Abstract
The cation-independent mannose-6-phosphate receptor (CI-MPR) follows a highly regulated sorting itinerary to deliver hydrolases from the trans-Golgi network (TGN) to lysosomes. Cycling of CI-MPR between the TGN and early endosomes is mediated by GGA3, which directs TGN export, and PACS-1, which directs endosome-to-TGN retrieval. Despite executing opposing sorting steps, GGA3 and PACS-1 bind to an overlapping CI-MPR trafficking motif and their sorting activity is controlled by the CK2 phosphorylation of their respective autoregulatory domains. However, how CK2 coordinates these opposing roles is unknown. We report a CK2-activated phosphorylation cascade controlling PACS-1- and GGA3-mediated CI-MPR sorting. PACS-1 links GGA3 to CK2, forming a multimeric complex required for CI-MPR sorting. PACS-1-bound CK2 stimulates GGA3 phosphorylation, releasing GGA3 from CI-MPR and early endosomes. Bound CK2 also phosphorylates PACS-1Ser278, promoting binding of PACS-1 to CI-MPR to retrieve the receptor to the TGN. Our results identify a CK2-controlled cascade regulating hydrolase trafficking and sorting of itinerant proteins in the TGN/endosomal system.
Keywords: CI-MPR, CK2, endosome, GGA3, PACS-1
Introduction
The localization and trafficking of itinerant membrane cargo proteins within the trans-Golgi network (TGN)/endosomal system relies upon canonical sorting motifs within their cytosolic domains, which are recognized by components of the vesicular trafficking machinery (Robinson, 2004). These motifs include tyrosine (Yxxφ)- and dileucine ([D/E]xxxL[L/I])-based signals, which bind to the heterotetrameric adaptors (APs), acidic-dileucine (DxxLL)-based motifs, which bind to GGAs and acidic cluster-based motifs, which bind to PACS proteins. The cytosolic domain of one membrane protein, the cation-independent mannose-6-phosphate receptor (CI-MPR), requires motifs that bind to each of these three groups of sorting molecules to localize to the TGN and to efficiently sort cathepsin D to lysosomes (Chen et al, 1997; Wan et al, 1998; Meyer et al, 2000; Puertollano et al, 2001a; Ghosh et al, 2003). The GGAs sort the CI-MPR into clathrin-coated vesicles at the TGN and may also mediate CI-MPR trafficking between endosomal compartments (Doray et al, 2002b; Mattera et al, 2003; Puertollano and Bonifacino, 2004). By contrast, PACS-1 and AP-1, which mediate endosome-to-TGN retrieval, are required to localize CI-MPR to the TGN (Wan et al, 1998; Meyer et al, 2000; Crump et al, 2001). In addition, other sorting molecules including TIP47, Retromer and EpsinR also function in the endosome-to-TGN retrieval of CI-MPR (Diaz and Pfeffer, 1998; Arighi et al, 2004; Saint-Pol et al, 2004; Seaman, 2004), supporting the highly regulated and complex trafficking pathway followed by this multifunctional receptor.
We identified PACS-1 through its binding to the protein kinase CK2 (CK2)-phosphorylated acidic cluster (…EECPpSDpSEEDE…) on the furin cytosolic domain (Figure 1A and Wan et al, 1998). The 140 amino acid PACS-1 cargo-binding region (FBR, Figure 1A) contains an eight-amino-acid segment ETELQLTF175 that binds AP-1, and is required for correct subcellular localization of furin and CI-MPR to the TGN (Crump et al, 2001). PACS-1 also binds to acidic cluster motifs on several additional itinerant cellular proteins (Thomas, 2002), including proprotein convertase 6B (Xiang et al, 2000), polycystin-2 (Köttgen et al, 2005) and VAMP4 (Hinners et al, 2003), as well as the viral proteins HCMV gB (Crump et al, 2003) and HIV-1 Nef (Piguet et al, 2000). Studies using dominant negative-, siRNA- or antisense-based methods show PACS-1 is required for the TGN localization of each of these proteins, suggesting a broad role for PACS-1 in cellular homeostasis and disease.
Figure 1.
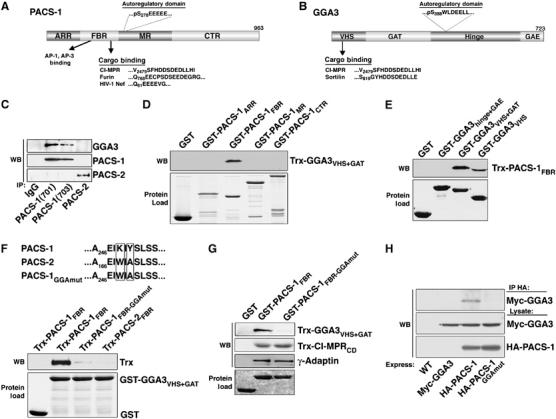
PACS-1 binds to GGA3. (A) Diagram of PACS-1 showing the atropin-1-related region (ARR), cargo-binding region (FBR), which interacts with cargo and AP-1/AP-3 adaptor complexes (Wan et al, 1998; Crump et al, 2001), the middle region (MR), which contains the autoregulatory acidic cluster and Ser278 (Scott et al, 2003), and the C-terminal region (CTR) and PACS-1 cargo. (B) Diagram of GGA3 showing the VHS (Vps27, Hrs, Stam) domain, which binds to cargo proteins, the GAT (GGA and TOM) domain, which binds to ARF1, the hinge segment, which contains the autoregulatory acidic-dileucine motif and Ser388, and the GAE (γ-adaptin ear) domain (Bonifacino, 2004) and GGA3 cargo. (C) Endogenous PACS-1 was immunoprecipitated from rat brain (lower panel) using anti-PACS-1 701 or 703, anti-PACS-2 834 or control IgG and co-precipitating GGA3 analyzed by SDS–PAGE and Western blot (upper panel). Immunoprecipitated PACS-1 and PACS-2 are shown by Western blot (bottom panel). (D–G) The indicated GST-fusion proteins were incubated with the indicated Trx-fusion proteins or with purified AP-1, isolated with glutathione sepharose and analyzed by Western blot using anti-Trx or anti-γ-adaptin antibody (upper panels). Each GST-protein is also shown (lower panel). The Trx-PACS-1FBR (residues 117–294) band is shifted lower in (E) because Trx-PACS-1FBR migrates at the same size as GST-GGA3VHS. GST-GGA3VHS−GAT captured ∼1% of Trx-PACS-1FBR input, and GST-PACS-1FBR (residues 117–294) captured ∼1%, 0.5% and 1% of the Trx-GGA3VHS−GAT, γ-adaptin and Trx-CI-MPRcd input, respectively. Binding assays were conducted as described in Materials and methods, except 4% NP40 was used. (H) A7 cells infected with wild-type (WT) AV or AV expressing Myc-GGA3, Myc-GGA3 and HA-PACS-1, or Myc-GGA3 and HA-PACS-1GGAmut were harvested and HA-tagged proteins immunoprecipitated and co-precipitating myc-GGA analyzed by Western blot (upper panel). Lower panels show myc-GGA3 and HA-PACS-1 expression.
Similar to furin, binding of PACS-1 to the CI-MPR cytosolic domain (CI-MPRCD) requires the CK2 phosphorylatable acidic cluster …DDpSDEDLLHI, located at the CI-MPRCD C-terminus (Wan et al, 1998). Interestingly, the three GGA family members (1–3) also bind to this phosphorylated motif on the CI-MPRCD but require the dileucine motif for binding, which furin lacks (Puertollano et al, 2001a). The GGAs contain three principal domains including the VHS domain, which binds to cargo proteins, the GAT domain, which binds to ARF1, a hinge segment, which binds clathrin and contains an autoregulatory acidic-dileucine motif (GGA1 and 3 only) and the GAE domain, which binds to several accessory proteins (Figure 1B and Bonifacino, 2004). Through these interactions, the GGAs function as monomeric clathrin adaptors that link itinerant cargo directly to clathrin (Puertollano et al, 2001b). However, why GGAs and PACS-1 share overlapping binding sites on the CI-MPRCD is not known.
The functional similarities shared by PACS-1 and GGAs extend to regulation of their cargo binding. The sorting activity of PACS-1 is regulated by the CK2- and PP2A-controlled phosphorylation of an autoregulatory domain (Scott et al, 2003). Phosphorylation of PACS-1Ser278 within the PACS-1 autoregulatory domain activates cargo binding and is required for the endosome-to-TGN transport of furin, CI-MPR and HIV-1 Nef. Similar to PACS-1, GGA1 and GGA3 binding to cargo proteins is regulated by CK2 phosphorylation of an autoregulatory domain within the GGA1 and GGA3 hinge segment (Doray et al, 2002a; Ghosh and Kornfeld, 2003). Phosphorylation of GGA1Ser355 (which corresponds to GGA3Ser388, see Figure 1B) within the GGA1 autoregulatory domain inhibits binding to CI-MPR. Therefore, CK2 phosphorylation of the PACS-1 autoregulatory domain promotes cargo binding (Scott et al, 2003), whereas CK2 phosphorylation of GGA1 or three autoregulatory domains inhibits cargo binding (Doray et al, 2002a).
CK2 is a ubiquitous protein kinase with more than 300 putative polypeptide substrates and is a heterotetramer composed of two catalytic subunits (αα, αα′, or α′α′) and two regulatory β subunits (Meggio and Pinna, 2003). The regulation of this basally active kinase has long remained enigmatic, although the binding of the regulatory β subunit to polyamines or substrate proteins can increase kinase activity three-fold (Litchfield, 2003). The requirement for CK2 phosphorylation for the regulation of PACS-1, GGA1 and GGA3 action led us to determine how this kinase may control the PACS-1 and GGA3-mediated trafficking of CI-MPR. We report that PACS-1 binds to GGA3 and recruits CK2, forming a multimeric complex, which regulates PACS-1/GGA3-mediated sorting of CI-MPR between the TGN and early endosomes. Together our results describe a novel cellular mechanism for the phospho-regulation of membrane protein traffic through the TGN/endosomal system.
Results
PACS-1 binds to GGA3
Despite regulating opposing CI-MPR trafficking steps, PACS-1 and GGAs share several biochemical functions. These include binding to the CI-MPRCD at a C-terminal acidic cluster and the regulation of their binding to membrane cargo by the CK2 phosphorylation of an autoregulatory domain (Wan et al, 1998; Puertollano et al, 2001a; Doray et al, 2002a; Scott et al, 2003). These common properties led us to ask if GGA3 and PACS-1 associate in vivo. Accordingly, we immunoprecipitated PACS-1 from rat brain using two different PACS-1 antibodies and found that GGA3 co-precipitated with PACS-1 (Figure 1C). By contrast, GGA3 did not co-precipitate with PACS-2, which is a PACS-1 homologue that mediates ER/mitochondria trafficking (Simmen et al, 2005). To determine if PACS-1 bound directly to GGA3 and to identify the GGA3-binding region of PACS-1, we used glutathione-S-transferase (GST)-tagged PACS-1 fusion proteins corresponding to predicted domains of PACS-1 (Figure 1A) to capture Thioredoxin (Trx)-tagged GGA3VHS+GAT (Figure 1D). Only GST-PACS-1FBR, which binds to cargo including CI-MPRCD, was able to precipitate Trx-GGA3VHS+GAT. Reciprocal mapping experiments using purified GST-GGA3 constructs (Figure 1B) showed that the GGA3 VHS domain, which binds the CI-MPRCD, was sufficient to bind Trx-PACS-1FBR (Figure 1E). These results demonstrate a direct interaction between PACS-1 and GGA3 through their cargo-binding regions.
To further define the GGA3-binding site on the PACS-1 FBR, we took advantage of the fact that through the FBR, PACS-1 and PACS-2 are 75% identical and 83% homologous. Serial mutation of nonhomologous amino acids was used to identify residues in the PACS-1 FBR required for binding GGA3. Using this approach, we found that mutation of PACS-1 FBR residues K249IY to the corresponding PACS-2 residues (W171IA; hereafter PACS-1 FBR-GGAmut) disrupted GGA3 binding (Figure 1F). In addition, we found that protein-binding studies showed that the K249Y251 → WA substitution had no effect on GST-PACS-1FBR binding to purified AP-1 or Trx-tagged CI-MPRCD (Figure 1G). Therefore, we introduced the K249Y251 → WA mutation into full-length PACS-1 (hereafter PACS-1GGAmut), and compared the ability of hemagglutinin (HA)-tagged PACS-1 and HA-PACS-1GGAmut to co-immunoprecipitate co-expressed myc-GGA3 (Figure 1H). In agreement with our in vitro binding studies, we found that myc-GGA3 co-immunoprecipitated with HA-PACS-1, but not with HA-PACS-1GGAmut. Thus, we identified a PACS-1 mutant that fails to bind GGA3 but is unaffected for binding cargo and AP-1.
Blocking the PACS-1/GGA3 interaction disrupts CI-MPR and GGA3 localization
We expressed PACS-1GGAmut in cells to determine if PACS-1 binding to GGA3 is required for the steady-state localization of their mutual cargo protein: CI-MPR. In control cells or PACS-1-expressing cells, CI-MPR demonstrated a paranuclear staining pattern that overlapped with TGN46 (Figure 2A). However, in PACS-1GGAmut-expressing cells, CI-MPR showed a pronounced redistribution to an endosomal population that overlapped with the early endosomal marker EEA1. As a control, we asked whether PACS-1GGAmut disrupted the localization of furin, which requires PACS-1 for endosome-to-TGN retrieval (Wan et al, 1998), but lacks the canonical D/ExxLL GGA-binding motif (Figure 2B). We found that expression of PACS-1 or PACS-1GGAmut failed to affect the TGN localization of FLAG-furin, suggesting that PACS-1GGAmut selectively disrupts the trafficking of itinerant cargo that depend on binding to both PACS-1 and GGAs. Because GGA3 distributes between the TGN and early endosomes (Puertollano and Bonifacino, 2004), we also examined the localization of GGA3 in PACS-1GGAmut-expressing cells (Figure 2C). We found that expression of PACS-1GGAmut, but not PACS-1, caused a striking redistribution of GGA3 from a paranuclear localization to a dispersed endosome population that overlapped with the redistributed CI-MPR. In addition, we tested the effect of a interfereing mutant PACS-1 molecule, PACS-1Admut, that fails to bind AP-1 and redistributes the CI-MPR and furin from the TGN (Crump et al, 2001), on the localization of GGA3. We observed no effect of PACS-1Admut expression on the localization of GGA3 (Figure 2C), suggesting that redistribution of GGA3 to endosomal compartments induced by PACS-1GGAmut results from the inability of PACS-1GGAmut to interact with GGA3. These findings suggest the PACS-1/GGA3 interaction is required for CI-MPR retrieval and for release of GGA3 from endosomal membranes.
Figure 2.
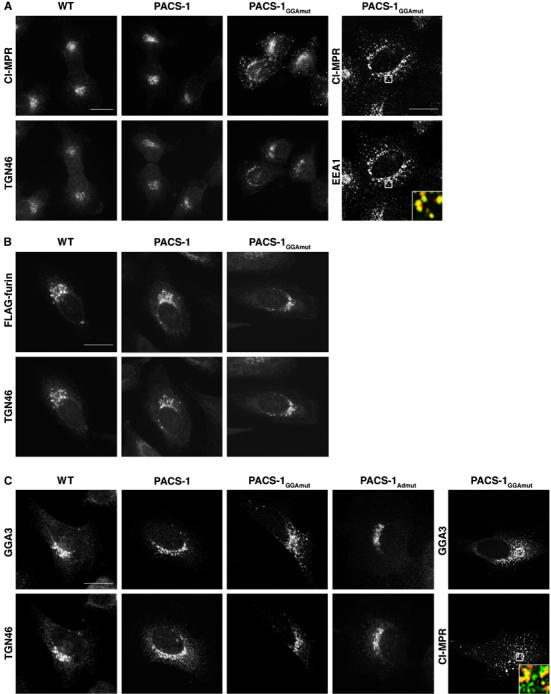
PACS-1GGAmut disrupts CI-MPR trafficking. (A) A7 cells infected with WT vaccinia virus (VV) or VV recombinants expressing PACS-1 or PACS-1GGAmut were stained with antibodies to detect CI-MPR, TGN46 or EEA1 as indicated. Inset: colocalization of CI-MPR (green) and EEA1 (red) from the boxed area. CI-MPR staining outside the TGN area increased from 11±5 and 9±7% in the WT and PACS-1-expressing cells, respectively, to 40±10% for PACS-1GGAmut-expressing cells. (B) A7 cells expressing FLAG-furin were treated as in (A) and stained with anti-FLAG and anti-TGN46. (C) A7 cells infected with VV:WT or with VV expressing PACS-1, PACS-1Admut or PACS-1GGAmut and then costained with anti-GGA3 and anti-TGN46 or anti-CI-MPR. Inset: Colocalization of GGA3 (green) and CI-MPR (red) from the boxed area. GGA3 staining outside the TGN area increased from 11±6, 8±4 and 9±5% in the WT, PACS-1- and PACS-1Admut- expressing cells, respectively, to 38±7% for PACS-1GGAmut-expressing cells. Scale bars=20 μm.
PACS-1 is required for CI-MPR function
To better understand how PACS-1 and GGA3 might cooperate to direct trafficking of CI-MPR, we conducted protein–protein binding assays to define the PACS-1-binding site on the CI-MPRCD. Previously, we found that truncation of the last 10 amino acids (…DDpS2484DEDLLHI) of the CI-MPRCD, which contain a CK2 phosphorylatable acidic cluster and constitute a DxxLL GGA-binding motif, abolished binding to the FBR region of PACS-1 (Wan et al, 1998). First, we sought to determine if, similar to the interaction of PACS-1 and furin (Wan et al, 1998), as well as the CI-MPR with GGA3 (Kato et al, 2002), phosphorylation of CI-MPR Ser2484 would enhance binding to the PACS-1 FBR (Figure 3A). We tested the binding of Trx-PACS-1FBR to GST-CI-MPRCD phosphorylated by CK2 or to GST-CI-MPRCD mutants containing a phosphomimic Ser2484 → Asp or nonphosphorylatable Ser2484 → Ala substitution. We found that both preincubation of GST-CI-MPRCD with CK2 and the Ser2484 → Asp substitution enhanced binding to Trx-PACS-1FBR, indicating that like other PACS-1 cargo proteins, CK2 phosphorylation of Ser2484 within the CI-MPR acidic cluster enhanced binding to PACS-1. Second, we conducted an alanine scan of each of the acidic residues from Asp2482 to Asp2487 and found that alanine mutation of any of the acidic residues reduced binding to Trx-PACS-1FBR (Figure 3B). Finally, we found that Leu2488 → Ala and Leu2489 → Ala mutations had no effect on Trx-PACS-1FBR binding, whereas these mutations completely blocked Trx-GGA3VHS−GAT binding, as previously reported (Figure 3C and Puertollano et al, 2001a). Thus, PACS-1 and GGA3 share overlapping but not identical CI-MPR-binding sites.
Figure 3.
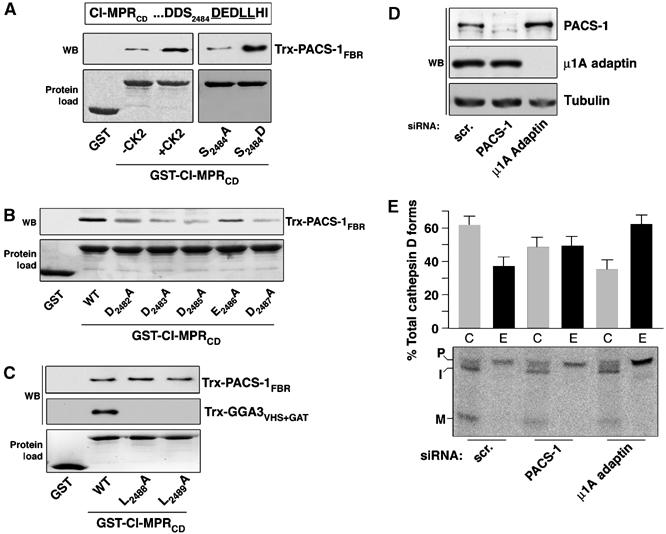
PACS-1 is required for CI-MPR function. (A–C) GST, GST-CI-MPRCD preincubated or not with CK2, or GST-CI-MPRCD containing the indicated mutations was incubated with Trx-PACS-1FBR (residues 117–294) or Trx-GGA3VHS+GAT, isolated with glutathione sepharose, washed three times with GST-binding buffer containing 4% NP-40 and analyzed by Western blot using anti-Trx (upper panels). Input of each GST-protein is shown (lower panel). GST-CI-MPRCD pulled down ∼1% of the Trx-PACS-1FBR. (D) A7 cells were treated with scrambled (scr.) or PACS-1 siRNAs and cell lysates analyzed by Western blot using anti-PACS-1 or anti-tubulin. (E) A7 cells were treated with the indicated siRNA and Cathepsin D pulse chase experiments performed. Cellular and secreted fractions were immunoprecipitated with anti-cathepsin D and analyzed by fluorography. Precursor (P), intermediate (I) and mature (M) forms of cathepsin D are shown (lower panel). The percentage of missorted (secreted) cathepsin D compared to the processed form is shown (n=3, P=0.01).
The importance of CI-MPR Asp2485, which is required for GGA binding and sorting of lysosomal enzymes (Chen et al, 1997; Puertollano et al, 2001a), for binding to PACS-1, as well as the requirement of PACS-1 for the TGN localization of CI-MPR (Wan et al, 1998; Simmen et al, 2005), led us to determine if PACS-1 is required for CI-MPR function. Therefore, we investigated the effect of PACS-1 depletion on the sorting of lysosomal enzymes by CI-MPR. The sorting and maturation of cathepsin D, a ligand of CI-MPR, to lysosomes was followed in metabolically labeled cells. The intracellular (C) and extracellular (E) forms of cathepsin D were immunoprecipitated from both the cells and medium after pulse-chase in the presence of mannose-6-phosphate. We found that siRNA depletion of PACS-1 (Figure 3D), which redistributes CI-MPR from the TGN (Simmen et al, 2005), caused an ∼20% increase in secreted cathepsin D and a corresponding ∼20% decrease in intracellular cathepsin D compared to control cells (Figure 3E). As a positive control, and in agreement with previous studies, we found that siRNA depletion of the μ1A subunit of AP-1 caused ∼50% of the newly synthesized procathepsin D to be released into the culture medium. Additionally, we observed no change in the half-life of CI-MPR in PACS-1-depleted cells (data not shown), indicating that this increased secretion of cathepsin D does not result from decreased CI-MPR stability.
PACS-1 binds to and activates CK2
The overlapping PACS-1- and GGA3-binding sites on CI-MPR, as well as the requirement for binding of PACS-1 to GGA3 to control the TGN localization of CI-MPR, suggested that the interaction between PACS-1, GGA3 and CI-MPR is tightly regulated. One clue to the underlying mechanism controlling the PACS-1/GGA3-dependent sorting of CI-MPR is the prominent role CK2 phosphorylation plays in the regulation of each protein (Doray et al, 2002a; Scott et al, 2003). Although earlier studies demonstrated that an AP-1-associated CK2 activity could phosphorylate GGA1 (Doray et al, 2002b), we speculated that a more direct association of CK2 with PACS-1 and GGA3 might afford greater signaling efficacy. Accordingly, we immunoprecipitated PACS-1 from rat brain and assayed the bound material for co-precipitating CK2 activity (Figure 4A). We observed a ∼14-fold increase in PACS-1-associated CK2 activity compared to the control, which was blocked by the CK2-specific inhibitor TBB, but not the PKA inhibitor PKI. To identify the region of PACS-1 that associates with CK2, we used GST-PACS-1 segments (see Figure 1A) to capture CK2α from rat brain cytosol (Figure 4B). Similar to our analysis of GGA3 binding (Figure 1), we found that CK2α was captured solely by GST-PACS-1FBR. We more precisely identified PACS-1 FBR residues required for CK2 binding by testing a battery of GST-PACS-1FBR truncations and substitutions for their ability to capture CK2α from rat brain cytosol (Figure 4C). We found that an 18-amino acid segment of the PACS-1 FBR between L194 and A212 was required to capture CK2α (Figure 4D). Next, we conducted an alanine scan of this PACS-1 segment and found that an R196RKRY → AAAAA substitution (hereafter called PACS-1FBR–CKmut) blocked CK2α association with GST-PACS-1FBR, whereas alanine substitution of adjacent five-amino-acid segments, including K201NRTI → AAAAA and L206GYKT → AAAAA, did not. As a control, we observed no difference between the binding of GST-PACS-1FBR or GST-PACS-1FBR−CKmut to purified AP-1, Trx-CI-MPRCD or Trx-GGA3VHS+GAT (Figure 4E). Thus, PACS-1 associates with CK2 in vivo and the PACS-1 FBR-CKmut substitution specifically blocks the CK2/PACS-1 interaction.
Figure 4.
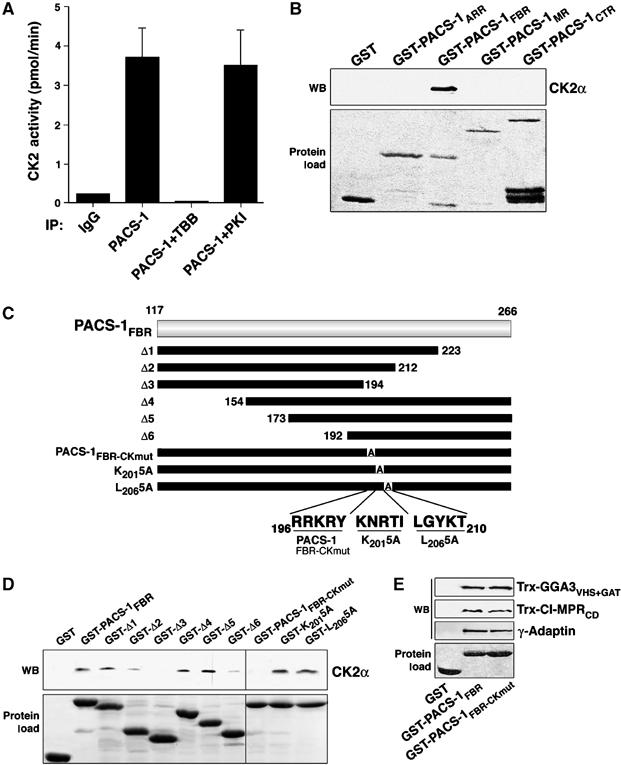
PACS-1 associates with CK2. (A) Rat brain cytosol was incubated with affinity-purified anti-PACS-1 or control IgG to immunoprecipitate endogenous PACS-1, and co-precipitating CK2 activity was measured with an in vitro kinase assay in the absence or presence of 40 μM TBB (CK2 inhibitor) or 400 μM PKI (PKA inhibitor). Error bars represent mean and s.d. of three independent experiments. (B–D) GST-PACS-1 ARR, FBR (residues 117–266), MR or CTR, or the indicated PACS-1 FBR truncations or alanine substitutions (see Figure 1A and D) were incubated with rat brain cytosol, captured with glutathione sepharose and analyzed by Western blot using anti-CK2α (panel B, D, upper panel). Input of each GST-protein is also shown (panel B, D, lower panel). GST-PACS-1FBR captured ∼3% of the input CK2α. Relative to GST-PACS-1FBR, the interaction of CK2 with GST-Δ2 and GST-Δ6 was reduced 60 and 75%, respectively (n=3). (E) GST, GST-PACS-1FBR (residues 117–266) or GST-PACS-1FBR−CKmut was incubated with Trx-GGA3VHS+GAT, Trx-CI-MPRCD or purified AP-1, captured with glutathione sepharose and analyzed by Western blot using anti-Trx or anti-γ-Adaptin antibody (upper panels). GST-PACS-1FBR (residues 117–266) pulled down 9% of the Trx-CI-MPRCD. Input of each GST-protein is shown (lower panel).
To determine which CK2 subunit associates with PACS-1, we conducted a yeast-two-hybrid analysis (Figure 5A). We found that yeast expressing PACS-1 FBR and CK2β, but not CK2α or CK2α′, supported growth under histidine selection. Moreover, cotransformation of PACS-1FBR–CKmut with CK2β failed to support cell growth, further indicating that PACS-1 R196RKRY is required for the interaction between the PACS-1 FBR and CK2β. To determine if the PACS-1 FBR binds directly to CK2β, we conducted a protein–protein binding assay, and found that Trx-CK2β bound directly to GST-PACS-1FBR but not GST-PACS-1FBR–CKmut (Figure 5B). Finally, to confirm the effect of the CKmut substitution in the context of full-length PACS-1, we expressed full-length HA-PACS-1 or HA-PACS-1CKmut in cells, immunoprecipitated the PACS-1 proteins and examined co-precipitating endogenous CK2α and β by Western blot (Figure 5C). In agreement with the in vitro protein capture studies, we found that HA-PACS-1, but not HA-PACS-1CKmut, co-precipitated CK2.
Figure 5.
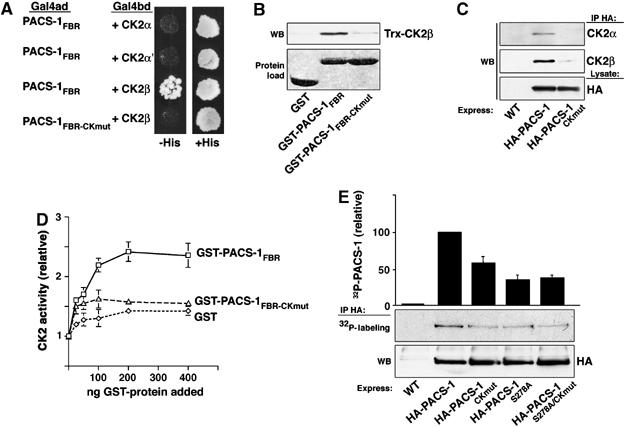
PACS-1 binding to CK2β activates CK2. (A) Yeast transformed with the indicated Gal4 activation and DNA-binding domain (Gal4ad and Gal4bd) constructs were screened for growth on His+ and His− media. (B) GST, GST-PACS-1FBR (residues 117–266) or GST-PACS-1FBR–CKmut was incubated with Trx-CK2β, isolated with glutathione sepharose, washed twice with GST-binding buffer, once with GST-binding buffer containing 1% deoxycholate and analyzed by Western blot using anti-Trx (upper panel). Input of each GST-protein is shown (lower panel). GST-PACS-1FBR captured 2.5% of the input Trx-CK2β. (C) A7 cells infected with VV:WT or VV expressing HA-PACS-1 or HA-PACS-1CKmut were lysed, immunoprecipitated with HA antibody and any co-immunoprecipitating CK2α and CK2β detected by Western blot using subunit-specific antisera (upper panels). HA-PACS-1 expression is shown (bottom panel). (D) In vitro CK2 holoenzyme activity assayed in the absence or presence of purified GST, GST-PACS-1FBR (residues 117–266) or GST-PACS-1FBR–CKmut. Activity is normalized to a parallel sample assayed in the absence of added protein. Error bars represent mean and s.d. of three independent experiments. (E) A7 cells were infected with VV:WT or VV expressing HA-PACS-1, HA-PACS-1CKmut, HA-PACS-1S278A or HA-PACS-1S278A/CKmut and metabolically labeled with 32Pi. HA-proteins were immunoprecipitated with mAb HA.11, resolved by SDS–PAGE and analyzed by autoradiography (upper panel). HA-PACS-1 expression is shown (bottom panel). Error bars represent mean and s.d. of three independent experiments.
One characteristic property of CK2 is the three-fold activation observed upon binding of polycationic molecules or proteins containing clusters of basic amino acids to a patch of acidic residues in the regulatory β subunit (Bonnet et al, 1996; Leroy et al, 1997). As the R196RKRY cluster of basic amino acids in the PACS-1 FBR is required for binding to CK2β, we tested the effect of PACS-1 on CK2 activity levels using an in vitro kinase assay. Purified bovine CK2 holoenzyme was preincubated with increasing concentrations of GST, GST-PACS-1FBR or GST-PACS-1FBR–CKmut, and CK2 activity was scored as incorporation of 32P into a peptide substrate (Figure 5D). GST-PACS-1FBR stimulated CK2 activity ∼2.5-fold, whereas GST or GST-PACS-1FBR–CKmut had a lesser (∼0.5-fold) effect on CK2 activity. Thus, PACS-1 FBR binding stimulates the activity of the CK2 holoenzyme.
We previously determined that CK2 phosphorylation of Ser278 within the PACS-1 autoregulatory domain activates cargo binding and accounts for ∼50% of the incorporated phosphate on PACS-1 (Scott et al, 2003). Thus, our finding that PACS-1 bound and activated CK2 suggested that this interaction may be critical for regulating the phosphorylation state of PACS-1. To test this possibility, we metabolically labeled replicate plates of cells expressing full-length HA-PACS-1, HA-PACS-1CKmut or HA-PACS-1S278A with 32Pi, and quantified the amount of radiolabel incorporated into each protein (Figure 5E). We observed ∼40% less 32P incorporation into HA-PACS-1CKmut compared to HA-PACS-1, whereas HA-PACS-1S278A exhibited ∼60% less 32P incorporation compared to HA-PACS-1. This indicated that the PACS-1/CK2 interaction is required for efficient PACS-1 phosphorylation, but does not reduce PACS-1 phosphorylation to the level observed by Ser278 → Ala substitution. Therefore, to gauge whether the CKmut substitution affects PACS-1 Ser278 phosphorylation, we examined the 32P incorporation into a PACS-1S278A/CKmut double mutant. We predicted that if CK2 that is bound to PACS-1 phosphorylates only Ser278, then PACS-1S278A/CKmut would exhibit equal 32P incorporation compared to PACS-1S278A. Conversely, if CK2 bound to PACS-1 primarily phosphorylates residues other than Ser278, then PACS-1S278A/CKmut would incorporate less 32P than PACS-1S278A. We observed no difference between the 32P incorporation of HA-PACS-1S278A and HA-PACS-1S278A/CKmut, suggesting that CK2 binding to PACS-1 is required for efficient phosphorylation of Ser278 and thus the ability of PACS-1 to bind cargo.
PACS-1-bound CK2 inactivates GGA3 to retrieve CI-MPR to the TGN
The inhibitory effects of the CKmut substitution suggested that PACS-1CKmut may interfere with the PACS-1-dependent sorting of membrane cargo. To test this possibility, we expressed PACS-1CKmut in cells and determined any effect on the TGN localization of CI-MPR and furin. Similar to PACS-1GGAmut, PACS-1CKmut caused CI-MPR to redistribute to an EEA1-positive compartment (Figure 6A). We also found that PACS-1CKmut caused furin to redistribute from the TGN, suggesting that PACS-1 binding to CK2 is required for the sorting of all PACS-1 cargo (Figure 6B). To determine whether PACS-1CKmut blocked PACS-1-dependent trafficking solely because this mutant cannot bind CK2 to phosphorylate Ser278, we expressed a double mutant, PACS-1S278D/CKmut, in cells (Figure 6A). We previously showed that the phosphomimic construct PACS-1S278D had no effect on PACS-1-dependent sorting when expressed in cells, and could rescue the disruption of endosome-to-TGN trafficking caused by depletion of PACS-1 in a cell-free assay (Scott et al, 2003). Therefore, based on our determination that CK2 bound to PACS-1 is required for efficient phosphorylation of Ser278 (Figure 5), we predicted that the PACS-1S278D/CKmut double mutant would override the disruption of CI-MPR localization caused by PACS-1CKmut. Accordingly, we found that expression of PACS-1S278D/CKmut had no effect on the localization of CI-MPR (Figure 6A). Together, these results suggest that PACS-1 recruits CK2 and activates the kinase to promote cargo binding by phosphorylating Ser278 in the PACS-1 autoregulatory domain.
Figure 6.
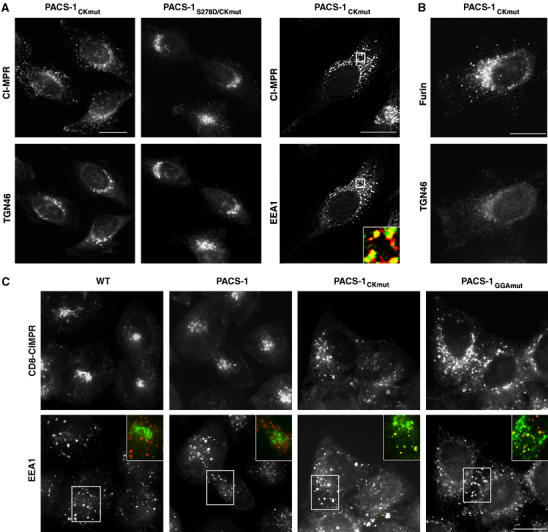
PACS-1CKmut blocks trafficking of CI-MPR and furin. (A) A7 cells infected with VV expressing PACS-1CKmut or PACS-1CKmut/S278D, then stained with anti-CI-MPR, anti-TGN46 or anti-EEA1. Inset: colocalization of CI-MPR (green) and EEA1 (red). CI-MPR staining outside the TGN area was 13±6 and 10±5% in the WT and PACS-1-expressing cells (Figure 2A), respectively, compared with 39±9% for PACS-1CKmut-expressing cells and 11±7% in PACS-1CKmut/S278D-expressing cells. (B) A7 cells transfected with FLAG-furin, infected with VV expressing PACS-1CKmut and stained with anti-FLAG and anti-TGN46. FLAG-furin staining outside the TGN area increased from 9±6 and 11±4% in the WT and PACS-1-expressing cells (Figure 2B), respectively, to 36±5% in PACS-1CKmut-expressing cells. (C) HeLa:CD8-CIMPR cells infected with WT VV or VV expressing PACS-1, PACS-1CKmut or PACS-1GGAmut were incubated with 4 μg/ml anti-CD8 for 1 h at 37°C, fixed, and then incubated with anti-EEA1 and stained with subtype-specific secondary antibodies. Inset: Staining of CD8-CIMPR (green) and EEA1 (red).
Next, we asked whether PACS-1CKmut or PACS-1GGAmut interferes with the retrieval of internalized CI-MPR to the TGN. Due to a lack of mAbs that can be used to monitor the trafficking endogenous CI-MPR following endocytosis, we chose to study the effect of the PACS-1 mutants on the trafficking of internalized CD8-CIMPR, a chimera containing the CD8 lumenal domain fused to the CI-MPR transmembrane and cytosolic domains (Seaman, 2004). Accordingly, we found that PACS-1Ckmut and PACS-1GGAmut, but not PACS-1, blocked the retrieval of CD8-CIMPR to the paranuclear region and caused the reporter to accumulate in an EEA1-positive compartment (Figure 6C).
The ability of PACS-1CKmut and PACS-1GGAmut to redistribute CI-MPR to EEA1-positive endosomes (Figure 6A and C) suggested that, as for PACS-1GGAmut, PACS-1CKmut might disrupt the steady-state localization of GGA3. Accordingly, we found that PACS-1CKmut caused GGA3 to redistribute with CI-MPR to an endosome population (Figure 7A). Because CK2 phosphorylation of GGA3 blocks cargo binding (Doray et al, 2002a), we next asked whether the ability of PACS-1 to bind GGA3 may affect the efficiency of GGA3 phosphorylation by CK2. Therefore, we metabolically labeled cells expressing HA-PACS-1 or HA-PACS-1CKmut with 32Pi and quantified the amount of immunoprecipitated, 32P-labeled, endogenous GGA3 (Figure 7B). We found that HA-PACS-1 expression increased the amount of 32P-GGA3, whereas expression of HA-PACS-1CKmut reduced by ∼45% the amount of 32P-GGA3. These results suggest that PACS-1 recruits CK2 to GGA3, enabling CK2 to phosphorylate and inactivate the binding of GGA3 to CI-MPR. To further test this possibility, we determined whether PACS-1 could form a ternary complex with GGA3 and CK2β in vitro and found that GST-GGA3VHS+GAT could capture Trx-CK2β only in the presence of Trx-PACS-1FBR (Figure 7C). Together, our results suggest that PACS-1 recruits CK2 to phosphorylate both PACS-1 and GGA3, thereby inactivating GGA3 and activating PACS-1, thus causing PACS-1 to bind CI-MPR and direct its retrieval to the TGN.
Figure 7.
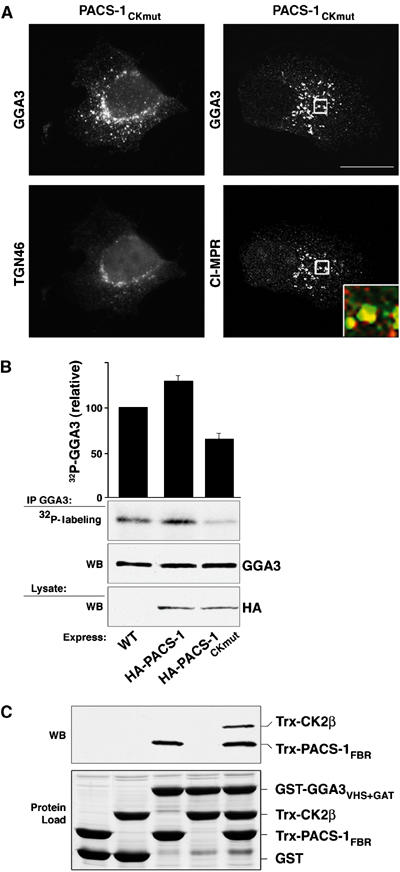
PACS-1Ckmut redistributes GGA3 and controls GGA3 phosphorylation. (A) A7 cells infected with VV expressing PACS-1CKmut, then stained with anti-GGA3 and anti-TGN46 or anti-CI-MPR. Inset: Colocalization of GGA3 (green) and CI-MPR (red). GGA3 staining outside the TGN area increased from 11±6 and 8±4% in the WT and PACS-1-expressing cells (Figure 2), respectively, to 36±5% in PACS-1CKmut-expressing cells. Scale bars=20 μm. (B) A7 cells were infected with WT:VV or VV expressing HA-PACS-1 or HA-PACS-1CKmut, metabolically labeled with 32Pi, and endogenous GGA3 was immunoprecipitated and analyzed by 12% SDS–PAGE and autoradiography. Error bars represent mean and s.d. of three independent experiments normalized to the PACS-1 sample. (C) GST-GGA3VHS+GAT or GST alone was incubated with Trx-CK2β or Trx-PACS-1FBR (residues 117–294) or both, isolated with glutathione sepharose using 4% NP40 and analyzed with anti-Trx mAb (upper panel). Input of each fusion protein is shown (lower panel). Trx-CK2β (∼0.1%) input was captured in the GST-GGA3VHS−GAT/Trx-PACS-1FBR/CK2β ternary complex.
Discussion
The results presented here show that PACS-1, GGA3 and CK2 form a multimeric complex to regulate the endosomal sorting and TGN retrieval of CI-MPR. We found that PACS-1 FBR binds directly to the cargo-binding VHS domain of GGA3 (Figure 1). Substitution of PACS-1 residues K249Y251 (PACS-1GGAmut) blocked binding of PACS-1 to GGA3, but had no effect on adaptor, cargo protein or CK2 binding (Figure 1 and data not shown). Likewise, mutation of the R196RKRY basic amino-acid cluster (PACS-1CKmut) blocked binding of PACS-1 to CK2β, but did not affect cargo, adaptor or GGA3 binding (Figures 4 and 5). Expression of PACS-1GGAmut or PACS-1CKmut caused the redistribution of CI-MPR and GGA3 from the TGN to an early endosomal compartment and also blocked the ability of internalized CD8-CIMPR to traffic to the TGN, causing this CI-MPR reporter to also accumulate in EEA1-positive endosomes (Figures 2 and 6). However, only PACS-1CKmut disrupted TGN localization of furin (Figure 6), suggesting that the interaction of PACS-1 with GGA3 is specifically required for the trafficking of a subset of cargo proteins that bind to both PACS-1 and GGAs. Furthermore, binding of PACS-1 to CK2β stimulated CK2 activity (Figure 5), and was required for the phosphorylation of both PACS-1Ser278 and GGA3, which forms a ternary complex with PACS-1 and CK2β (Figures 5 and 7). Considering the known requirement for CK2 phosphorylation to regulate cargo binding of PACS-1 and GGA3 (Doray et al, 2002a; Scott et al, 2003), one interpretation of these findings is that PACS-1 directs CK2 to GGA3 to initiate the retrieval of CI-MPR from endosomes to the TGN.
Previously, we and others reported that CI-MPR requires an acidic cluster/PACS-1-dependent retrieval step to localize to the TGN (Wan et al, 1998), and function for lysosomal enzyme sorting (Chen et al, 1997). Here we show that CK2 phosphorylation of Ser2484, within the CI-MPR acidic cluster, enhances binding to PACS-1, and that each of the acidic residues within the CI-MPR acidic cluster contributes to PACS-1 binding (Figure 3). By contrast, binding of PACS-1 to CI-MPR is not affected by mutation of LL2489, which are required for GGA3 binding (Figure 3 and Puertollano et al, 2001a). Thus, PACS-1 and GGA3 bind to overlapping but distinct motifs on the CI-MPR. In addition, siRNA depletion of PACS-1 disrupted maturation and lysosomal delivery of cathepsin D, but had no effect on CI-MPR stability. These results differ from those observed with depletion of the retromer subunit Vps26 (Arighi et al, 2004) or TIP47 (Diaz and Pfeffer, 1998), molecules that function in retrieving CI-MPR from Hrs-coated maturing endosomal intermediates or late endosomes, respectively, to the TGN and whose depletion results in a dramatic reduction of CI-MPR half-life. This suggests that PACS-1 functions upstream of or in a separate pathway from retromer and TIP47 for sorting CI-MPR. Interestingly, the cytosolic domain of sortilin, which also sorts lysosomal cargo, binds to GGAs (Nielsen et al, 2001; Lefrancois et al, 2003) and PACS-1 (our unpublished data), and contains a cluster of acidic residues nearly identical to that found on the CI-MPR. Thus, the mechanism described here for control of CI-MPR trafficking may be common to other acidic-dileucine containing receptors. Possibly, the interaction of PACS-1 with CK2 and GGA3 prolongs movement of CI-MPR through endosomes, representing a timing mechanism to aid ligand uncoupling before retrieval of the receptor to the TGN. Alternatively, PACS-1 and GGA3 may combine to retrieve non-ligated CI-MPR to the TGN, whereas ligated receptor would continue to the prelysosomal compartment to release cargo to lysosomes and then be retrieved to the TGN by a retromer- or TIP47-based pathway. The association of PACS-1 and GGA3 with nonligated receptor in early endosomes may explain why depletion of PACS-1 or AP-1 has no effect on CI-MPR stability, whereas disruption of retromer or TIP47 decreases the CI-MPR half-life (Diaz and Pfeffer, 1998; Meyer et al, 2000; Arighi et al, 2004; Seaman, 2004).
Our observation that the R196RKRY polybasic segment is required for PACS-1 to bind and activate CK2 in vitro and in vivo (Figures 4 and 5) provides a mechanism for localizing this kinase to phosphorylate regulatory sites on PACS-1 and GGA3. In particular, our demonstration that blocking CK2β binding to PACS-1 prevented activation of the CK2 holoenzyme, caused a 40% decrease in PACS-1 phosphorylation (Figure 5), and disrupted the TGN localization of CI-MPR, furin and GGA3 (Figures 6 and 7), suggests that localization of CK2 to PACS-1 is required for activation of PACS-1 cargo binding. Exactly how PACS-1 stimulates CK2 activity remains unknown, but may occur in a similar way as spermine or FGF-2, which are proposed to bind the acidic groove of CK2β, causing a conformational change in the CK2 holoenzyme that correlates with an increase in CK2 activity (Leroy et al, 1995, 1997; Bonnet et al, 1996). Additionally, our finding that PACS-1, GGA3 and CK2β form a ternary complex in vitro (Figure 7), and that expression of PACS-1CKmut disrupted the phosphorylation and localization of GGA3 suggests that CK2 controls GGA3 phosphorylation through an interaction with PACS-1.
The disruption of CI-MPR trafficking we observed with expression of PACS-1GGAmut or PACS-1CKmut (Figure 6) is similar to our findings in cells lacking PACS-1 (Wan et al, 1998; Simmen et al, 2005), or expressing dominant negative PACS-1 molecules that cannot bind cargo or AP-1 (Crump et al, 2001; Scott et al, 2003), and further supports the role of PACS-1 in the endosome-to-TGN retrieval step of acidic cluster containing cargo proteins as determined using a cell-free assay (Scott et al, 2003). Notably, the redistributed steady-state concentration of CI-MPR and GGA3 from the TGN to an EEA1-positive compartment we observed with expression of PACS-1GGAmut or PACS-1CKmut is reminiscent of that observed with overexpression of Rabaptin5, which shifts the localization of endogenous GGA1 and CI-MPR to enlarged early endosomes (Mattera et al, 2003). However, expression of PACS-1Admut, which does not bind AP-1, caused the redistribution of the CI-MPR from the TGN (Crump et al, 2001), but had no effect on GGA3 localization (Figure 2), suggesting a temporal ordering of molecular interactions such that PACS-1 is required downstream of GGA3 in the endosomal sorting of CI-MPR and also recruits AP-1 to retrieve this itinerant receptor to the TGN. These results suggest that PACS-1 must bind GGA3 for GGA3 to release from an early endosome compartment and that PACS-1 does not utilize GGA3 as a clathrin adaptor, rather AP-1 or -3 must also be present (Crump et al, 2001). We do not know how the block of PACS-1 binding to GGA3 traps GGA3 in this endosomal compartment, but it is possible that PACS-1 is required to receive the CI-MPR from GGA3 before GGA3 can release from this compartment, or perhaps PACS-1 affects the ability of GGA3 to interact with the early endosomal membrane, possibly by directing CK2 phosphorylation.
Several reports have now identified phosphorylation sites on the GGAs (Doray et al, 2002a; McKay and Kahn, 2004; Kametaka et al, 2005), including two CK2 sites thought to regulate GGA3 function: Ser355 of GGA1 (which corresponds to Ser388 in GGA3) and Ser372 of GGA3. Phosphorylation of GGA1 Ser355 promotes an intramolecular interaction of the GGA DxxLL autoregulatory domain with the cargo-binding VHS domain (Doray et al, 2002a), resulting in a conformational change that correlates with an inhibition of CI-MPR binding (Ghosh and Kornfeld, 2003). CK2 phosphorylation of GGA3 Ser372 is required for EGF-stimulated phosphorylation of GGA3 Ser368 by an unidentified kinase, which causes a conformational change in GGA3 that correlates with reduced association with membranes (Kametaka et al, 2005). Our finding that expression of PACS-1 interfering mutants that cannot bind GGA3 or CK2, respectively, shifted the steady-state distribution of GGA3 with CI-MPR from the TGN to an EEA1-positive compartment (Figures 2 and 7) may represent the manifold effect of phosphorylation of GGA3 Ser272 and Ser388 on binding to cargo and membranes. Whether phosphorylation of PACS-1 at sites in addition to Ser278 control cargo and membrane association warrant further investigation. Nonetheless, our results suggest that a CK2-initiated phosphorylation cascade controls a novel cellular mechanism for regulating the dynamic movement of membrane protein traffic through the TGN/endosomal system.
Materials and methods
Cell lines and recombinant virus
A7 and HeLa:CD8-CIMPR (from M Seaman, University of Cambridge) cells were cultured as previously described (Wan et al, 1998). Viral recombinants were constructed using standard methods (Blagoveshchenskaya et al, 2002). Vaccinia virus (VV) recombinants expressing human HA-PACS-1, HA-PACS-1Admut and HA-PACS-1S278A were previously described (Crump et al, 2001; Scott et al, 2003). VV and adenovirus (AV) recombiants expressing HA-PACS-1CKmut, HA-PACS-1GGAmut HA-PACS-1S278A/CKmut and HA-PACS-1S278D/CKmut and myc-GGA3 (long form) were constructed using techniques previously described (Blagoveshchenskaya et al, 2002).
DNA constructs
pGEX3x plasmids expressing PACS-1 segments ARR, FBR (residues 117–266 or 117–294 as indicated in figure legends), MR and CTR, the PACS-1 FBR truncations Δ1, Δ2, Δ4, Δ5, Δ6 and pET32-PACS-1FBR (residues 117–294) expressing His/thioredoxin (Trx)-fusion proteins were previously described (Crump et al, 2001). PGEX3x plasmids expressing PACS-1 FBR (residues 117–266 or residues 117–294 as indicated in figure legends), Δ3, K2015A, L2065A, GGAmut and CKmut, GST-CI-MPRCD mutants and pVP16 PACS-1FBR–CKmut were generated by standard PCR methods and subcloned into pGEX3x or pVP16. There are no qualitative differences on binding of cargo proteins to FBR117–266 and FBR117–294 but binding to FBR117–294 is quantitatively reduced as described in legends to figures. pET32-CK2β was constructed by subcloning CK2β from pGEX3x-CK2β. pGEX3x-CK2β, pGBT9-CK2α, pGBT9-CK2α′ and pGBT9-CK2β were provided by D Litchfield. PCR3.1GGA3 as well as pGEX constructs expressing the VHS, VHS+GAT, and Hinge+GAE domains of GGA3 were provided by J Bonifacino. pET32-GGA3VHS+GAT was subcloned from pGEX3xGGA3VHS+GAT using standard techniques. pVP16-PACS-1FBR and pGEX3xCI-MPRCD were previously described (Wan et al, 1998).
Yeast-two-hybrid
HF7c yeast were transformed with pVP16-PACS-1FBR (Wan et al, 1998) as well as pGBT9-CK2α, pGBT9-CK2α′ or pGBT9-CK2β, then grown on media lacking histidine supplemented with 1 mM 3-aminotriazole according to standard methods (Clontech).
Protein purification
pGEX or pET32-Trx vectors were transformed into BL21 (DE3) pLysS cells (Novagen). GST- or Trx-fusion proteins were purified using glutathione sepharose (Amersham-Pharmacia) or Ni-NTA-agarose (Qiagen), respectively, according to the manufacturer's protocol. Purified porcine AP-1 was provided by S Tooze. Native CK2 was purified from bovine testicles as described (Litchfield et al, 1990).
Metabolic labeling
A7 cells were infected with VV expressing HA-tagged PACS-1 proteins (m.o.i.=3) for 16 h, washed and incubated with phosphate-free media for 1 h after which 0.5 mCi/ml 32Pi (NEN) was added for 2 h. The labeled cells were washed with PBS and lysed in labeling buffer (PBS with 1% TX-100, 50 mM NaF, 80 mM β-glycerol phosphate and 0.1 μM orthovanadate). HA-PACS-1 proteins were immunoprecipitated with mAb anti-HA.11 (IgG1, Covance no. MMS-101P), separated by SDS–PAGE and analyzed by autoradiography. To label endogenous GGA3, A7 cells were infected with VV expressing HA-PACS-1 proteins (m.o.i.=10) for 4 h, processed as above, and endogenous GGA3 was immunoprecipitated from the cell lysate using mAb anti-GGA3 (IgG1, BD no. 612310), separated by SDS–PAGE and GGA3 phosphorylation determined by autoradiography.
Co-immunoprecipitation
Endogenous PACS-1 or PACS-2 was immunoprecipitated using affinity-purified rabbit anti-PACS-1 701 (Simmen et al, 2005) or purified rabbit anti-PACS-1 703 (raised against PACS-1 residues KERQLSKPLSERTNSSD530) or purified rabbit anti-PACS-2 834 (raised against PACS-2 residues RITKTESLVIPSTRSE425) from freshly isolated rat brain. The immunoprecipitates were separated by 10% SDS–PAGE and analyzed by Western blot with mAb GGA3 (IgG1, BD no. 612310). For co-immunoprecipitation of expressed proteins, HA-PACS-1 was immunoprecipitated with mAb anti-HA.11 (IgG1, Covance no. MMS-101P), and co-immunoprecipitating proteins were analyzed by Western blot with mAb anti-myc clone9E10 (IgG1, Santa Cruz no. sc-40), mAb anti-CK2β (IgG1, Abcam no. ab15848) or rabbit anti-CK2α (Upstate no. 06–873).
Cathepsin sorting
A7 cells were treated twice with siRNA specific for PACS-1 (CUCAGUGGUCAUCGCUGUG), μ1A (AAGGCAUCAAGUAUCGGAAGA) or a control siRNA as described (Simmen et al, 2005). Protein expression was determined by Western blot using purified rabbit anit-PACS-1 701, rabbit anti-μ1A Adaptin (provided by L Traub) and mAb antiacetylated α-tubulin (IgG2b, Sigma no. T6793). Cathepsin D pulse-chase analysis was performed as described (Meyer et al, 2000). Briefly, siRNA-treated cells were labeled with 35S-Met/Cys (NEN) for 0.5 h, then chased with fresh medium containing excess methionine and 10 mM M6P for 4 h. Cathepsin D was immunoprecipitated from the media and cell extract using rabbit anti-cathepsin D (DAKO no. A0561), separated by SDS–PAGE and detected by fluorography.
Kinase assays
Endogenous PACS-1 was immunoprecipitated from rat brain using affinity-purified rabbit anti-PACS-1 701, washed twice with PBS and once with kinase buffer (20 mM MOPS, pH 7.2, 25 mM β-glycerol phosphate, 5 mM EGTA, 1 mM sodium orthovanadate and 1 mM DTT). Kinase assays were performed with 200 μM CK2 substrate (RRRDDDSDDD), 100 μM ATP, 15 mM MgCl2 and 5 μCi γ-32P-ATP in the absence or presence of 40 nM 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB; provided by L. Pinna) or 400 nM PKI (Upstate). Reactions were incubated for 10 min at 30°C, stopped with trichloroacetic acid, spotted on to P81 paper, washed with 0.75% phosphoric acid, and counted. Kinase assays to determine the activation level of CK2 were performed as above using 40 ng native bovine CK2 and increasing amounts of GST, GST-PACS-1FBR (residues 117–266) or GST-PACS-1FBR–CKmut
GST-protein-binding assays
Lysate interactions. 3 μg of each purified PACS-1 fragment was incubated with 250 μl rat brain cytosol for 2 h at 4°C, followed by incubation with glutathione agarose for 30 min. Glutathione beads were pelleted, washed with in vitro binding buffer (25 mM Hepes pH 7.2, 250 mM KCl, 2.5 mM MgOAc, 100 mM NaCl) and analyzed by Western blot with rabbit anti-CK2α (Upstate no. 06–873).
Direct interactions. Except where indicated, 1 μg of Trx-protein was incubated with 3 μg of GST-protein in GST-binding buffer (20 mM Tris pH 7.5, 200 mM NaCl, 1 mM MgCl2, 1% NP40) for 2 h at RT, followed by incubation with glutathione agarose for 30 min. Glutathione beads were pelleted, washed three times with GST-binding buffer, and analyzed by Western blot with mAb anti-Thioredoxin (Trx; IgG1, Invitrogen no. R920–25). GST-PACS-1 pulldown of purified AP-1 was performed as described (Crump et al, 2001).
Ternary complex. GST-GGA3VHS+GAT (3 μg) was preincubated with 3 μg Trx-PACS-1FBR in GST-binding buffer containing 4% NP40 for 2 h at RT, followed by the addition of 3 μg Trx-CK2β for 2 h at RT, then glutathione agarose for 30 min. Glutathione beads were pelleted by centrifugation, washed with GST-binding buffer containing 4% NP40, and analyzed by Western blotting with mAb anti-Thioredoxin (Trx; IgG1, Invitrogen no. R920–25).
Immunofluorescence microscopy
A7 cells or HeLa:CD8-CIMPR cells grown to 80% confluency were infected with VV expressing PACS-1, PACS-1GGAmut, PACS-1CKmut, PACS-1Admut or PACS-1S278D/CKmut (m.o.i.=10) or transfected with pcDNA FLAG-furin. Cells were fixed and processed for immunofluorescence as previously described (Crump et al, 2001). mAb anti-CI-MPR (clone 2G11, IgG2a, obtained from S Pfeffer, 1:2 (Dintzis et al, 1994)), mAb anti-GGA3 (IgG1, BD no. 612310, 1:50), mAb anti-EEA1 (IgG1, BD no. 610457, 1:100), mAb anti-CD8 (clone UCHT-4, IgG2a, Sigma no. C7423, 4 μg/ml for antibody uptake studies), mAb anti-FLAG tag (M1, Kodak no. IB13006, 1:300) and rabbit anti-TGN46 (Abcam no. ab16052, 1:100) were used as primary antibodies to localize antigens. As indicates in the figure legends, following incubation with species- and subtype-specific fluorescently labeled secondary antisera (Molecular Probes), images were captured at RT using a × 60 oil immersion objective on an Olympus Fluo-View FV300 confocal laser scanning microscope and processed with the NIH Image J program or a × 63 oil immersion objective on a Leica DM-RB microscope and Hamamatsu C4742-95 digital camera and processed with the scion image 1.62 program. CI-MPR, Flag-furin and GGA3 redistribution was quantified morphometrically by comparing the staining area and pixel intensity of these molecules for each construct/marker pair (25 cells from three independent experiments), relative to the corresponding TGN stain (TGN46), according to the following formula: (Mean pixel intensity)O × AreaO/(Mean pixel intensity)T × AreaT, where O=outside the TGN and T=whole cell. Background was set to the spurious signal intensity observed in the nuclear area. Quantification of immunofluorescence images was carried out using NIH Image J. Values for each morphometric analysis are provided in the respective figure legend. Antibody uptake experiments were conducted as described previously (Scott et al, 2003).
Acknowledgments
We thank Q Justmann for preliminary experiments and members of the Thomas lab, C Enns, J Bonifacino and S Kaech Petrie (CROET Imaging Center) for helpful discussions. We thank J Bonifacino, D Litchfield, S Tooze, S Pfeffer, L Pinna, M Seaman and L Traub for reagents. The authors declare no financial conflict of interest with the described work. This work was supported by NIH grants AI49793 and DK37274 (to GT).
References
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS (2004) Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol 165: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Thomas L, Fellciangell SF, Hung CH, Thomas G (2002) HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111: 853–866 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS (2004) The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Bonnet H, Filhol O, Truchet I, Brethenou P, Cochet C, Amalric F, Bouche G (1996) Fibroblast growth factor-2 binds to the regulatory beta subunit of CK2 and directly stimulates CK2 activity toward nucleolin. J Biol Chem 271: 24781–24787 [DOI] [PubMed] [Google Scholar]
- Chen HJ, Yuan J, Lobel P (1997) Systematic mutational analysis of the cation-independent mannose 6-phosphate/insulin-like growth factor II receptor cytoplasmic domain. An acidic cluster containing a key aspartate is important for function in lysosomal enzyme sorting. J Biol Chem 272: 7003–7012 [DOI] [PubMed] [Google Scholar]
- Crump CM, Hung CH, Thomas L, Wan L, Thomas G (2003) Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J Virol 77: 11105–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Xiang Y, Thomas L, Gu F, Austin C, Tooze SA, Thomas G (2001) PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J 20: 2191–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E, Pfeffer SR (1998) TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell 93: 433–443 [DOI] [PubMed] [Google Scholar]
- Dintzis SM, Velculescu VE, Pfeffer SR (1994) Receptor extracellular domains may contain trafficking information. Studies of the 300-kDa mannose 6-phosphate receptor. J Biol Chem 269: 12159–12166 [PubMed] [Google Scholar]
- Doray B, Bruns K, Ghosh P, Kornfeld SA (2002a) Autoinhibition of the ligand-binding site of GGA1/3 VHS domains by an internal acidic cluster-dileucine motif. Proc Natl Acad Sci USA 99: 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Ghosh P, Griffith J, Geuze HJ, Kornfeld S (2002b) Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297: 1700–1703 [DOI] [PubMed] [Google Scholar]
- Ghosh P, Griffith J, Geuze HJ, Kornfeld S (2003) Mammalian GGAs act together to sort mannose 6-phosphate receptors. J Cell Biol 163: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Kornfeld S (2003) Phosphorylation-induced conformational changes regulate GGAs 1 and 3 function at the trans-Golgi network. J Biol Chem 278: 14543–14549 [DOI] [PubMed] [Google Scholar]
- Hinners I, Wendler F, Fei H, Thomas L, Thomas G, Tooze SA (2003) AP-1 recruitment to VAMP4 is modulated by phosphorylation-dependent binding of PACS-1. EMBO Rep 4: 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Mattera R, Bonifacino JS (2005) Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol Cell Biol 25: 7988–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Misra S, Puertollano R, Hurley JH, Bonifacino JS (2002) Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism. Nat Struct Biol 9: 532–536 [DOI] [PubMed] [Google Scholar]
- Köttgen M, Benzing T, Simmen T, Tauber R, Buchholz B, Feliciangeli S, Huber TB, Schermer B, Kramer-Zucker A, Hèopker K, Simmen KC, Tschucke CC, Sandford R, Kim E, Thomas G, Walz G (2005) Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J 24: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois S, Zeng J, Hassan AJ, Canuel M, Morales CR (2003) The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J 22: 6430–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy D, Herichâe JK, Filhol O, Chambaz EM, Cochet C (1997) Binding of polyamines to an autonomous domain of the regulatory subunit of protein kinase CK2 induces a conformational change in the holoenzyme. A proposed role for the kinase stimulation. J Biol Chem 272: 20820–20827 [DOI] [PubMed] [Google Scholar]
- Leroy D, Schmid N, Behr JP, Filhol O, Pares S, Garin J, Bourgarit JJ, Chambaz EM, Cochet C (1995) Direct identification of a polyamine binding domain on the regulatory subunit of the protein kinase casein kinase 2 by photoaffinity labeling. J Biol Chem 270: 17400–17406 [DOI] [PubMed] [Google Scholar]
- Litchfield DW (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J 369: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield DW, Lozeman FJ, Piening C, Sommercorn J, Takio K, Walsh KA, Krebs EG (1990) Subunit structure of casein kinase II from bovine testis. Demonstration that the alpha and alpha' subunits are distinct polypeptides. J Biol Chem 265: 7638–7644 [PubMed] [Google Scholar]
- Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS (2003) Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J 22: 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay MM, Kahn RA (2004) Multiple phosphorylation events regulate the subcellular localization of GGA1. Traffic 5: 102–116 [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J 17: 349–368 [DOI] [PubMed] [Google Scholar]
- Meyer C, Zizioli D, Lausmann S, Eskelinen EL, Hamann J, Saftig P, von Figura K, Schu P (2000) mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J 19: 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Madsen P, Christensen EI, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, Petersen CM (2001) The sortilin cytoplasmic tail conveys Golgi–endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J 20: 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D (2000) HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol 2: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS, Cell B (2001a) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS (2004) Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol 6: 244–251 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS (2001b) The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell 105: 93–102 [DOI] [PubMed] [Google Scholar]
- Robinson MS (2004) Adaptable adaptors for coated vesicles. Trends Cell Biol 14: 167–174 [DOI] [PubMed] [Google Scholar]
- Saint-Pol A, Yâelamos B, Amessou M, Mills IG, Dugast M, Tenza D, Schu P, Antony C, McMahon HT, Lamaze C, Johannes L (2004) Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev Cell 6: 525–538 [DOI] [PubMed] [Google Scholar]
- Scott GK, Gu F, Crump CM, Thomas L, Wan L, Xiang Y, Thomas G (2003) The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J 22: 6234–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN (2004) Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol 165: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G (2005) PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J 24: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G (2002) Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3: 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Molloy SS, Thomas L, Liu G, Xiang Y, Rybak SL, Thomas G (1998) PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94: 205–216 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Molloy SS, Thomas L, Thomas G (2000) The PC6B cytoplasmic domain contains two acidic clusters that direct sorting to distinct trans-Golgi network/endosomal compartments. Mol Biol Cell 11: 1257–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]


