Abstract
Surrogate light chain, which escorts the mu heavy chain to the cell surface, is a critical component of the pre-B cell receptor complex. The two proteins that comprise the surrogate light chain, VpreB and λ5/14.1, contain both unique regions and Ig-like domains. The unique regions have been postulated to function in the assembly of the surrogate light chain. However, by using transient transfection of COS7 cells, we show that deletion of the unique regions of both proteins did not inhibit the assembly of surrogate light chain. Instead, in vivo folding studies showed that the unique region of λ5/14.1 acts as an intramolecular chaperone by preventing the folding of this protein when it is expressed in the absence of its partner, VpreB. The Ig domains of both λ5/14.1 and VpreB are atypical. The one in VpreB lacks one of the canonical β strands whereas the one in λ5/14.1 has an extra β strand. Deletion of the extra β strand in λ5/14.1 completely abrogated the formation of the surrogate light chain, demonstrating that complementation of the incomplete Ig domain in VpreB by the extra β strand in λ5/14.1 was necessary and sufficient for the folding and assembly of these proteins. Our studies reveal two novel mechanisms for regulating surrogate light chain formation: (i) the presence of an intramolecular chaperone that prevents folding of the unassembled subunit but that remains part of the mature assembled protein, and (ii) splitting an Ig domain between two proteins to control their folding and assembly.
The pre-B cell receptor (pre-BCR) complex is transiently expressed at low cell density on B cell precursors and functions as a critical checkpoint in B cell development (reviewed in refs.1 and 2). However, the factors that control the expression of this complex are not well understood. Like the mature BCR, the pre-BCR is assembled in the endoplasmic reticulum (ER), a process that involves binding of the surrogate light chain to the heavy chain, displacement of the ER chaperone Ig heavy chain binding protein (BiP) from the heavy chain, and association of the Igα/Igβ signal-transducing heterodimer (3, 4). However, assembly of the pre-BCR has an added degree of complexity. Unlike conventional light chains that are comprised of two Ig domains encoded by a single protein, the surrogate light chain is made up of two proteins, VpreB and λ5/14.1 (λ5 in the mouse), each of which consists of a single Ig domain that must noncovalently assemble with its partner (5–8).
VpreB and λ5/14.1 will assemble and transverse the secretory pathway as a complex even in the absence of heavy chain expression (9, 10). However, our previous studies have suggested that VpreB is unstable when expressed in the absence of normal λ5/14.1 and that λ5/14.1 folds inefficiently in COS7 cells in the absence of VpreB (11). This was somewhat surprising because conventional light chain domains fold independently of each other both in vitro (12) and in vivo (13). These observations raise questions about the structural components of the surrogate light chain proteins that control their stability and their ability to assemble with each other and with mu heavy chain.
VpreB and λ5/14.1 have several unusual features that distinguish them from conventional light chains. Both proteins have atypical ER-targeted signal peptide sequences. In VpreB, the 19-amino acid signal peptide has an uncharacteristic proline at the −1 position (9, 14, 15). Mature VpreB protein is comprised of a 102-amino acid sequence with homology to a conventional variable region sequence; however, conventional variable domains include a J region sequence and the complete domain folds into a 9 β strand structure. VpreB lacks the region with homology to the J region and therefore the folded domain would be expected to include only 8 β strands. The carboxyl-terminal portion of VpreB consists of a unique sequence of 24 amino acids without homology to known Ig domains.
The signal peptide for λ5/14.1 is 44 amino acids long, which is significantly longer than the typical 19- to 24-amino acid signal sequence found in most secreted proteins; this feature could affect the cleavage of the signal peptide and therefore the folding of the λ5/14.1 molecule. A second unusual feature of λ5/14.1 is the 50-amino acid unique region without homology to known proteins, which is found immediately carboxyl-terminal to the signal peptide. It has been proposed that this region of λ5/14.1 might interact with the unique region of VpreB in the assembly of the surrogate light chain complex (9); alternatively, the unique regions of both VpreB and λ5/14.1 might form a ligand binding domain for the extracellular pre-BCR. The unique region of λ5/14.1 is followed by 119 amino acids with homology to both the Jλ region and the constant region of λ light chain. As most Ig constant domains are 100–110 amino acids long and fold into a 7 β strand domain, the J region sequence, separated from its typical upstream sequence, might adversely affect the folding of the remaining portion of this protein. Finally, although there is 85% identity between the Ig domain of λ5/14.1 and the constant region of conventional λ light chain, the differences between the two sequences might influence the folding of λ5/14.1.
In the present study, we investigated the role of the various regions of VpreB and λ5/14.1 in controlling the folding, assembly, and secretion of these two molecules. Our data demonstrated that neither the unusual signal sequences nor the primary structure of the Ig-like domains contribute to the instability or inadequate folding of VpreB or λ5/14.1. We found that the J region homology sequence in the λ5/14.1 protein was essential for assembly of this molecule with VpreB. In contrast to previous suggestions (9), the unique regions of these proteins did not play a critical role in assembly of the surrogate light chain complex. Finally, our studies revealed that the unique region of λ5/14.1 has the unusual function of inhibiting the folding and secretion of this molecule when it is expressed in the absence of VpreB.
MATERIALS AND METHODS
Cell Culture and DNA Transfections.
COS7 monkey fibroblasts were maintained in DMEM supplemented with 10% fetal bovine serum and 2 mM l-glutamine. Subconfluent monolayers of COS7 cells were transfected by Lipofectamine (GIBCO/BRL) as described (11).
Construction of Expression Vectors.
Construction of wild-type VpreB and λ5/14.1 cDNAs in the expression vector pcDNA3 (Invitrogen) has been described (11). PCR was used to amplify a conventional λ light chain by using cDNA from peripheral blood lymphocytes as template. This λ light chain containing Vλ1–13 variable region, Jλ2 joining region, and Cλ2 constant region (16, 17) was cloned into pcDNA3. The mutants and the chimeras were constructed from these three vectors by using recombinant PCR mutagenesis. All of the PCR amplified fragments were sequenced by using dideoxy chain termination before transfection.
Metabolic Labeling and Immunoprecipitations.
Forty hours after transfection, COS7 cells were metabolically labeled with 0.2 mCi/ml (1 Ci = 37 GBq) of [35S]Translabel for 3 hr. Labeled cells were lysed in buffer consisting of 1% Triton X-100, 10 mM Tris (pH 8.0), 140 mM NaCl, 1% bovine hemoglobin, 0.2 unit/ml aprotinin, 10 μg/ml leupeptine, 1 mM phenylmethylsulfonyl fluoride, 50 μg/ml 1-chloro-3-tosylamido-7-amino-2-heptanone either with or without 20 mM N-ethylmaleimide (NEM). Lysates and supernatants were incubated with murine anti-VpreB mAb (18) or goat anti-λ antibody (Sigma) and immune complexes were precipitated with anti-mouse Ig Sepharose (Zymed) or protein G Sepharose, respectively. Immunoprecipitates were electrophoresed on 12.5% SDS/polyacrylamide mini-gels with or without 20 mM 2-mercaptoethnol and visualized by fluorography as described (11).
RESULTS
λ5/14.1 Is Not Secreted or Folded When Expressed Without VpreB.
To determine the requirements for surrogate light chain assembly and secretion, we began by transiently expressing a conventional λ light chain or λ5/14.1 and VpreB in COS7 cells. As expected, the conventional λ light chain and the assembled surrogate light chain proteins were readily secreted during the 3 hr labeling period (Fig. 1A). By contrast, λ5/14.1 was not secreted when expressed alone. Because quality control measures operate in the ER to ensure that improperly folded proteins are retained, we examined the folding status of λ5/14.1 by analyzing the migration of this protein in an SDS/polyacrylamide gel under reducing and nonreducing conditions. Once an Ig domain folds, the characteristic disulfide bond is formed and the oxidized protein migrates more rapidly through the gel (19).
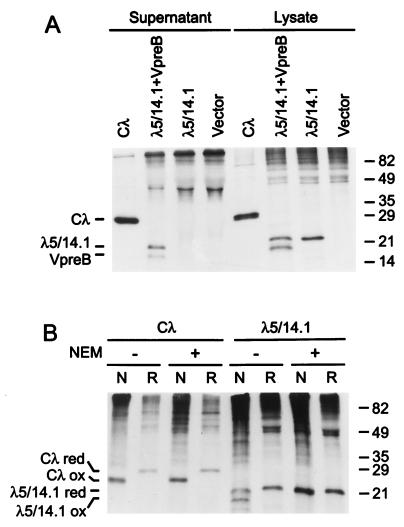
Figure 1.
Secretion and folding of conventional λ light chain (Cλ), surrogate light chain, and λ5/14.1 (A) COS7 cells were transfected with expression vectors as indicated. After metabolic labeling, both lysates and supernatants were immunoprecipitated with goat anti-λ antiserum and separated in a 12.5% SDS/PAGE gel under reducing conditions. The migration of molecular mass markers is indicated on the right margin (kDa). (B) COS7 cells were transfected with the vectors indicated then lysed in the absence (−) or presence (+) of NEM. Proteins were immunoprecipitated with goat anti-λ and analyzed under nonreducing (N) or reducing (R) conditions.
The conventional λ light chain and λ5/14.1 were transiently expressed in COS7 cells. As reported (13), the λ light chain migrated more rapidly under nonreducing conditions than reducing conditions (Fig. 1B). Addition of the alkylating agent NEM to the cells before lysis had no effect on the migration of this protein, because it is rapidly and fully oxidized in these cells. By contrast, only a portion of λ5/14.1 migrated as a folded protein under nonreducing conditions (Fig. 1B). When NEM was added to prevent postlysis oxidation, the nonreduced and reduced λ5/14.1 proteins migrated with the same mobility, demonstrating that when λ5/14.1 protein is expressed alone, it is unable to fold in vivo.
Construction of λ5/14.1 Mutants and Chimeras.
To identify the region(s) of λ5/14.1 that influence its folding and assembly, we made a series of constructs that either contained various regions of the λ5/14.1 sequence juxtaposed to regions of the conventional λ light chain or simply deleted regions of the λ5/14.1 sequence (Fig. 2). One construct, λ5(CλL), replaced the leader sequence of λ5/14.1 with that of the conventional light chain; a second, λ5ΔU, deleted the 50-amino acid unique region of λ5/14.1; a third, λ5Δβ, deleted the extra β strand with homology to a Jλ region. Finally, to examine the possibility that the Ig homologous regions of the λ5/14.1 protein were intrinsically inefficient in folding, we substituted the corresponding regions from the conventional λ light chain [λ5(CλC)].
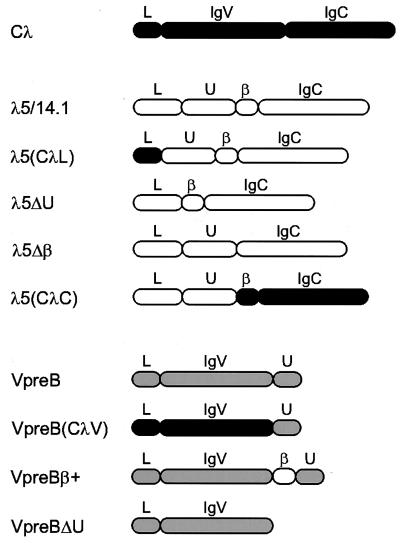
Figure 2.
Schematic diagram of wild-type and mutant proteins included in this study. The domain structure of conventional λ light chain (■), λ5/14.1 (□), and VpreB (░⃞) are indicated. Wild-type λ5/14.1 has four cysteines, one in the unique region, two that form the disulfide bridge in the Ig domain and one that is required for binding to the mu heavy chain; VpreB has only two cysteines both of which lie within the Ig-like domain. The λ5(CλL) mutant has the 19-amino acid leader sequence of conventional λ light chain and the169-amino acid mature λ5/14.1 polypeptide. The λ5(CλC) mutant contains the 44-amino acid leader peptide and 50-amino acid unique region of λ5/14.1 and the 119-amino acid Jλ2 and Cλ2 sequence of conventional λ light chain. Mutants λ5ΔU and λ5Δβ lack the 50-amino acid unique region (codon 45–94) and 14-amino acid β strand (codon 95–108), respectively. The VpreB(CλV) contains the leader sequence and variable region of λ light chain (codon 1–114 of Vλ1–13) without the ninth β strand; like VpreB, it does include the 24 amino acids of the unique region of VpreB (codon 122–145 of VpreB). The mutant VpreBβ+ has the 14-amino acid Jλ region from λ5/14.1 (codon 95–108 of λ5/14.1) inserted between the variable region and the unique region of VpreB, completing the nine β-strand Ig variable domain. VpreBΔU lacks carboxyl-terminal 24-amino acid unique region. The domain structure of the wild-type and the mutants are shown as follows: L, leader peptide; U, unique region; IgV, Ig variable domain; IgC, Ig constant domain; β, β strand homologous to Ig J region.
The Unique Region of λ5/14.1 Inhibits Its Folding and Secretion.
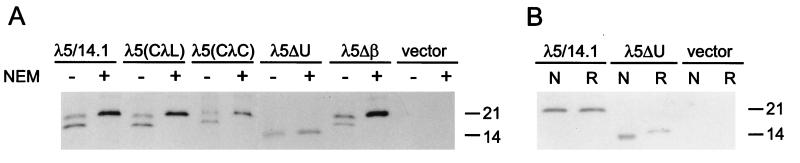
Wild-type λ5/14.1 and the various λ5/14.1 mutants and chimeras were expressed in COS7 cells, and the folding status of these proteins was determined by lysing transfectants in the presence or absence of NEM and analyzing the labeled λ5/14.1 proteins on nonreducing gels (Fig. 3A). The data revealed that neither the replacement of the signal sequence with a more conventional one or the removal of the extra β strand that constitutes the Jλ-like region influenced the migration of λ5/14.1, indicating that these regions do not contribute to the poor folding of λ5/14.1. Furthermore, substitution of the conventional λ constant region domain for the Ig domain in λ5/14.1 did not improve the folding of this molecule. Only the unique region-deleted λ5/14.1 protein displayed an altered pattern in this analysis. It had the same migration pattern in the presence and absence of NEM. This could mean either the λ5ΔU protein was completely unable to fold even in the presence of postlysis oxidation, or that this protein was efficiently folded in the cell. To distinguish between these possibilities, we lysed the proteins in the presence of NEM to “lock” them as they were in the cell and then examined them under both reducing and nonreducing conditions. Because the wild-type λ5/14.1 protein has not folded and formed its intradomain disulfide bond in the cell, it migrates the same under both reducing and nonreducing conditions (Fig. 3B). However, the λ5ΔU protein migrates faster under nonreducing conditions, demonstrating that it has folded and formed its intradomain disulfide bond. Thus, the unique region is responsible for inhibiting the folding of the λ5/14.1 protein when it is expressed in the absence of VpreB. This conclusion is further supported by the observation that NEM does not bind cysteines in λ5ΔU and cause a modest retardation in its mobility, whereas this effect is observed with λ5/14.1 and the other 3 constructs that have not formed their intradomain disulfide bond (Figs. 3A and 1B).
Figure 3.
Comparison of folding of wild-type and mutant λ5/14.1 (A) COS7 cells were transfected with wild-type or mutant λ5/14.1 as indicated. After metabolic labeling, cells were lysed in the absence (−) or presence (+) of NEM. The lysates were immunoprecipitated with goat anti-λ antiserum and separated under nonreducing conditions. (B) Immunoprecipitation of labeled COS7 transfectants was performed as described above in the presence of NEM. The samples were separated under nonreducing (N) or reducing (R) conditions.
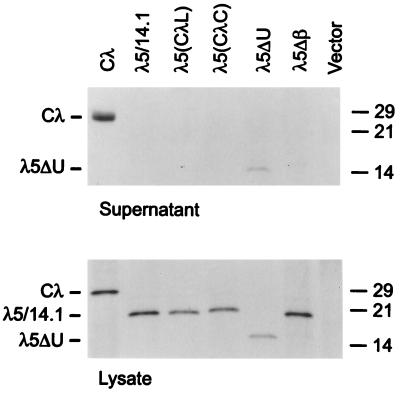
As a more rigorous test of the appropriate folding of the various constructs, we evaluated their ability to be secreted. As expected, the λ light chain was secreted and the λ5/14.1 protein was not. (Fig. 4). In keeping with their inability to fold properly, the λ5(CλL), the λ5(CλC), and the λ5Δβ proteins were not secreted from the cell although all these proteins were made at levels equivalent to those of the conventional light chain. Only λ5ΔU was secreted from these cells, demonstrating that removing the unique region from λ5/14.1 was sufficient to produce a protein that was adequately folded to pass the stringent ER quality control.
Figure 4.
Secretion of λ5/14.1 mutants COS7 cells were transfected with the vectors indicated. Culture supernatants were harvested and cell lysates were prepared in the presence of NEM. Both lysates and supernatants were immunoprecipitated with goat anti-λ antiserum and samples were separated under reducing conditions.
VpreB Protein Is Inefficiently Folded in Vivo.
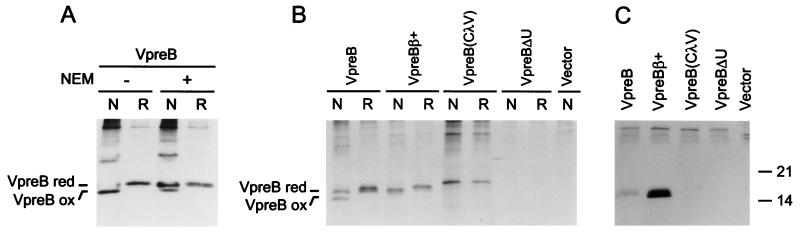
Our previous studies had shown that VpreB can be secreted in the absence of λ5/14.1; however, these same studies demonstrated that VpreB was unstable when expressed in B cell precursors with a mutant λ5/14.1 (11). This finding suggests that VpreB may require a normal λ5/14.1 protein for efficient folding. This possibility was tested by analyzing cell lysates from VpreB transfected COS7 cells under reducing and nonreducing conditions and in the presence and absence of NEM. As shown in Fig. 5A, although all of the VpreB migrated more rapidly under nonreducing conditions in the absence of NEM, only a fraction of the VpreB migrated more rapidly under nonreducing conditions in the presence of NEM. These findings indicate that VpreB folds more efficiently than λ5/14.1 but not as well as λ light chain.
Figure 5.
Folding and secretion of wild-type and mutant VpreB (A) COS7 cells were transfected with VpreB and metabolically labeled cells were lysed in the absence (−) or presence (+) of NEM. Proteins were immunoprecipitated with anti-VpreB and analyzed under nonreducing (N) or reducing (R) conditions. (B) COS7 cells were transfected with the vectors indicated, and labeled cells were lysed in the presence of NEM, immunoprecipitated with anti-VpreB, and separated under nonreducing (N) or reducing (R) conditions. (C) The supernatants from the experiment shown in B were immunoprecipitated with anti-VpreB mAb and separated under reducing conditions.
Construction of VpreB Mutants.
Three VpreB constructs were produced to evaluate the importance of the various regions of this molecule in protein folding and in assembly with λ5/14.1 (Fig. 2). In the first, the signal sequence and variable region domain of VpreB were replaced with those of λ light chain [VpreB(CλV)]. Like VpreB, this construct did not include the ninth β strand of the variable region of λ light chain, but it did include the 24 amino acids of the unique region of VpreB. To determine whether the addition of a ninth β strand to the region of VpreB with homology to a variable region of λ light chain would affect the folding of this protein or its ability to bind λ5/14.1, the 14 amino acids from the Jλ homologous region of λ5/14.1 were inserted between the eighth β strand and the unique region of VpreB (VpreBβ+). In the last construct, the 24 amino acids of the unique region were deleted (VpreBΔU).
Addition of Ninth β Strand Induces Efficient Folding and Secretion of VpreB.
Wild-type and mutant VpreB constructs were expressed in COS7 cells to evaluate the folding and secretion of these proteins. Cells were lysed in the presence of NEM and the lysates were immunoprecipitated with a mAb to VpreB and analyzed on SDS/polyacrylamide gels under reducing and nonreducing conditions (Fig. 5B). The results demonstrated that neither the unusual leader sequence nor the Ig variable homology domain of VpreB could account for the poor folding of VpreB; however, when the ninth β strand was added to the variable region domain of VpreB, all of the protein was readily oxidized. The role of the unique region of VpreB could not be evaluated in this experiment because the mAb to VpreB apparently binds to this portion of the molecule. Examination of the supernatants from the COS7 cells transfected with the VpreB constructs demonstrated that there was a strong correlation between formation of the intradomain disulfide bond and secretion. Although the wild-type VpreB protein was secreted into the supernatant, the VpreBβ+ construct was secreted much more efficiently and the VpreB(CλV) was not secreted at all (Fig. 5C).
Binding of λ5/14.1 to VpreB.
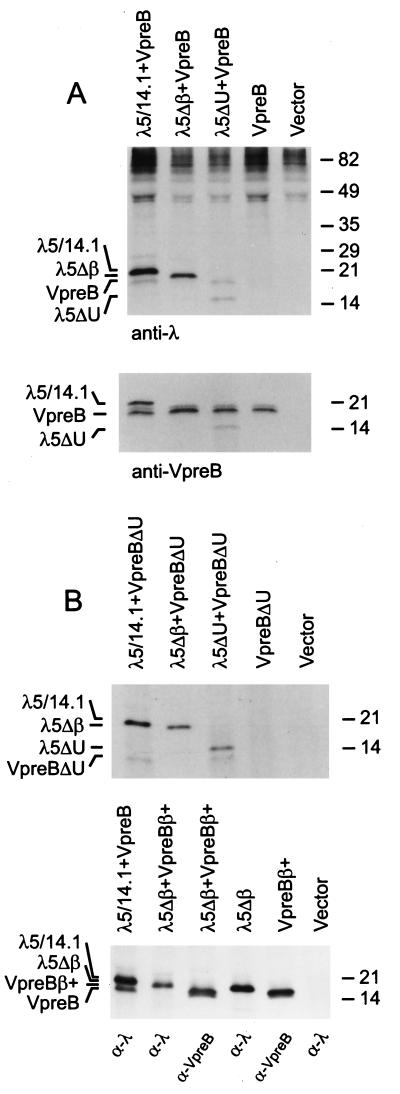
To explore the requirements for surrogate light chain assembly, we transiently expressed VpreB and the various mutants of λ5/14.1 in COS7 cells. Equivalent amounts of VpreB were coprecipitated with both wild-type λ5/14.1 and the λ5ΔU mutant by using anti-λ antiserum (Fig. 6A Upper). However, VpreB was not coprecipitated with λ5Δβ, although approximately equal amounts of VpreB were expressed in these cells (Fig. 6A Lower). When the samples were immunoprecipitated with anti-VpreB, the wild-type λ5/14.1 and the λ5ΔU were coprecipitated but not the λ5Δβ. These results suggested that the unique region of λ5/14.1 was not essential for the binding of λ5/14.1 to VpreB; however, the first β strand preceding the Ig constant region domain of the λ5/14.1 was needed for this interaction.
Figure 6.
Assembly of VpreB and λ5/14.1 COS7 cells were transfected with the vectors indicated, lysates were made in the presence of NEM and the proteins were immunoprecipitated with (A) anti-λ or anti-VpreB. (B) Upper, anti-λ; Lower, anti-λ in lanes 1, 2, 4, and 6 or anti-VpreB in lanes 3 and 5. The expected positions of the recombinant proteins are shown on the left. Molecular mass markers are shown on the right.
The vector expressing VpreBΔU mutant was then cotransfected with wild-type and mutant λ5/14.1 constructs. As expected from the previous experiments, VpreBΔU was not coprecipitated with the λ5Δβ mutant. However, VpreBΔU could be immunoprecipitated with either wild-type λ5/14.1 or λ5ΔU (Fig. 6B Upper). Thus, in the absence of the unique regions of both λ5/14.1 and VpreB, these two proteins were able to assemble efficiently.
As a final step to identify a role for the unique regions of VpreB and λ5/14.1 in the formation of the surrogate light chain, COS7 cells were transfected with wild-type VpreB and λ5/14.1 or with the VpreB construct containing the ninth β strand (VpreBβ+) and the λ5/14.1 construct lacking the extra β strand (λ5Δβ). The mutant VpreBβ+ could not be coimmunoprecipitated with anti-λ antisera and the mutant λ5Δβ could not be coimmunoprecipitated with anti-VpreB (Fig. 6B Lower). These results demonstrate that the unique regions of λ5/14.1 and VpreB cannot induce the binding of these two proteins in the absence of the Ig complementation. Further, when coupled with the studies shown in the top panel of Fig. 6B, the results indicate that complementation of the incomplete Ig domain in VpreB with the extra β strand, Jλ homology region, in λ5/14.1 is both necessary and sufficient for assembly of the surrogate light chain complex.
DISCUSSION
A variety of quality control mechanisms ensure that only properly folded and assembled molecules exit the ER (20). Our studies describe two novel mechanisms that are used to regulate the folding, assembly and transport of the components of the surrogate light chain, VpreB and λ5/14.1. Although both VpreB and λ5/14.1 encode proteins with Ig domain-like motifs, neither of the motifs is completely characteristic of those seen in members of the Ig superfamily. Typical Ig domains consist of 7 or 9 β strands that fold independently into the prototypical Ig domain structure (21, 22). The VpreB Ig domain is incomplete in that it lacks the J region sequence that typically constitutes the ninth and final β strand in a variable region Ig domain. In the absence of this ninth β strand, VpreB folds inefficiently. On the other hand, λ5/14.1 has a complete constant region Ig domain with 7 β strands preceded by an extra β strand with homology to J region sequence. The ability of the J region sequence in λ5/14.1 to complement the incomplete Ig domain in VpreB allows the efficient folding of VpreB and the assembly of VpreB and λ5/14.1 to form the surrogate light chain complex. A second novel mechanism is used to prevent the folding and secretion of λ5/14.1 in the absence of VpreB. The 50-amino acid unique region at the amino-terminal end of λ5/14.1 inhibits the folding and the secretion of this molecule if it is expressed in the absence of VpreB. When the unique region is deleted, λ5/14.1 folds and successfully exits the secretory pathway. This mechanisms ensures that λ5/14.1 is not secreted independently of VpreB.
Our studies provide support for some aspects of the three-dimensional models of the surrogate light chain proposed by Kudo et al. (23) and Guelpa-Fonlupt et al (9). Based on sequence data, both groups suggested that the Jλ homologous β strand in λ5/14.1 might interdigitate with VpreB to form a complete Ig domain. Our data show that the transfer of the Jλ homologous β strand from λ5/14.1 to VpreB enhanced the ability of VpreB to fold and exit the secretory pathway; however, this modified VpreB no longer assembled with λ5/14.1. Thus, splitting the Ig domain between VpreB and λ5/14.1 controlled both the folding and assembly of the surrogate light chain. A second aspect of the earlier models for the surrogate light chain deals with the unique regions of VpreB and λ5/14.1. It was suggested that the unique region of VpreB and λ5/14.1 might interact with each other, based on their proposed proximity in space and the potential for salt bridge formation between the acidic and basic residues of these two regions (9). Although we cannot rule out the possibility that the two unique regions interact in normal B cell precursors, our mutants demonstrate that this interaction is not required to stabilize the VpreB and λ5/14.1 proteins, nor is it sufficient in the absence of interdigitation of the two domains to support their association.
If the unique region of VpreB does not bind the unique region of λ5/14.1, one must account for the mechanism by which VpreB relieves the inhibition of λ5/14.1 folding imposed by the unique region of λ5/14.1. One possible explanation is that the Ig domain of VpreB may be able to successfully compete with the unique region of λ5/14.1 for binding to the Jλ homology region. In the absence of VpreB, the unique region might interact with the Jλ homology region as first proposed by Melchers et al. (1) and in some way inhibit the folding of the remaining portion of λ5/14.1; however, deletion of the extra β strand did not relieve this inhibition. Alternatively, the λ5/14.1 unique region could directly associate with any of the remaining β strands in λ5/14.1 and alter their folding and prevent oxidation. In these models, the unique region of λ5/14.1 acts as intramolecular chaperone that prevents folding of the λ5/14.1 constant region domain until it can assemble with the VpreB domain. The concept of assembly-dependent folding is supported by recent data demonstrating that the CH1 domain of free heavy chains is not folded while bound to BiP and requires light chain assembly to displace BiP and allow the domain to fold (R. Hellman, J. W. Brewer, Y. K. Lee, L.M.H., unpublished work). However, the mechanism by which this is achieved in the case of the surrogate light chain is quite different. The λ5/14.1 molecule uses an intramolecular or cis chaperone rather than the professional, trans chaperone BiP used by heavy chains.
Intramolecular chaperones have been described in some secreted proteins, including subtilisin, bovine pancreatic trypsin inhibitor and type β1 transforming growth factor (24–26). These proteins contain amino-terminal pro regions that are required for the correct folding of the functional domain. However, in contrast to the unique region of λ5/14.1, these pro regions are removed from the mature protein. It has been proposed that the pro regions may stabilize the transitional state for the correct folding pathway of the mature protein or they may destabilize nonproductive folding intermediates (27). The unique region of λ5/14.1 may play a similar role.
Three well-defined mechanisms used to regulate the assembly and transport of multimeric complexes were first demonstrated in studies on proteins with Ig domains. (i) The CH1 Ig domain of mu heavy chain binds the 70-kDa heat shock protein chaperone BiP, causing the retention of mu in the ER until the surrogate light chain or the conventional light chain can replace BiP (28, 29). (ii) The transmembrane form of mu heavy chain has two hydrophilic amino acids that must be masked by the transmembrane domains of the heterodimeric signal transducing complex, Igα-Igβ, before mu can be expressed on the cell surface. (iii) The penultimate cysteine residue found on the secreted form of mu is bound by a thiol-mediated retention mechanism in the ER until the J chain and the monomeric subunits assemble.
The fact that the Ig domain is so well characterized has allowed us to define the two additional mechanisms controlling folding and assembly described in this paper. Studies on the folding characteristics of the Ig domain (12, 13, 20, 29) permitted us to identify defective folding of both the VpreB and λ5/14.1 Ig domains and correction of those defects by the addition of an extra β strand in VpreB or the removal of the unique region, the intramolecular chaperone, from λ5/14.1. Finally, although a very large number of Ig domains have been described, and it is well recognized that Ig domains tend to homo- or heterodimerize (30), our studies are the first to show that splitting an Ig domain between two proteins can be used to control the folding and assembly of those proteins. It is likely that as additional proteins are analyzed, similar examples of inhibitory intramolecular chaperones or split domains will be described.
Acknowledgments
These studies were supported by National Institutes of Health Grants AI25129 (M.E.C.) and GM54068 (L.M.H.), the Assisi Foundation, National Cancer Institute Grant P30 CA21765; March of Dimes FY97-0384; American Lebanese Syrian Associated Charities; and by funds from the Federal Express Chair of Excellence.
ABBREVIATIONS
- BCR
B cell receptor
- BiP
Ig heavy chain binding protein
- ER
endoplasmic reticulum
- NEM
N-ethylmaleimide
Footnotes
A Commentary on this article begins on page 2571.
References
- 1.Melchers F, Karasuyama H, Haasner D, Bauer S, Kudo A, Sakaguchi N, Jameson B, Rolink A. Immunol Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 3.Reth M, Hombach J, Wienands J, Campbell K S, Chien N, Justement L B, Cambier J C. Immunol Today. 1991;12:196–201. doi: 10.1016/0167-5699(91)90053-V. [DOI] [PubMed] [Google Scholar]
- 4.Lassoued K, Illges H, Benlagha K, Cooper M D. J Exp Med. 1996;183:421–429. doi: 10.1084/jem.183.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai S, Baltimore D. Nature (London) 1987;329:172–174. doi: 10.1038/329172a0. [DOI] [PubMed] [Google Scholar]
- 6.Kerr W G, Cooper M D, Feng L, Burrows P D, Hendershot L M. Int Immunol. 1989;1:355–361. doi: 10.1093/intimm/1.4.355. [DOI] [PubMed] [Google Scholar]
- 7.Karasuyama H, Kudo A, Melchers F. J Exp Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsubata T, Reth M. J Exp Med. 1990;172:973–976. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guelpa-Fonlupt V, Bossy D, Alzari P, Fumoux F, Fougereau M, Schiff C. Mol Immunol. 1994;31:1099–1108. doi: 10.1016/0161-5890(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 10.Bornemann K D, Brewer J W, Perez E, Doerre S, Sita R, Corley R B. J Immunol. 1997;158:2551–2557. [PubMed] [Google Scholar]
- 11.Minegishi Y, Coustan-Smith E, Wang Y-H, Cooper M D, Campana D, Conley M E. J Exp Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto Y, Hamaguchi K. J Mol Biol. 1982;156:911–926. doi: 10.1016/0022-2836(82)90147-4. [DOI] [PubMed] [Google Scholar]
- 13.Hendershot L M, Wei J, Gaut J R, Melnick J, Aviel S, Argon Y. Proc Natl Acad Sci USA. 1996;93:5269–5274. doi: 10.1073/pnas.93.11.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo A, Melchers F. EMBO J. 1987;6:2267–2272. doi: 10.1002/j.1460-2075.1987.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasicek T J, Leder P. J Exp Med. 1990;172:609–620. doi: 10.1084/jem.172.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits J L, Wang J, Shimizu N. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y H, Nomura J, Faye-Petersen O M, Cooper M D. J Immunol. 1998;161:1132–1139. [PubMed] [Google Scholar]
- 19.Braakman I, Helenius J, Helenius A. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond C, Helenius A. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 21.Amzel L M, Poljak R J. Annu Rev Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- 22.Bork P, Holm L, Sander C. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 23.Kudo A, Bauer S, Melchers F. In: Progress in Immunology VII. Melchers F, editor. Berlin: Springer; 1989. pp. 339–347. [Google Scholar]
- 24.Zhu X, Ohta Y, Jordan F, Inouye M. Nature (London) 1989;339:483–484. doi: 10.1038/339483a0. [DOI] [PubMed] [Google Scholar]
- 25.Weissman J S, Kim P S. Cell. 1992;71:841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- 26.Gray A M, Mason A J. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- 27.Silen J L, Agard D A. Nature (London) 1989;341:462–464. doi: 10.1038/341462a0. [DOI] [PubMed] [Google Scholar]
- 28.Haas I G, Wabl M. Nature (London) 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- 29.Hendershot L M. J Cell Biol. 1990;111:829–837. doi: 10.1083/jcb.111.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]