Abstract
The type II transforming growth factor (TGF)-β receptor gene (TGFBR2) is often mutated in nucleotide repeat sequences in colorectal cancers that are replication error positive (RER+). These mutations are thought to be selected for escape from growth inhibition by TGF-β rather than representing bystander events because of an increased mutation rate. We investigated the role of TGFBR2 mutations in 12 colorectal cancer cell lines. Six of these were RER+, and these were shown to have homozygous TGFBR2 mutations. All cell lines then were tested for changes in proliferation in response to TGF-β stimulation. Despite homozygous mutation of the type II TGF-β receptor, two RER+ cell lines, Lovo and SW48, showed statistically significant growth inhibition when stimulated by TGF-β1 in serum-free conditions. This shows that the type II TGF-β receptor can be bypassed in certain cases to maintain growth inhibition. We next investigated whether there was any alternative mode through which TGFBR2 mutation may give a selective advantage, such as a change in adhesion molecule expression. All cell lines were stimulated with TGF-β1 and adhesion molecules detected by ELISA. No consistent changes were identified between the RER+ and the RER− cell lines, although changes in E-cadherin, β-catenin, and γ-catenin were identified in individual cell lines. We conclude that (i) type II TGF-β receptor activity can be bypassed and thus TGFBR2 mutations in RER+ cancers may, at least sometimes, be just “bystander” events and (ii) TGF-β can affect adhesion molecule expression so that TGFBR2 mutation may give rise to a selective advantage through an effect other escape from growth inhibition.
Human colorectal carcinomas (CRCs) arise from normal epithelium through a process of somatic evolution involving an accumulation of mutations that confer a selective advantage to the cells in which they occur(1–3). It is possible, however, that occasionally, mutations that give neither a selective advantage nor disadvantage are incorporated in a cancer by chance. Such “bystander” mutations would normally be expected to be rare, as the mutation rate in most cancers, at least initially, is low (4). A subgroup of CRCs, however, lose DNA mismatch-repair function during tumor development, one consequence of which is an increase in the mutation rate (5, 6). Loss of mismatch repair renders a cell prone to insertion/deletion-type mutations occurring especially at sites of nucleotide repeat sequences during replication (7). These replication errors (RERs) frequently result in the generation of multiple mutant alleles at certain nucleotide repeat sequences known as microsatellites. Tumors with multiple new alleles at microsatellite loci are said to show microsatellite instability and to be replication error-positive (RER+) (8).
Microsatellites are nearly always intronic, noncoding sequences with no known function, and mutations of these can probably be regarded as neutral (i.e., without selective advantage or disadvantage). In this case, the mutant microsatellite alleles, which accumulate by chance as a result of an increased mutation rate in RER+ cancers, can be regarded as true bystander mutations.
Short runs of mono- or dinucleotide repeat sequences can also be found in the coding regions of certain tumor suppressor genes (e.g., Bax, E2F4, insulin-like growth factor (IGF) 2 receptor, and type II TGF-β receptor). Insertion/deletion mutations have been found in the repeat sequences of these genes and have been shown to occur significantly more frequently in RER+ CRCs than in RER− CRCs (9–12). Because these are frameshift mutations resulting in an altered gene product, they have always been assumed to give a selective advantage. It is equally possible, however, that, as in the case of microsatellite instability, these represent nothing more than bystander mutations.
The TGF-β superfamily of growth and differentiation factors influences a wide range of cellular processes, including epithelial cell proliferation, differentiation, extracellular matrix production, and migration (13). These factors are also involved in embryonic morphogenesis, tissue differentiation, and wound healing. The physiology of TGF-β activity is extremely complicated and is influenced by a multitude of factors, including levels of TGF-β and presence of other growth factors. At least nine different cellular TGF-β binding proteins and five different TGF-β receptors have been identified that show different patterns of expression in different tissues (13). The interactions of these receptors with TGF-β with other members of the TGF-β superfamily and with each other is not completely clear. It appears that during TGF-β activation, the ligand binds initially to the type II receptor, which then heterodimerizes with, and phosphorylates, the type I receptor, which by itself is not thought to bind the ligand directly (14).
The type II receptor gene (TGFBR2) contains an A (10) tract in exon 3 and GT (3) tracts in exons 5 and 7 (15, 16). These repeat sequences are prone to replication errors and TGFBR2 mutations occur in a very high proportion of RER+ CRCs. These mutations might be expected to give a selective advantage by providing a means of escaping TGF-β-mediated growth control. However, given the complex pattern of TGF-β signaling, it is quite possible that there is some built-in redundancy. The TGFBR2 mutations may therefore represent bystander events consequent to the RER+ phenotype.
In this study we investigated whether mutations in TGFBR2 do actually give rise to resistance to TGF-β1. We demonstrated that, in certain cases, the type II receptor can be bypassed to allow some growth inhibition by TGF-β. We also showed that TGF-β can stimulate changes in adhesion molecule expression, raising the possibility that, if TGFBR2 mutations do give a selective advantage, it may be through an effect other than growth inhibition.
MATERIALS AND METHODS
Cell Culture.
Twelve cell lines were studied in total (Table 1), and references for these are available from the authors. The cells were grown routinely in DMEM (Imperial Cancer Research Fund) with 10% fetal calf serum at an atmospheric CO2 content of 10%. Stocks of cells were maintained in 10-cm Petri dishes and passaged when the cells had reached near-confluence.
Table 1.
Genotype of cell lines
| Cell line | RER status | LOH at 3p22 |
TGFBR2 mutation
|
||
|---|---|---|---|---|---|
| Exon 3 | Exon 5 | Exon 7 | |||
| LS174T | + | No | −1/−1 | no | no |
| HCA7 | + | No | −1/−1 | no | no |
| LOVO | + | No | −1/−2 | no | no |
| SW48 | + | No | −1/−1 | no | no |
| DLD1 | + | No | −1/wt | CTG > CCG (codon 452) | no |
| HCT116 | + | No | −1/−1 | no | no |
| COLO201 | − | No | no | no | no |
| COLO205 | − | No | no | no | no |
| COLO320 | − | No | no | no | no |
| HCA46 | − | No | no | no | no |
| HT29 | − | No | no | no | no |
| CACO2 | − | No | no | no | no |
LOH, loss of heterozygosity; wt, wild type.
Cell Line Genotyping.
DNA was extracted from approximately 107 cells by using the Nucleon Bacc2 kit (Scotlab, Lankashire, U.K.), and all cell lines were genotyped as follows.
Microsatellite instability and loss of heterozygosity.
Nine microsatellite loci (D3S3613, D3S3510, D3S2338, D3S3473, D5S82, D5S346, D18S84, D16S520, and BAT-26) were examined. Details of these are given in Table 2. Primers were dye-labeled, taking into account the size of the microsatellite markers to allow several PCR products to be analyzed simultaneously. About 2 μl of pooled PCR product was mixed with 0.5 μl of Rox350 size standard (Applied Biosystems) and 1 μl of formamide loading buffer, heat-denatured, and electrophoresed on a 6% acrylamide sequencing gel on an ABI Prism 377 semiautomated sequencer for 2–4 hours. Data were analyzed by using genescan 2.0.2 software (Perkin–Elmer). Loci showing multiple alleles differing by 2 base pairs were labeled as having microsatellite instability, and the RER+ phenotype was defined as microsatellite instability at two or more of these loci.
Table 2.
Details of primers used
| Marker | GenBank database | Distance to TGFBR2, centimorgans |
|---|---|---|
| D3S3613 | 609720 | 0.09 |
| D3S3510 | 598896 | 0.09 |
| D3S2338 | 343612 | 0.52 |
| D3S3473 | 594438 | 0.09 |
| D5S82 | 180445 | N/A |
| D5S346 | 181171 | N/A |
| D18S84 | 190574 | N/A |
| D16S520 | 200145 | N/A |
| BAT-26 | 9834505 | N/A |
| TGFBR2 exon 3 | Ref. 16 | N/A |
| TGFBR2 exon 5 | Ref. 16 | N/A |
| TGFBR2 exon 7 | Ref. 16 | N/A |
Cell lines were typed for allelic loss of the TGFBR2 gene by using the markers D3S3613, D3S3510, D3S2338, and D3S3473, which are tightly linked to the TGFBR2 locus (mapping to 3p22). In the absence of control normal DNA for these cell lines, we argued that homozygosity at all four markers was probably an indication of allelic loss (the odds of this happening by chance would be about 1 in 1,000).
Sequence analysis of exons 3,5, and 7 of TGFBR2.
All of the cell lines were also examined for mutations in the TGFBR2 gene. This gene contains nucleotide repeat sequences that are prone to mutation in RER+ cancers. By using previously described primers (Table 2; refs.15 and 16), we examined these exons by direct sequencing of PCR products. PCR was performed for each exon by using the appropriate reaction conditions, and products were separated from oligonucleotides and unincorporated dNTPs by using a Sephadex G50 spin column (Amersham Pharmacia). Five microliters of purified PCR product was used in a thermocycle sequencing reaction with the Ready Reaction Dye Terminator Cycle Sequencing kit (Applied Biosystems). PAGE was performed by using standard conditions on a 377 Prism fluorescence-based, semiautomated DNA sequencer (Applied Biosystems). Sequence analysis was performed by using sequencher software (Gene Codes, Ann Arbor, MI).
TGF-β1 Stimulation.
Once the cell lines had been genotyped, they were tested for response to TGF-β1 (Promega) stimulation at a concentration of 2.5 ng/ml. This number was deemed to produce the optimal response in a series of experiments in which the cell line HCA 46, which is known to respond to TGF-β, was tested for changes in carcinoembryonic antigen expression on stimulation with different concentrations of TGF-β1 (data not shown).
All experiments were carried out in quadruplicate in 96-well plates, and TGF-β stimulation was tested in growth media containing 10% fetal calf serum (FCS), 2% FCS, and serum-free medium. Cells from stock plates were trypsinized and washed, and eight wells were plated for each cell line, with approximately 105 cells per well in 100 μl of the appropriate growth medium. The cells were allowed to settle for 24 hours, and TGF-β1 was added to four wells of each cell line at a final concentration of 2.5 ng/ml (= 100 pmol). Cells were allowed to incubate with TGF-β1 for 24 hours and were then analyzed with the MTS assay (see below) for cell proliferation and with ELISA for adhesion molecule expression. In those cases in which the experiments were performed in medium containing 2% FCS or serum-free medium, the cells in the stock plates were cultured in medium containing 2% FCS for 24 hours prior to trypsinization.
MTS Assay for Cell Proliferation.
After incubation with TGF-β1, cell proliferation was measured by using the CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega) in accordance with the manufacturer’s instructions. This assay is referred to here as the MTS assay and comprises a metabolic colorimetric test that has been shown to correlate precisely with cell number. Absorption at λ = 490 nm was measured on a Titertek Multiscan PLUS (Labsystems, Chicago) plate scanner and the pooled readings of the four wells of stimulated cells were compared with the four wells of unstimulated cells by using the two-tailed t test.
To define growth curves for the cell lines SW48 and Lovo, MTS assays were performed at 12, 24, and 36 hours. These cell lines also underwent antibody inhibition experiments in which the TGF-β1 was mixed with an equal volume of a solution of anti-TGF-β1 antibody (Promega) before adding to the cells. The antibody solution had been diluted to give a 2:1 molar ratio of antibody/TGF-β.
Cloning of PCR Products and Sequence Analysis of Exon 3 TGFBR2.
Because the RER+ cell lines SW48 and Lovo had responded to TGF-β stimulation, it was necessary to exclude the possibility of contamination with wild-type TGFBR2. PCR products for exon 3 of TGFBR2 from these cell lines were cloned into the commercially available pMOSBlue vector (Amersham Pharmacia). Pooled products from three PCR reactions from each case were run on a 1% Tris acetate–EDTA (TAE) gel, a slice containing the band was cut out under UV light, and PCR products were retrieved from the gel slices by using the Nucleon GX kit (Scotlab). Purified products were treated with pk enzyme (supplied with the pMOSBlue kit) and ligated overnight into dephosphorylated pMOSBlue vector. Ligated product was transfected into pMOSBlue-competent cells by the “heat shock” method and plated onto agar plates containing 50 μg/ml ampicillin and 15 μg/ml tetracycline. Thirty minutes before plating the cells, 50 μl of a solution containing 25 mg/ml 5-bromo-4-chloro-3-indolyl β-galactoside (X-Gal), and 50 mM isopropyl B-d-thiogalactopyranoside (IPTG) was applied to the plates. Plated cells were incubated overnight at 37°C, and white colonies were picked and cultured overnight in 5 ml of growth medium containing 50 μg/ml ampicillin. The picked colonies also underwent direct PCR for exon 3 to check for the presence of the insert and, of the “PCR-positive” clones, 20 were selected from each cell line and purified by using the Qiagen (Chatsworth, CA) Miniprep kit. Automated sequencing was then performed as described above by using exon 3 primers and 1 μl of a 1:10 dilution of the miniprep as template.
Single-Strand Conformation Polymorphism (SSCP).
PCR products for exon 3 of TGFBR2 from SW48 and Lovo cells were also subjected to SSCP analysis to exclude contamination with wild-type TGFBR2. The banding patterns for SW48 and Lovo, which are homozygous for exon 3 mutations, were compared with DLD1 (which is heterozygous for mutations) and a single human random control (wild-type only). For SSCP analysis, 3 μl of PCR product was mixed with 4 μl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol), denatured, and immediately placed on ice. The mixture was run overnight at 20 mA on a 6% polyacrylamide gel containing 10% glycerol. The gel was fixed in a solution containing 10% ethanol and 0.5% acetic acid and stained by soaking for 15 minutes in a 0.1% solution of silver nitrate. After two quick washes in distilled water, the gel was incubated in a solution of 1.5% sodium hydroxide and 0.1% formaldehyde to visualize the bands. The gel was dried and analyzed for comparative banding patterns.
ELISA.
Expression of a number of adhesion molecules was measured by using an ELISA assay. Details of the antibodies used are given in Table 3. After incubation with TGF-β1, medium was removed and the cells were washed once in PBS and fixed with 0.02% glutaraldehyde in PBS (20 minutes at room temperature). Cells were incubated overnight in 0.1% gelatin (Sigma) in PBS to prevent nonspecific attachment of antibody and, after two quick washes with 0.2% casein in PBS, were incubated for 1 hour with 50 μl of primary antibody at the appropriate dilution (Table 3). Next, the cells were washed for a total of 30 minutes in a solution containing 0.2% casein/0.2% Tween 20 in PBS with at least 10 changes of washing solution. The secondary antibody was a horseradish peroxidase-conjugated rabbit anti-mouse (Dako), and 50 μl of a 1:1,000 dilution of this was added to each well. After a 1-hour incubation, the cells were washed as in the last washing step, and 100 μl of o-phenylenediamine dihydrochloride (OPD)/H2O2 (SigmaFast; Sigma) was added as the chromogen. Absorption readings at λ = 492 were taken every 5 min, and after 15–20 minutes, 50 μl of 3 M HCl was added, and a final reading was taken. The pooled readings of the four wells of stimulated cells were compared with the four wells of unstimulated cells by using an unpaired two-tailed t test. To ensure that any changes identified were not false changes that had become apparent as a result of changes in cell proliferation, all cell lines were tested for changes in proliferation by the MTS assay in parallel with the ELISA assay. Care was taken to ensure that the same batch of FCS was used to supplement the growth medium in all parallel proliferation/adhesion experiments.
Table 3.
Details of antibodies used
| Antibody | Target | Dilution | Source |
|---|---|---|---|
| HECD-1 | E-cadherin | neat supernatant | ICRF |
| α-catenin | α-catenin | 1:500 | Affinity (Nottingham, U.K.) |
| β-catenin | β-catenin | 1:1000 | Affinity |
| γ-catenin | γ-catenin | 1:1000 | Affinity |
| Ali-28 | APC (N-terminal) | 1:50 | ICRF, ref. 20 |
| PR1A3 | CEA | 1:1000 | ICRF, ref. 21 |
APC, antigen-presenting cell; CEA, carcinoembryonic antigen; ICRF, Imperial Cancer Research Fund.
RESULTS
Genotyping of Cell Lines.
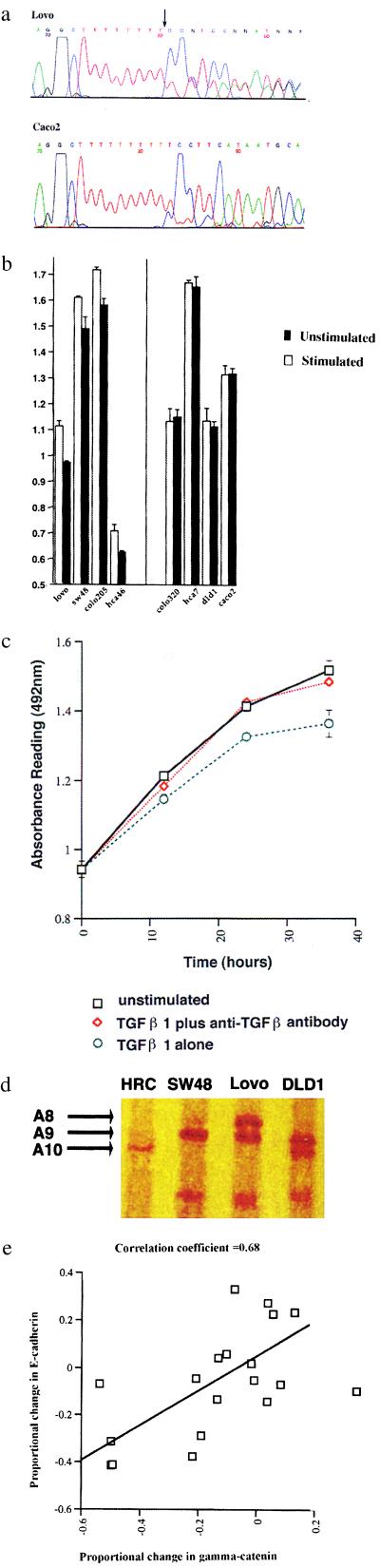
Details of the genotypic analysis of the cell lines are shown in Table 1. All six cell lines that had previously been designated as RER+ were confirmed as showing microsatellite instability, and all were shown to contain homozygous TGFBR2 mutations (Fig. 1a). A single missense mutation was found in exon 5 in the cell line DLD1, while all of the remaining mutations were either single- or double-base pair deletions in the poly(A) tract of exon 3 (Fig. 1a). No mutations in these regions were found in the RER− cell lines. None of the cell lines showed allelic loss at the TGFBR2 locus.
Figure 1.
(a) Reverse sequence analysis for exon 3 of TGFBR2 in the cell lines Lovo (Upper) and Caco2 (Lower). Lovo shows both a single- and double-base pair deletion in the A10 tract (arrow). Caco2 shows the corresponding wild-type sequence. (b) Histogram showing the changes in absorbance reading in response to TGF-β stimulation in the four cell lines (Lovo, SW48, Colo205, and Hca46) that were responsive (Left) and four cell lines (Colo320, HCA7, DLD1, andCaco2) which were unresponsive (Right). Each experiment was performed in quadruplicate, and means thus derived were used for comparison. Two of the responsive cell lines (Lovo and SW48) were RER+ with TGFBR2 mutations and two (Colo205 and HCA46) were RER−, without evidence of TGFBR2 mutations. All four responsive cell lines showed a significant reduction in cell mass when cultured in the presence of TGF-β (two-tailed t test): Lovo, P = 0.0007; SW48, P = 0.02; Colo205, P = 0.002; HCA46, P = 0.01. Error bars indicate standard deviation. (c) Growth curve for Lovo over a 36-hour period, plotted at 0, 12, 24, and 36-hour time points. Cells were grown in the absence of TGFβ1 (a); in the presence of TGF-β1 and anti-TGFβ1 antibody (b); or in the presence of TGF-β1 only (c). Growth inhibition in response to TGF-β1 was seen in Lovo after 12 hours (P = 0.009). The addition of anti-TGF-β1 antibody to the culture negated the growth-inhibitory effects of TGF-β1, thus demonstrating the specificity of the response. Error bars indicate standard deviation. (d) SSCP analysis of exon 3 in TGFBR2 for a human random control (HRC), SW48, Lovo, and DLD-1. This exon has a poly(A) tract consisting of A10 (wild-type) in HRC, A9 in SW48, A8 and A9 in Lovo, and A9 and A10 in DLD1. Each allele showed specific band mobility, and double bands were seen in those cell lines containing two different alleles. All four samples had unique banding patterns, and there was no trace of an A10 (wild-type) allele in SW48 and Lovo (the two responsive RER+ cell lines). (e) Scatterplot of proportional changes in E-cadherin and γ-catenin in response to TGF-β1 stimulation in growth medium supplemented with 10% FCS. The correlation coefficient was 0.68.
Proliferative Responses to TGF-β1 Stimulation.
Initially, the proliferative response of the cell lines to TGF-β1 stimulation proved to be highly variable and inconsistent. There was variation of response with variations of both the batch and the concentration of FCS used. These variations were obviously caused by the presence of growth factors in the FCS interacting either directly with the cells or modulating the activity of the added TGF-β1. We concluded that the effects of the added TGF-β1 could only be isolated if the experiments were performed in serum-free medium. Under these conditions, four of the cell lines showed statistically significant growth inhibition of up to 13% in response to stimulation with TGF-β1 for 24 hours (Fig. 1b). Two of these cell lines were RER+ (SW48 and Lovo) and two were RER− (HCA46 and Colo 205).
Because the two RER+ cell lines had homozygous mutation of the TGFBR2 gene and ought to have been nonresponsive, they were investigated further. The results were confirmed on at least three separate occasions by using a different batch of TGF-β1 each time and with fresh batches of cell lines retrieved from frozen stores. Growth curves for the cell lines were then plotted over a 36-hour period, measuring cell proliferation at four time points (0, 12, 24, and 36 hours) (Fig. 1c). Because the cells were maintained in serum-free conditions, the growth curve experiments were not pursued beyond 36 hours. Interestingly, a significant growth-inhibitory response to TGF-β1 in Lovo was first observed after 12 hours, whereas the response in SW48 became evident after 24 hours. This may be because of the fact that Lovo is a faster grower and a more robust cell line. To ascertain the specificity of the response, anti-TGF-β1 antibody was added to the ligand, which negated its growth-inhibitory effects (Fig. 1c). This confirmed that the response was caused by TGF-β1 rather than any contaminant in the preparation.
Cloning and SSCP Analysis of Exon 3 of TGFBR2.
To further exclude the possibility of the presence of wild-type receptor, PCR products for exon 3 of TGFBR2 from SW48 and Lovo were cloned, and 20 clones from each cell line were sequenced. This should reveal contamination at a 5% level, but there was no evidence of the wild-type sequence. In addition, exon 3 of TGFBR2 for SW48 and Lovo was analyzed by SSCP and compared with DLD1 and normal DNA from a human random control. The length of the poly(A) tract influenced the migration of the bands, and although all of the samples had unique banding patterns, there was no evidence of a band representing an allele with an A10 tract in SW48 and Lovo (Fig. 1d). This method has been predicted to have a sensitivity level as low as 5% (17), showing, as did the cloning, that if there is contamination, it is below this level.
Changes in Adhesion Molecule Expression in Response to TGF-β1 Stimulation.
As with the proliferative responses, the changes in adhesion molecule expression varied with variation in batch and concentration of FCS. It was found that, in those experiments performed in serum-free medium or medium with 2% FCS, there was loss of large numbers of cells because of the frequent washing steps. For this reason only, the results obtained in medium with 10% FCS were further analyzed, although this is a higher concentration of FCS than would be preferred. All of the experiments that produced a significant change in adhesion molecule expression were repeated, and some consistent changes were observed even with different batches of FCS. The cell line HCA 46 was the most responsive of all of the cell lines, and this showed a significant increase in E-cadherin (P = 0.0039), β-catenin (P = 0.0005), and γ-catenin (P = 0.0006) [(with a decrease in cell proliferation (P = 0.023)]. The increase in E-cadherin expression was also observed in SW48 (P = 0.006).
It was noticed that, on the whole, the E-cadherin and γ-catenin changed together and that, although the direction of the change varied between cell lines, both of these molecules usually changed in the same direction. When the proportional change of all of the cell lines was analyzed (including repeats), a distinct correlation between E-cadherin and γ-catenin (correlation coefficient = 0.68) became apparent (Fig. 1e).
DISCUSSION
Among the manifold activities of TGF-β is the ability to inhibit cell growth and induce differentiation. The high frequency of TGFBR2 mutations in RER+ CRCs has been assumed to represent mutation selection for escape from TGF-β-mediated growth inhibition. If this were the case, then it would be counterintuitive to expect cells with TGFBR2 mutations to show growth inhibition in response to TGF-β stimulation. Our results, however, show that, in fact, TGF-β response does not correlate absolutely with TGFBR2 mutation. This implies that, at least in certain cases, the type II receptor can be bypassed. These results were obtained in cells grown in serum-free medium, and therefore the effects are most likely to be caused by the activity of TGF-β1 alone. This supports the idea that TGF-β1 can bypass TGFBR2, presumably by acting through other receptors such as the type V receptor. This receptor has been shown to allow TGF-β1-mediated growth inhibition in Mink lung cell lines that have homozygous mutations of the type II receptor (18, 19). Alternatively, TGF-β1 may also be able to stimulate receptors of other members of the TGF-β superfamily (such as activin, BMP4, and inhibin).
Although the MTS assay is generally a reliable indicator of cell numbers, it is a metabolic assay, and so it is possible that the changes observed were not caused by actual growth inhibition but by some metabolic change. Whether or not this is the case, the data still support type II receptor bypass by TGF-β1.
Our data clearly raise questions about the role of TGFBR2 mutations during the development of CRCs. There are several possible explanations for our findings. It may be that TGFBR2 mutations give no selective advantage and may arise as bystander mutations in RER+ tumors because the A10 tract is hypermutable. It is possible that our experimental conditions do not reflect the situation in vivo and that, in the presence of normal human serum, growth inhibition usually does occur. In our experiments, we saw a growth inhibition of up to 13% in 24 hours, which is of uncertain biological significance. This is likely to be an underestimate, given the fact that serum-free conditions will slow growth and induce apoptosis. It is, however, possible that most growth inhibition is mediated through the type II receptor and that a small amount of growth inhibition through alternative receptor activation is a price that the tumor can afford to pay if the majority of growth inhibition is removed by TGFBR2 mutation.
Another explanation of our results is that TGFBR2 mutations may be selected for effects other than growth inhibition, such as expression of adhesion molecules. With this question in mind, we investigated changes in the expression of six adhesion molecules in response to TGF-β1 stimulation. The methodology used limited reliable interpretation to those data accrued from experiments carried out in growth medium containing 10% FCS. An overall difference between the RER+ and RER− cell lines did not emerge. The changes were variable between cell lines and between different batches of FCS. This observation is most likely attributable to differences in the pattern of somatic mutations between the cell lines. There were, however, some consistent reproducible changes; in particular, HCA 46 showed an increase in carcinoembryonic antigen, E-cadherin, β-catenin, and γ-catenin expression, whereas SW48 showed an increase in E-cadherin expression. Given the wide range of activities within the repertoire of TGF-β function, these changes are not unexpected. An interesting observation was the correlation between E-cadherin and γ-catenin changes. Both molecules can be found in Adherens junctions, and γ-catenin can be found in desmosomal junctions.
In conclusion, in comparing six RER+ cell lines with proven homozygous TGFBR2 mutations with six RER− cell lines, we showed, in serum-free conditions, that the growth of some of the RER+ cell lines could be inhibited and undergo growth inhibition by TGF-β1 and that TGF-β1 could also induce changes in the expression of certain adhesion molecules. It is thus possible that TGFBR2 mutations are neutral bystander events or that these mutations give a selective advantage through alteration in the pattern of adhesion molecule expression rather than direct growth inhibition.
Acknowledgments
We thank Reg Boone for technical assistance and A. Rowan for assistance with the ELISA studies.
ABBREVIATIONS
- TGF
transforming growth factor
- CRC
colorectal carcinoma
- RER
replication error
- SSCP
single-strand conformation polymorphism
References
- 1.Bodmer W F. J R College Physicians Lond. 1996;31:82–89. [PMC free article] [PubMed] [Google Scholar]
- 2.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Ilyas M, Tomlinson I P. Histopathology. 1996;28:389–399. doi: 10.1046/j.1365-2559.1996.339381.x. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson I P, Novelli M R, Bodmer W F. Proc Natl Acad Sci USA. 1996;93:14800–14833. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peltomaki P, Aaltonen L A, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Green J S, Jass J R, Weber J L, Leach F S, et al. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 6.Eshleman J R, Lang E Z, Bowerfind G K, Parsons R, Vogelstein B, Willson J K, Veigl M L, Sedwick W D, Markowitz S D. Oncogene. 1995;10:33–37. [PubMed] [Google Scholar]
- 7.Modrich P. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 8.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 9.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 10.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 11.Souza R F, Appel R, Yin J, Wang S, Smolinski K N, Abraham J M, Zou T T, Shi Y Q, Lei J, Cottrell J, et al. Nat Genet. 1996;14:255–257. doi: 10.1038/ng1196-255. [DOI] [PubMed] [Google Scholar]
- 12.Souza R F, Yin J, Smolinski K N, Zou T T, Wang S, Shi Y Q, Rhyu M G, Cottrell J, Abraham J M, Biden K, et al. Cancer Res. 1997;57:2350–2353. [PubMed] [Google Scholar]
- 13.Alevizopoulos A, Mermod N. BioEssays. 1997;19:581–591. doi: 10.1002/bies.950190709. [DOI] [PubMed] [Google Scholar]
- 14.Wrana J L, Attisano L, Wieser R, Ventura F, Massague J. Nature (London) 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 15.Takenoshita S, Hagiwara K, Nagashima M, Gemma A, Bennett W P, Harris C C. Genomics. 1996;36:341–344. doi: 10.1006/geno.1996.0471. [DOI] [PubMed] [Google Scholar]
- 16.Takenoshita S, Tani M, Nagashima M, Hagiwara K, Bennett W P, Yokota J, Harris C C. Oncogene. 1997;14:1255–1258. doi: 10.1038/sj.onc.1200938. [DOI] [PubMed] [Google Scholar]
- 17.Wu J K, Ye Z, Darras B T. Am J Hum Genet. 1993;52:1273–1275. [PMC free article] [PubMed] [Google Scholar]
- 18.Leal S M, Liu Q, Huang S S, Huang J S. J Biol Chem. 1997;272:20572–20576. doi: 10.1074/jbc.272.33.20572. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Huang S S, Huang J S. J Biol Chem. 1997;272:18891–18895. doi: 10.1074/jbc.272.30.18891. [DOI] [PubMed] [Google Scholar]
- 20.Efstathiou J A, Noda M, Rowan A, Dixon C, Chinery R, Jawhari A, Hattori T, Wright N A, Bodmer W F, Pignatelli M. Proc Natl Acad Sci USA. 1998;95:3122–3127. doi: 10.1073/pnas.95.6.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richman P I, Bodmer W F. J Pathol. 1988;156:197–211. doi: 10.1002/path.1711560305. [DOI] [PubMed] [Google Scholar]