Abstract
The import motor of the mitochondrial translocase of the inner membrane (TIM23) mediates the ATP-dependent translocation of preproteins into the mitochondrial matrix by cycles of binding to and release from mtHsp70. An essential step of this process is the stimulation of the ATPase activity of mtHsp70 performed by the J cochaperone Tim14. Tim14 forms a complex with the J-like protein Tim16. The crystal structure of this complex shows that the conserved domains of the two proteins have virtually identical folds but completely different surfaces enabling them to perform different functions. The Tim14–Tim16 dimer reveals a previously undescribed arrangement of J and J-like domains. Mutations that destroy the complex between Tim14 and Tim16 are lethal demonstrating that complex formation is an essential requirement for the viability of cells. We further demonstrate tight regulation of the cochaperone activity of Tim14 by Tim16. The first crystal structure of a J domain in complex with a regulatory protein provides new insights into the function of the mitochondrial TIM23 translocase and the Hsp70 chaperone system in general.
Keywords: chaperone, mitochondria, protein translocation
Introduction
The biogenesis of mitochondria requires the import of many hundreds of different proteins that are synthesized as preproteins in the cytosol. This process is mediated by several preprotein translocases present in the mitochondrial membranes (Endo et al, 2003; Koehler, 2004; Rehling et al, 2004; Mokranjac and Neupert, 2005). The majority of preproteins carry an N-terminal matrix targeting signal (MTS) and use the translocase of the outer membrane (TOM) complex in the outer membrane and the translocase of the inner membrane (TIM23) complex in the inner membrane for concerted translocation across outer and inner membranes. The TOM translocase is the common entry gate for these preproteins and facilitates recognition and initial translocation of the MTS.
The TIM23 translocase accepts the preproteins directly from the TOM complex. It consists of 10 known components, which are highly conserved throughout the eukaryotic kingdom and are, with two exceptions, essential for cell viability. The TIM23 complex can be operationally divided into a membrane integrated channel unit and an import motor that is associated with the channel at its matrix face. The channel unit recognizes the MTS of the preproteins as they appear at the outlet of the TOM pore and subsequently mediates their insertion into and translocation across the inner mitochondrial membrane. This latter process requires the electrical membrane potential ΔΨ across the inner membrane. Further translocation into the matrix is performed by the import motor in a reaction driven by ATP hydrolysis. The central component of the motor is mitochondrial Hsp70 (mtHsp70).
Hsp70 chaperones are involved in folding of newly synthesized proteins, prevention of protein aggregation and protein translocation. Like all members of the Hsp70 family of chaperones, mtHsp70 consists of an N-terminal nucleotide-binding domain and a C-terminal peptide-binding domain. Hsp70 chaperones use the energy of ATP hydrolysis at the nucleotide-binding domain to drive conformational changes of the peptide-binding domain, thereby altering its affinity for substrates (Bukau and Horwich, 1998; Young et al, 2004; Jiang et al, 2005; Vogel et al, 2006). In the case of mtHsp70, the substrates are the precursor polypeptides that are entering the mitochondrial matrix. The ATP-hydrolysis-driven cycle allows Hsp70 chaperones to associate with unfolded protein substrates in the ATP form and then bind them tightly upon hydrolysis to ADP. Release of ADP and rebinding of ATP triggers the release of the bound substrate.
Progression through the ATPase cycle of Hsp70s is generally helped by two types of cochaperones, nucleotide exchange factors and J proteins, named after the founding member of the family, Escherichia coli cochaperone DnaJ. The mtHsp70 in the import motor requires Mge1, a member of the GrpE family which catalyzes the nucleotide exchange reaction and a specific J protein, Tim14/Pam18, which stimulates the ATPase activity (D'Silva et al, 2003; Mokranjac et al, 2003; Truscott et al, 2003). In addition, the import motor contains at least two more essential proteins, Tim16/Pam16 and Tim44. Tim16 is a J-like protein as it contains a domain with sequence similarity to the J domain of Tim14 but lacks the invariant tripeptide motif His-Pro-Asp (HPD), a signature of J proteins (Kelley, 1998; Frazier et al, 2004; Kozany et al, 2004). Tim16 forms a subcomplex with Tim14 in the import motor and appears to be necessary for the recruitment of Tim14 to the translocase. In in vitro experiments, Tim16 did not stimulate the ATPase activity of mtHsp70. Rather, it inhibited the stimulatory activity of Tim14 (Li et al, 2004; D'Silva et al, 2005; Mokranjac et al, 2005). Tim44 can be viewed as a central organizer of the import motor. On the one hand, it binds to the channel unit of the TIM23 complex and on the other hand, it interacts with mtHsp70 thereby recruiting the chaperone to the translocase. In addition, Tim44 is necessary to link the Tim14–Tim16 subcomplex to the translocase.
The exact mechanism of the import motor is still a matter of debate (Matouschek et al, 2000; Neupert and Brunner, 2002). It is, however, clear that multiple cycles of precursor binding to and release from mtHsp70 lead to a stepwise translocation of the unfolded polypeptide chain into the matrix. A deep understanding of the structure and function of the import motor is greatly hampered by the lack of a high-resolution structure of its components.
Here we report the crystal structure of the Tim14–Tim16 subcomplex of the import motor. It reveals a unique arrangement of J and J-like domains. Our results show that the cochaperone activity of Tim14 is not sufficient for its function within the import motor, but that the regulation of its activity is equally important. We show that the regulation is mediated by Tim16 and provide insights into the regulatory process based on the structure of the heterodimer. Furthermore, we present evidence for a higher oligomeric state of the complex. In the end, we propose a model as to how Tim14 and Tim16 interact in a dynamic manner with each other and with Tim44 to couple the ATP hydrolysis by mtHsp70 to the vectorial movement of preproteins into the mitochondria.
Results
Domain structure of Tim14 and Tim16
Tim14 is anchored in the inner mitochondrial membrane by a single transmembrane domain and exposes a conserved, C-terminal domain into the matrix (Figure 1A, left panel). The J domain, necessary for the function of Tim14, is a part of this conserved domain. Yeast Tim14, in addition, exposes an N-terminal domain into the intermembrane space. In order to address the importance of these domains for the function of Tim14, we analyzed the ability of truncation mutants to support growth of yeast cells in the absence of the wild-type protein. In the first mutant described here the intermembrane space domain was deleted, whereas in the second mutant only the conserved domain in the matrix was present. As the mitochondrial targeting information of Tim14 is located in its transmembrane domain and a segment directly following it, the second mutant of Tim14 was targeted to the mitochondrial matrix by adding the presequence of subunit 9 of the FOF1 ATPase. Deletion of the N-terminal segment of Tim14 had virtually no effect on the function of Tim14 in yeast, in agreement with the lack of its conservation through evolution (Figure 1A, right panel). Interestingly, this domain was recently proposed to be essential for the recruitment of the import motor to the membrane embedded part of the TIM23 translocase (Chacinska et al, 2005). However, the mutant lacking the entire intermembrane space domain of Tim14 grew like wild-type yeast under all conditions tested, including both fermentable and nonfermentable carbon sources (data not shown). Even the combined deletion of the intermembrane and transmembrane domains did not have a strong effect on the functionality of Tim14. The mutant containing only the conserved matrix domain was still able to support growth of cells and could functionally replace wild-type Tim14. This domain consists of the J domain and additional 10 residues at its N-terminal side.
Figure 1.
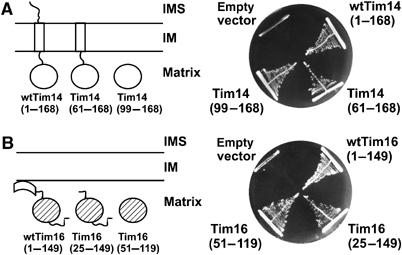
Domain analysis of Tim14 and Tim16. (A) Schematic representation of Tim14 domain structure and of truncation mutants (left panel). A haploid deletion strain of TIM14 harboring a wild-type copy of TIM14 on the URA plasmid was transformed with centromeric plasmids carrying either wild-type Tim14 or the indicated Tim14 mutants. Cells were plated on medium containing 5-fluoroorotic acid, which selects for cells that have lost the URA plasmid. Plasmids carrying wild-type Tim14 or an empty plasmid were used as positive and negative controls, respectively (right panel). (B) The same analysis was performed with Tim16. IM, inner membrane of mitochondria; IMS, intermembrane space.
Tim16 does not contain a predicted transmembrane segment, still it is firmly attached to the inner membrane (Figure 1B, left panel). It contains two conserved domains, one at the very N-terminus and one towards the C-terminus of the protein. They are connected by a linker whose sequence is not conserved. A further extension, not conserved in sequence and length, is present at the C terminus. The N-terminal domain consists of about 25, mostly hydrophobic amino-acid residues. This domain is most likely responsible for the membrane association of Tim16. The conserved J-like domain is predicted to have the characteristics of a J domain but has a DKE/S motif instead of HPD. The importance of the various domains for the function of Tim16 was analyzed in the same way as described above for Tim14. In a first mutant, the N-terminal 24 amino acids were deleted. A second mutant contained only the conserved J-like domain. The targeting signal for import of Tim16 is present in the N-terminal domain (our unpublished data). Therefore, both mutants were targeted to the mitochondrial matrix using the presequence of subunit 9 of the FOF1 ATPase. Both mutants were able to support growth of cells lacking wild-type Tim16 (Figure 1B, right panel). We therefore conclude that the J-like domain is the only essential part of Tim16.
Crystallization and structure determination of the Tim14–Tim16 complex
To understand assembly, architecture, subunit interactions and functions of Tim14 and Tim16, we set out to determine the crystal structure of the essential parts of these proteins. When expressed separately in E. coli cells, the essential domains of both Tim14 and Tim16 aggregated and were mostly found in inclusion bodies. We therefore coexpressed Tim14 and Tim16 from a single vector. Both proteins were found soluble in the E. coli cytosol under these conditions (data not shown). Furthermore, when a His6 tag was placed at the N-terminus of Tim14, both proteins were efficiently copurified on an NiNTA-agarose column. They formed a complex in E. coli as they do in mitochondria. The essential parts of Tim14 and Tim16 are obviously sufficient for complex formation.
The Tim14 (99–168)–Tim16 (54–119) complex crystallized from 1.65 M sodium citrate, pH 7.0. The crystals were well ordered and diffracted to 2.0 Å resolution (see Table I). The crystal structure was determined by single-wavelength anomalous diffraction phasing method using a K2OsCl6–Tim14–Tim16 heavy metal derivative. The refined structure revealed a well-defined electron density for the whole Tim14–Tim16 complex, except for the last two amino-acid residues of Tim16, which were structurally disordered. The asymmetric unit contained eight Tim14–Tim16 heterodimers, which were virtually identical. The overlay of the core structures of eight Tim14 and eight Tim16 molecules showed r.m.s. deviations for Cα positions of <0.3 Å and <0.6 Å in Tim14 and Tim16, respectively. In successive rounds of model building and refinement, the model was completed and included 921 water molecules and four citrate molecules. Temperature factors were refined with restraints between bonded atoms and between noncrystallographic symmetry-related atoms yielding R-values of Rfree=25.2% and Rcrys=20.5%.
Table 1.
Data collection and refinement statistics
| Tim14–Tim16 | Tim14–Tim16–K2OsCl6 | |
|---|---|---|
| Crystal parameters | ||
| Space group | P212121 | P43212 |
| Cell constants | a=112 Å; b=115 Å; c=162 Å | a=b=114Å; c=163 Å |
| Molecules per AUa | 16 | 8 |
| Heavy metal | — | 7 |
| Data collection | ||
| Beam line | BW6, DESY | BW6, DESY |
| Wavelength (Å) | 1.05 | Pk 1.1402 |
| Resolution range (Å)b | 99–2.0 (2.03–2.00) | 99–3.5 (3.52–3.47) |
| No. of observations | 4 022 553 | 879 765 |
| No. of unique reflectionsc | 137 971 | 26 166 |
| Completeness (%)b | 97.7 (96.9) | 98.4 (100) |
| Rmerge (%)b,d | 8.8 (48.4) | 7.27 (27.0) |
| I/σ (I)b | 10.5 (2.6) | 54.8 (16.1) |
| Refinement (REFMAC5) | ||
| Resolution range (Å) | 15–2.0 | |
| No. of reflections working set | 121 692 | |
| No. of reflections test set | 6405 | |
| No. of non-hydrogen | 8846 | |
| Solvent water | 921 | |
| No. of citrates | 4 | |
| Rwork/Rfree (%)e | 20.5/25.2 | |
| Rwork/Rfree (2.03–2.00 Å) (%)e | 27.4/33.9 | |
| R.m.s.d. bond lengths (Å)/(deg)f | 0.02/1.75 | |
| Average B-factor (Å2) | 32.04 | |
| Ramachandran plot (%)g | 98.7/1.3/0.0 | |
| aAsymmetric unit. | ||
| bThe values in parentheses of resolution range, completeness, Rmerge and I/σ (I) correspond to the last resolution shell. | ||
| cFriedel pairs were treated as different reflections. | ||
| dRmerge(I)=∑hkl∑j ∣[I(hkl)j−I(hkl)] ∣/[∑hklIhkl, where I(hkl)j is the jth measurement of the intensity of reflection hkl, and 〈I(hkl)〉 is the average intensity. | ||
| eR=∑hkl∣∣Fobs∣−∣Fcalc∣∣/∑hkl∣Fobs∣, where Rfree is calculated without a sigma cutoff for a randomly chosen 10% of reflections, which were not used for structure refinement, and Rwork is calculated for the remaining reflections. | ||
| fDeviations from ideal bond lengths/angles. | ||
| gNumber of residues in favored region/allowed region/outlier region. | ||
Structures of individual Tim14 and Tim16 subunits
In the Tim14–Tim16 complex, the J domain of Tim14 follows the typical fold of J domains with the characteristic arrangement of three helices (I, II and III). It lacks, however, helix IV (Figure 2A, left panel). Helices II and III form an antiparallel hairpin connected by a short linker, which contains the invariant HPD motif. The hairpin structure is supported by helix I from the backside. The HPD motif is well defined in the electron density map and adopts an identical conformation in all eight subunits, suggesting that it is part of a rather rigid structure. The first 10 amino acids of the Tim14 construct are not part of the J domain structure and form an arm in front of helix I (see below).
Figure 2.
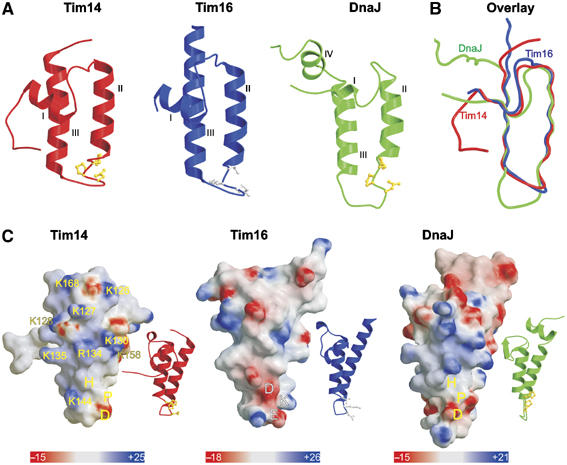
Topology and charge patterns of Tim14 and Tim16. (A) Ribbon presentations of the J domain of Tim14 (left panel) and the J-like domain of Tim16 (central panel). For comparison, the J domain of DnaJ from E. coli is included (right panel). Helices I–III are indicated. HPD motifs in the J domains are presented as balls-and-sticks models and colored in yellow. Corresponding DKE motif in Tim16 is colored in gray. As compared to the J domain of DnaJ, Tim14 and Tim16 lack helix IV. (B) Superposition of the J and J-like domains of Tim14, Tim16 and DnaJ. (C) Surface representations of Tim14, Tim16 and DnaJ. Ribbon presentations of the same orientation are shown as insets. The domains are rotated clockwise by 80° relative to the representations in (A) to bring helix II to the front. Surface colors indicate positive (intense blue) and negative electrostatic potentials (intense red) with the scale bar giving the actual potentials in kT/e. Lys and Arg residues of Tim14 contributing to the positive surface charge distribution are indicated.
The J-like domain of Tim16, in complex with Tim14, has all the structural characteristics of J domains described above (Figure 2A, middle panel). The loop connecting helices II and III is highly ordered. However, instead of the HPD motif it contains a conserved DKE/S motif.
Superposition of J and J-like domains of Tim14 and Tim16, respectively, shows a high structural consensus (Figure 2B). The only difference appears to be in the length of helix III, which is shorter in Tim14. Also, when Tim14 and Tim16 are compared to DnaJ, notable differences are visible only in the loop connecting helices II and III; this loop is longer in DnaJ (Figure 2B). All three structures diverge at the end of helix III.
NMR perturbation mapping and mutational analyses have located the binding site of J domains to their Hsp70 partners at one surface of the J domain (Greene et al, 1998; Genevaux et al, 2002). This binding surface, which is positively charged, is mostly formed by the HPD loop and helix II. The surface charge model shows that a similar positively charged surface is present on the corresponding face of the J domain of Tim14 (Figure 2C). Interestingly, such clustering of positive charges is not present on the corresponding face of Tim16 (Figure 2C).
Structure of the Tim14–Tim16 heterodimer
The Tim14–Tim16 complex is an elongated molecule with the two subunits arranged back-to-back in a way that the loops between helices II and III face opposite sides of the heterodimer (Figure 3A). The complex has a pseudo-two-fold symmetry. Numerous contacts between Tim14 and Tim16 arise from complementarity in both shape and polarity. The buried surface area at the interface between Tim14 and Tim16 subunits is about 1060 Å2. This corresponds to 20% of total surface of each Tim14 and Tim16. There are two major areas of interaction.
Figure 3.
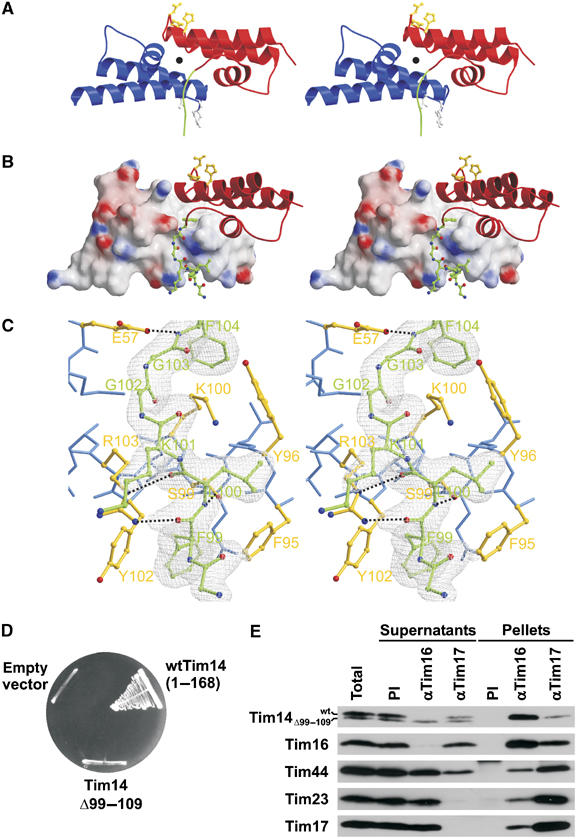
N-terminal arm of Tim14 embraces Tim16. (A) Ribbon model of Tim14–Tim16 heterodimer given in stereo representation. Tim14 is shown in red, Tim16 in blue. The HPD and DKE motifs are highlighted in yellow and gray, respectively, and shown as balls-and-sticks models. The pseudo-two-fold symmetry axis is indicated by a black full circle. The N-terminal arm of Tim14 is highlighted in green. (B) Stereo representation of interacting parts of Tim14 and Tim16. Tim 14 is shown as ribbon model, Tim16 as surface model. The N-terminal arm of Tim14 that embraces helix III of Tim16 is represented as balls-and-sticks model, carbon atoms are colored in green, oxygen atoms in red and nitrogen atoms in blue. (C) Stereo representation of a Tim14–Tim16 interacting section around the arm of Tim14. The electron density map (colored in gray), contoured from 1σ, is only displayed for the arm of Tim14 with 2FO−FC coefficients. Temperature factor refinement indicates full occupancies of the whole arm. The residues of the arm of Tim14 are numbered in green according to sequence numbering. Residues of Tim16, which are in close contact with the arm of Tim14, are colored in yellow and represented as balls-and-sticks, whereas remaining residues of Tim16 are colored in blue. Hydrogen bonds between Tim14 and Tim16 are shown as black dotted lines. (D) A Tim14 mutant which contains an intact J domain but lacks the arm region cannot support growth of yeast cells in the absence of wild-type Tim14. The analysis was carried out as described in the legend to Figure 1A. (E) A Tim14 mutant which lacks the arm region does not interact with Tim16 in vivo. Mitochondria were isolated from wild-type yeast cells transformed with a plasmid carrying a Tim14 mutant lacking the arm region. They were solubilized with digitonin and subjected to immunoprecipitation with affinity-purified antibodies to Tim16 and Tim17 or preimmune serum (PI) as control. Supernatants and pellets were analyzed by SDS–PAGE and immunodecoration with the indicated antibodies.
First, the N-terminal arm of Tim14 embraces helix III of Tim16 and, in addition, makes contacts with helices I and II (Figure 3B). These contacts are both polar and hydrophobic. Flanking residues of the Tim14 arm, Phe99 and Phe104, protrude into two distinct hydrophobic pockets formed predominantly by Tim16 (Figure 3C). They fix Tim14 by formation of a clamp. Two consecutive residues of Tim16, Phe95 and Tyr96 fix the Tim14 arm from the Tim16 side (for the detailed description of Tim14–Tim16 contacts, see Supplementary data).
Mutations in the HPD motif of Tim14 abolish its function as a J protein and have a lethal effect on yeast cells (D'Silva et al, 2003; Mokranjac et al, 2003). The arm that forms contacts with Tim16 is not part of the J domain and its deletion should not abolish the function of Tim14 as a cochaperone for mtHsp70. We therefore asked whether a mutant form of Tim14 that lacks the arm region is able to function in the import motor. Deletion of the arm region had a deleterious effect on the function of Tim14, as this mutant was not able to support cell growth at all (Figure 3D). Apparently, also mutations outside of the J domain can lead to a nonfunctional Tim14.
In order to test whether the arm region is necessary for complex formation between Tim14 and Tim16, the mutant form of Tim14 was expressed in wild-type yeast cells, mitochondria were isolated and complex formation was assessed by co-immunoprecipitation. The mutant form of Tim14 did not interact with Tim16 as it was absent from the precipitate of the Tim16 antibody and apparently remained in the supernatant (Figure 3E). It was also not recruited to the translocation unit as it was completely absent from the precipitate of Tim17 antibody. In contrast, antibodies to Tim16 depleted both Tim16 and wild-type Tim14 from the mitochondrial lysate, showing that these two proteins form a stable complex (Kozany et al, 2004 and Figure 3E), part of which was also recruited to the membrane embedded unit of the translocase as shown by partial co-precipitation with antibodies to Tim17. Taken together, complex formation is an essential property of Tim14 and Tim16. Interestingly, a mutagenesis analysis performed in order to narrow down the interaction to the level of single amino-acid residues in Tim14 and Tim16 showed that single and even double exchanges are tolerated at lower temperatures, indicating that the interaction network relies on many contacts and is therefore rather robust (Supplementary Figure 1). Still, these mutations lead to severe growth defects and are not tolerated at all at 37°C.
The second interface of Tim14 and Tim16 is a continuation of the hydrophobic groove formed around Phe104 of Tim14 and is responsible for the back-to-back arrangement of the two subunits (for details, see Supplementary data part II). A particularly interesting part of this interface is formed by the HPD loop of Tim14 and a groove made up by helices I and II of Tim16 (Figure 4A). The conserved Gly145 and Gly146 residues of Tim14, neighbors of the HPD motif at the C-terminal side, are deeply inserted in this groove. A network of interactions involving the backbone and side chains of Tim14 is responsible for this tight interaction (Figure 4B). The amide group in the side chain of Asn140 of Tim14, which flanks the HPD motif at the N-terminal side, is hydrogen bonded to the amide group in the side chain of Asn87 in Tim16. The carbonyl oxygen of Lys145 at the C-terminal side of the HPD motif in Tim14 makes a strong hydrogen bond to the hydroxyl group of Tyr82 in Tim16. Carbonyl oxygen of Gly145 is hydrogen bonded to the side chain of Arg79 in Tim16. Finally Ser147, the first residue of Tim14 helix III, is linked via its hydroxyl group to carbonyl oxygens of Ile61 and Lys60 in Tim16. The combination of these interactions and strong hydrophobic interactions between the helices of Tim14 and Tim16 lead to a severely constrained conformation of the HPD loop and a firm attachment to Tim16.
Figure 4.
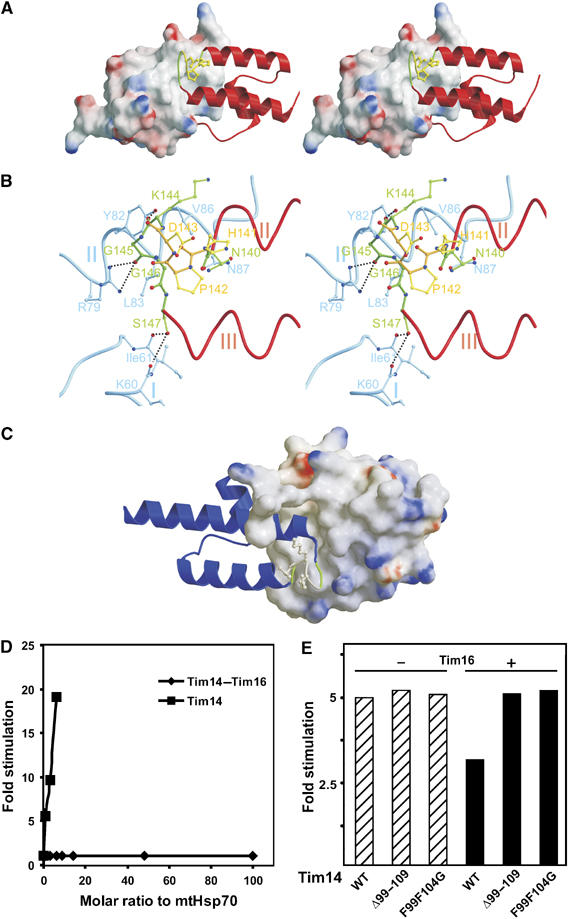
The HPD loop of Tim14 is projecting into a groove formed by Tim16. (A) Stereo representation of Tim14 as ribbon model projecting into a groove of Tim16 represented as surface model. The HPD containing loop connecting helices II and III is embedded into a groove made up by residues of helices I and II of Tim16. (B) Network of hydrogen bonds between the HPD loop of Tim14 and helices I and II of Tim16 leads to a constrained conformation of the HPD loop. (C) DKE loop in Tim16 (ribbon model) is similarly inserted into a Tim14 groove (surface model). (D) The Tim14–Tim16 complex does not stimulate the ATPase activity of mtHsp70 in vitro, in contrast to Tim14 alone. The factor of stimulation was determined by measuring the ATPase activity of mtHsp70 in the presence of either various amounts of Tim14–Tim16 complex or of Tim14 and relating it to intrinsic activity of mtHsp70. (E) The arm region of Tim14 is critical for the regulation of Tim14 by Tim16. Stimulation of mtHsp70's ATPase activity by wild-type Tim14 (WT) or its two mutant forms, Tim14Δ99–109 and Tim14F99F104G, was determined in the absence (−) or presence (+) of Tim16. A typical experiment is shown.
On the other hand, the DKE loop of Tim16 is in close contact with a groove made up by helices I and II of Tim14 (Figure 4C). This is a reflection of the pseudo-symmetry of the heterodimer. As in the corresponding contacts of Tim14 with Tim16, Asn87 of Tim16 is hydrogen bonded to Asn140 in Tim14, while the first amino-acid residue of helix III, Ser94 of Tim16 interacts with Gln115 and Ile116 in the helix I of Tim14. The DKE loop of Tim16 is, however, not as tightly inserted into the Tim14 groove as is the HPD loop in the Tim16 groove. Only the carbonyl oxygen of Gly92 in Tim16 is hydrogen bonded to the side chain of Asn118 in Tim14.
It seems questionable whether Tim14 in the described constrained conformation can function as an active cochaperone for mtHsp70. We therefore compared the ability of free Tim14 to stimulate the ATPase activity of mtHsp70 with that of Tim14 in the complex with Tim16. The chaperone was incubated with radiolabeled ATP at 25°C in the presence of Mge1 and either Tim14 alone or Tim14–Tim16 complex. Whereas Tim14 alone stimulated the ATPase activity of mtHsp70, Tim14 bound to Tim16 displayed no activity (Figure 4D). Apparently, even though the HPD loop and the positively charged surface of helix II are exposed in the complex, Tim16 blocks the cochaperone activity of Tim14. We assessed the stimulatory activities of two Tim14 mutants, Tim14Δ99–109 and Tim14F99F104G, which were not able to support growth of yeast at all or only poorly, respectively, and found virtually no difference as compared to the wild-type Tim14 protein (Figure 4E). However, addition of Tim16 did not inhibit their stimulatory activities, in contrast to the situation when Tim16 was added to the wild-type Tim14 (Figure 4E). This further confirms the importance of the Tim14 arm for the complex formation and the regulation of the import motor activity.
Higher order structure of the Tim14–Tim16 heterodimer
The asymmetric unit cell in the crystal structure contains an octamer of Tim14–Tim16 heterodimers. We asked whether higher oligomers are present in vivo and are therefore of physiological significance. We expressed His-tagged forms of Tim14 or Tim16 in wild-type yeast cells. We have previously shown that these forms are functional as they fully support growth of yeast cells lacking the respective wild-type protein (Mokranjac et al, 2005). Mitochondria were isolated from cells expressing either the His-tagged Tim14 or His-tagged Tim16 and from wild type as a control. They were lysed with digitonin and incubated with NiNTA-agarose beads. Proteins bound to the affinity matrix and those which remained in the supernatant fraction were analyzed by SDS–PAGE and immunoblotting (Figure 5A). In case of mitochondria containing His-tagged Tim14, both Tim16 and Tim44 were bound to the NiNTA-agarose beads together with the His-tagged Tim14. In addition, a significant amount of wild-type Tim14 was recovered in the bound fraction. As we have previously shown that the total amount of Tim14 present in mitochondria is bound to Tim16 (Kozany et al, 2004; Mokranjac et al, 2005), this result suggests that two Tim14 molecules are present in the complex. Furthermore, wild-type Tim16 was retained on the NiNTA-agarose beads together with the His-tagged Tim16. Thus, the complex appears to represent, at least, a tetramer.
Figure 5.
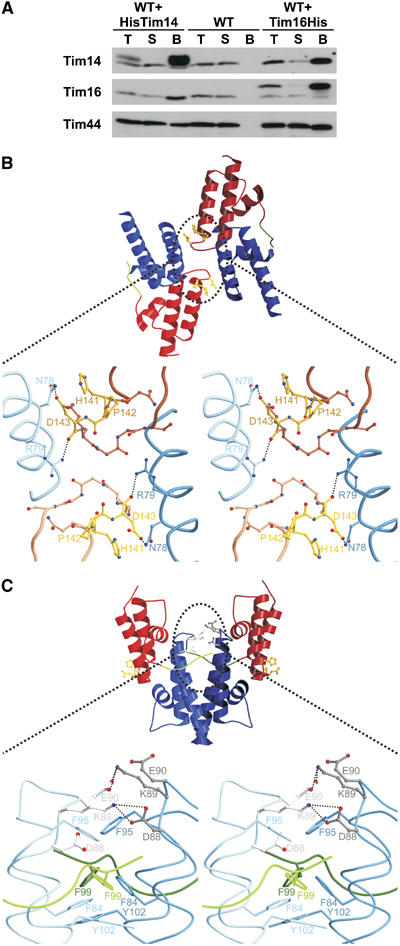
The Tim14–Tim16 complex is organized as a tetramer. (A) His-tagged versions of Tim14 or of Tim16 were expressed in wild-type yeast cells. Mitochondria were isolated, solubilized with digitonin and the detergent extracts were incubated with NiNTA-agarose beads. Untransformed wild-type cells were used as a control. Fractions were analyzed by SDS–PAGE and immunodecoration using antibodies against Tim14, Tim16 and Tim44 as indicated. Total (T) and supernatant (S) fractions represent 20% of the material present in the bound fractions. (B) Ribbon model of type I of Tim14–Tim16 heterodimer interactions observed in the crystal. In this type of tetramer, the HPD motifs of both Tim14 are masked. The enlargement of the contact area in the stereo representation shows that the contact is made essentially by two pairs of amino-acid residues in Tim14 and Tim16. (C) Ribbon model of type II of Tim14–Tim16 heterodimer interactions in the crystal structure. The contact is made by dimerization of the Tim16 subunits of two heterodimers. The stereo representation highlights the network of hydrogen bonds. In addition, a number of hydrophobic interactions stabilize this contact.
There are two types of tetrameric contacts in the crystal structure described here (Figures 5B and C). The tetramer shown in Figure 5B contains two heterodimers that are rotated about 60° relative to each other. The molecules are oriented in a way that each Tim16 contacts the HPD motif in Tim14 of the opposite heterodimer. Interactions between two heterodimers in this tetramer comprise essentially only two pairs of amino-acid residues. Arg79 of Tim16 from one heterodimer forms a hydrogen bond with the carbonyl oxygen of Asp143 in Tim14 from the other heterodimer. The other interaction occurs between Tim16 Asn78 and Asp143 in the HPD motif of Tim14 of the other heterodimer. In order to investigate whether these interactions represent a physiologically important contact, we exchanged Arg79 of Tim16 by Ala. This mutation neither produced a phenotype in vivo nor is it conserved in the Tim16 of various species. This suggests that this interaction is a crystallographic rather than a functional contact.
In the other type of tetramer, shown in Figure 5C, contact occurs via extensive interactions between two Tim16 molecules. Interestingly, two Tim16 molecules form hydrogen bonds between their two DKE motifs. Interactions are asymmetric. Lys89 from one Tim16 is hydrogen-bridged to Asp88 of the second Tim16, whereas Lys89 from the second Tim16 is hydrogen-bridged to Glu90 from the first Tim16. These tetramers are further stabilized by a network of van der Waals interactions between Phe84, Phe95 and Tyr102 from both heterodimers, and, interestingly, Phe99 from Tim14. In this tetramer, the proposed Hsp70-interacting surfaces of both Tim14 molecules are surface exposed.
Discussion
We describe here a high-resolution structure of the complex of Tim14 and Tim16, an essential part of the import motor of the TIM23 translocase, which drives translocation of preproteins into mitochondria. Our data reveal that the conserved domain of Tim14, part of which is its J domain, and the J-like domain of Tim16 form a stable 1:1 complex. In the complex, both proteins have the typical fold of J domains. Superposition of the polypeptide backbones reveals remarkably high similarity between Tim14 and Tim16 but also with the J domain of DnaJ. Tim16, however, lacks the signature motif of J domains, the HPD tripeptide in the loop connecting helices II and III, qualifying it ‘only' as a J-like protein. Furthermore, the surfaces of the two proteins are different. As a typical J protein, Tim14 contains a highly positively charged surface, which has been proposed to play a crucial role in the interaction with the Hsp70 partners (Greene et al, 1998; Genevaux et al, 2002). In contrast, the corresponding surface of Tim16 is mostly neutral or negatively charged. This offers an explanation why introducing simply an HPD motif did not convert Tim16 into a functional J protein (Li et al, 2004). These characteristics are shared by all Tim14 and Tim16 homologues (Figure 6). Furthermore, essentially all residues involved in the heterodimer formation are strictly conserved, suggesting that Tim16 binds its Tim14 partner in the same manner in all organisms (Figure 6).
Figure 6.
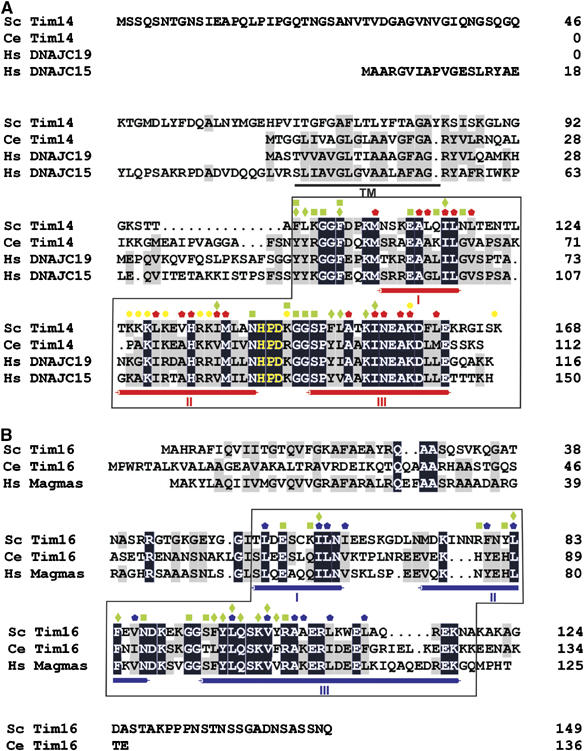
Evolutionary conservation of Tim14 and Tim16. Sequence alignment of Tim14 (A) and Tim16 (B) with their homologues. Identical residues are shown as white letters on black background and similar residues are shaded in gray. The characteristic α helices of the J domain fold are indicated below the alignments. The HPD motif is shown in yellow. Predicted transmembrane domain (TM) of Tim14 is underlined. Red/blue pentagons—Tim14/Tim16 residues forming the hydrophobic cores of the J/J-like domains; Yellow circles—Tim14 residues which form the positively charged surface for interaction with mtHsp70; green squares/diamonds—residues involved in polar/hydrophobic interactions between Tim14 and Tim16. The parts of Tim14 and Tim16, which were crystallized, are boxed.
The high structural similarity between Tim14 and Tim16 suggests that the latter protein evolved from the functional J protein. During evolution, Tim16 apparently lost the HPD motif and therefore the ability to stimulate the ATPase activity of Hsp70 chaperones. Instead, it acquired other functions. Interestingly, analysis of the yeast genome revealed the presence of two other J-like proteins (Walsh et al, 2004), not homologous to Tim16. It will be interesting to see for which functions they have specialized during evolution.
The Tim14–Tim16 heterodimeric complex comprises a novel, pseudo-symmetrical arrangement of J folds. A network of interactions is responsible for the tight packing of Tim14 and Tim16. First, an arm in Tim14, present in front of its J domain, embraces Tim16. We showed that the arm is indispensable for the activity of Tim14. Therefore, the J domain alone is not sufficient for making a functional J protein of the import motor. Second, helices I, II and III are tightly packed together. Such tight packing results in constrained conformations of the loops between helices II and III of both subunits predicting that, in this conformation, Tim14 cannot interact properly with mtHsp70. Indeed, we found that Tim14 in the complex with Tim16 is not able to stimulate the ATPase activity of mtHsp70. This is also in agreement with previous findings which demonstrated the stimulation of the ATPase activity of mtHsp70 by purified Tim14 and inhibition of this stimulation by addition of isolated Tim16 (Li et al, 2004; Mokranjac et al, 2005). Apparently, correct positioning and integration, in space and probably time, of the J domain is required for its activity in the import motor clearly showing that the regulation of the cochaperone activity of Tim14 is as important as the activity itself. Also, the exchange of the J domain of Tim14 by that of Mdj1, the J protein partner of mtHsp70 in protein folding in the mitochondrial matrix, is not tolerated (data not shown). This further illustrates the necessity for a specific J protein for the stimulation of the ATPase activity of mtHsp70 in the import motor. Interestingly, exchanges of J domains between two J proteins frequently result in nonfunctional chimeras (Hennessy et al, 2005). Furthermore, the genome of Saccharomyces cerevisiae encodes 22 different J proteins and only 14 Hsp70 chaperones (Walsh et al, 2004). A similar disproportion prevails in higher eukaryotes. This clearly shows that it is the J protein which determines the biochemical process for which Hsp70 is needed and then, directly or indirectly, recruits the chaperone to a specific site of action. The regulatory mechanisms as to when and where Hsp70s are activated remain largely obscure. It will be most interesting to see if other J-like proteins are present in complexes with functional J proteins and thereby have similar regulatory activities as Tim16.
J domains were reported to change their conformation upon binding to Hsp70s (Huang et al, 1999; Berjanskii et al, 2002; Landry, 2003; Hennessy et al, 2005). The NMR structure of the J domain of DnaJ shows a high degree of flexibility, particularly in the HPD loop (Szyperski et al, 1994). Fluctuations in the HPD loop of SV40 T large antigen (Tag) are correlated with the reorientation of the positively charged residues on helix II (Berjanskii et al, 2002). These fluctuations are produced by a crankshaft motion of the two consecutive glycine residues following the HPD motif. Intriguingly, the HPD loops of yeast Tim14 and SV40 Tag are identical in sequence, HPDKGG (in some Tim14 homologues the lysine residue is replaced by an arginine). This suggests that a similar rearrangement can occur in the HPD loop of Tim14. Tim14 is an active cochaperone of mtHsp70 (D'Silva et al, 2003; Mokranjac et al, 2003; Truscott et al, 2003) and therefore the flexible conformation of the HPD loop of Tim14 is likely to be induced via a change in the conformation of the Tim14–Tim16 complex. A major determinant of the conformation of both proteins in the complex likely resides in its oligomeric nature. Biochemical and crystallographic data presented here favor the existence of Tim14–Tim16 as tetramers or even higher oligomers. They are also in agreement with previous results demonstrating the ability of Tim16 to form homodimers (Kozany et al, 2004; D'Silva et al, 2005). The tetramers are formed by interactions between two Tim16 molecules of two heterodimers. Tim16–Tim16 interactions occur through the surfaces, which correspond to the ones in Tim14 which are involved in the interaction with mtHsp70. The DKE motifs from two Tim16 molecules are fixed via asymmetric hydrogen bridges. At the same time, these DKE motifs are part of the loops that interact with the binding grooves on Tim14. Interestingly, the DKE loops of Tim16 contain two consecutive and invariant glycine residues, which can undergo conformational changes as described above for SV40 Tag. Notably, Phe99 of Tim14, the first residue of the arm, is also part of the hydrophobic network, which stabilizes the tetramers.
Based on previous mechanistic insights and the results presented here, we put forward a working hypothesis for the function of the J and J-like proteins in the import motor. Tim44 recruits both mtHsp70 and the Tim14–Tim16 complex to the protein conducting channel of the TIM23 translocase (Schneider et al, 1994; Mokranjac et al, 2003; Kozany et al, 2004). When a precursor polypeptide appears at the outlet of the channel, it is recognized by and loosely bound to Tim44 (Schneider et al, 1994). We propose that as a result of precursor binding, Tim44 undergoes a conformational change which is sensed by the Tim16–Tim16 interface of the tetramer. This will result in loosening of the Tim14 arm and, as a consequence, to a change in the conformation of the Tim14–Tim16 heterodimer. Tim14, in a more flexible conformation, is now able to stimulate the ATPase activity of mtHsp70. MtHsp70 in the ADP form can then exert a strong grip on the precursor allowing transfer of a segment of the polypeptide into the mitochondrial matrix.
Elucidation of the exact steps of the mitochondrial import motor will require extensive biochemical (Liu et al, 2003) and genetic analyses combined with determination of intricate crystal structures, such as the complex of Tim44 with mtHsp70, Tim14 and Tim16, in different nucleotide states of the chaperone and in the presence/absence of the unfolded polypeptide chain.
Materials and methods
Yeast strains and plasmids
Wild-type yeast strain YPH499 was used (Sikorski and Hieter, 1989). Deletion strains of TIM14 and TIM16 that carry the corresponding wild-type genes on a URA plasmid were described before (Mokranjac et al, 2005). Tim14 and Tim16 mutants were cloned into the centromeric yeast plasmid pRS314 under the control of their respective promoters and 3′UTRs, and subsequently transformed into the respective deletion strains. Functionality of mutant proteins was assessed on 5-fluoroorotic acid-containing medium, which selects for cells which lost the URA plasmid carrying the wild-type copy of the gene (Boeke et al, 1987). A Tim14 mutant lacking amino-acid residues 99–109 was also transformed into YPH499. Plasmids for expression of His-tagged versions of Tim14 and Tim16 in yeast were described before (Mokranjac et al, 2005), with the difference that the respective 3′UTRs were added after the stop codon. They were transformed into YPH499. Mitochondria were isolated from yeast cells grown at 30°C in the synthetic medium lacking tryptophan and containing 0.1% glucose and 2% lactate as carbon sources (Sambrook et al, 1989).
Expression and purification of the Tim14–Tim16 complex
The nucleotide sequence coding for an N-terminal TEV cleavage site and amino-acid residues 99–168 of Tim14 was cloned into the multiple cloning site 1 of pETDuet-1 (Novagen) between BamHI and PstI sites to produce His6-TEV-Tim14 (99–168). This plasmid was digested with NdeI and XhoI to introduce a fragment encoding Tim16 (54–119) into the multiple cloning site 2 of the same plasmid. The resulting plasmid enabled expression of the Tim14 fragment with a cleavable His-tag and the non-tagged Tim16 fragment from two distinct T7 promoters. The E. coli strain BL21 (DE3) harboring the plasmid was grown in LB medium supplemented with 100 μg/ml ampicillin at 37°C to an OD600∼0.6. Expression of recombinant proteins was induced with 1 mM isopropyl-β-D-thiogalactopyranoside for 3 h at 37°C. The Tim14–Tim16 complex was purified on a NiNTA-agarose column (Qiagen) according to the manufacturer's instructions. After cleavage of the His-tag with TEV protease, the Tim14–Tim16 complex was loaded onto a Superdex-75 gel filtration column (GE Healthcare) equilibrated with 20 mM Tris–HCl, 300 mM NaCl, pH 8.0. Protein containing fractions were concentrated using a Microsep 3K Omega centrifugal device (Pall Life Sciences) and used for crystallization.
Crystallization and structure determination
Crystals of Tim14–Tim16 were grown at 20°C within 4 days to a final size of 200 × 200 × 200 μm3 by using the hanging drop vapor diffusion method. The drops contained equal volumes of protein (400 mg/ml) and reservoir solution (1.65 M sodium citrate, pH 7.0). Before exposure to X-rays, crystals were soaked in a solution of 1.65 M sodium citrate, pH 7.0, and 5% glycerol for 30 s and subsequently frozen in a stream of cold nitrogen gas at 95 K (MSC-XStream).
Native data to 2.0 Å resolution were collected on the BW6 beamline at DESY, Hamburg using MarCCD detector (Table I). The data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). The space group of Tim14–Tim16 crystals was P212121 with unit cell dimensions of a=111.7 Å, b=114.8 Å, c=162.2 Å. The self-rotation function calculated using MOLREP (Vagin and Teplyakov, 1997) indicated possible tetragonal pseudo-symmetry of the native crystals. Indeed, after testing a variety of Tim14–Tim16 crystals that were incubated with various heavy metal atom solutions, it turned out that crystals treated with K2OsCl6 for 6 h changed the space group to P43212 with unit cell dimensions of a=b=114.1 Å, c=163 Å. SAD data were collected at 3.5 Å resolution using absorption peak wavelength at 1.1402 Å (see Table I). Seven Os4+ sites were localized using SHELXD (Schneider and Sheldrick, 2002). Subsequent phasing with MLPHARE (The CCP4 suite: programs for protein crystallography) and solvent flattening with DM (Cowtan and Main, 1996) resulted in an interpretable electron density map showing four Tim14 and four Tim16 subunits in the asymmetric unit, which was modelled by polyalanine. Next, we transferred the model to the orthorhombic cell using MOLREP. After rigid body refinement and preliminary positional refinement with REFMAC (Murshudov et al, 1997), 16 chains (eight Tim14 and eight Tim16 subunits) in the asymmetric unit of the orthorhombic crystal form were re-traced and phases extended to 2.0 Å resolution using ARP-WARP (Perrakis et al, 1999). The model has been completed using the interactive three-dimensional graphics programs MAIN (Turk, 1992) and refined with REFMAC5. Temperature factors were refined with restraints between bonded atoms and between noncrystallographic symmetry-related atoms. The final model comprises fully defined Tim14, whereas in Tim16 the two C-terminal amino acids are structurally disordered. The data of Tim14–Tim16 have been deposited in the RCSB Protein Data bank: PDB ID code 2GUZ.
ATPase assay
ATPase assays were performed according to Sichting et al (2005) and Mokranjac et al (2005), using as the J protein either free Tim14 (as MBP-Tim14) or the one bound to Tim16 (as the Tim14–Tim16 complex). In the inhibition experiments, wild-type or mutant forms of Tim14 (as MBP fusion proteins) were preincubated with His6Tim16 (25–149) for 5 min at 25°C before the ATPase assay was performed.
Miscellaneous
Co-immunoprecipitation and NiNTA pull down assays were previously described (Mokranjac et al, 2005). Protein concentration was determined according to Bradford (1976) using bovine plasma gamma globulin as a standard.
Supplementary Material
Supplementary data
Acknowledgments
We are grateful to Dr Alexey Rak for helpful discussions and to Marica Malesic, Heiko Germeroth and Ulrike Gärtner for expert technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 594, the Fonds der Chemischen Industrie, the German–Israeli Foundation and by the German–Israeli Project Cooperation DIP F.5.1.
References
- Berjanskii M, Riley M, Van Doren SR (2002) Hsc70-interacting HPD loop of the J domain of polyomavirus T antigens fluctuates in ps to ns and micros to ms. J Mol Biol 321: 503–516 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Main P (1996) Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D 52: 43–48 [DOI] [PubMed] [Google Scholar]
- D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA (2003) J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci USA 100: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva PR, Schilke B, Walter W, Craig EA (2005) Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc Natl Acad Sci USA 102: 12419–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P (2004) Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol 11: 226–233 [DOI] [PubMed] [Google Scholar]
- Genevaux P, Schwager F, Georgopoulos C, Kelley WL (2002) Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 162: 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA 95: 6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40–Hsp70 interactions. Protein Sci 14: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Ghose R, Flanagan JM, Prestegard JH (1999) Backbone dynamics of the N-terminal domain in E. coli DnaJ determined by 15N- and 13CO-relaxation measurements. Biochemistry 38: 10567–10577 [DOI] [PubMed] [Google Scholar]
- Jiang J, Prasad K, Lafer EM, Sousa R (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell 20: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem Sci 23: 222–227 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K (2004) The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol 11: 234–241 [DOI] [PubMed] [Google Scholar]
- Landry SJ (2003) Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry 42: 4926–4936 [DOI] [PubMed] [Google Scholar]
- Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W (2004) The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 279: 38047–38054 [DOI] [PubMed] [Google Scholar]
- Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA (2003) Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300: 139–141 [DOI] [PubMed] [Google Scholar]
- Matouschek A, Pfanner N, Voos W (2000) Protein unfolding by mitochondria. The Hsp70 import motor. EMBO Rep 1: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W (2005) Protein import into mitochondria. Biochem Soc Trans 33: 1019–1023 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K (2003) Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22: 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Popov-Celeketic D, Berg A, Hell K, Neupert W (2005) The import motor of the yeast mitochondrial TIM23 preprotein translocase contains two different J proteins, Tim14 and Mdj2. J Biol Chem 280: 31608–31614 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neupert W, Brunner M (2002) The protein import motor of mitochondria. Nat Rev Mol Cell Biol 3: 555–565 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin V (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004) Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5: 519–530 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Laboratory Press: Cold Spring Harbor, New York, USA [Google Scholar]
- Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W (1994) Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371: 768–774 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Sichting M, Mokranjac D, Azem A, Neupert W, Hell K (2005) Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J 24: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2–108) containing the highly conserved J domain. Proc Natl Acad Sci USA 91: 11343–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Muller H, Sickmann A, Meyer HE, Meisinger C, Guiard B, Rehling P, Pfanner N (2003) A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol 163: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D (1992) Improvement of a programme for molecular graphics and manipulation of electron densities and its application for protein structure determination. Technische Universitaet Muenchen: Munich
- Vagin AA, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vogel M, Bukau B, Mayer MP (2006) Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell 21: 359–367 [DOI] [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 5: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU (2004) Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 5: 781–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


