Abstract
Genomic disorders are conditions that result from DNA rearrangements, such as deletions or duplications. The identification of the dosage-sensitive gene(s) within the rearranged genomic interval is important for the elucidation of genes responsible for complex neurobehavioral phenotypes. Smith-Magenis syndrome is associated with a 3.7-Mb deletion in 17p11.2, and its clinical presentation is caused by retinoic acid inducible 1 (RAI1) haploinsufficiency. The reciprocal microduplication syndrome, dup(17)(p11.2p11.2), manifests several neurobehavioral abnormalities, but the responsible dosage-sensitive gene(s) remain undefined. We previously generated a mouse model for dup(17)(p11.2p11.2), Dp(11)17/+, that recapitulated most of the phenotypes observed in human patients. We have now analyzed compound heterozygous mice carrying a duplication [Dp(11)17] in one chromosome 11 along with a null allele of Rai1 in the other chromosome 11 homologue [Dp(11)17/Rai1– mice] in order to study the relationship between Rai1 gene copy number and the Dp(11)17/+ phenotypes. Normal disomic Rai1 gene dosage was sufficient to rescue the complex physical and behavioral phenotypes observed in Dp(11)17/+ mice, despite altered trisomic copy number of the other 18 genes present in the rearranged genomic interval. These data provide a model for variation in copy number of single genes that could influence common traits such as obesity and behavior.
Introduction
Genomic rearrangements including submicroscopic deletions, duplications, and inversions are being recognized as important mutational mechanisms involved in several aspects of genome evolution, susceptibility to traits, and human genomic disorders (1–6). Although the segmental aneuploidy usually involves several genes, only a small subset of these genes convey phenotypes as a function of copy number alteration (i.e., variation from the normal 2 copies with 1 present on each chromosome homologue); such genes are referred to as dosage-sensitive genes. Genomic disorders (6) are frequent conditions (~1 per 1,000 births) (7) that result from genomic imbalance due to DNA rearrangements. The resulting phenotypes can be ascribed to 1 or more dosage-sensitive gene(s) present in the rearranged interval. The rearrangements that are large enough to be visualized by high-resolution cytogenetic techniques and comprise multiple unrelated contiguous genes are referred to as contiguous gene syndromes (CGSs). Such conditions often present complex phenotypes that can include specific neurobehavioral traits (8, 9). However, establishing a correlation between each phenotypic feature and the particular dosage-sensitive gene has often been a major challenge due to the usually large number of genes mapping within or around the rearranged genomic interval. Murine models have proven to be extremely valuable for the identification of the predominant responsible gene(s) (10).
New duplication syndromes continue to be described (11), but in virtually none of these has the exact dosage-sensitive gene or genes been elucidated. In some cases a unique dosage-sensitive gene within the rearranged interval can give a phenotype when either present in 3 copies or haploinsufficient. This is the case for 2 reciprocal rearrangement syndromes, Charcot-Marie-Tooth disease type 1A (CMT1A; associated with a 1.4-Mb microduplication in 17p12) and hereditary neuropathy with liability to pressure palsies (HNPP; associated with the reciprocal microdeletion), that are both dysmyelinating peripheral neuropathies that result from an altered dosage of a single gene within a rearranged genomic fragment: PMP22, which encodes peripheral myelin protein (12). In contrast, it is possible that more than 1 dosage-sensitive gene within the rearranged region or different genes can convey a distinct phenotype with duplication versus deletion or with parent-of-origin effects when deletions are inherited from the mother versus the father. In Angelman syndrome (associated with maternal deletion of 15q11–q13) and Prader-Willi syndrome (resulting from deletion of the equivalent paternal genomic interval), the responsible dosage-sensitive gene is not the same, despite the fact that the same genomic region is involved. E6-associated ubiquitin–protein ligase gene, UBE3A, is responsible for the Angelman phenotype, while it has been proven that it is not the dosage-sensitive gene that causes the Prader-Willi phenotype (13).
Smith-Magenis syndrome (SMS) and dup(17)(p11.2p11.2) represent reciprocal CGSs, associated with a deletion and duplication of a 3.7-Mb interval within chromosome 17 band p11.2, respectively. The SMS (~1 per 25,000 births) phenotype includes craniofacial abnormalities, brachydactyly, obesity, behavioral abnormalities, seizures, sleep abnormalities, and mental retardation. While a commonly deleted region of approximately 3.7 Mb is present in the majority of SMS patients (>70%–80%) (14), an approximately 1.1-Mb SMS critical region was defined, and it includes 23 genes (15). Recently, haploinsufficiency of 1 of these 23 genes, retinoic acid inducible 1 (RAI1), was associated with most features of SMS (16–18), suggesting that it is the dosage-sensitive gene within this genomic region responsible for SMS. Whereas the responsible dosage-sensitive gene in individuals with a deletion can be identified through rare individuals with frameshift or nonsense alleles, finding the dosage-sensitive gene for a duplication syndrome poses additional challenges. The dup(17)(p11.2p11.2) clinical presentation includes learning disabilities, attention deficit disorder, autistic and obsessive-compulsive behaviors, short stature (below the 50th percentile), reduced body weight, malocclusion, and dental abnormalities (19). The dosage-sensitive gene(s) responsible for this syndrome remain unidentified.
Human chromosome 17p11.2 is syntenic to the 32- to 34-cM region of murine chromosome 11, in which 19 genes are conserved in order and orientation (Figure 1) (15). Using chromosome engineering (20), we generated a mouse model for dup(17)(p11.2p11.2) syndrome, Dp(11)17/+, and a mouse model for SMS, Df(11)17/+, heterozygous for either a duplication or deletion (deficiency), respectively, of an approximately 3-Mb mouse genomic interval that is syntenic to the SMS critical region (21). Dp(11)17/+ mice are significantly leaner than wild-type mice (21), hyperactive, and exhibit impaired contextual fear conditioning (22). Df(11)17/+ mice manifest craniofacial abnormalities, marked obesity, seizures, hypoactivity, and circadian rhythm alteration (21, 22). Normal gene dosage in this specific genomic interval in Dp(11)17/Df(11)17 compound heterozygous mice was able to rescue the obesity, craniofacial, and seizure phenotypes (21) (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI28953DS1). The goals of the present study were to determine (a) whether the behavioral phenotypes observed in Dp(11)17/+ mice are due to a gene dosage effect; and (b) whether there is a specific gene whose copy number variation is responsible for the phenotype observed in Dp(11)17/+ mice.
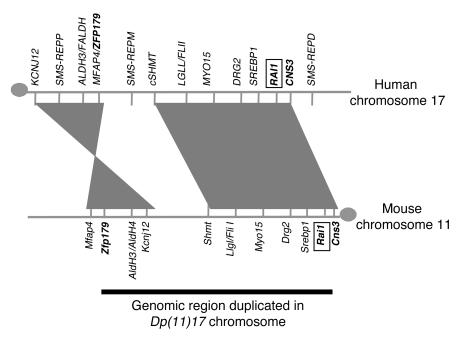
Figure 1. Representation of the genomic region duplicated in Dp(11)17.
A schematic representation of the 17p11.2 region of human chromosome 17 and the 32- to 34-cM interval of the syntenic region of mouse chromosome 11 is shown. The gray circles depict the centromeres. The horizontal black line (bottom) denotes the genomic interval encompassing the Dp(11)17 duplication generated by chromosomal engineering. Note that the numbers, orientations, and relative orders of the genes in these syntenic genomic intervals are extremely conserved. Three low-copy repeats are present in the human region: SMS-REPD (distal); SMS-REPM (middle); SMS-REPP (proximal). The mouse Rai1 and human RAI1 genes are indicated in boxes.
Results
Correcting genomic balance normalizes Dp(11)17 phenotypic traits.
We explored the possibility that a gene-dosage effect may cause the behavioral phenotypes we observed in our chromosome-engineered animal models by normalizing the gene copy numbers and then examining the resultant phenotype. Compound heterozygous Dp(11)17/Df(11)17 animals were bred. These mice contain the normal disomic copy number of each of the 19 genes within the rearrangement interval. We previously showed that the underweight phenotype observed in Dp(11)17/+ animals and the obesity observed in Df(11)17/+ animals were rescued in the Dp(11)17/Df(11)17 mice (21). Thus, normalizing the copy number of the genes within the rearranged interval normalizes the weight phenotype, which is consistent with the hypothesis that the complex phenotype in the animals with segmental aneuploidy is the result of gene dosage. To simplify the analysis of the data, all the behavioral studies were performed in male mice, since gender specificity was previously observed for Dp(11)17/+ mice (22).
We were able to rescue many of the behavioral phenotypes observed in the male animals with segmental aneuploidies by normalizing the gene copy number in Dp(11)17/Df(11)17 mice (Figures 2 and 3). The vertical activity and center/total distance ratio determined by the open-field test — used to assesses exploratory activity and providing some measure of anxiety levels, which were abnormal in Dp(11)17/+ mice — were normalized in the Dp(11)17/Df(11)17 animals (Figure 2, A and B). Surprisingly, reconstitution of the normal gene dosage in Dp(11)17/Df(11)17 animals did not normalize the total distance traveled by Dp(11)/+ mice (Figure 2C). Potential explanations for the results presented in Figure 2C include: Rai1 position effects, the presence of a cis-regulatory element near or within the rearranged genomic interval that regulates another gene affecting this phenotype, or limitations of the behavioral testing paradigm when evaluating relatively small numbers of animals. The percent freezing to context as determined by the conditioned fear test assessing learning and memory, which was abnormal in Dp(11)17/+ animals, was corrected when the mice were balanced genomically (Figure 3A). No significant difference was found in percent freezing to the sound cue among the different genotypes (Figure 3B).
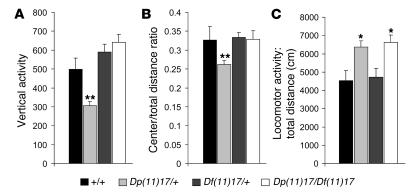
Figure 2. Genomic balance restores normal behavior.
The results of 3 different parameters that were measured in the open-field paradigm (vertical activity, center/total distance, and total distance) are represented as bar graphs for each of the 4 genotypes tested. (A) Vertical activity [F(3, 46) = 26.9; P < 0.0001]. (B) Center/total distance ratio [F(3, 46) = 6.740; P = 0.001]. (C) Total distance (cm) [F(3, 46) = 4.260; P = 0.01]. Note that the abnormal vertical activity and center/total distance ratio observed in Dp(11)17/+ animals was normalized after restoration of genomic balance [i.e., Dp(11)17/Df(11)17]. Black bars, wild type; light gray bars, Dp(11)17/+; dark gray bars, Df(11)17/+; white bars, Dp(11)17/Df(11)17. Values represent mean ± SEM. Actual values are given in Supplemental Table 2. *P < 0.05, **P < 0.01 compared with wild-type littermate.
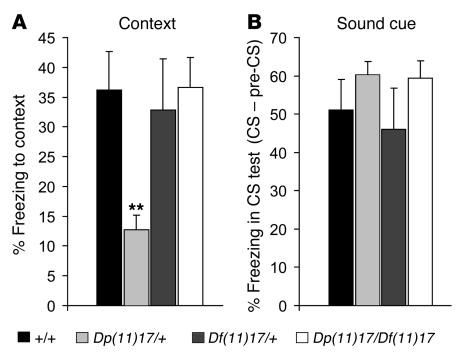
Figure 3. Genomic balance corrects learning and memory deficits.
The percentages of freezing to context [F(3, 46) = 10.847, P < 0.0001] (A) and the sound cue (P > 0.05) (B) are represented for each of the 4 genotypes tested. Note that the abnormal freezing of Dp(11)17/+ animals was corrected in Dp(11)17/Df(11)17 mice. Black bars, wild type; light gray bars, Dp(11)17/+; dark gray bars, Df(11)17/+; white bars, Dp(11)17/Df(11)17. The mean ± SEM values are presented. Actual values are given in Supplemental Table 2. **P < 0.01 compared with wild type.
Correcting Rai1 copy number in Dp(11)17/+ mice.
We next investigated Rai1 copy number to determine whether it was the dosage-sensitive gene responsible for the physical and behavioral phenotypes in Dp(11)17/+ mice. We have previously generated mice carrying a null mutation in Rai1 (Rai1+/– mice), and these animals manifest craniofacial abnormalities and are obese (23). By crossing Dp(11)17/+ mice with heterozygous Rai1+/–mice, we generated animals carrying a heterozygous duplication of 3 Mb containing Rai1, along with 18 other genes on one chromosome, and a null allele of Rai1 on the other chromosome [Dp(11)17/Rai1– mice] (Figure 4A). Therefore the Dp(11)17/Rai1–mice, like the Dp(11)17/+ mice, retain 3 copies of each of those 18 other genes but differ in that they now have the normal disomic (n = 2) copy number of Rai1.
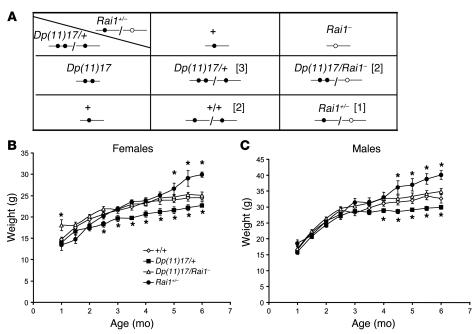
Figure 4. Mating scheme to normalize Rai1 and the correlation of Rai1 copy number changes and body weight.
(A) Expected genotypes of the progeny from the mating of Dp(11)17/+ and Rai1+/– mice. Rai1 alleles are represented by a line with circles on it. The filled circles represent normal Rai1 gene, whereas the open circles represent the null copy of Rai1. Rai1 gene copy number is further indicated in brackets for each genotype. The 4 different genotypes were obtained at expected Mendelian ratios. Mice for each of the 4 genotypes were weighed every 2 weeks until 6 months of age. (B) Weight curves for female wild-type (open diamonds), Dp(11)17/+ (filled squares), Dp(11)17/Rai1–(open triangles), and Rai1+/–(filled circles) mice. (C) Data for male mice are shown. Each point represents the average of weights from at least 10 mice, and error bars are indicated. *P < 0.05.
Body weight normalization in Dp(11)17/Rai1– mice.
Dp(11)17/+ mice are significantly underweight when compared with wild-type littermates (21). To explore the influence of Rai1 copy number on this particular phenotype, mice of the different genotypes were weighed biweekly from 1 to 6 months of age (Figure 4, B and C). Remarkably, Dp(11)17/Rai1– mice, carrying 2 copies of the Rai1 gene but 3 copies of the other 18 genes in the chromosome-engineered duplication interval, showed body weight similar to that of the wild-type animals, and their weight was significantly different from that of the Dp(11)17/+ and Rai1+/– mice. These data indicate that normalization of Rai1 gene copy number in the context of Dp(11)17 is sufficient to restore body weight to normal, in spite of all the other genes remaining trisomic in this genomic interval. This finding suggests that duplication of Rai1 is responsible for the reduced body weight phenotype of the Dp(11)17/+ mice. In addition, we have previously shown that homozygous Dp(11)17/Dp(11)17 animals (carrying 4 copies of the Rai1 gene), are even leaner than the heterozygous mice and display significant growth retardation (21). Furthermore, heterozygous Rai1+/– mice (23) are obese, as are mice in which this genomic interval was deleted to different extents, with the deletions always encompassing the Rai1 gene (21, 24). These data in aggregate indicate that Rai1 copy number and gene dosage are critical to body weight regulation.
Behavioral characterization as a function of Rai1 copy number variation.
Dup(17)(p11.2p11.2) patients also have neurobehavioral abnormalities, including mild to borderline mental retardation, attention deficit disorder, hyperactivity, and autistic features (19). Despite the impossibility of recapitulating the complexity of human neurobehavioral phenotypes in mice, many of these phenotypes are associated with intermediate traits (also called endophenotypes) that can be analyzed in animal models (25). Such endophenotypes may result from chromosomal abnormalities or single gene defects or represent complex traits. In fact, Dp(11)17/+ mice exhibit elevated levels of anxiety, learning and memory deficits, and hyperactivity (22). Therefore, we next assessed the neurobehavioral phenotypic consequences of reconstituting Rai1 copy number to the normal disomic dosage in male mice.
Partial normalization of behaviors assayed by open-field testing.
We subjected the Dp(11)17/Rai1– mice to the open-field test, used to assess exploratory activity and anxiety-related responses in a novel arena.
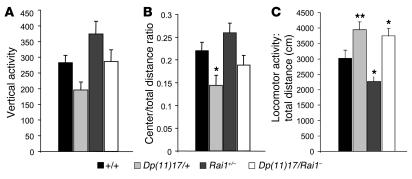
Dp(11)17/+ mice displayed less rearing behavior or vertical activity (Figure 5A) in the open field than wild-type mice.Although this difference in rearing was not quite statistically significant (P = 0.065), it is consistent with the statistically significant difference shown in Figure 2A. In contrast, Rai1+/– mice displayed more rearing than wild-type mice (P = 0.055), and Dp(11)17/Rai1– mice displayed rearing responses similar to those in wild-type mice (P = 0.459). Together these findings indicate that correcting Rai1 gene copy number normalized the rearing behavioral response of Dp(11)17/+ mice in the genetic backgrounds analyzed in the present study. Anxiety-related responses in the open field as measured by the center/total distance ratio (Figure 5B) clearly indicate that Dp(11)17/+ mice spend a lower proportion of their exploration in the center of the open field compared with wild-type mice (P < 0.05). The decreased center/total ratio for the Dp(11)17/+ mice replicates our previous findings of increased anxiety-related behaviors in Dp(11)17/+ animals (22) and is consistent with those data shown for the Dp(11)17/+ mice in Figure 2. Interestingly, the Dp(11)17/Rai1– mice displayed a similar center/total ratio compared with wild-type littermates, indicating that the increased anxiety-related behavior in the open field in the Dp(11)17/+ mice was attenuated by normalization of the Rai1 copy number. Rai1+/– mice also had a center/total ratio similar to that of wild-type littermates. Finally, replicating our previous reported results (22), and similar to the results presented in Figure 2, Dp(11)17/+ animals traveled significantly farther compared with wild-type littermates (P = 0.008) (Figure 5C). Consistent with the Dp(11)17/Df(11)17 mice, Dp(11)17/Rai1– mice were also more active than wild-type littermates (P < 0.05), indicating that this abnormality may not be totally related to copy number of Rai1. Although normalizing copy number of Rai1 did not significantly alter the response of Dp(11)17/+ mice, Rai1+/– mice were significantly less active (P < 0.05) compared with wild-type mice, suggesting that reduced Rai1 copy number does contribute to this behavioral response.
Figure 5. Correcting Rai1 gene dosage rescues behavioral deficits.
Different parameters measured in the open field are represented for each of the 4 genotypes tested. (A) Vertical activity [F(3, 73) = 4.458; P = 0.06]. (B) Center/total distance ratio [F(3, 71) = 4.893; P < 0.004]. (C) Total distance (cm) [F(3, 73) = 9.498; P < 0.001]. Black bars, wild type; light gray bars, Dp(11)17/+; dark gray bars, Rai1+/–; white bars, Dp(11)17/Rai1–. Values represent mean ± SEM. Actual values are given in Supplemental Table 3. *P < 0.05, **P < 0.01, significant differences compared with wild-type littermates.
The light-dark exploration test, typically used to assess anxiety-related responses, and the prepulse inhibition test, used to assess sensorimotor gating, did not show any overall significant differences (P > 0.05) for any of the mice reported in this article (data not shown). A decrease in light-dark transitions and in startle response was previously reported for Dp(11)17/+ mice (22). As mice of different genetic backgrounds were used in the 2 studies, it is possible that there were modifiers present that affected the outcome of Rai1 copy number variation.
Learning and memory is normalized in Dp(11)17/Rai1– mice.
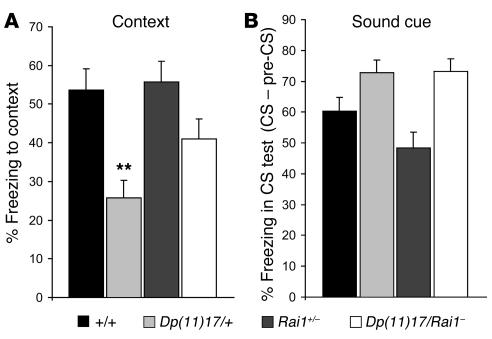
We assessed learning and memory using a conditioned fear test based on the Pavlovian paradigm. As previously reported (22), Dp(11)17/+ mice displayed significantly less freezing during the context test than wild-type mice (P = 0.0005) (Figure 6A). However, the percentage of freezing observed for Dp(11)17/Rai1– mice was similar to that observed for wild-type mice (P > 0.05), clearly indicating that the contextual fear impairment is directly related to Rai1 copy number. Levels of freezing during the pre–conditioned stimulus (pre-CS) and CS phases were not significantly different among the various genotypes (P > 0.05) (Figure 6B).
Figure 6. Normalizing Rai1 gene dosage rescues learning and memory deficits.
The percentages of freezing to the context [F(3, 73) = 6.573, P = 0.001] (A) and the sound cue (B) are represented for each of the 4 genotypes. Black bars: wild type; light gray bars: Dp(11)17/+; dark gray bars, Rai1+/–; white bars: Dp(11)17/Rai1–. Note that the abnormal freezing of Dp(11)17/+ animals is totally corrected in Dp(11)17/ Rai1– mice. The mean ± SEM values are presented. Actual values are given in Supplemental Table 3. **P < 0.001, significant difference compared with wild type.
Discussion
We present essentially 2 experimental paradigms in this report: study 1 compared Dp(11)17/+ and Df(11)17/+ mice with Dp(11)17/Df(11)17 mice; study 2 compared Dp(11)17/+ and Rai1+/– mice with Dp(11)17/Rai1– mice. It is important to note the remarkable similarities between the data for the 2 studies, which strengthen our hypothesis that gene copy number is a critical factor underlying several complex behavioral responses and body weight control in the Dp(11)17/+ mouse model. Genetic background can directly influence the behavioral phenotype of mutant mice; however, the results of the studies were similar despite differences in genetic background, consistent with the idea that the Rai1 gene copy number plays a more prominent role than a possible modifier locus in these particular genetic backgrounds. Our data indicate that Rai1 is the gene responsible for most features of the Dp(11)17/+ phenotype and by extension for the dup(17)(p11.2p11.2) syndrome. However, the role of the other 18 genes in the duplication interval was not specifically addressed, so that their potential influence on phenotype cannot be ruled out by this study.
Although our experiments were performed in an animal model for a relatively rare human condition, the body weight and behavioral traits studied can certainly often represent very common but complex traits. The identification of genetic factors influencing obesity is of broad interest in terms of public health, since they might provide some clues to the genetic determinants of obesity-related conditions such as cardiovascular diseases and diabetes. Also startling and perhaps having greater implications are our findings relating Rai1 gene copy number to behavioral traits. We have verified herein that Rai1 is a dosage-sensitive gene implicated in complex traits that control exploratory rearing behaviors and anxiety-related responses in the open field and conditioned fear. Our findings demonstrate the recovery of the wild-type phenotype solely by means of correction of Rai1 gene copy number in a Dp(11)17 genetic background. The human RAI1 gene has been implicated in neurobehavioral traits; polymorphic variation at a trinucleotide repeat causing polyglutamine expansion can modify a specific form of spinocerebellar ataxia (SCA2) (26). RAI1 is also linked to neuroleptic response in patients with schizophrenia (27). In addition, the SMS neurobehavioral traits are manifested in patients with RAI1 loss-of-function alleles (16–18, 28). However, the phenotypic effects of varying RAI1 gene copy number have never before to our knowledge been systematically evaluated.
Our findings that Rai1 copy number variation causes obesity and behavioral traits have implications beyond rare syndromes and Mendelian conditions. Most CGS patients present with unique behavioral features (8, 9, 29), suggesting that gene copy number alteration, either loss or gain of copies inside the rearranged genomic interval, may be critical to the manifestation of such behavioral traits. The microdeletions causing such syndromes are often flanked by segmental duplications, or low-copy repeats (LCRs), that mediate genomic deletion and duplication rearrangements via nonallelic homologous recombination (NAHR) (30). Genomic segmental duplications constitute approximately 5% of the human genome (31), and there is a growing body of evidence suggesting that such architecture of the genome may not only be related to susceptibility to rearrangements causing genomic disorders but also may be involved in gene copy number variation in normal individuals (32). The existence of gene copy number variation in the human genome (32–34) can potentially predispose individual behavioral phenotypes or specific personality characteristics and perhaps influence population-specific behaviors. Our results suggest that Rai1 may be one such gene.
Methods
Animals.
Dp(11)17/+ and Df(11)17/+ mice were in a C57BL/6 Tyrc-Brd genetic background (N8 and N7, respectively), while Rai1+/–mice were in a mixed C57BL/6 Tyrc-Brd × 129S5/SvEvBrd genetic background (F2). Depending on the experiments, heterozygous Dp(11)17/+ mice were crossed with Df(11)17/+ or with Rai1+/–mice, and the resulting strains were genotyped by Southern blotting or PCR as described previously (21, 23). Mice were housed 2–5 per cage in a room with a 12-hour light/12-hour dark cycle (lights on at 6 am, off at 6 pm) with access to food and water ad libitum.
For the Dp(11)17/Df(11)17 study, 10 Dp(11)17/Df(11)17, 7 Df(11)17/+, 21 Dp(11)17/+, and 12 wild-type male mice were evaluated. For the Dp(11)17/Rai1–study, 21 Dp(11)17/Rai1–, 18 Dp(11)17/+, 17 Rai1+/–, and 21 wild-type littermate control male mice were evaluated. The battery of behavioral tests consisted of: (a) an open-field exploration test; (b) a light-dark test; (c) an assessment of acoustic startle response and prepulse inhibition of the startle response; and (d) the Pavlovian conditioned fear test. The selection of tests for the battery was based on our previous studies (22). The behavioral experiments were run with multiple batches of 5–10 mice. In general, behavioral testing was performed between 9 am and 2 pm. Experiments were conducted by an experimenter blinded to the genotypes of the mice. All the mice were 2 months of age at the beginning of the behavioral testing. All behavioral testing procedures were approved by the Baylor Institutional Animal Care and Use Committee and followed NIH guidelines.
Locomotor activity in the open field.
Locomotor activity was evaluated by placing a mouse into the center of a clear Plexiglas (40 × 40 × 30 cm) open-field arena and allowing it to explore for 30 minutes. Overhead incandescent lights provided room lighting that measured approximately 800 lx inside the test arenas. In addition, white noise was present at approximately 55 db inside the test arenas. Activity in the open field was quantitated by a computer-operated Digiscan optical animal activity system (RXYZCM [ref. 16]; AccuScan Instruments Inc.) containing 16 photoreceptor beams on each side of the arena, which divides the arena into 256 equally sized squares. Total distance (locomotor activity), movement time (in seconds), movement speed (cm/s), vertical activity (rearing measured by number of photobeam interruptions), and center distance (the distance traveled in the center of the arena) were recorded. The center distance was also divided by the total distance to obtain a center distance/total distance ratio. The center distance/total distance ratio can be used as an index of anxiety-related responses (35). Data were collected in 2-minute intervals over the 30-minute test session. Open-field activity data for the total 30-minute test were analyzed using 1-way (genotype) ANOVA.
Light-dark exploration.
One to 3 days later, mice were tested in the light-dark exploration test, which consists of a polypropylene chamber (44 × 21 × 21 cm) unequally divided into 2 chambers by a black partition containing a small opening. The large chamber was open and brightly illuminated (800 lx), while the small chamber was closed and dark. White noise was present in the room at approximately 55 db in the test chamber. Mice were placed into the illuminated side and allowed to move freely between the 2 chambers for 10 minutes. The time to enter the dark and the total number of transitions were recorded. Data were analyzed using 1-way (genotype) ANOVA.
Startle and prepulse inhibition of the startle.
One to 3 days later, mice were tested for prepulse inhibition of acoustic startle responses using the SR Lab System (SD Instruments), as previously described (36). At the beginning of a test session, a mouse was placed in the Plexiglas cylinder, where it was left undisturbed for 5 minutes. A test session consisted of 7 trial types. One trial type involved a 40-ms, 120-db sound burst used as the startle stimulus. There were 5 different types of trials involving acoustic prepulse plus acoustic startle stimulus: the 20-ms prepulse sounds, presented 100 ms before the startle stimulus, were at 74, 78, 82, 86, or 90 db. Finally, there was a trial where no stimulus was presented, to measure baseline movement in the cylinders. Six blocks of the 7 trial types were presented in pseudorandom order such that each trial type was presented once within a block of 7 trials. The average intertrial interval was 15 seconds (range, 10–20 s). The startle response was recorded for 65 ms (with the response measured every 1 ms) starting with the onset of the startle stimulus. The background noise level in each chamber was approximately 70 db. The maximum startle amplitude recorded during the 65-ms sampling window was used as the dependent variable.
The following formula was used to calculate percent prepulse inhibition of a startle response: 100 – [(startle response in acoustic prepulse + startle stimulus trials/startle response in startle stimulus alone trials) × 100]. Thus, a high-percentage prepulse inhibition value indicates good prepulse inhibition; i.e., the subject showed a reduced startle response when a prepulse stimulus was presented compared with when the startle stimulus was presented alone. Conversely, a low-percentage prepulse inhibition value indicates poor prepulse inhibition; i.e., the startle response was similar with and without the prepulse. Acoustic response amplitude data were analyzed using 1-way ANOVA. Prepulse inhibition data were analyzed using 2-way ANOVA with repeated measures.
Pavlovian conditioned fear.
One to 3 days later, performance in a conditioned fear paradigm was measured using a test chamber (26 × 22 × 18 cm) made of clear Plexiglas (front wall) and stainless steel (back wall and 2 side walls). The bottom of the test chamber was a grid floor used to deliver a mild electric foot shock. The test chamber was placed inside a sound-attenuated chamber (internal dimensions: 56 × 38 × 36 cm; Med Associates Inc.). Mice were observed through windows in the front of the sound-attenuated chamber. A mouse was placed in the test chamber (house lights on) and allowed to explore freely for 2 minutes. White noise (80 db), which served as the CS, was then presented for 30 seconds, followed by a mild (2 s, 1.0 mA) foot shock, which served as the unconditioned stimulus (US). Two minutes later, another CS-US pairing was presented. The mouse was removed from the chamber 15–30 seconds later and returned to its home cage. Freezing behavior was recorded using the standard interval sampling procedure every 10 seconds. Freezing was operationally defined as no movement with the exception of those movements associated with breathing. Responses (run, jump, and vocalize) to the foot shock were also recorded. If a mouse did not respond to the foot shock, it was excluded from the analysis. Twenty-four hours later, the mouse was placed back into the test chamber for 5 minutes, and the presence of freezing behavior was recorded every 10 seconds (context test). One to 2 hours later, the mouse was tested for its freezing to the auditory CS. Environmental and contextual cues were changed for the auditory CS test: a black Plexiglas triangular insert was placed in the chamber to alter its shape and spatial cues, red house lights replaced the white house lights, the wire grid floor was covered with black Plexiglas, and vanilla extract was placed in the chamber to alter the smell. Finally, the sound-attenuated chamber was illuminated with red house lights. There were 2 phases during the auditory CS test. In the first phase (pre-CS), freezing was recorded for 3 minutes without the auditory CS. In the second phase, the auditory CS was turned on, and freezing was recorded for another 3 minutes. The number of freezing intervals was converted to a percentage freezing value. Context and CS test data were analyzed using 1-way (genotype) ANOVA.
Statistics.
The data obtained from the open-field, light-dark, acoustic startle, and conditioned fear tests were analyzed using 1-way (genotype) ANOVA. Prepulse inhibition data were analyzed using 2-way (genotype × prepulse sound level) ANOVA with repeated measures. Differences were considered significant at P < 0.05. Based on our previous findings (21), we were able to employ independent planned comparisons to compare the data from the wild-type animals with those data from the other 3 genotypes [i.e., Dp(11)17/+; Rai1+/–; and either Dp(11)17/Df(11)17, in which the gene dosage for the interval is normalized, or Dp(11)17/Rai1–, in which Rai1 copy number is specifically normalized] for the 2 separate experiments [i.e., Dp(11)17/+ with Df(11)17/+ and Dp(11)17/+ with Rai1+/–].
Supplementary Material
Acknowledgments
We thank J.I. Young for critical review of the manuscript. This work was supported in part by National Institute of Child Health and Development grant P01 HD39420 and National Institute of Dental and Craniofacial Research grant R01 DE015210 to J.R. Lupski, the Neurobehavioral Core of the Baylor Mental Retardation and Developmental Disabilities Research Center (P30 HD24064), and NIH research grant R03 TW07536 funded by the Fogarty International Center (to K. Walz). The Centro de Estudios Científicos (CECS) receives institutional support from Empresas CMPG, the Millenium Scientific Initiative, Fundación Andes, the Tinker Foundation, and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) grant 1061067 (to K. Walz).
Footnotes
Nonstandard abbreviations used: CGS, contiguous gene syndrome; CS, conditioned stimulus; RAI1, retinoic acid inducible 1; SMS, Smith-Magenis syndrome.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:3035–3041 (2006). doi:10.1172/JCI28953
References
- 1.Cheng Z., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- 2.Johnson M.E., et al. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 3.Courseaux A., Nahon J.L. Birth of two chimeric genes in the Hominidae lineage. Science. 2001;291:1293–1297. doi: 10.1126/science.1057284. [DOI] [PubMed] [Google Scholar]
- 4.Stefansson H., et al. A common inversion under selection in Europeans. Nat. Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez E., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. . Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 6.Lupski J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 7.Shaffer L.G., Lupski J.R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 8. Shaffer, L.G., Ledbetter, D.H., and Lupski, J.R. 2001. Molecular cytogenetics of contiguous gene syndromes: mechanisms and consequences. InThe metabolic and molecular bases of inherited diseases. Volume 227. C.R. Scriver, et al., editors. McGraw-Hill. New York, New York, USA. 6077–6096. [Google Scholar]
- 9.Inoue K., Lupski J.R. Genetics and genomics of behavioral and psychiatric disorders. Curr. Opin. Genet. Dev. 2003;13:303–309. doi: 10.1016/s0959-437x(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 10.Walz K., Fonseca P., Lupski J.R. Animal models for human contiguous gene syndromes and other genomic disorders. Genet. Mol. Biol. 2004;27:305–320. [Google Scholar]
- 11.Somerville M.J., et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lupski, J.R., and Chance, P.F. 2005. Hereditary motor and sensory neuropathies involving altered dosage or mutation ofPMP22 : the CMT1A duplication and HNPP deletion. Chapter 70. InPeripheral neuropathy . P.J. Dyck and P.K. Thomas, editors. Elsevier Science. Philadelphia, Pennsylvania, USA. 1659–1680. [Google Scholar]
- 13.Burger J., Horn D., Tonnies H., Neitzel H., Reis A. Familial interstitial 570 kbp deletion of the UBE3A gene region causing Angelman syndrome but not Prader-Willi syndrome. Am. J. Med. Genet. 2002;111:233–237. doi: 10.1002/ajmg.10498. [DOI] [PubMed] [Google Scholar]
- 14.Potocki L., Shaw C.J., Stankiewicz P., Lupski J.R. Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. Genet. Med. 2003;5:430–434. doi: 10.1097/01.gim.0000095625.14160.ab. [DOI] [PubMed] [Google Scholar]
- 15.Bi W., et al. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 2002;12:713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slager R.E., Newton T.L., Vlangos C.N., Finucane B., Elsea S.H. Mutations in RAI1 associated with Smith-Magenis syndrome. . Nat. Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- 17.Bi W., et al. Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum. Genet. 2004;115:515–524. doi: 10.1007/s00439-004-1187-6. [DOI] [PubMed] [Google Scholar]
- 18.Girirajan S., Elsas L.J., Devriendt K., Elsea S.H. RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. . J. Med. Genet. . 2005;42:820–828. doi: 10.1136/jmg.2005.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potocki L., et al. Molecular mechanism for duplication 17p11.2 — the homologous recombination reciprocal of the Smith-Magenis microdeletion [letter]. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Solis R., Liu P., Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 21.Walz K., et al. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol. Cell. Biol. 2003;23:3646–3655. doi: 10.1128/MCB.23.10.3646-3655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walz K., et al. Behavioral characterization of mouse models for Smith-Magenis syndrome and dup(17)(p11.2p11.2). Hum. Mol. Genet. 2004;13:367–378. doi: 10.1093/hmg/ddh044. [DOI] [PubMed] [Google Scholar]
- 23.Bi W., et al. Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. . Hum. Mol. Genet. 2005;14:983–995. doi: 10.1093/hmg/ddi085. [DOI] [PubMed] [Google Scholar]
- 24.Yan J., et al. Reduced penetrance of craniofacial anomalies as a function of deletion size and genetic background in a chromosome engineered partial mouse model for Smith-Magenis syndrome. Hum. Mol. Genet. 2004;13:2613–2624. doi: 10.1093/hmg/ddh288. [DOI] [PubMed] [Google Scholar]
- 25.Seong E., Seasholtz A.F., Burmeister M. Mouse models for psychiatric disorders. Trends Genet. 2002;18:643–650. doi: 10.1016/s0168-9525(02)02807-x. [DOI] [PubMed] [Google Scholar]
- 26.Hayes S., et al. GAG repeat length in RAI1 is associated with age at onset variability in spinocerebellar ataxia type 2 (SCA2). . Hum. Mol. Genet. 2000;9:1753–1758. doi: 10.1093/hmg/9.12.1753. [DOI] [PubMed] [Google Scholar]
- 27.Joober R., et al. Analysis of 14 CAG repeat-containing genes in schizophrenia. Am. J. Med. Genet. 1999;88:694–699. [PubMed] [Google Scholar]
- 28.Bi W., et al. RAI1 point mutations, CAG repeat variation, and SNP analyses in nondeletion Smith-Magenis syndrome. . Am. J. Med. Genet. 2006 doi: 10.1002/ajmg.a.31510. In press. [DOI] [PubMed] [Google Scholar]
- 29.Dykens E.M. Measuring behavioral phenotypes: provocations from the “new genetics.”. Am. J. Ment. Retard. 1995;99:522–532. [PubMed] [Google Scholar]
- 30.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 31.Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp A.J., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sebat J., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 34.Iafrate A.J., et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 35.Swanson J.M., et al. Attention-deficit hyperactivity disorder and hyperkinetic disorder. Lancet. 1998;351:429–433. [PubMed] [Google Scholar]
- 36.Weaver D.R. The suprachiasmatic nucleus: a 25-year retrospective. J. Biol. Rhythms. 1998;13:100–112.. doi: 10.1177/074873098128999952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.