Abstract
The 26S proteasome is a multisubunit protease responsible for regulated proteolysis in eukaryotic cells. It is composed of one catalytic 20S proteasome and two 19S regulatory particles attached on both ends of 20S proteasomes. Here, we describe the identification of Adrm1 as a novel proteasome interacting protein in mammalian cells. Although the overall sequence of Adrm1 has weak homology with the yeast Rpn13, the amino- and carboxyl-terminal regions exhibit significant homology. Therefore, we designated it as hRpn13. hRpn13 interacts with a base subunit Rpn2 via its amino-terminus. The majority of 26S proteasomes contain hRpn13, but a portion of them does not, indicating that hRpn13 is not an integral subunit. Intriguingly, we found that hRpn13 recruits UCH37, a deubiquitinating enzyme known to associate with 26 proteasomes. The carboxyl-terminal regions containing KEKE motifs of both hRpn13 and UCH37 are involved in their physical interaction. Knockdown of hRpn13 caused no obvious proteolytic defect but loss of UCH37 proteins and decrease in deubiquitinating activity of 26S proteasomes. Our results indicate that hRpn13 is essential for the activity of UCH37.
Keywords: deubiquitinating enzyme, proteasome, Rpn13, ubiquitin, UCH37
Introduction
The ubiquitin–proteasome system is the main non-lysosomal route for intracellular protein degradation in eukaryotes (Glickman and Ciechanover, 2002). Short-lived proteins as well as abnormal proteins in cells are recognized by the ubiquitin system and are marked with ubiquitin chains as a degradation signal. Polyubiquitinated proteins are then recognized and degraded by 26S proteasomes. The 26S proteasome is a huge protein complex of approximately 2.5 MDa composed of one proteolytically active 20S proteasome and two 19S regulatory particles (RP), each attached to one end of the 20S proteasome (Baumeister et al, 1998). The 20S proteasome is a barrel-shaped complex formed by the axial stacking of four rings made up of two outer α-rings and two inner β-rings, which are each made up of seven structurally similar α- and β-subunits, respectively, being associated in the order of α1−7β1−7β1−7α1−7 (Coux et al, 1996). The interior of the cavity composed of β-rings is responsible for its proteolytic activity. However, the entry of substrates into the cavity of 20S proteasomes is restricted by the narrow gate of α-rings, which is closed in itself. The 19S RP plays an important role in the degradation of ubiquitinated proteins. The 19S RP can be divided into two subcomplexes, known as ‘base' and ‘lid' (Glickman et al, 1998). The base is made up of six ATPases (Rpt1–Rpt6) and two large regulatory components, Rpn1 and Rpn2, functioning as presumptive receptor(s) of ubiquitin-like proteins (Leggett et al, 2002), whereas the lid contains multiple non-ATPase subunits (Rpn3, Rpn5–9, Rpn11–13, and Rpn15). The base complex binds to the outer α-ring of the 20S proteasome and opens a narrow gate in an ATP-dependent manner (Smith et al, 2005). In addition, the ATPase subunits supply energy for unfolding target proteins, so that they can be translocated into the β-ring cavity of 20S proteasomes where active sites are located. The role of the lid complex is less unraveled. Among the lid subunits, Rpn11 is known as a metalloprotease that cleaves the peptide bonds between the substrate and the most proximal ubiquitin of the polyubiquitin chains (Verma et al, 2002; Yao and Cohen, 2002). Rpn10 is thought to lie between the base and the lid complex and serve as one of the ubiquitin receptors (Verma et al, 2004).
In addition to the genuine proteasome subunits, several molecules that associate with proteasomes and play auxiliary roles have been identified (Verma et al, 2000; Leggett et al, 2002). Most of the proteasome studies have been carried out using yeast cells, especially budding yeasts, and less is known about mammalian proteasomes and some of the counterparts of yeast proteasome subunits have not yet been identified. Here, we show that Adrm1, which was previously reported as a membrane glycoprotein (Shimada et al, 1991, 1994), is an ortholog of yeast Rpn13 and identify it as a novel interacting protein of mammalian 26S proteasomes. Furthermore, we reveal that it recruits UCH37, a deubiquitinating enzyme (DUB) associated with proteasomes (Lam et al, 1997; Li et al, 2000; Stone et al, 2004).
Results
Adrm1 is a mammalian ortholog of yeast Rpn13
To identify proteins involved with mammalian proteasomes, we searched for cellular proteins that physically associate, directly or indirectly, with 26S proteasomes in mammalian cells. For this purpose, the human ortholog of Rpn10 (hRpn10: hereafter, ‘h' is used as a prefix to indicate a human ortholog of any yeast proteasome subunit) with a carboxyl-terminal Flag tag was expressed in HEK293 cells and immunoprecipitated from cell lysates with anti-Flag antibody. The immunoprecipitates were eluted with Flag peptides, digested with Lys-C endopeptidase, and analyzed using a highly sensitive direct nano-flow liquid chromatography/tandem mass spectrometry (LC–MS/MS) (Natsume et al, 2002). Following a database search, 30 peptides were assigned to MS/MS spectra for Flag-hRpn10-associated complexes (Supplementary Figure 1). These data identified almost all the subunits of 26S proteasomes. In addition, we identified a molecule with yet unknown relevance to proteasomes, called Adrm1.
Adrm1 was previously reported as a membrane glycoprotein with a molecular mass of 110 kDa and involved in cell adhesion (Shimada et al, 1994; Simins et al, 1999; Natsume et al, 2002). Exploration of the Proteome BioKnowledge Library (https://www.proteome.com/proteome/Retriever/index.html) database indicated that Adrm1 is weakly homologous to Rpn13, a subunit of budding yeast proteasomes (Verma et al, 2000). The overall sequence of Adrm1 showed 24.9% homology with that of Rpn13, whereas it exhibited a high homology of 60.2% with the amino-termini of Adrm1 (residues 22–111) and Rpn13 (residues 7–103). We also identified 74.4% homology between the C-termini of Adrm1 (residues 366–407) and Rpn13 (residues 114–156) (Figure 1). Accordingly, we renamed the molecule hRpn13.
Figure 1.
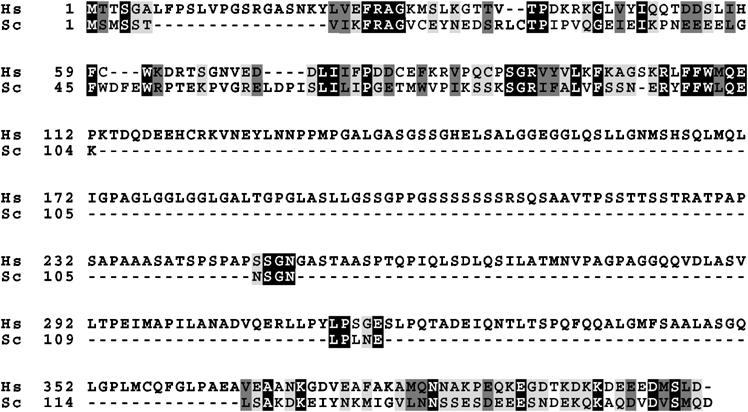
Sequence alignment of human Adrm1 and budding yeast Rpn13. The amino-acid sequences were aligned by the DIALIGN-T program. Amino acids that are identical, strongly similar, and weakly similar between the two sequences are boxed in black, dark gray, and light gray, respectively. Hs: Homo sapiens; Sc: Saccharomyces cerevisiae.
Identification of hRpn13 as a novel proteasome interacting protein in mammals
To verify that hRpn13 is a human counterpart of yeast Rpn13, we tested whether hRpn13 is incorporated into 26S proteasomes. Extracts of 293T cells were fractionated by glycerol gradient centrifugation, and each fraction was subjected to immunoblotting with anti-hRpn13 antibody. Almost all hRpn13 proteins co-sedimented with 26S proteasomes that were detected by succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-MCA)-hydrolyzing activity and sedimentation of a genuine proteasome subunit hRpn1 (Figure 2A). Free forms of hRpn13 were not apparently observed (Figure 2A). 26S proteasomes were further purified from fractions 18–20 by immunoprecipitation using anti-hRpt6 antibody, and then the immunoprecipitates were subjected to two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). Immunoblot for hRpn13 detected a spot that had an isoelectric point (pI) and molecular mass that corresponded to the predicted values of pI 4.8 and 42.1 kDa, respectively (Figure 2B). Tandem mass spectrometric analysis identified this spot as Adrm1 (data not shown). Adrm1 has been reported as a glycosylated membrane protein with a molecular mass of 110 kDa (Shimada et al, 1994), which is in conflict with the notion that Adrm1, that is, hRpn13, is a subunit of proteasomes. Immunostaining of HeLa cells using anti-hRpn13 and anti-hRpn1 antibodies showed that the distribution of hRpn13 was mainly in the nucleus and partially in the cytosol, a pattern quite similar to the distribution of hRpn1 (Figure 2C). These results were in agreement with those reported previously showing that proteasomes are predominantly present in the nuclei of rapidly proliferating mammalian cells (Kumatori et al, 1990). Thus, in our hands, there was no evidence that hRpn13 is a membrane protein.
Figure 2.
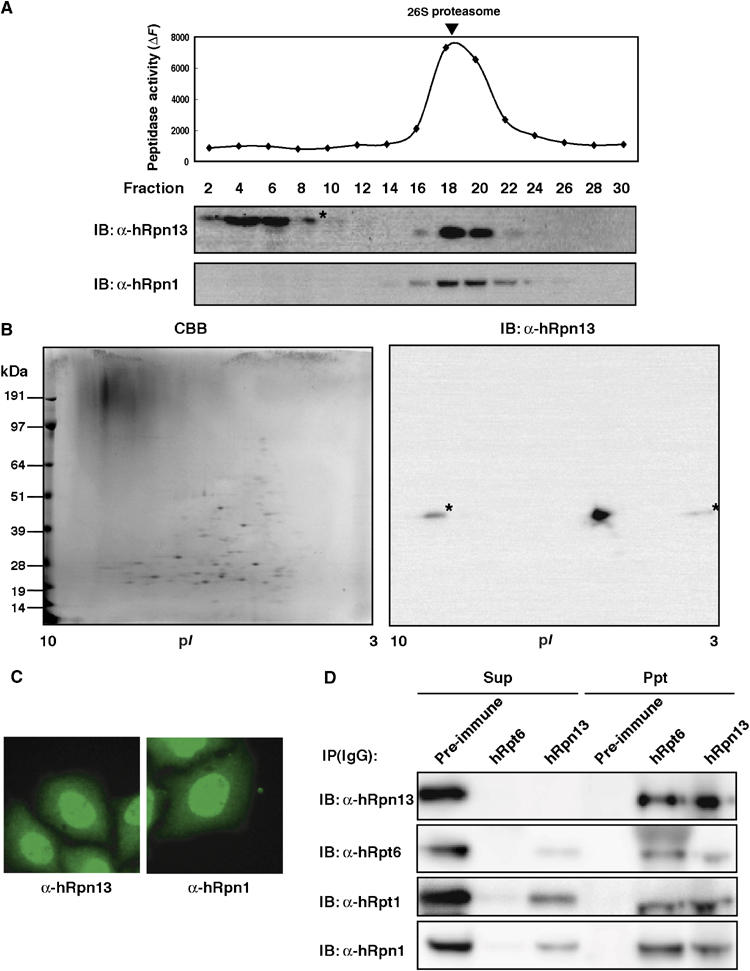
hRpn13 is a subunit of mammalian 26S proteasomes. (A) Sedimentation velocity analysis. Extracts of HEK293T cells were fractionated by 10–40% glycerol gradient centrifugation into 32 fractions from the top. An aliquot of each fraction was subjected to the assay of Suc-LLVY-MCA-hydrolyzing activity (upper panel). Immunoblot analysis of each fraction was performed using antibodies against hRpn1 and hRpn13 (lower panels). Arrowhead indicates the peak fraction of 26S proteasomes. Asterisk indicates artifact bands. (B) Affinity purification of human proteasomes. Fractions 18–20 in panel A were subjected to immunoprecipitation using anti-hRpt6 antibody. The precipitates were eluted with glycine–HCl and resolved by 2D-PAGE, followed by Coomassie brilliant blue (CBB) staining (left panel) and immunoblot with anti-hRpn13 antibody (right panel). (C) Intracellular distribution of hRpn13 in HeLa cells. hRpn13 (left panel) and hRpn1 (right panel) were detected with anti-hRpn13 or anti-hRpn1 antibody and visualized with Alexa488-conjugated anti-rabbit IgG antibody. (D) Immunodepletion analysis. Fractions 18–20 in panel A were pooled and immunoprecipitated with anti-hRpt6 antibody, anti-hRpn13 antibody, or preimmune serum. The unbound fractions and immunoprecipitates were subjected to SDS–PAGE, followed by immunoblotting for hRpn13, hRpt6, hRpt1, and hRpn1.
Next, we examined whether all 26S proteasomes contain hRpn13. The 26S proteasome fractions (fractions 18–20 in Figure 2A) were immunodepleted using anti-hRpt6 antibody, anti-hRpn13 antibody, or preimmune serum. Anti-hRpt6 antibody completely immunodepleted hRpn13 and other 19S RP subunits such as hRpt1 and hRpn1. In contrast, anti-hRpn13 antibody did not remove all the 19S RP subunits, with about 10% input of 19S RP being left in the unbound fraction, whereas it completely depleted hRpn13 (Figure 2D). These results indicate that hRpn13 is incorporated into the majority of 26S proteasomes, but a portion of 26S proteasomes does not contain hRpn13. Therefore, we conclude that hRpn13 is one of the near-stoichiometric proteasome interacting proteins (PIPs), like Ubp6 and Ecm29 in budding yeast (Leggett et al, 2002), and is not an integral subunit of 26S proteasomes.
The conserved N-terminal region of hRpn13 is required for association with proteasomes
Based on the sequence alignment, we postulated that hRpn13 is composed of three regions. The N- and C-terminal regions are conserved between budding yeast and human, and the internal region that lies between the N- and C-terminal regions is not found in the budding yeast Rpn13 (Figure 1). We tentatively divided hRpn13 into three portions. The ‘N' portion encompassed the conserved N-terminal region (i.e. amino acids 1–132) and the ‘C' portion corresponded to amino acids 269–407 that included the conserved C-terminal region. The portion between ‘N' and ‘C' was designated ‘M' portion (Figure 3A). In the next step, we determined the portion required for incorporation into proteasomes. Various deletion constructs encoding wild-type and mutant hRpn13 with Flag tag were expressed in HEK293T cells and immunoprecipitated with anti-Flag antibody. The hRpn13 mutants that lacked the C portion or encoded solely the N portion precipitated various proteasome subunits such as hα6, hRpn10, and hRpn1, as did full-length hRpn13 (Figure 3B). In contrast, hRpn13 that lacked the N portion did not precipitate these subunits (Figure 3B). These results indicate that the N portion, that is, the conserved N-terminal region of hRpn13, is essential and sufficient for its association with proteasomes.
Figure 3.
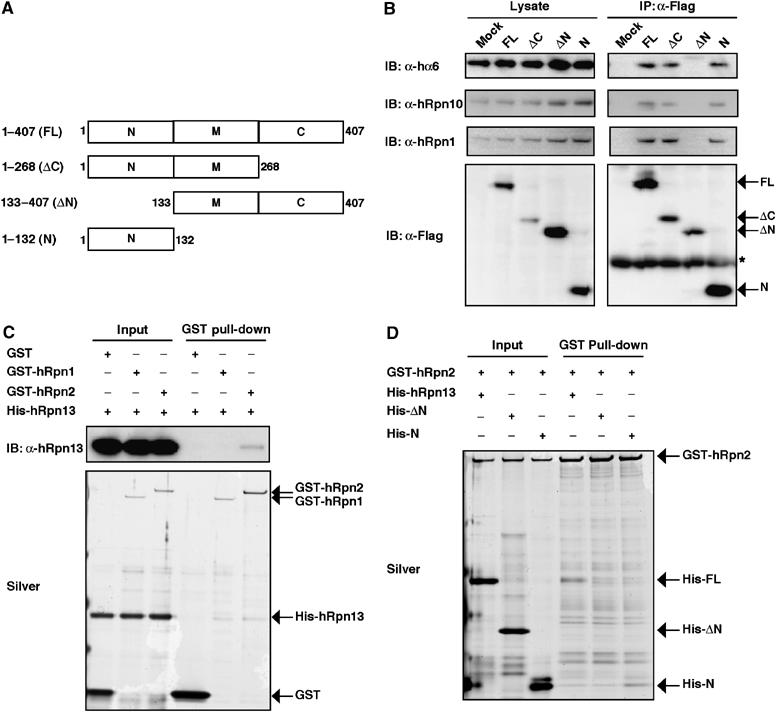
hRpn13 associates with proteasomes via its N-terminal region. (A) Schematic representation of constructs for Rpn13 and its dissected mutants used. (B) Flag-tagged plasmids encoding hRpn13 and its deletion mutants depicted in panel A were transfected into HEK293T cells. The cell lysates were immunoprecipitated with anti-Flag antibody, followed by immunoblotting for hα6, hRpn10, hRpn1, and Flag epitopes. (C) GST pull-down analysis of recombinant proteins. GST, GST-hRpn1, and GST-hRpn2 were incubated with 6xHis-hRpn13 for 1 h at 4°C and then precipitated with glutathione Sepharose. The bound proteins were analyzed by SDS–PAGE, followed by silver staining and immunoblotting with anti-hRpn13 antibody. (D) GST pull-down analysis of recombinant proteins. GST-hRpn2 was incubated with 6xHis-hRpn13 or its mutant forms and then pulled down with glutathione Sepharose followed by silver staining.
hRpn13 directly interacts with a base subunit hRpn2
The next set of experiments determined the subunit of 26S proteasomes associated with hRpn13. For this purpose, we tested the interaction between hRpn13 and each subunit of mammalian 19S RP by yeast two-hybrid analysis. The results identified hRpn1 and hRpn2 as possible interacting subunits with hRpn13 (data not shown). Furthermore, comprehensive interactive proteome analysis in budding yeast showed interaction of Rpn13 with Rpn2 (Ito et al, 2001). Therefore, we purified recombinant proteins of hRpn1, hRpn2, and hRpn13 and performed in vitro binding analysis (Figure 3C). hRpn13 was pulled down by glutathione S-transferase (GST)-hRpn2 but not by GST-hRpn1, indicating that hRpn13 is directly associated with hRpn2. Next, we tested whether the N-terminal region of hRpn13 is required for the association with hRpn2. As shown in Figure 3D, the N portion of hRpn13 was required and sufficient for interaction with GST-hRpn2, which was consistent with the results shown in Figure 3B. Based on these results, we conclude that hRpn13 is associated with 26S proteasomes via hRpn2, with which the conserved N-terminal region of hRpn13 interacts.
Knockdown of Rpn13 does not cause proteolytic defects in proteasomes
In budding yeast, deletion of Rpn13 is not lethal, unlike most other proteasome subunits, but causes defect in the degradation of a ubiquitin-fusion-degradation (UFD) substrate (Verma et al, 2000). To elucidate the function of hRpn13 in mammalian cells, we knocked down hRpn13 by small interfering RNA (siRNA) in HEK293T cells. As a positive control for proteasome dysfunction, knockdown of hRpt2 was also performed. In hRpt2 knockdown cells, a decrease in hRpt2 protein levels resulted in a concomitant loss of hRpn13 as well as other genuine proteasome subunits such as hRpn1 and hα6. Consequently, hRpt2-knockdown cells showed cell death (data not shown) and accumulation of polyubiquitinated proteins (Figure 4A), which are hallmarks of proteasome dysfunction. These results indicate that hRpt2 is required for the integrity of proteasome structure and function and that hRpn13 is stably expressed in the presence of normal proteasomes. In hRpn13-knockdown cells, the expression of hRpn13 was almost abrogated, but the expression of other proteasome subunits was not changed (Figure 4A). The cells showed normal growth (data not shown) and no accumulation of polyubiquitinated proteins (Figure 4A). These features were in contrast to the phenotypes of hRpt2 knockdown. When the extracts from HEK293T cells were fractionated by glycerol-density gradient centrifugation, an active enzyme that catalyzes the degradation of the fluorogenic substrate Suc-LLVY-AMC was sedimented with a sedimentation coefficient of approximately that of 26S, but low activity was found in the slowly sedimenting fractions corresponding to the sedimentation position of the purified 20S proteasome (Figure 4B, upper panel). The addition of 0.05% SDS, which is a potent artificial activator of the latent 20S proteasome, caused marked activation of the enzyme sedimenting like the 20S proteasome (Figure 4B, lower panel). As shown in Figure 4B, the peptide-hydrolyzing activities of both 20S and 26S proteasomes remained unchanged irrespective of hRpn13 knockdown. Next, we tested the activities of protein degradation in vitro (Figure 4C and D) and in vivo (Figure 4E). Assay of antizyme (AZ)-dependent ornithine decarboxylase (ODC) degradation, which measures ATP-dependent and ubiquitin-independent proteolytic activity of 26S proteasomes (Murakami et al, 1992), did not show decreased proteolytic activity. Rather, the activity was increased 1.4-fold in the hRpn13-knockdown cells (Figure 4C). We also examined ubiquitin-dependent proteolytic activity using in vitro ubiquitinated cIAP1 (inhibitor of apoptosis-1), which is a ubiquitin ligase that catalyzes its own ubiquitination for degradation (Yang et al, 2000). As shown in Figure 4D, extracts of hRpn13-knockdown cells showed normal proteolytic activity in the degradation of ubiquitinated cIAP1 proteins. IκBα is also a well-known substrate of 26S proteasomes that is rapidly ubiquitinated and degraded in response to TNF-α (Suzuki et al, 2000). To check proteasome activities in vivo, we monitored the degradation rate of IκBα. hRpn13-knockdown cells showed the same degradation efficiency as control cells (Figure 4E). Considered together, the above results suggest that hRpn13 is not essential for overall protein degradation and viability of mammalian cells.
Figure 4.
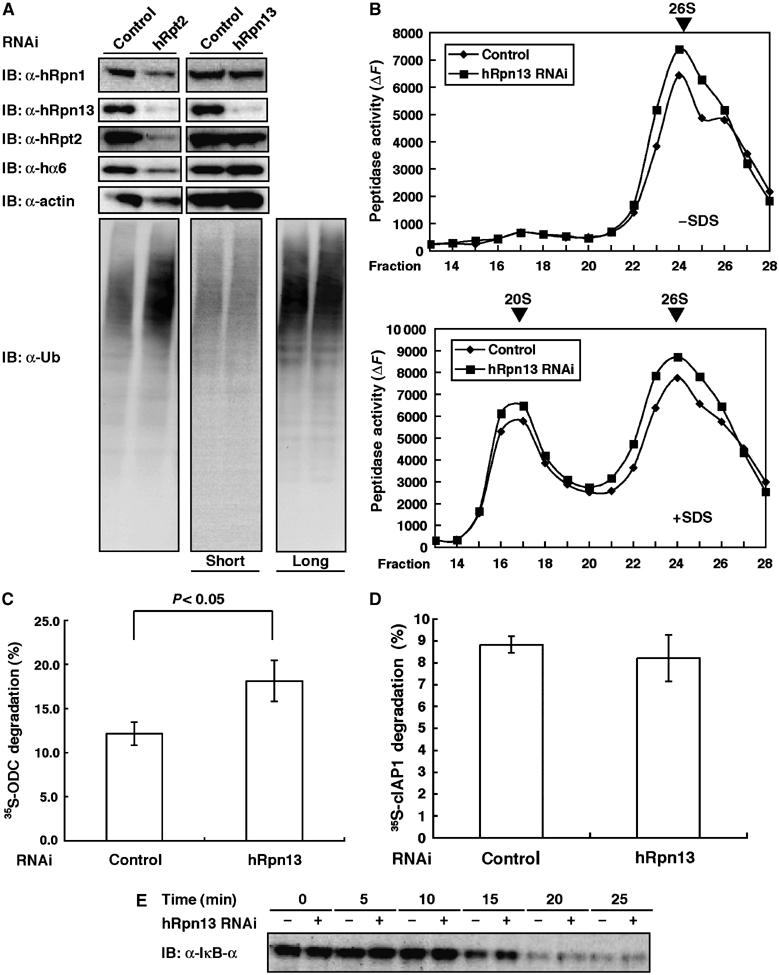
siRNA-mediated knockdown of hRpn13 does not cause proteolytic defects in proteasomes. (A) HEK293T cells were transfected with siRNA against hRpn13 or hRpt2. After 48 h for hRpt2 knockdown and 96 h for hRpn13 knockdown, cell extracts were subjected to SDS–PAGE, followed by immunoblotting with the indicated antibodies. (B) Extracts of control and Rpn13-knockdown cells were fractionated by 8–32% glycerol gradient centrifugation into 32 fractions from the top. Suc-LLVY-AMC hydrolysis activities were measured in the absence (left) or presence (right) of 0.05% SDS. 20S: 20S proteasome; 26S: 26S proteasome. (C) Ubiquitin-independent protein-degrading activity of proteasome. ATP- and AZ-dependent degradation of 35S-labeled ODC protein was assayed. Knockdown cells showed significantly increased activity (P<0.05, one-way analysis of variance). Data are mean±s.e.m. values of three independent experiments. (D) Ubiquitinated cIAP1-degrading activity of proteasomes. Ubiquitin-dependent degradation of ubiquitinated 35S-labeled cIAP1 was assayed. Data are mean±s.e.m. values of three independent experiments. (E) Effect of hRpn13 knockdown on TNF-α-induced degradation of IκBα in vivo. HEK293T cells were treated with TNF-α for the indicated times in the presence of cycloheximide. The levels of IκBα proteins were analyzed by immunoblotting.
hRpn13 interacts with UCH37 via its C-terminus
Whereas the N-terminal region of hRpn13 is essential for association with 26S proteasomes (Figure 3), the role of the conserved C-terminal region is still unknown. To elucidate its role, we again used the proteomic approach. Flag-tagged hRpn13ΔN that did not interact with proteasomes (Figure 3) was expressed in HEK293 cells and immunoprecipitated from the cell lysate with anti-Flag antibody, followed by LC–MS/MS analysis. The peptides most abundantly identified were those of UCH37 (also called UCHL5) (Figure 5A), which is reported to be associated with proteasomes in fission yeast, fly, and mammals (Lam et al, 1997; Holzl et al, 2000; Li et al, 2000, 2001; Stone et al, 2004). We noticed that both hRpn13 and UCH37 had KEKE motifs at their C-terminal extremities. The conserved C-terminal region of hRpn13 is composed of one typical KEKE at its C-terminus (hereafter referred to as KE2) and one KE-rich region adjacent to KE2, which does not fit the definition of KEKE motif, as proposed previously (Realini et al, 1994), but is rich in lysine and glutamic acid residues (hereafter referred to as KE1) (Figure 5B, left panel). A proline residue known as a potent α-helical and β-sheet structure breaker (MacArthur and Thornton, 1991) was located between KE1 and KE2. KEKE motifs are known to mediate protein–protein interactions (Realini et al, 1994), and we hypothesized that the interaction between these two molecules was KEKE motif dependent. To test this notion, we expressed Flag-tagged hRpn13, hRpn13ΔKE2 lacking typical KEKE motif, and hRpn13ΔKE1+2, which lacked both KE1 and KE2 and thus the entire conserved C-terminal region, in HEK293T cells and immunoprecipitation was achieved with anti-Flag antibody (Figure 5B, right panel). Although the three constructs precipitated nearly the same amount of hα6 and hRpn1, hRpn13ΔKE2 precipitated less amount of UCH37 compared with the wild-type hRpn13, and hRpn13ΔKE1+2 did not precipitate UCH37. These results indicate that the conserved C-terminal region is required for the interaction with UHC37.
Figure 5.
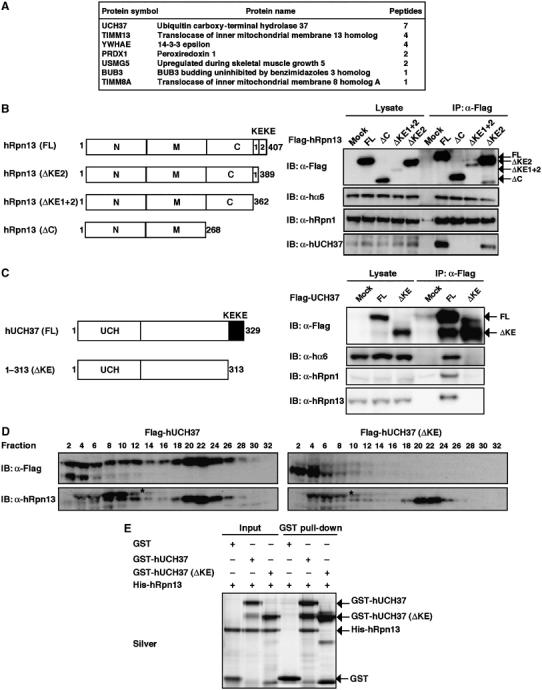
UCH37 interacts with hRpn13. (A) A list of proteins detected in four independent hRpn13ΔN immunoprecipitations by LC–MS/MS analysis. The number of identified peptides of each protein is shown. (B) Flag-tagged plasmids encoding hRpn13 and its deletion mutants depicted in the left panel were transfected into HEK293T cells. The cell lysates were immunoprecipitated with anti-Flag antibody, followed by immunoblotting for hα6, UCH37, hRpn1, and Flag (right panel). (C) Schematic representation of the structures of wild-type UCH37 (FL) and KEKE domain deletion mutant (left panel). HEK293T cells stably expressing Flag-tagged UCH37 (FL), ΔKE, and an empty vector were lysed and subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblot for Flag, hα6, hRpn1, and hRpn13 (right panel). (D) The extracts of cells stably expressing Flag-UCH37 and Flag-UCH37ΔKE were fractionated by 10–40% glycerol gradient centrifugation. Immunoblot analysis was performed for each fraction using antibodies against Flag and hRpn13. Asterisks indicate artifact bands. (E) GST pull-down analysis of recombinant proteins. GST, GST-UCH37, or GST-ΔKEKE were incubated with 6xHis-Rpn13 and precipitated as in Figure 3C, followed by silver staining.
Next, we examined the role of the KEKE motif of UCH37. As shown in Figure 5C, UCH37 that lacked the KEKE motif (UCH37ΔKE) did not associate with hRpn13 and other proteasome subunits, whereas full-length UCH37 did. Glycerol-density gradient analysis of the extracts showed that a large portion of full-length UCH37 co-sedimented with 26S proteasomes, whereas UCH37ΔKE was observed exclusively in much lighter fractions, presumably as a free form, and that overexpression of UCH37ΔKE did not affect the association of hRpn13 with 26S proteasomes (Figure 5D). In in vitro experiments, GST-UCH37 pulled down hRpn13, but GST-UCH37ΔKE did not (Figure 5E), verifying the results depicted in Figure 5C and D. These results indicate that the KEKE motif of UCH37 is necessary for interaction with hRpn13, and hence, with 26S proteasomes.
Knockdown of hRpn13 causes loss of UCH37 proteins
Next, we examined the relationship between hRpn13 and UCH37 in knockdown experiments. We also performed knockdown of USP14, a human ortholog of yeast Ubp6, which is another proteasome-associated DUB (Leggett et al, 2002), to examine the relative contributions of UCH37 and USP14 in deubiquitinating activities of 26S proteasomes. Intriguingly, knockdown of hRpn13 caused marked reduction of total cellular UCH37 proteins but not USP14 proteins. On the other hand, knockdown of UCH37 did not affect the level of hRpn13 proteins (Figure 6A). The observed phenotypes of UCH37 knockdown were similar to those of hRpn13 knockdown with regard to cell growth and levels of polyubiquitinated proteins, which were almost the same as the control (Figure 6B and C). Glycerol-density gradient analysis of the extracts of hRpn13-knockdown cells again showed loss of UCH37 proteins in both free and 26S proteasome fractions, with unchanged distributions of proteasome subunits such as hRpn1 and α6 (Figure 6D). Next, we examined the mechanism of decrease in UCH37 proteins in hRpn13-knockdown cells. Control and hRpn13-knockdown cells were treated with various protease inhibitors such as epoxomycin, which is highly specific for proteasomes, MG132, which inhibits both proteasomes and lysosomal enzymes, and E-64d and pepstatin A, which are specific to lysosomal cathepsins. Although these agents worked effectively in inhibiting the relevant proteases (as monitored by the accumulation of polyubiquitinated proteins for proteasome inhibition and accumulation of lipid-conjugated forms of LC3 proteins for inhibition of lysosomal enzymes; Komatsu et al, 2005), UCH37 was not increased by any of the inhibitors (Figure 6E). Pulse–chase experiments using HEK293T cells that stably expressed Flag-tagged UCH37 showed almost the same half-life of UCH37 proteins in knockdown cells and control cells (Figure 6F). Semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) showed almost the same expression levels of UCH37 mRNA in hRpn13-knockdown cells and control cells (Figure 6G). Considered together, these results indicate that hRpn13 is required for maintaining normal protein levels of UCH37, and that loss of UCH37 proteins in hRpn13 is not due to metabolic instability of UCH37 proteins or due to repression of transcription of UCH37 mRNA. At present, the mechanism is not clear.
Figure 6.
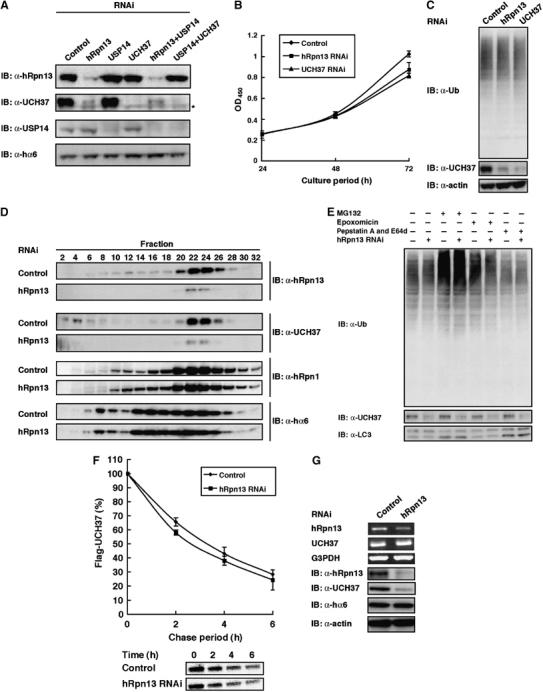
Knockdown of hRpn13 causes loss of UCH37 proteins. (A) HEK293T cells were transfected with siRNA against hRpn13, UCH37, or USP14. Where indicated, cells were transfected with a mixture of siRNAs. After 96 h, cell extracts were subjected to SDS–PAGE, followed by immunoblotting with the indicated antibodies. (B) Growth rates of hRpn13 knockdown, UCH37 knockdown, and mock cells. The cells were seeded in triplicate in 96-well dishes on day 0 (72 h after transfection), cultured in normal growing medium, and their proliferation was measured every 24 h. Data are mean±s.d. values. (C) Cell extracts of hRpn13 knockdown, UCH37 knockdown, and mock cells were subjected to SDS–PAGE, followed by immunoblotting with the indicated antibodies. (D) Samples fractionated by 10–40% glycerol gradient centrifugation were immunoblotted with the indicated antibodies. (E) Lack of accumulation of UCH37 proteins in hRpn13-knockdown cells following inhibition of the proteasome or lysosomal cathepsins. HEK293T cells transfected with siRNA were treated with MG132 (50 μM, 2 h), epoxomycin (1 μM, 12 h), or E-64 and pepstatin A (10 μg/ml, 12 h). The cells were lysed and subjected to immunoblot. (F) Pulse–chase analysis of Flag-UCH37. HEK293T cells stably expressing Flag-UCH37 were pulse-labeled for 30 min with 35S-labeled methionine and chased for the indicated time periods. After immunoprecipitation with M2 agarose, samples were separated by SDS–PAGE and autoradiographed (bottom panels). The upper panel shows the results of band quantitative analysis. Data are mean±s.d. values of three independent experiments. (G) hRpn13 knockdown does not alter UCH37 mRNA transcription. Semiquantitative RT–PCR (the upper three panels) and immunoblotting (the lower three panels) were performed using total RNA and proteins extracted from control and hRpn13-knockdown cells 48 h after transfection with siRNAs.
Knockdown of hRpn13 decreases deubiquitinating activities of 26S proteasomes
Finally, we tested the deubiquitinating activities of knockdown cells shown in Figure 6A. As there are abundant DUBs that are not associated with proteasomes, we partially purified complexes of 26S proteasomes and proteasome-associated DUBs by glycerol-density gradient centrifugation, and the 26S proteasome fractions identified by Suc-LLVY-MCA-hydrolyzing activities were used in the following experiments. As shown in Figure 7A, the 26S proteasome fraction of each knockdown cell contained proteasome subunits at a comparable level to each other. Proteins of hRpn13, UCH37, and USP14 were almost completely lost in 26S proteasomes of the cells transfected with siRNAs against hRpn13, UCH37, and USP14, respectively. The deubiquitinating activities of these samples were assayed using ubiquitin-AMC as a substrate. Notably, the deubiquitinating activities of hRpn13- and UCH37-deficient proteasomes were approximately only one-third of those of the control. In contrast, knockdown of USP14 did not significantly reduce the activity. Concomitant knockdown of USP14 with knockdown of hRpn13 or UCH37 did not have additive effects. These results clearly indicate that UCH37 is the dominant DUB associated with mammalian 26S proteasomes and that recruitment of UCH37 by hRpn13 is required for this activity.
Figure 7.
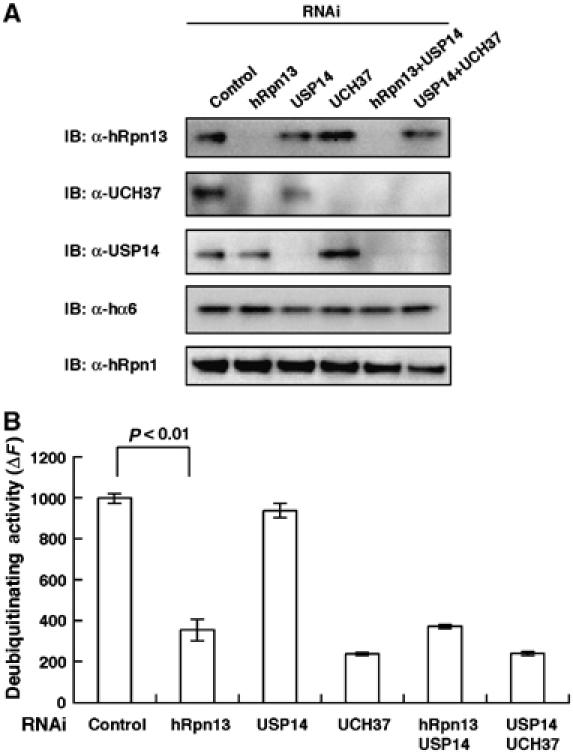
Knockdown of hRpn13 reduces deubiquitinating activities of 26S proteasomes. (A) hRpn13 and proteasome associating DUBs in knockdown cells. Cell extracts shown in Figure 6A were fractionated by 10–40% glycerol gradient centrifugation into 32 fractions. Immunoblot analysis of 26S proteasome fractions determined by Suc-LLVY-hydrolyzing activity was performed using the indicated antibodies. (B) The deubiquitinating activities of 26S proteasome fractions shown in panel A were measured using ubiquitin-AMC as a substrate. hRpn13- and UCH37-knockdown cells showed significantly reduced deubiquitinating activities (P<0.01). Data are mean±s.e.m. values of three independent experiments.
Discussion
Adrm1 was originally described as a heavily glycosylated membrane protein of molecular mass 110 kDa (Shimada et al, 1991). However, recent studies showed that Adrm1 was hardly, if any, glycosylated and most of it could be detected as a 42 kDa protein (Simins et al, 1999; Hasegawa et al, 2001; Lamerant and Kieda, 2005). Likewise, the antibody against Adrm1 we raised in this report did not detect the 110 kDa form of Adrm1. Moreover, immunocytochemical analysis of HeLa cells using this antibody indicated that Adrm1 is a soluble protein distributed both in the cytosol and nucleus, which is quite similar to the distribution of proteasomes, as revealed by staining for hRpn1 (Figure 1). Database search analysis suggested that Adrm1 is a human ortholog of yeast Rpn13 subunit. Indeed, Adrm1 was identified in the purified 26S proteasomes at nearly stoichiometric amount, and so we designated it hRpn13. While this manuscript was in preparation, Jorgensen et al (2006) reported Adrm1 as a novel proteasome-associated factor (Jorgensen et al, 2006). We also identified hRpn2 as an hRpn13-interacting subunit (Figure 3). A previous report mapped the location of p37A (Drosophila ortholog of UCH37) in Drosophila 26S proteasomes to the interface between the base and the lid, by electron microscopy using gold-labeled ubiquitin-aldehyde bound to the p37A/UCH37 (Holzl et al, 2000), which is consistent with our results.
However, immunodepletion analysis showed that not all the 26S proteasomes contain hRpn13 (Figure 2D). In this regard, it is better not to call hRpn13 a subunit of proteasomes but rather regard it as one of the PIPs. Even in yeast, there is no definite evidence to indicate that Rpn13 is a constitutive subunit of proteasomes. In a quantitative mass spectrometric analysis of budding yeast 26S proteasomes, Rpn13 was identified by much smaller number of peptides, compared to authentic proteasome subunits (Guerrero et al, 2006). The relatively low homology between Adrm1 and yeast Rpn13 may reflect this fact, as genuine proteasome subunits, but not PIPs, have much higher similarities between human and yeast subunits. Knockdown experiments further revealed that hRpn13 is not essential for proteolysis or viability of mammalian cells, whereas knockdown of hRpt2, a 19S ATPase subunit, was fatal (Figure 4). It is noteworthy that all subunits of RP, except Rpn9, Rpn10, and Rpn15/Sem1, are essential for proliferation of the budding yeast examined so far. Considered together, these results suggest that hRpn13 plays an auxiliary role in the proteasome-dependent protein degradation.
hRpn13 has no known functional motifs and does not seem to be necessary for the structural integrity of proteasomes. Then, what is the role of hRpn13 in proteasomes? hRpn13 can be divided into three functional regions: the N-terminal, C-terminal, and the inserted regions. The former two regions are well conserved from budding yeast to human, whereas the latter one is not found in budding yeast (Figure 1). We proved that the conserved N-terminal region is required for association with proteasomes (Figure 3). We also examined the biological role of the C-terminal region, and our results revealed that it serves as an acceptor for UCH37 (Figure 5). Moreover, loss of hRpn13 proteins caused concurrent loss of UCH37 proteins, indicating that hRpn13 recruits UCH37 to proteasomes and at the same time it is required for maintenance of UCH37 protein levels (Figure 6). As the half-life of UCH37 protein in hRpn13-knockdown cells was similar to that of control cell, the role of hRpn13 in maintaining UHC37 protein levels does not seem to be the stabilization of UCH37 protein. Consistent with this notion, neither the use of a proteasome inhibitor nor a lysosomal inhibitor resulted in accumulation of UCH37 proteins in knockdown cells. The amount of UCH37 mRNA was also unaltered in knockdown cells. At present, we do not know the reason for loss of UCH37 protein in knockdown cells, and the role of the insertion region is yet to be determined. No ortholog of UCH37 is found in budding yeast. Evolutionarily, UCH37 orthologs have emerged synchronously with the insertion region of Rpn13 orthologs. This region may have some role in the relationship with UCH37 and/or other proteins yet to be identified.
Ubp6 (USP14 in mammals) is another well-known DUB that associates with proteasomes. A mutation in USP14 in mice causes neurological disorders, demonstrating the importance of USP14 in mammals. Low expression of USP14 in mice is associated with reduced levels of free ubiquitin, suggesting its role in recycling ubiquitins (Anderson et al, 2005), as is also suggested in studies in budding yeast (Guterman and Glickman, 2004). However, the deubiquitinating activity of proteasomes is mainly attributed to UCH37 in fission yeast (Stone et al, 2004). Our present study also demonstrates that the deubiquitinating activities of 26S proteasomes are affected more profoundly by loss of UCH37 than USP14, indicating that UCH37 is the dominant DUB over USP14 in mammalian proteasomes. Recruitment of UCH37 by hRpn13 is essential for the activity of UCH37, as hRpn13 knockdown resulted in loss of UCH37, followed by a decrease in the deubiquitinating activity of 26S proteasomes at a level comparable to UCH37 knockdown. Further studies are required to distinguish the roles of USP14 (Ubp6) and UCH37 in organisms that have both molecules. It is plausible that the two molecules have distinct roles.
Despite the significant reduction of deubiquitinating activity of hRpn13-deficient proteasomes, they efficiently degraded substrate proteins such as ODC, ubiquitinated c-IAP, and IκBα (Figure 4). These observations seem to be in marked contrast to the case of yeast Rpn13, whose deletion caused a complete defect in degradation of a UFD substrate (Verma et al, 2000). However, as yeasts that lack Rpn13 are viable (Winzeler et al, 1999), the proteolytic defect is probably specific to UFD substrates. Rpn13 may recruit a component essential for degradation of UFD substrates in yeast. The precise role of hRpn13 and UCH37 still remains elusive. Curiously, knockdown of hRpn13 significantly increased the degrading activity of ODC but not that of ubiquitinated cIAP1 protein in vitro and IκBα in vivo. As sole knockdown of UCH37 did not increase the degrading activity of ODC (data not shown), the results observed in hRpn13 knockdown are not simply due to loss of UCH37. hRpn13 may influence access of protein to the channel of ATPase rings of proteasomes by sitting in the space between the base and lid (Holzl et al, 2000), or may recruit proteins that are relevant to proteolysis other than UCH37. At least from these results, both hRpn13 and UCH37 do not seem to be generally required for protein degradation by 26S proteasomes. It is assumed that UCH37 disassembles polyubiquitin chains from the distal end, shortening it such that the attached proteins can be released from the proteasome if there is a delay in efficient degradation (Stone et al, 2004). hRpn13 and UCH37 may be important in some specific situations. Further studies are required to determine the specific functions of hRpn13 and UCH37.
Materials and methods
Plasmids and cloning
The complementary DNAs (cDNAs) used in the present study were obtained by RT–PCR from total RNA isolated from HeLa cells or mouse livers using Superscript III (Invitrogen, San Diego, CA) and Pyrobest DNA polymerase (Takara Shuzo, Ohtsu, Japan). All amplified fragments were cloned into pcDNA3.1 vector (Invitrogen) and sequenced for confirmation. Deletion mutants (hRpn13ΔC, ΔN, N, ΔKE1 and ΔKE1+2, and UCH37ΔKE) were generated by PCR from wild-type hRpn13 and UCH37 and the Flag tag was introduced at the N-terminus of the constructs. For expression of GST and 6xHis fusion proteins, the cDNAs were subcloned into pGEX6P-1 (Amersham Life Science, Buckinghamshire, UK) and pET-28a (Novagen, Madison, WI) vector, respectively.
Immunological analysis
For immunoprecipitation analysis, HEK293T cells were transfected with plasmids using Fugene 6 (Roche, Mannheim, Germany). After 36 h, the cells were lysed with ice-cold phosphate-buffered saline (PBS) containing 1% Nonidet P-40 (NP-40) and centrifuged at 20 000 g for 10 min at 4°C. The supernatant was added with M2-agarose (Sigma Chemical Co., St Louis, MO) and rotated for 1 h at 4°C. The immunoprecipitates were washed five times with ice-cold PBS containing 0.5% NP-40 and then boiled in SDS sample buffer in the presence of β-mercaptoethanol (β-ME). Samples were subjected to SDS–PAGE, transferred to polyvinylidene fluoride membrane, and analyzed by immunoblotting with anti-Flag (M2; Sigma). Polyclonal antibodies against hRpn1, hRpn10, hRpt2, hRpn13, UCH37, and USP14 were raised in rabbits using recombinant proteins expressed in and purified from BL21RIL strain (Novagen) as GST fusion proteins: mouse Rpn1 (full length), mouse Rpn10 (residues 1–251), hRpt2 (residues 1–82), hRpn13 (residues 361–407), mouse UCH37 (residues 228–329), and human USP14 (full length). Anti-LC3 antibody was described previously (Komatsu et al, 2005). The antibodies for polyubiquitin and actin were purchased (FK2; MBL, Ina, Japan, Chemicon International Inc., Temecula, CA). All experimental protocols described in this study were approved by the Ethics Review Committee for Animal Experimentation of Tokyo Metropolitan Institute of Medical Science.
GST pull-down assay
Recombinant GST- or 6xHis-tagged proteins were produced in Escherichia coli and purified with glutathione Sepharose 4B (Amersham) or Ni-NTA Sepharose (Qiagen, Hilden, Germany). After elution of proteins from the beads, the preparations were dialyzed against buffer A (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM β-ME, and 10% glycerol). In GST pull-down analysis, 5 μg of each sample was mixed in 600 μl of buffer A and constantly rotated for 1 h at 4°C, and then 30 μl of glutathione Sepharose 4B was added and further rotated for 1 h. After washing with buffer A, bound proteins were eluted with 10 mM glutathione and subjected to SDS–PAGE, followed by immunoblot and silver staining (Wako Pure Chemical Industries, Osaka, Japan).
RNAi experiments
siRNAs targeting hRpt2, hRpn13, UCH37, and USP14 with the following 25-nucleotide sequences were purchased from Invitrogen: Rpt2, 5′-GGAGUACGAUGUGUAAGUGCCCAUU-3′; Rpn13, 5′-GGAGGGUCUACGUGCUGAAGUUCAAA-3′; UCH37, 5′-ACCGAGCTCATTAAAGGATTCGGTT-3′; USP14, 5′-UCAGCAUCGUAACACCAGAAGAUAU-3′. siRNAs were transfected into HEK293T cells with Lipofectamine 2000 (Invitrogen) at a final concentration of 2 nM in six-well dishes. The cells were analyzed 96 h (hRpn13, UCH37, USP14, hRpn13+USP14, and USP14+UCH37) or 48 h (hRpt2) after transfection. For protease inhibition assay, HEK293T cells transfected with siRNA (96 h) were cultured in the presence or absence of protease inhibitors (50 μM MG132 for 2 h, 10 μg/ml E64d and 10 μg/ml pepstatin A for 12 h, or 1 μM epoxomycin for 12 h). Cell growth was measured using Cell Counting Kit-8 (Wako) according to the instructions provided by the manufacturer. Briefly, cells were seeded at 1 × 103 cells/well in 96-well plates. Absorbance was measured using Microplate reader (Bio-Rad).
Glycerol-density gradient analysis
Mouse livers were homogenized in a Potter-Elvehjem Homogenizer in buffer B (in mM, 25 Tris–HCl (pH 7.5), 2 ATP, 5 MgCl2, and 1 dithiothreitol). HEK293T cells were lysed in buffer B containing 0.2% NP-40. The homogenates and lysates were clarified by centrifugation at 20 000 g and subjected to 10–40% (v/v) or 8–32% (v/v) linear glycerol-density density gradient centrifugation (22 h, 83 000 g) as described previously (Hirano et al, 2005).
In vitro assay of proteasome activity
Proteasome peptidase activity was measured using a peptide substrate, succinyl-Leu-Leu-Val-Tyr-7-amino-4-methyl-coumarin (Suc-LLVY-MCA), and the degradation of the recombinant 35S-labeled ODC was assayed in the presence of ATP, an ATP-regenerating system, and AZ, as described previously (Hirano et al, 2005). For the assay of cIAP1 degradation, cDNAs encoding Flag-cIAP1 subcloned into pcDNA3.1 were transcribed in vitro, translated, and radiolabeled as described previously (Hirano et al, 2005). The 35S-labeled Flag-cIAP1 was purified using M2-agarose (Sigma) and eluted with Flag-peptide (Sigma). For ubiquitination of cIAP1, 3 000 000 c.p.m. of 35S-labeled cIAP1, 0.25 μg of E1, 0.9 μg of UbcH5, and 33 μg of ubiquitin (Sigma) were mixed and incubated in a volume of 80 μl for 90 min at 30°C, as described previously (Murata et al, 2001). Finally, 2.5 μl of the ubiquitination mixture was added to 10 μl of cell lysates in the presence of 2 mM ATP, incubated at 37°C for 20 min, and then radioactivities of trichloroacetic acid-soluble fractions were measured.
TNF--dependent IκB degradation
HEK293T cells transfected with siRNA were treated with 100 μg/ml cycloheximide (Sigma) for 10 min, and then human TNF-α (Genzyme, Cambridge, MA) was added at a final concentration of 10 ng/ml. Changes in the protein levels of endogenous IκBα after treatment with TNF-α were analyzed by immunoblotting with anti-IκBα (c-21) (Santa Cruz Biotechnology, Santa Cruz, CA).
Deubiquitination assay
For ubiquitin-7-amino-4-methylcoumarin (ubiquitin-AMC) (Boston Biochem) hydrolysis assays, 10 μl of 26S proteasome fraction separated by glycerol gradient centrifugation was incubated with 0.25 μM ubiquitin-AMC for 15 min at 37°C. The release of AMC was measured fluorometrically.
Pulse–chase analysis
HEK293T cells stably expressing Flag-UCH37 were transfected with siRNA for hRpn13 or control siRNA. Pulse–chase experiments were performed as described previously (Hirano et al, 2005).
RT–PCR analysis
Total RNA (2.5 μg) was reverse transcribed using SuperScript III (Invitrogen) and oligo(dT)20 primers. Specific primers for each gene were as follows: 5′-AAGGATCCATGAGCATCCTGGCCACGATGAAC G-3′ and 5′-TTCTCGAGTCAGTCCAGGCTCATGTCCTCC-3 ′ for hRpn13, 5′-AAGGATCCATGACGGGCAACGCCGGGGAG-3′ and 5′-TTCTCGAGTCATTTGGTTTCCTGAGCTTTC-3 ′ for UCH37, and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for G3PDH.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank Y Murakami for providing the ODC degradation assay system and K Furuyama for technical support. This work was supported in part by a grant to SM from JST and grants to SM and KT from the Ministry of Education, Science and Culture of Japan.
References
- Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, Wilson SM (2005) Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem 95: 724–731 [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 [DOI] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65: 801–847 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94: 615–623 [DOI] [PubMed] [Google Scholar]
- Guerrero C, Tagwerker C, Kaiser P, Huang L (2006) An integrated mass spectrometry-based proteomic approach. Mol Cell Proteomics 5: 366–378 [DOI] [PubMed] [Google Scholar]
- Guterman A, Glickman MH (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem 279: 1729–1738 [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Sakurai N, Kinoshita T (2001) Xoom is maternally stored and functions as a transmembrane protein for gastrulation movement in Xenopus embryos. Dev Growth Differ 43: 25–31 [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S (2005) A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437: 1381–1385 [DOI] [PubMed] [Google Scholar]
- Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, Baumeister W (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol 150: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JP, Lauridsen AM, Kristensen P, Dissing K, Johnsen AH, Hendil KB, Hartmann-Petersen R (2006) Adrm1, a putative adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol 360: 1043–1052 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, Tachikawa T, Shin S, Ichihara A (1990) Abnormally high expression of proteasomes in human leukemic cells. Proc Natl Acad Sci USA 87: 7071–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385: 737–740 [DOI] [PubMed] [Google Scholar]
- Lamerant N, Kieda C (2005) Adhesion properties of adhesion-regulating molecule 1 protein on endothelial cells. FEBS J 272: 1833–1844 [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507 [DOI] [PubMed] [Google Scholar]
- Li T, Duan W, Yang H, Lee MK, Bte Mustafa F, Lee BH, Teo TS (2001) Identification of two proteins, S14 and UIP1, that interact with UCH37. FEBS Lett 488: 201–205 [DOI] [PubMed] [Google Scholar]
- Li T, Naqvi NI, Yang H, Teo TS (2000) Identification of a 26S proteasome-associated UCH in fission yeast. Biochem Biophys Res Commun 272: 270–275 [DOI] [PubMed] [Google Scholar]
- MacArthur MW, Thornton JM (1991) Influence of proline residues on protein conformation. J Mol Biol 218: 397–412 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A (1992) Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360: 597–599 [DOI] [PubMed] [Google Scholar]
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K (2001) CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T (2002) A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem 74: 4725–4733 [DOI] [PubMed] [Google Scholar]
- Realini C, Rogers SW, Rechsteiner M (1994) KEKE motifs. Proposed roles in protein–protein association and presentation of peptides by MHC class I receptors. FEBS Lett 348: 109–113 [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa M, Schlom J, Greiner JW (1991) Identification of a novel tumor-associated Mr 110 000 gene product in human gastric carcinoma cells that is immunologically related to carcinoembryonic antigen. Cancer Res 51: 5694–5703 [PubMed] [Google Scholar]
- Shimada S, Ogawa M, Takahashi M, Schlom J, Greiner JW (1994) Molecular cloning and characterization of the complementary DNA of an M(r) 110 000 antigen expressed by human gastric carcinoma cells and upregulated by gamma-interferon. Cancer Res 54: 3831–3836 [PubMed] [Google Scholar]
- Simins AB, Weighardt H, Weidner KM, Weidle UH, Holzmann B (1999) Functional cloning of ARM-1, an adhesion-regulating molecule upregulated in metastatic tumor cells. Clin Exp Metast 17: 641–648 [DOI] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL (2005) ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell 20: 687–698 [DOI] [PubMed] [Google Scholar]
- Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C (2004) Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol 344: 697–706 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K (2000) Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J Biol Chem 275: 2877–2884 [DOI] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR III, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11: 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin–proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD (2000) Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 288: 874–877 [DOI] [PubMed] [Google Scholar]
- Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


