Abstract
The Hdm2 oncoprotein inhibits p53 functions by two means: (i) it blocks p53’s transactivation activity and (ii) it targets p53 for degradation in a proteasome-dependent manner. Recent data indicate that Hdm2 shuttles between the nucleus and the cytoplasm and that the regulation of p53 levels by Hdm2 requires its nuclear export activity. Two different models are consistent with these observations. In the first, Hdm2 binds to p53 in the nucleus and shuttles p53 from the nucleus to the cytoplasm, and then it targets p53 to the cytoplasmic proteasome. Alternatively, Hdm2 and p53 could be exported separately from the nucleus and then associate in the cytoplasm, where Hdm2 promotes the degradation of p53. To distinguish between these two models, several Hdm2 mutants were employed. Hdm2NLS lacks the ability to enter the nucleus, whereas Hdm2NES is deficient in nuclear export. Hdm2NLS, Hdm2NES, or the combination of both mutants were unable to promote p53 degradation in the cotransfected 2KO cells (which were null for both the p53 and mdm2 genes), although wild-type Hdm2 efficiently reduced p53 levels under the same conditions. This observation is not a result of the differences in expression levels or stability between Hdm2 and these mutants. Moreover, coexpression of these mutants had no effect on wild-type Hdm-2-induced p53 destabilization. Thus, Hdm2 must shuttle p53 from the nucleus to the cytoplasm to target it for degradation in the cytoplasm.
The p53 tumor suppressor is a key component of cellular mechanisms that serve to maintain genomic integrity in response to an array of cellular stress (1, 2). P53 is a short-lived protein that is maintained at low levels in normal cells. On stimulation by cellular stress such as DNA damage, p53 is transiently stabilized (3) and activated as a transcriptional factor in the nucleus, where it acts to induce the transcription of a set of genes that includes p21WAF1/CIP1, bax, and hdm2 (mdm2 in mouse) (4–9). Whereas the products of p21WAF1/CIP1 and bax mediate the antiproliferative function of p53 by blocking cell cycle progression and provoking cell elimination (4, 7), hdm2 encodes a cellular negative regulator of p53 and acts as an oncogene (10–14). The Hdm2 oncoprotein directly binds to the transactivation domain of p53 and abrogates the transactivation activity of p53 (11, 15, 16). In addition, Hdm2 promotes the degradation of p53 (17, 18). Thus, Hdm2 forms an autoregulatory feedback loop with p53, in which p53 can control both its own levels and its own activity by inducing the expression of Hdm2 (17–19). However, the mechanism by which Hdm2 mediates p53 degradation remains enigmatic.
It has been shown that Hdm2-mediated p53 degradation occurs through a proteasome-dependent pathway in the cytoplasm (17, 18). Honda et al. (20) reported that Hdm2 could function as an E3 ubiquitin ligase. Recent data have shown that Hdm2 consistently shuttles between the nucleus and the cytoplasm (21). Moreover, inhibition of nuclear export of Hdm2 results in the accumulation of p53 (21, 22), suggesting that Hdm2 shuttles p53 from the nucleus to the cytoplasm and then targets p53 for degradation in the cytoplasm. However, these data do not preclude another model in which p53 itself shuttles from the nucleus to the cytoplasm in an Hdm2-independent manner, and Hdm2 only acts in the cytoplasm to induce the degradation of p53 (J. Stommel and G. Wahl, personal communication). Employing a number of mutants of Hdm2 that are defective in either nuclear import or nuclear export, we demonstrate in this report that the nucleocytoplasmic shuttling of Hdm2 is essential for its ability to promote p53 degradation. Therefore, Hdm2 must shuttle p53 from the nucleus to the cytoplasm to target p53 to the cytoplasmic proteasome. These results point to a novel mechanism by which protein levels are regulated, and this could provide a new therapeutic strategy to restore p53 levels in the cancer cells in which Hdm2 is overexpressed.
MATERIALS AND METHODS
Plasmids.
pCMVhdm2 (encoding wild-type Hdm2) (11) was mutated within the putative nuclear localization signal (NLS)-coding region by using the QuikChange kit for site-directed mutagenesis (Stratagene) and two complementary oligonucleotides, 5′-CTGGTGAACGACAAACAAAACGCCACAAA-TCTGATAG-3′ and 5′-CTATCAGATTTGTGGAGTTTTGTTTGTCGTT-CACCAG-3′. This resulted in mutations of amino acids R181 and R183 to threonine and leucine, respectively. The expression plasmids for Hdm2NES, Hdm2Δ150–230, Hdm2G58A, Hdm2D68A, and the parental plasmid pCMV-neo-bam3 (pCMV) were described previously (11, 21, 23). p53 expression plasmid was pCMV-neo-bam-p53 (encoding human wild-type p53).
Cells and Transfection.
2KO cell line was derived from mouse embryo fibroblasts that lacked the p53 and mdm2 genes (provided by Guillermina Lozano). Both 2KO and H1299 cell lines were maintained in DMEM containing 10% FBS and transfected by using Lipofectamine (GIBCO/BRL) according to the manufacturer’s instructions.
Immunoblotting.
2KO cells (6 × 105) were cotransfected with 1 μg of pCMV-neo-bam-p53 and 5 μg of pCMVhdm2 or the Hdm2 mutant expression plasmid(s). Cells were collected 24 hours after transfection. Proteins were separated on 8% SDS/polyacrylamide gels (50 μg per lane) and then transferred to nitrocellulose membranes (Amersham Pharmacia). The p53 and Hdm2 proteins were detected by usng PAb421 and 4B11 monoclonal hybridoma supernatants respectively. Immunoblotting was performed as described (24).
Immunofluorescence.
Two micrograms of plasmid(s) encoding wild-type Hdm2 or its mutants was (co)transfected into 2KO or H1299 cells in a 35-mm dish. The transfected cells were washed with PBS and fixed with 100% methanol 24 hours after transfection. The Hdm2 protein and its mutants were detected by 2A10 monoclonal hybridoma supernatant as described (22). Cellular staining was observed by using a fluorescent microscope.
RESULTS
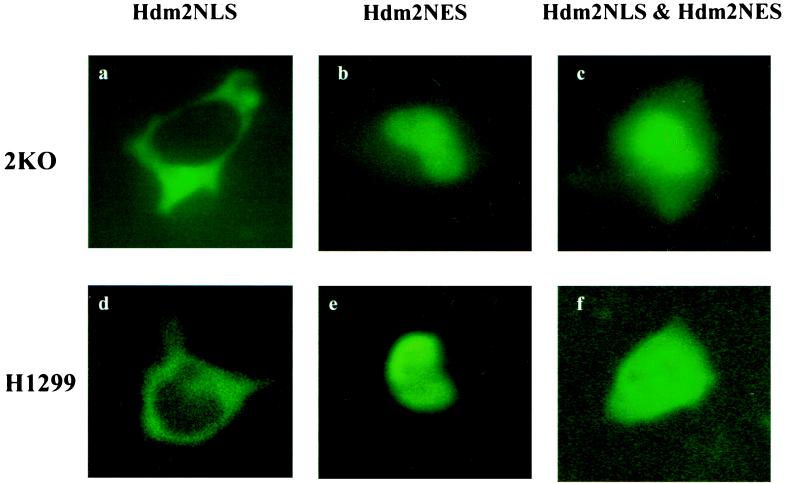
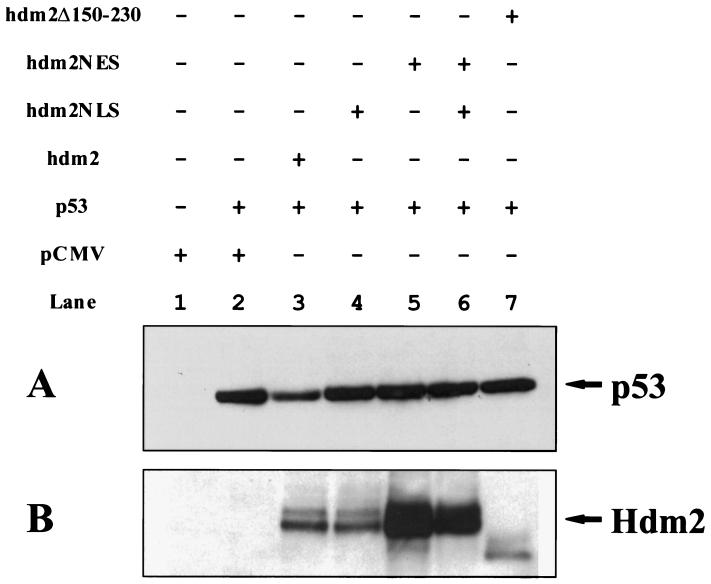
The Hdm2 oncoprotein contains a number of functional domains (25), including a p53-binding domain (10), a leucine-rich nuclear export signal (NES) (21), and a NLS (13). Whereas the NES is necessary and sufficient to direct nuclear export of Hdm2 (21), the NLS has been shown to mediate the transport of Hdm2 into the nucleus (11). It has been shown that both the p53-binding domain and the NES are required for Hdm2’s ability to induce p53 degradation (17, 18, 21). Blocking nuclear export of Hdm2 stabilized p53 in the nucleus (21, 22), suggesting that Hdm2 must associate with p53 and migrate from the nucleus to the cytoplasm to carry out this function. If p53 shuttles from the nucleus to the cytoplasm independently of Hdm2 (J. Stommel and G. Wahl, personal communication) and Hdm2 only acts in the cytoplasm to promote p53 degradation, then the cytoplasmic-localized Hdm2 should be able to perform this function. To test this possibility, an Hdm2 mutant, termed Hdm2NLS, was generated by introducing two point mutations into the codons for the NLS of Hdm2 to change two amino acids (R181 and R183) to threonine and leucine, respectively (Fig. 1). Because such basic amino acids are conserved among the NLSs of many nuclear proteins and are crucial for nuclear localization of those molecules (Fig. 1) (26), these mutations are expected to abolish Hdm2’s ability to enter the nucleus. Cellular localization of Hdm2NLS was determined by using immunofluorescence. As expected, Hdm2NLS is exclusively located in the cytoplasm of both 2KO (null for p53 and Mdm2) and H1299 cells (null for p53) (Fig. 2 a and d), whereas wild-type Hdm2 is largely localized to the nucleus (data not shown), indicating that R181 and R183 are essential for nuclear import of Hdm2. Hdm2NES, another mutant of Hdm2 (21), contains point mutations within the NES that eliminate its ability to be exported from the nucleus to the cytoplasm and is localized entirely in the nucleus (Fig. 2 b and e). Coexpression of Hdm2NLS and Hdm2NES does not alter their cellular localization (Fig. 2 c and f). To avoid ambiguity in the interpretation of experimental results that might be imposed by endogenous Hdm2, activity of the Hdm2 mutants in triggering p53 degradation was determined in the 2KO cell line that was obtained by passaging on a 3T3 schedule primary mouse fibroblasts derived from a mouse null for both the mdm2 and p53 genes. 2KO cells were cotransfected with an expression plasmid encoding human wild-type p53 and an empty vector (pCMV), a pCMV-hdm2 vector, or its mutant expression plasmids separately or a combination of the plasmids for Hdm2NLS and Hdm2NES (Fig. 3). Twenty-four hours after transfection, steady-state levels of the p53 protein in the transfected cells were determined by using Western blot analysis. As shown in Fig. 3A, the amount of p53 was markedly reduced in cells cotransfected with the plasmid encoding wild-type Hdm2 (Fig. 3A, compare lanes 2 and 3). On the contrary, coexpression of either Hdm2NLS or Hdm2NES alone or both of them with p53 did not affect the detectable amounts of p53 in these cells (Fig. 3A, compare lanes 2 with 4, 5, and 6). Similarly, Hdm2Δ150–230, a deletion mutant of Hdm2 lacking both NLS and NES, localized to the cytoplasm (11) and lost the ability to accelerate p53 degradation (Fig. 3A, compare lanes 2 and 7). In addition, two Hdm2 mutants (G58A and D68A) that did not bind to p53 (23) also failed to promote p53 degradation in the cotransfected 2KO cells (data not shown).
Figure 1.
The nuclear localization sequence in Hdm2. A part of the Hdm2 primary structure that resembles the NLSs of various nuclear proteins is aligned with these known NLSs. The basic amino acid residues that are critical for NLS function are shown in bold. Numbers refer to the position of the amino acids in the proteins. A mutant Hdm2 (Hdm2NLS) was generated with the amino acid changes (R181T, R183L).
Figure 2.
Cellular localization of Hdm2 mutants. 2KO and H1299 cells were (co)transfected with the expression plasmid(s) encoding Hdm2 mutants as indicated above the pictures. Cellular localization of Hdm2 mutants in 2KO (a–c) or H1299 (d–f) cells was determined by using immunofluorescence 24 hours after transfection as described in Materials and Methods.
Figure 3.
Effect of wild-type Hdm2 and its mutants on p53 stability. 2KO cells were cotransfected with p53 expression plasmid and the plasmids encoding either wild-type Hdm2 or its mutants as indicated above the figure. At 24 hours after transfection, steady-state levels of p53 (A) and Hdm2 (B) were determined by immunoblotting as described in Materials and Methods. Two species of the Hdm2 protein with different molecular weights were detected (B). This may be a result of different levels of phosphorylation or some other posttranslational modification. The apparent molecular weight of Hdm2Δ150–230 is lower than that of the full-length Hdm2 proteins because of the deletion (B, lane 7).
To preclude the possibility that lower expression levels or higher instability of Hdm2NLS and Hdm2NES in the transfected cells resulted in loss of their ability to target p53 for degradation, steady-state levels of these mutants and wild-type Hdm2 transfected into cells were determined by using immunoblotting. Fig. 3B shows that levels of the Hdm2 mutants were similar to or even greater than that of wild-type Hdm2 (Fig. 3B, compare lanes 3 with 4, 5, and 6).
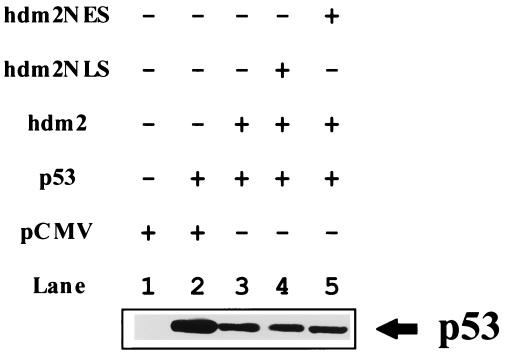
Another trivial explanation for these data is that when Hdm2NLS and Hdm2NES were coexpressed, Hdm2NES might act as a dominant suppressor of Hdm2NLS by sequestering p53 in the nucleus. If so, Hdm2NES should be able to attenuate Hdm2’s ability to induce p53 degradation. To test this possibility, the wild-type p53 plasmid was cotransfected with either wild-type Hdm2 plasmid alone or wild-type Hdm2 and Hdm2NES or Hdm2NLS plasmids together into 2KO cells. p53 levels were then determined (Fig. 4). Immunodetection showed that coexpression of Hdm2NES or Hdm2NLS did not affect wild-type Hdm2-induced reduction of p53 levels (Fig. 4). Thus, the mutant Hdm2 proteins do not act in a transdominant fashion. Taken together, these data demonstrate that nucleocytoplasmic shuttling of Hdm2 is required for its ability to trigger p53 degradation in the cytoplasm. It is not sufficient to have Hdm2 in the nucleus and cytoplasm, as it is unable to shuttle p53 between these compartments.
Figure 4.
Coexpression of either Hdm2NLS or Hdm2NES with wild-type Hdm2 has no effect on Hdm2’s ability to promote p53 degradation. 2KO cells were cotransfected with p53 expression plasmid and the Hdm2 plasmid alone or with the plasmid encoding its mutant (as indicated). At 24 hours after transfection, steady-state levels of p53 was determined by using immunoblotting as described in Materials and Methods.
DISCUSSION
These data eliminate the model in which p53 and Hdm2 independently shuttle from the nucleus to the cytoplasm and assemble there, and then p53 is degraded. This was formally possible because J. Stommel and G. Wahl have recently shown that p53 monomers/dimers could shuttle from the nucleus to the cytoplasm of a cell (J. Stommel and J. Wahl, personal communication). Why must the Hdm2–p53 heteromolecular complex itself shuttle from the nucleus to the cytoplasm to efficiently degrade p53? There are several possibilities: (i) the complex must exit from the nucleus via the nucleopore, which in turn is linked directly to the proteasome, (ii) delivery of the Hdm2–p53 complex is done by a protein complex assembled in the nucleus which is required for functioning in the cytoplasm, or (iii) Hdm2 modification of p53 occurs first in the nucleus (ubiquitin) and then acts (as a docking protein to the cytoplasmic proteasome) again in the cytoplasm. The ideas generated from the results reported here are testable and should continue to elucidate the mechanisms that regulate p53 levels in a cell.
Acknowledgments
We thank J. Stommel and J. Wahl for communication of unpublished data, Guillermina Lozano for 2KO cell line, and D. Freedman for helpful discussion. This work was supported by a grant from the National Cancer Institute (Pol-CA41086) to A.J.L.; W.T. was supported by the National Institute of Health Cancer Training Grant.
ABBREVIATIONS
- NLS
nuclear localization signal
- NES
nuclear export signal
References
- 1.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 2.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 5.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 6.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. Nature (London) 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 8.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Lin J, Levine A J. Mol Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Wu X, Lin J, Levine A J. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fakharzadeh S S, Trusko S P, George D L. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlay C A. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 16.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Nature (London) 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 20.Honda, R., Tanaka, H. & Yasuda, H. (1997) FEBS Lett.420. [DOI] [PubMed]
- 21.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman D A, Levine A J. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman D A, Epstein C B, Roth J C, Levine A J. Mol Med. 1997;3:248–259. [PMC free article] [PubMed] [Google Scholar]
- 24.Tao W, Kurschner C, Morgan J I. J Biol Chem. 1997;272:15547–15552. doi: 10.1074/jbc.272.24.15547. [DOI] [PubMed] [Google Scholar]
- 25.Piette J, Neel H, Marechal V. Oncogene. 1997;15:1001–1010. doi: 10.1038/sj.onc.1201432. [DOI] [PubMed] [Google Scholar]
- 26.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]