Abstract
Orf virus, a member of the poxvirus family, produces a pustular dermatitis in sheep, goats, and humans. The lesions induced after infection with orf virus show extensive proliferation of vascular endothelial cells, dilation of blood vessels and dermal swelling. An explanation for the nature of these lesions may lie in the discovery that orf virus encodes an apparent homolog of the mammalian vascular endothelial growth factor (VEGF) family of molecules. These molecules mediate endothelial cell proliferation, vascular permeability, angiogenesis, and lymphangiogenesis via the endothelial cell receptors VEGFR-1 (Flt1), VEGFR-2 (KDR/Flk1), and VEGFR-3 (Flt4). The VEGF-like protein of orf virus strain NZ2 (ORFV2-VEGF) is most closely related in primary structure to VEGF. In this study we examined the biological activities and receptor specificity of the ORFV2-VEGF protein. ORFV2-VEGF was found to be a disulfide-linked homodimer with a subunit of ≈25 kDa. ORFV2-VEGF showed mitogenic activity on bovine aortic and human microvascular endothelial cells and induced vascular permeability. ORFV2-VEGF was found to bind and induce autophosphorylation of VEGFR-2 and was unable to bind or activate VEGFR-1 and VEGFR-3, but bound the newly identified VEGF165 receptor neuropilin-1. These results indicate that, from a functional viewpoint, ORFV2-VEGF is indeed a member of the VEGF family of molecules, but is unique, however, in that it utilizes only VEGFR-2 and neuropilin-1.
Vascular endothelial growth factor (VEGF) is a homodimeric glycoprotein that has mitogenic activity specific for endothelial cells (1, 2) and the ability to induce vascular permeability (3). VEGF has important regulatory functions in the formation of new blood vessels during embryonic vasculogenesis and in angiogenesis during adult life (reviewed in ref. 4). The significance of the role played by VEGF has been demonstrated in studies showing that inactivation of a single VEGF allele results in embryonic lethality because of failed development of the vasculature (5, 6). VEGF is also an important mediator of angiogenesis in a number of pathological conditions including tumor formation. Furthermore, numerous inhibitors of the VEGF/VEGF receptor system have been shown to prevent tumor growth via an antiangiogenic mechanism (7, 8).
Alternative mRNA splicing of a single VEGF gene gives rise to five isoforms of VEGF (2, 9–11). In addition, several proteins related in primary structure to VEGF have been reported. The VEGF family now includes placenta growth factor (PlGF)(12), VEGF-B (13), VEGF-C (14), and VEGF-D (15), each of which shows between 30% and 45% amino acid sequence identity with VEGF. The VEGF family members share a VEGF homology domain that contains the six cysteine residues that form the cystine knot motif. Functional characteristics of the VEGF family include varying degrees of mitogenicity for endothelial cells, induction of vascular permeability, and angiogenic and lymphangiogenic properties (16–18).
The VEGF family members are ligands for a set of mammalian tyrosine kinase receptors (VEGF receptors, VEGFRs). VEGF binds and activates both VEGFR-1 (Flt1)(19) and VEGFR-2 (Flk1 or KDR)(20), whereas PlGF and VEGF-B bind only VEGFR-1 (21, 22). VEGF-C and VEGF-D bind VEGFR-2 in addition to VEGFR-3 (Flt4) (14, 15). At least VEGF and PlGF-2 also appear to interact with a neuronal cell-guidance receptor, neuropilin-1(NP-1) (23, 24). Gene targeting studies have demonstrated the absolute requirement of VEGFR-1, VEGFR-2, and VEGFR-3 for embryonic development (25–27). These studies show that VEGFR-1 plays a role in vascular endothelial tube formation, VEGFR-2 is important for endothelial/hematopoietic cell differentiation and mitogenesis, and VEGFR-3 is involved in regulation of vascular remodeling and the formation of large vessels (reviewed in refs. 16 and 17).
We have identified VEGF-like proteins encoded by two isolates of orf virus, NZ2 and NZ7 (28). Both proteins show significant amino acid sequence similarity to VEGF. In recent years there have been numerous reports of viral virulence factors that appear to be derived from captured host genes (for reviews see refs. 29–31), but orf virus is the only virus reported to encode a VEGF-like protein. Orf virus is the type species of the parapoxvirus genus (32). It causes a highly contagious pustular dermatitis in sheep and goats and is readily transmittable to humans. The dermal lesions are remarkable for the extensive vascular proliferation and dilation seen in histological sections (33, 34). Generally, orf virus infections resolve in a few weeks, but severe infections that fail to resolve without surgical intervention are seen in immune-impaired individuals (35, 36). The large proliferative lesions seen in these cases have a tumor-like appearance. Expression of an orf virus gene able to stimulate angiogenesis may provide an explanation for these histological observations. We report here that the purified VEGF-like gene product from orf virus strain NZ2 is mitogenic for endothelial cells and induces vascular permeability. We also show that it binds to VEGFR2 and NP-1, inducing tyrosine kinase phosphorylation of VEGFR-2, but does not bind to VEGFR-1 or VEGFR-3. This receptor-binding specificity is unique among the VEGF family of growth factors.
MATERIALS AND METHODS
Expression of ORFV2-VEGF in COS Cells and Purification by Affinity Chromatography.
A DNA fragment containing nucleotides 4–401 of the VEGF-like gene of orf virus strain NZ2, was prepared by PCR using pVU89 as a template (28). This fragment was inserted into the pEFBOS-I-Flag expression vector (obtained from Clare MacFarlane, Walter and Eliza Hall Institute, Melbourne), immediately upstream from the DNA sequence encoding the Flag octapeptide (IBI/Kodak). Protein synthesis would, therefore, give rise to a secreted orf virus VEGF-like polypeptide that was tagged with the Flag octapeptide at its C terminus. This protein was designated ORFV2-VEGF. Flag-tagged proteins were expressed transiently in COS cells by using the DEAE-dextran method (37). After 3 days of incubation, a portion of the transfected COS cells were metabolically labeled as described (38). The remaining culture was incubated for a total of 7 days. Conditioned cell culture medium was collected and clarified by centrifugation before Flag-tagged proteins were recovered by affinity chromatography by using M2-gel (anti-Flag; IBI/Kodak).
SDS/PAGE and Immunoblotting.
Purified proteins or washed immunoprecipitates were combined with SDS/PAGE sample buffer either with or without 2% 2-mercaptoethanol, boiled, and resolved by SDS/PAGE (39). When required, proteins were transferred to nitrocellulose and blotted with M2 antibody as described (15).
Bioassay to Monitor Binding to the Extracellular Domain of VEGFR-2.
A bioassay was performed by using Ba/F3 cells, which express a chimeric receptor containing the extracellular domain of the mouse VEGFR-2 and the transmembrane and cytoplasmic domains of the mouse erythropoietin receptor (EpoR) (15). The cells were maintained in DMEM containing 10% FBS, 50 mM l-glutamine, 50 μg/ml gentamicin, and 10% WEHI-3D-conditioned medium as a source of interleukin-3 (IL-3). Cells expressing the VEGFR-2-EpoR chimeric receptor were washed three times in PBS and once in complete medium lacking IL-3. Cells (104) were distributed into 96-well microtiter plates containing dilutions of the test reagent or medium alone. Cells were incubated for 48 h at 37°C in a humidified atmosphere of 10% CO2. Cell proliferation was quantitated by the addition of 1 μCi (1 Ci = 37 GBq) of [3H]thymidine (Amersham Pharmacia) for 4 h before harvesting. Incorporated [3H]thymidine was determined by using a cell harvester (Tomtec, Orange, CT) and β counting.
Binding Assays with Soluble VEGFR Extracellular Domains.
293 Epstein–Barr virus-encoded nuclear antigen (EBNA) cells were transfected with plasmids encoding the soluble receptor-Ig fusion proteins VEGFR-1-Ig (E. Korpelainen, Haartman Institute, Helsinki), VEGFR-2-Ig (Y. Gunji, Haartman Institute, Helsinki), VEGFR-3-Ig (K. Pajusola, Biotechnology Institute, Helsinki) (15) and NP-1-Ig. All Ig fusion proteins are human VEGFRs. Cells were incubated for 24 h after transfection, washed with DMEM containing 0.2% BSA, and serum starved for 24 h. The fusion proteins were then precipitated from the clarified, conditioned medium by using protein A-Sepharose beads. Beads were then combined with 100 μl of 10× binding buffer (5% BSA, 0.2% Tween 20, and 10 μg/ml heparin) and 900 μl of conditioned medium from 293EBNA cells that had been transfected with expression plasmids encoding ORFV2-VEGF, VEGF, VEGF-DΔNΔC (mutant that contains the VEGF homology domain of VEGF-D) (15), VEGF-C156S (40), or a vector control, and biosynthetically labeled with [35S]Cys/Met for 4–16 h. After 2.5 h at room temperature, the Sepharose beads were washed three times with binding buffer at 4°C, once with PBS and boiled in SDS/PAGE sample buffer before resolving the proteins by SDS/PAGE under reducing conditions. Radiolabeled proteins were detected by using a PhosphorImager analyzer (Molecular Dynamics).
Ligand-Stimulated Receptor Autophosphorylation.
ORFV2-VEGF, VEGF165, and VEGF-CΔNΔC were diluted in DMEM containing 0.2% BSA and used to stimulate porcine aortic endothelial cells (PAE) expressing VEGFR-2 or NIH/VEGFR-3. Stimulation of cells and analysis of phosphorylated receptors were carried out as described (14).
Mitogenesis Assays with Endothelial Cells.
Mitogenic assays were carried out with bovine aortic endothelial cells (BAEs) and human microvascular endothelial cells (HMVEC). In brief, cells grown in RPMI 1640 medium plus 10%FBS and supplements or Epstein–Barr medium (EBM)2, 5% FBS plus growth supplements, respectively, were removed with trypsin, washed, and distributed at 103 cells per well in a 96-well plate or 5 × 103 cells per 24-well plate. Cells were allowed to adhere for 6–16 h at 37°C before samples of growth factor, diluted in medium with reduced serum and without growth supplements, were added. After 72 h of growth at 37°C, cell proliferation was quantitated by using either the substrate (3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), or counting.
Miles Assay for Vascular Permeability.
The Miles vascular permeability assay was performed using anesthetized Guinea pigs as described (41). Animals were given an intracardiac injection of 500 μl of 0.5% Evans blue dye in PBS to introduce the dye into the bloodstream. Test samples (100–150 μl) were injected intradermally into a shaved area on the back of each animal. After 30 min, animals were sacrificed and an area of skin excised. For quantification of extravasated dye, samples of skin were incubated for 3–4 days in formamide at room temperature. The amount of dye extracted was determined by using OD 620 nm.
RESULTS
Alignment of ORFV2-VEGF and VEGF Family Members.
Alignment of the predicted amino acid sequence of ORFV2-VEGF with members of the VEGF family demonstrates that ORFV2-VEGF has a high degree of sequence homology with the VEGF homology domain (VHD) of this family of proteins (Fig. 1). ORFV2-VEGF contains all six cysteine residues of the cystine-knot motif that are absolutely conserved among family members. ORFV2-VEGF does not contain the extended N-and C-terminal regions seen in VEGF-C and VEGF-D. Overall, ORFV2-VEGF is 43.3%, 34.3%, 25.4%, 26.9%, and 33.6% identical to human VEGF165, VEGF-B, VEGF-C, VEGF-D, and PlGF, respectively. This sequence similarity of ORFV2-VEGF to the mammalian VEGFs raises the question of whether the structural relatedness extends to receptor binding and biological function.
Figure 1.
Comparison of ORFV2-VEGF with other members of the VEGF family of growth factors. The deduced amino acid sequence of ORFV2-VEGF was aligned with the sequences of VEGF121 (10), VEGF165 (49), PlGF (12), VEGF-B167 (13), and truncated sequences of VEGF-C (50) and VEGF-D (15). The residues that show identity with ORFV2-VEGF are boxed. The conserved cysteine residues of the cystine knot motif are indicated with an asterisk. The signal sequence, as determined by N-terminal sequencing, is indicated by the line above the sequence. The potential sites of N-and O-linked glycosylation are indicated by a bracket and dashed line, respectively. The VEGF homology domain is indicated by arrows.
Expression and Purification of ORFV2-VEGF.
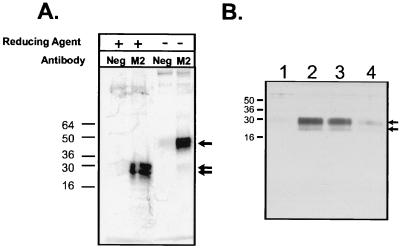
To determine the function of ORFV2-VEGF, the coding region of the VEGF-like gene was cloned into a eukaryotic expression vector such that the expressed product would have the Flag octapeptide at the C terminus. This protein (ORFV2-VEGF) was transiently expressed in COS cells and biosynthetically labeled with [35S]Cys/Met. The labeled Flag fusion protein in the conditioned cell culture medium was precipitated with mAb M2 (anti-Flag gel) and analyzed by using SDS/PAGE (Fig. 2a). Under nonreducing conditions, a band of ≈44–48 kDa was observed, whereas under reducing conditions two faster-migrating bands of ≈23–26 kDa were seen (Fig. 2a). The bands detected are consistent with ORFV2-VEGF being a disulfide-linked homodimer with a monomeric molecular mass of ≈25 kDa. This is larger than the predicted size of 13,456 Da for ORFV2-VEGF and suggests modification by glycosylation. Examination of the ORFV2-VEGF sequence reveals one potential N-linked glycosylation site (Asn85–Th87) and two potential O-linked glycosylation sites (Thr121–Thr125). N-glycanase treatment reduced the size of the ORFV2-VEGF monomer by ≈5 kDa (data not shown). The remaining size difference, we believe, is caused by O-linked glycosylation, the consensus sequences for which are present in the threonine/proline-rich C terminus of ORFV2-VEGF.
Figure 2.
Analysis and purification of expressed ORFV2-VEGF polypeptides. An ORFV2-VEGF-Flag was expressed in COS cells by transient transfection. (A) Cells were biosynthetically labeled with [35S]Cys/Met for 4 h. Conditioned medium from these cells was immunoprecipitated with either M2-gel or control beads, and the washed beads were eluted with SDS/PAGE sample buffer under reducing (2% 2-mercaptoethanol) or nonreducing conditions. The single arrow indicates the nonreduced form of ORFV2-VEGF, and the double arrows indicate the two species of the reduced form. (B) Conditioned medium from COS cells transfected with ORFV2-VEGF DNA was purified on M2-gel and eluted with free Flag peptide. The fractions (1, void; 2, 3, and 4, purified protein) eluted from the M2-gel were collected, combined with 2× SDS/PAGE sample buffer (reducing), boiled, and resolved by using SDS/PAGE (4–20% gradient). Proteins were identified by using silver staining. Molecular weight markers are indicated. Purified proteins are indicated by arrows.
Unlabeled ORFV2-VEGF was enriched from the conditioned medium of transfected COS cells by affinity chomatography with M2 resin followed by elution with Flag peptide. Analysis of this material by SDS/PAGE and silver staining (Fig. 2b) or Western blotting with anti-Flag mAbs (data not shown) demonstrated species of the same molecular mass as seen following biosynthetic labeling. N-terminal sequencing of the secreted purified protein demonstrated a single sequence identical to the deduced amino acid sequence from residue 21 to 43 (28) and confirmed the prediction that ORFV2-VEGF is a protein with a signal sequence of 20 aa.
ORFV2-VEGF Is a Ligand for VEGFR-2.
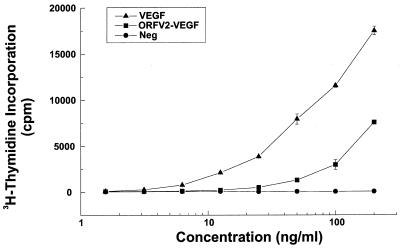
ORFV2-VEGF was tested in a bioassay, which detects ligands for VEGFR-2 (Fig. 3). The bioassay made use of Ba/F3 cells, into which we had introduced a chimeric receptor consisting of the extracellular domain of mouse VEGFR-2 and the transmembrane and cytoplasmic domains of the EpoR (15). Activation of the chimeric receptor rescues the cells from their dependence on IL-3 and causes the cells to proliferate in the absence of IL-3. VEGF, VEGF-CΔNΔC (the VEGF homology domain of VEGF-C), and VEGF-DΔNΔC (the VEGF homology domain of VEGF-D), which are all ligands for VEGFR-2, stimulate growth of this cell line in a specific and dose-dependent fashion (ref. 15; S.A.S, M. Achen, and K.A., unpublished data). Purified ORFV2-VEGF was able to induce detectable DNA synthesis in the bioassay cell line at a concentration of 25 ng/ml. By comparison VEGF, was able to induce DNA synthesis in the bioassay cell line from a concentration of 5 ng/ml. Overall, ORFV2-VEGF was about four-fold less potent in the bioassay compared with mouse VEGF. These results clearly demonstrate that ORFV2-VEGF can bind to and cross-link the extracellular domain of VEGFR-2 and induce a proliferative response.
Figure 3.
ORFV2-VEGF binds VEGFR-2 and activates a VEGFR-2 specific bioassay. Purified ORFV2-VEGF was tested for its ability to induce proliferation of Ba/F3 cells expressing a chimeric VEGFR-2. Bioassay cells were washed free of IL-3 and resuspended in dilutions of the ORFV2-VEGF, purified mouse VEGF164, or medium alone for 48 h at 37°C in a humidified atmosphere of 10% CO2. DNA synthesis was quantitated by [3H]thymidine incorporation and β counting. Values are expressed as mean ± SD and are representative of four experiments.
ORFV2-VEGF Binds to the Soluble VEGFR-2 and NP-1 Extracellular Domains.
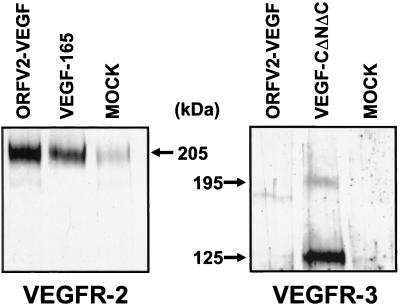
To further assess the interactions between ORFV2-VEGF and the VEGFRs, ORFV2-VEGF was tested for its capacity to bind to soluble Ig fusion proteins containing the extracellular domains of human VEGFR-1, VEGFR-2, and VEGFR-3. The fusion proteins, designated VEGFR-1-Ig, VEGFR-2-Ig, and VEGFR-3-Ig, were transiently expressed in 293 EBNA cells, precipitated from conditioned medium by using protein A-Sepharose, and incubated with metabolically labeled VEGF, VEGF-DΔNΔC, and ORFV2-VEGF. Labeled proteins that were bound to the Ig fusion proteins were analyzed by using SDS/PAGE (Fig. 4a). Polypeptides corresponding to the size of ORFV2-VEGF were precipitated by VEGFR-2-Ig from the medium of cells expressing ORFV2-VEGF. In contrast, VEGFR-1-Ig or VEGFR-3-Ig precipitated no proteins from this medium. As expected, a polypeptide of ≈24 kDa was precipitated by VEGFR-1-Ig and VEGFR-2-Ig from the medium of cells expressing mouse VEGF164 but was not precipitated by VEGFR-3-Ig. Also, as expected, a polypeptide of ≈22 kDa was precipitated by VEGFR-2-Ig and VEGFR-3-Ig from the medium of cells expressing VEGF-DΔNΔC but was not precipitated by VEGFR-1-Ig. No labeled polypeptides were precipitated by the three fusion proteins from the medium of cells transfected with the expression vector lacking sequences encoding VEGFs. ORFV2-VEGF also was tested for its ability to bind the NP-1 receptor, a recently reported ligand for VEGF (23) (Fig. 4b). The NP-1-Ig fusion protein was able to precipitate both VEGF165 and ORFV2-VEGF. Inclusion of 2 μg of unlabeled VEGF165 was able to abolish ORFV2-VEGF binding to NP-1, indicating that these factors have an overlapping site in the receptor (data not shown). Taken together, these data indicate that the ORFV2-VEGF can bind to VEGFR-2 and NP-1 but not to VEGFR-1 or VEGFR-3.
Figure 4.
VEGFR binding specificity of ORFV2-VEGF. Soluble fusion proteins consisting of the extracellular domain of VEGFRs and the Fc portion of human IgG1 were used to assess the receptor-binding specificity of ORFV2-VEGF. Biosynthetically labeled conditioned medium derived from 293EBNA cells transfected with ORFV2-VEGF, mouse VEGF164, human VEGF165, human VEGF-DΔNΔC, human VEGF-C156S, or vector alone were immunoprecipitated with VEGFR-1-Ig, VEGFR-2-Ig, VEGFR3-Ig, or NP-1 fusion proteins bound to protein A. Labeled growth factors immunoprecipitated with VEGFR-1-Ig, VEGFR-2-Ig, VEGFR3-Ig fusion proteins and M2-gel (anti-Flag) (A) or NP-1-Ig and VEGFR-1-Ig (B) were eluted from washed beads with SDS/PAGE sample buffer and resolved by using SDS/PAGE (15%). Note that only the ORFV2-VEGF and VEGF-DΔNΔC are Flag-tagged. Dried gels were visualized by using PhosphorImager analysis. The fusion protein used for each precipitation is listed above each panel.
ORFV2-VEGF Activates VEGFR-2.
The ability of ORFV2-VEGF to induce tyrosine phosphorylation of human VEGFR-2 and human VEGFR-3 was examined. For this purpose, ORFV2-VEGF was used to stimulate either PAE cells expressing VEGFR-2 or NIH 3T3 cells expressing VEGFR-3. After stimulation, cells were lysed, and VEGFR-2 or VEGFR-3 were immunoprecipitated and analyzed by Western blot analysis with phosphotyrosine-specific mAbs. ORFV2-VEGF stimulated tyrosine kinase phosphorylation of VEGFR-2 but not of VEGFR-3 (Fig. 5). As expected, the positive control proteins VEGF165 and VEGF-CΔNΔC were able to induce phosphorylation of VEGFR2 and VEGFR3, respectively (14, 15). These data demonstrate that ORFV2-VEGF can specifically induce the phosphorylation of VEGFR-2.
Figure 5.
Autophosphorylation of VEGFRs after stimulation by recombinant ORFV 2-VEGF. PAE cells expressing VEGFR-2 or NIH 3T3 cells expressing VEGFR-3 were made quiescent by starvation overnight in growth medium containing 0.2% BSA. The cells were stimulated with either ORFV 2-VEGF (100 ng/ml), VEGF165 (50 ng/ml), VEGF-CΔNΔC (100 ng/ml), or mock medium, lysed, and immunoprecipitated by using receptor-specific antibodies. The immunoprecipitates were analyzed by using phosphotyrosine immunoblotting. The apparent molecular weights of the tyrosyl phosphorylated VEGFR-2 and VEGFR-3 polypeptides are shown.
ORFV2-VEGF Is Mitogenic for Endothelial Cells.
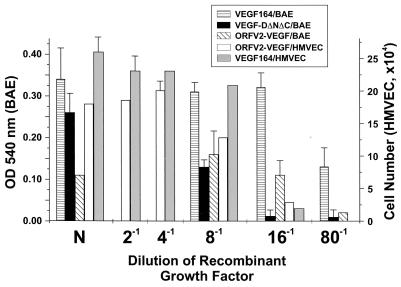
Members of the VEGF family of proteins show variable degrees of mitogenicity for endothelial cells. The mitogenic capacity of ORFV2-VEGF was tested by using BAEs and HMVECs. The cells were exposed to ORFV2-VEGF for 3 days before being assessed for growth. ORFV2-VEGF was able to stimulate an increase in the number of cells after 3 days compared with medium that did not contain added growth factor for both BAE and HMVEC (Fig. 6). Control proteins VEGF164 and VEGF-DΔNΔC also stimulated the endothelial cells. ORFV2-VEGF was therefore capable of inducing proliferation of both BAE and HMVEC cells with about a four-fold reduced potency to VEGF.
Figure 6.
Mitogenic activity of purified ORFV2-VEGF on endothelial cells. BAEs and HMVECs were treated with either purified ORFV2-VEGF, mouse VEGF164, or VEGF-DΔNΔC diluted in medium with reduced serum and without added growth factors (in the case of HMVEC). The factors were used either neat (N) or diluted 1:2, 1:4, 1:8, 1:16, or 1:80 prior to use. The neat concentration of factor was (VEGF164/BAE, 80 ng/ml; VEGF-DΔNΔC/BAE 80 ng/ml; ORFV2-VEGF/BAE, 80 ng/ml; ORFV2-VEGF/HMVEC 400 ng/ml; and VEGF164/HMVEC, 200 ng/ml). Note that the BAE have been tested at N, 1:8, 1:16, and 1:80 and the HMVEC cells at N, 1:2, 1:4, 1:8, and 1:16 only. After 72 h, the amount of cellular proliferation was quantitated by using either MTT substrate or counting. Values expressed (mean ± standard errors, representative of three experiments) are the cell number or proliferation above that were induced by the assay medium alone.
ORFV2-VEGF Induces Vascular Permeability.
Because orf virus lesions are characterized by swelling and fluid accumulation, the purified ORFV2-VEGF was tested for its ability to induce vascular permeability in a Miles assay. Guinea pigs were injected intradermally with aliquots of ORFV2-VEGF containing from 8 to 66 ng of factor (Fig. 7 a and b). In comparison to medium, alone there was detectable and dose-dependent permeability induced by the ORFV2-VEGF. ORFV2-VEGF is approximately five-fold less potent as a vascular permeability factor than mouse VEGF164.
Figure 7.
Vascular permeability activity of ORFV2-VEGF in the Miles assay. Anesthetized guinea pigs were given intracardiac injections of Evan’s Blue dye. Purified ORFV2-VEGF, mouse VEGF164, and appropriate controls were diluted in medium and applied (150 μl) intradermally to shaved areas on the back of the animal. After 30 min, the animals were sacrificed, and the skin was excised (A) and eluted in formamide, and the absorbance reading at 620 nm (B) was recorded. Values are expressed as mean ± SD and are representative of three experiments.
DISCUSSION
The pustular dermatitis induced by orf virus infection is characterized by dilation of blood vessels, swelling of the local area, and marked proliferation of endothelial cells lining the blood vessels. These features are seen in all species infected by orf (sheep, goats, and humans), and can result in the formation of a “tumor-like” growth or nodule caused by viral replication in epidermal cells. The discovery that orf viruses encode molecules with VEGF-like sequences (28) raises the important question of whether these proteins are capable of binding to mammalian VEGF-receptors and inducing characteristic VEGF-like effects such as mitogenesis of endothelial cells and vascular permeability, features that predominate the orf virus lesion. Our results demonstrate that ORFV2-VEGF is a biologically active member of the VEGF family. ORFV2-VEGF is mitogenic for endothelial cells and can induce vascular permeability with a potency that is similar to VEGF164. We have shown that ORFV2-VEGF is able to bind VEGFR-2 as a soluble binding domain or in the context of a cell-surface receptor (VEGFR-2 bioassay), where it can induce cross-linking of receptors to initiate downstream events that follow activation of the chimeric molecule. ORFV2-VEGF also is capable of specifically inducing activation by stimulating autophosphorylation of the VEGFR2 catalytic domain. The ability to induce receptor phosphorylation is an important step in the signaling mechanism of a receptor tyrosine kinase. Together, these data suggest that ORFV2-VEGF is capable of inducing activation of the VEGFR-2 signaling pathway analogous to VEGF stimulation. It would therefore be expected that ORFV2-VEGF would be capable of inducing the proliferation of endothelial cells, as VEGFR-2 appears to be a major mediator of such activity. The ability of ORFV2-VEGF to induce vascular permeability, combined with its restricted receptor binding specificity, implies that VEGFR-2 is a strong candidate for mediating vascular permeability in the VEGFR family, as has been previously suggested by analysis of VEGF-C mutants (40); the presence of a novel receptor-mediating permeability cannot, however, be formally excluded. Nevertheless, although ORFV2-VEGF binds VEGFR-2, it does not recognize the other tyrosine kinase receptors, VEGFR-1 and VEGFR-3. ORFV2-VEGF does, however, bind to the recently described VEGF receptor NP-1. This profile of receptor recognition by ORFV2-VEGF is unique among the VEGF family. Recent structural analyses of human VEGF identified residues thought to be important in binding to VEGFR-1 and VEGFR-2 (42–45). In light of the receptor-binding properties of ORFV2-VEGF, it is intriguing that the VEGF residues implicated as being critical in binding to VEGFR-1 are partially conserved in ORFV2-VEGF, whereas those involved in VEGFR-2 binding are not (42). Data from experiments to determine the crystal structure of VEGF and the predicted residues critical for binding (Phe-17, Ile-46, Glu-64, Gln-79, and Ile-83 for VEGFR-2; Asp-63 and Glu-64 for VEGFR-1) also suggest that some of the major residues required for VEGFR-1 interaction are conserved in ORFV2-VEGF, whereas residues important for VEGF interaction with VEGFR-2 are poorly conserved. The mechanism whereby ORFV2-VEGF binds to VEGFR-2 and NP-1 is clearly of interest; the lack of conservation of key residues suggests that the binding site for ORFV2-VEGF is different to that of VEGF. It will be of interest to see whether blocking the activity of ORFV2-VEGF will alter the lesion induced by orf virus infection.
Our original report of a VEGF-like gene in orf virus was based on sequence similarities with the isoforms of VEGF and with PlGF (28). Since that time, additional members of the VEGF family have been discovered. Analysis of these sequences shows that the highest level of sequence relatedness with VEGF family members is with VEGF itself (43%). This raises the possibility that ORFV2-VEGF is derived from the VEGF gene and that sequence divergence has resulted in loss of the capability to bind VEGFR-1. However, because another orf virus gene (a homolog of IL-10), which is likely to have been derived by capture of a host gene, shows 80% amino acid sequence identity to its mammalian counterpart (46), it is possible that the ORFV2-VEGF is derived from another as-yet-unidentified mammalian VEGF family member. These predictions are complicated by the presence of a surprisingly variant form of the viral VEGF in another strain of orf virus. Strain NZ7 encodes a protein that has only 23% amino acid identity with human VEGF and only 43% identity with ORFV2-VEGF (28). It has recently been shown that the VEGF-like protein from the NZ7 virus (VEGF-E) binds only to VEGFR2 (and not VEGFR1) and can induce mitogenesis and permeability (47). It is unclear whether the extensive sequence difference between the two viral proteins represents independent acquisition events from different sources or very extensive divergence from a single progenitor.
It seems likely that the biological activities of ORFV2-VEGF reported here contribute to the proliferative and highly vascularized nature of orf virus lesions. This is supported by our analysis of a recombinant orf virus in which the gene encoding ORFV2-VEGF has been deleted. Comparisons of lesions resulting from infection of sheep by wild-type and recombinant ORFV2-VEGF-deficient orf virus indicate that in the absence of ORFV2-VEGF, skin lesions are significantly less vascularized (unpublished data).
The identification of a viral VEGF protein that is capable of subverting mammalian VEGF receptors to aid in its viral infection also raises the possibility that other viruses may act in a similar fashion. A number of recently described viruses (Ebola, morbillivirus; ref. 48) produce lesions in which substantial changes to the integrity of blood vessels are observed. Analysis of other viral genomes for VEGF family members may broaden our understanding of endothelial cell activation during viral infection and provide a greater insight into the role of VEGF/VEGFR in pathological conditions.
Acknowledgments
We thank Clare MacFarlane for supplying expression vectors; R. Moritz for sequencing of ORFV2-VEGF; E. Korpelainen, Y. Gunji, and K. Pajusola for supplying plasmid constructs encoding VEGFR-Ig fusion proteins; Brendan Classon for identifying O-linked glycosylation sites; and Marc Achen, Margaret Hibbs, and Antony Burgess for critical reading of the manuscript.
ABBREVIATIONS
- KDR human VEGFR2
NP-1, neuropilin
- NZ2
New Zealand strain 2
- ORFV2-VEGF
VEGF-like protein from orf strain NZ2
- PlGF
placenta growth factor
- VEGF
vascular endothelial growth factor
- VEGFR
vascular endothelial growth factor receptor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Ferrara N, Henzel W J. Biochem Biophys Res Commun. 1989;161:851–858. doi: 10.1016/0006-291x(89)92678-8. [DOI] [PubMed] [Google Scholar]
- 2.Leung D W, Cachianes G, Kuang W J, Goeddel D V, Ferrara N. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 3.Senger D R, Galli S J, Dvorak A M, Perruzzi C A, Harvey V S, Dvorak H F. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- 4.Risau W. Nature (London) 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea K S, Powel-Braxton L, Hillan K J, Moore M W. Nature (London) 1996;380:439–443. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 6.Carmeliet P, Ferreira V, Breier G, Pollofeyt S, Keickens L, Gertenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Nature (London) 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim K J, Li B, Winer J, Armanini M, Gillett N, Phillips H S, Ferrara N. Nature (London) 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 8.Saleh M, Stacker S A, Wilks A F. Cancer Res. 1996;56:393–401. [PubMed] [Google Scholar]
- 9.Houck K A, Ferrara N, Winer J, Cachianes G, Li B, Leung D W. Mol Endocrinol. 1991;5:1806–1814. doi: 10.1210/mend-5-12-1806. [DOI] [PubMed] [Google Scholar]
- 10.Tischer E, Mitchell R, Hartman T, Silva M, Gospodarowicz D, Fiddes J C, Abraham J A. J Biol Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 11.Poltorak Z, Cohen T, Sivan R, Kandelis Y, Spira G, Vlodavsky I, Keshet E, Neufeld G. J Biol Chem. 1997;272:7151–7158. doi: 10.1074/jbc.272.11.7151. [DOI] [PubMed] [Google Scholar]
- 12.Maglione D, Guerriero V, Viglietto G, Delli Bovi P, Persico M G. Proc Natl Acad Sci USA. 1991;88:9267–9271. doi: 10.1073/pnas.88.20.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olofsson B, Pajusola K, Kaipainen A, von Euler G, Joukov V, Saksela O, Orpana A, Pettersson R F, Alitalo K, Eriksson U. Proc Natl Acad Sci USA. 1996;93:2576–2581. doi: 10.1073/pnas.93.6.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joukov V, Pajusola K, Kaipainen A, Chilov D, Lahtinen I, Kukk E, Saksela O, Kalkkinen N, Alitalo K. EMBO J. 1996;15:290–298. [PMC free article] [PubMed] [Google Scholar]
- 15.Achen M G, Jeltsch M, Kukk E, Mäkinen T, Vitali A, Wilks A F, Alitalo K, Stacker S A. Proc Natl Acad Sci USA. 1998;95:548–553. doi: 10.1073/pnas.95.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mustonen T, Alitalo K. J Cell Biol. 1995;129:895–898. doi: 10.1083/jcb.129.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achen M G, Stacker S A. Int J Exp Pathol. 1998;79:255–265. doi: 10.1046/j.1365-2613.1998.700404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risau W, Flamme I. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 19.De Vries C, Escobedo J A, Ueno H, Houck K, Ferrara N, Williams L T. Science. 1992;255:989–991. doi: 10.1126/science.1312256. [DOI] [PubMed] [Google Scholar]
- 20.Quinn T P, Peters K G, De Vries C, Ferrara N, Williams L T. Proc Natl Acad Sci USA. 1993;90:7533–7537. doi: 10.1073/pnas.90.16.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J E, Chen H H, Winer J, Houck K A, Ferrara N. J Biol Chem. 1994;269:25646–25654. [PubMed] [Google Scholar]
- 22.Olofsson B, Korpelainen E, Pepper M S, Mandriota S J, Aase K, Kumar V, Gunji Y, Jeltsch M M, Shibuya M, Alitalo K, Eriksson U. Proc Natl Acad Sci USA. 1998;95:11709–11714. doi: 10.1073/pnas.95.20.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soker S, Takashima S, Miao H Q, Neufeld G, Klagsbrun M. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 24.Migdal M, Huppertz B, Tessler S, Comforti A, Shibuya M, Reich R, Baumann H, Neufeld G. J Biol Chem. 1998;273:22272–22278. doi: 10.1074/jbc.273.35.22272. [DOI] [PubMed] [Google Scholar]
- 25.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Nature (London) 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 26.Fong G-H, Rossant J, Gertsenstein M, Breitman M L. Nature (London) 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 27.Dumont D J, Jussila L, Taipale J, Lymboussaki A, Mustonen T, Pajusola K, Breitman M, Alitalo K. Science. 1998;282:946–949. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 28.Lyttle D J, Fraser K M, Fleming S B, Mercer A A, Robinson A J. J Virol. 1994;68:84–92. doi: 10.1128/jvi.68.1.84-92.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry, M. & McFadden, G. (1997) Parasitology 115 Suppl., S89–S100. [DOI] [PubMed]
- 30.Smith G L, Symons J A, Khanna A, Vanderplasschen A, Alcami A. Immunol Rev. 1997;159:137–154. doi: 10.1111/j.1600-065x.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 31.Spriggs M K. Annu Rev Immunol. 1996;14:101–130. doi: 10.1146/annurev.immunol.14.1.101. [DOI] [PubMed] [Google Scholar]
- 32.Haig D M, Mercer A A. Vet Res. 1998;29:311–326. [PubMed] [Google Scholar]
- 33.Groves R W, Wilson Jones E, MacDonald D M. J Am Acad Dermatol. 1991;25:706–711. doi: 10.1016/0190-9622(91)70257-3. [DOI] [PubMed] [Google Scholar]
- 34.Wheeler C E, Cawely E P. Am J Pathol. 1956;32:535–545. [PMC free article] [PubMed] [Google Scholar]
- 35.Savage J, Black M M. Proc R Soc Med. 1972;65:766–768. doi: 10.1177/003591577206500918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan S T, Blake G B, Chambers S. Br J Plast Surg. 1991;44:465–467. doi: 10.1016/0007-1226(91)90209-3. [DOI] [PubMed] [Google Scholar]
- 37.Aruffo A, Seed B. Proc Natl Acad Sci USA. 1987;84:8573–8577. doi: 10.1073/pnas.84.23.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joukov V, Sorsa T, Kumar V, Jeltsch M, Claesson-Welsh L, Cao Y, Saksela O, Kalkkinen N, Alitalo K. EMBO J. 1997;16:3898–3911. doi: 10.1093/emboj/16.13.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Joukov V, Kumar V, Sorsa T, Arighi E, Weich H, Saksela O, Alitalo K. J Biol Chem. 1998;273:6599–6602. doi: 10.1074/jbc.273.12.6599. [DOI] [PubMed] [Google Scholar]
- 41.Miles A A, Miles E M. J Physiol (London) 1952;118:228–257. doi: 10.1113/jphysiol.1952.sp004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keyt B A, Nguyen H V, Berleau L T, Duarte C M, Park J, Chen H, Ferrara N. J Biol Chem. 1996;271:5638–5646. doi: 10.1074/jbc.271.10.5638. [DOI] [PubMed] [Google Scholar]
- 43.Muller Y A, Li B, Christinger H W, Wells J A, Cunningham B C, de Vos A M. Proc Natl Acad Sci USA. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller Y A, Christinger H W, Keyt B A, de Vos A M. Structure. 1997;5:1325–1338. doi: 10.1016/s0969-2126(97)00284-0. [DOI] [PubMed] [Google Scholar]
- 45.Wiesmann C, Fuh G, Christinger H W, Eigenbrot C, Wells J A, de Vos A M. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 46.Fleming S B, McCaughan C A, Andrews A E, Nash A D, Mercer A A. J Virol. 1997;71:4857–4861. doi: 10.1128/jvi.71.6.4857-4861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa S, Oku A, Sawano A, Yamaguchi S, Yazaki Y, Shibuya M. J Biol Chem. 1998;273:31273–31282. doi: 10.1074/jbc.273.47.31273. [DOI] [PubMed] [Google Scholar]
- 48.Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B, et al. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 49.Weindel K, Marme D, Weich H A. Biochem Biophys Res Commun. 1992;183:1167–1174. doi: 10.1016/s0006-291x(05)80313-4. [DOI] [PubMed] [Google Scholar]
- 50.Lee J, Gray A, Yuan J, Luoh S M, Avraham H, Wood W I. Proc Natl Acad Sci USA. 1996;93:1988–1992. doi: 10.1073/pnas.93.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]