Abstract
The structural requirements for generation of amyloid from the plasma protein transthyretin (TTR) are not known, although it is assumed that TTR is partly misfolded in amyloid. In a search for structural determinants important for amyloid formation, we generated a TTR mutant with high potential to form amyloid. We demonstrated that the mutant represents an intermediate in a series of conformational changes leading to amyloid. Two monoclonal antibodies were generated against this mutant; each displayed affinity to ex vivo TTR and TTR mutants with amyloidogenic folding but not to wild-type TTR or mutants exhibiting the wild-type fold. Two cryptic epitopes were mapped to a domain of TTR, where most mutations associated with amyloidosis occur and which we propose is displaced at the initial phase of amyloid formation, opening up new surfaces necessary for autoaggregation of TTR monomers. The results provide direct biochemical evidence for structural changes in an amyloidogenic intermediate of TTR.
Transthyretin (TTR) is a transport protein in plasma for thyroid hormone and forms a complex with retinol-binding protein. It has a potential to form amyloid fibrils and two major clinical forms are known. Senile systemic amyloidosis affects 25% of the individuals older than 80 years (1). Most cases of TTR-associated amyloidosis are linked to point mutations, of which more than 50 are known at present (2). One of the most common forms has a substitution of valine for methionine at position 30 of the 127-aa-long polypeptide, leading to widespread symptoms in the peripheral nervous system, known as familial amyloidosis with polyneuropathy.
Sixteen other proteins are known to form amyloid. Posttranslational modifications are observed in some cases associated with the formation of amyloid fibrils, including conformational changes and proteolytic cleavage (3). The role for these changes in self-aggregation is only partly understood. Analysis of amyloid fibrils of different origins indicates a common cross-β-pleated sheet conformation independent of the protein involved (4, 5). The three-dimensional structure of native TTR is established (6); it is a tetramer with four identical subunits, folding into a globular structure, each monomer having eight β-strands organized in two sheets. Thus, TTR has a predominance of β-structure, in contrast to several other amyloid-forming proteins with little β-structure, which has to be formed before aggregation starts. In the case of TTR-associated amyloid, it is not known whether the original conformation is preserved in the fibrils, although good evidence exists from in vitro experiments that the tetramers need to dissociate into alternatively folded monomers for amyloid to form (7). The package of the monomers into fibrils has been the subject of several studies and different, partly conflicting, models have been proposed (8–10).
Analysis of the distribution of mutations showed that they occur all along the polypeptide chain, although some areas seem to be spared. We previously described a broad area close to the “edge” of the molecule, i.e., around the β-strands designated C and D, with more frequent mutations leading to amyloidosis (11). It has been proposed that this area of the molecule is more flexible (12), and a current model proposes that this area bulges out from TTR when amyloid fibrils form (13).
Detailed x-ray diffraction studies with a resolution down to 1.7 Å of TTR V30M (TTR with the substitution V30M) has not given information concerning the mechanism for amyloid formation (14, 15). However, recent studies of the clinically aggressive L55P mutant suggested a possible organization of the fibrils based on the packing contacts in the crystal (16).
An amyloidogenic intermediate of TTR has been demonstrated, which might occur in a denaturing or degradation pathway (17). Such partly misfolded intermediates were isolated in a previous study from our laboratory (11, 18) by construction of mutants, in which the three amino acids of the D strand were either removed (TTRdel53–55) or substituted (TTR G53S, E54D, L55S, here designated TTRs53–55). These molecules rapidly formed aggregates, which gave a typical cross-β pattern in x-ray diffraction studies and a positive signal after staining with Congo Red or thioflavine T. Therefore, these mutants qualify as amyloid precursors in vitro and might carry structural determinants of intermediates in an in vivo pathway leading to amyloid formation.
In the present study we asked whether it would be possible to generate monoclonal antibodies against epitopes expressed only on amyloidogenic TTR mutants. Two such monoclonal antibodies are described here that provide direct biochemical evidence for amyloidogenic conformational changes in TTR and localize them in the edge area of the molecule.
MATERIALS AND METHODS
Expression of TTR.
TTR was expressed in Escherichia coli as previously described by using two different expression systems. In short, wild-type TTR, TTR V30M, TTRdel53–55, and TTRs53–55 were mutagenized, expressed after cloning into the pET3a vector, and purified by ion-exchange chromatography (11, 18–20). The TTR mutant carrying plasmids designated pINTR-RM, pINTR-10, pINTR-30, pTTR-10−30, pINTR-55, pINTR-60, pINTR-84, and pINTR-119 were used for expression, and TTR was extracted as described (21).
Cell Fusion.
Mice (BALB/c, female, 8–10 weeks old) were immunized with the recombinant mutant TTRs53–55 (11). After 6 weeks, sera were tested for antibodies against TTR. Mice with positive results were then given booster i.v. injections of 50 μg of protein. Fusion with mouse B cell myeloma cells (SP2/0) was performed 3 days later according to standard procedures and seeded in microtiter wells (200 μl per well). Selection for hybrid cells was done with RPMI medium 1640 containing 10% fetal calf serum and hypoxanthine/aminopterin/thymidine. Supernatants from wells with growing cells were screened for antibodies reactive with plasma wild-type TTR and/or TTRs53–55 bound to plates as follows.
ELISA.
TTR (wild-type or mutant protein) were bound to microtiter plates (Nunc) either directly or with a sandwich approach to a rabbit polyclonal antibody raised against human TTR (Dakopatts, Glostrup, Denmark). TTR was bound directly to wells by incubating plates overnight with 100 μl per well of TTR at a concentration of 5 μg/ml in PBS at 4°C. Wells were then washed with PBS containing 0.05% Tween 20 (Kebo Laboratory, Stockholm, Sweden), and unspecific binding sites were blocked by additional incubation at room temperature with PBS containing 0.5% skimmed milk.
With the sandwich ELISA, plates were first incubated overnight at 4°C with 100 μl per well of polyclonal antibody against human TTR (Dakopatts) at a concentration of 5 μg/ml in PBS. Blocking of unspecific binding sites was done as described above.
After the plates were coated, they were incubated with the TTR proteins at 37°C for 1 h. The plates were washed, and the supernatants were added to the wells, diluted 10 times in PBS. Bound antibodies were detected with horseradish peroxidase-labeled anti-mouse Ig antibodies (Dakopatts) according to standard procedures.
In competition experiments, horseradish peroxidase-labeled antibodies were added together with different concentrations of unlabeled antibodies.
Epitope Mapping.
Epitope mapping based on fusion of partial cDNA fragments of the TTRs53–55 gene to a phage protein was done with the NovaTope epitope-mapping system according the instructions of the supplier (Novagen; ref. 22).
Antibody Binding to Amyloid.
The ability of unlabeled antibody to compete with iodinated antibody for the consumption by ex vivo amyloid of labeled antibody was measured. The monoclonal antibodies used were iodinated with 125I according to the N-bromosuccinimide method. Vitreous amyloid from a patient with the TTR met30 mutation was dispersed through microtip sonication three times for 5 s each time at level 3 (Ultrasonic Processor XL, Heat Systems, Farmingdale, NY) and suspended in Tris-buffered saline, pH 7.4. The radiolabeled antibody was diluted to give a molar excess of TTR amyloid compared with the iodinated antibody and then mixed with various amounts of unlabeled mAb in dilutions ranging from 100 to 0 μg per tube. The total volume corresponded to 200 μl per tube. Aliquots (100 μl) of the dispersed vitreous TTR amyloid were added and incubated for 1 h and then centrifuged for 20 min at 12,000 × g. The supernatant (100 μl) was withdrawn and measured for radioactivity in a γ-counter (LKB model 1282). As a negative control, a mAb of the same isotype with irrelevant specificity was used.
Gel Electrophoresis.
Protein samples were separated on a 4–16% polyacrylamide gradient gel under native conditions, i.e., without SDS or reducing agent.
Immunoblotting.
Proteins were transferred to a poly(vinylidene difluoride)-plus membrane (Micron Separations, Westboro, MA), and the membrane was blocked with 5% skimmed milk. Immunodetection was performed with either a polyclonal rabbit anti-human TTR antibody (Dakopatts), and horseradish peroxidase-labeled donkey anti-rabbit IgG antibody (Amersham Pharmacia Biotech, Uppsala, Sweden), or monoclonal mouse anti-human antibodies, mAb 39–44 or mAb 56–61, and horse radish peroxidase-labeled sheep anti-mouse IgG antibody (Amersham). Detection was performed with enhanced chemiluminescence (ECL, Amersham).
Size-Exclusion HPLC.
Size-exclusion HPLC analysis was done on a Protein-Pak 125, 7.5- × 300-mm, Millipore column on a Waters chromatographic system with PBS as the eluent. The flow rate was 0.5 ml/min, and the sample volume was 250 μl. Column calibration was done with BSA, egg albumin, and soybean trypsin inhibitor.
RESULTS
The TTRs53–55 mutant used to immunize the BALB/c mice has been described (11, 18). It forms amyloid slowly at physiological pH and immediately at pH 4.5 and was proposed to represent an intermediate in a pathway leading to TTR amyloid. Four fusions were performed, and after screening with the same antigen to coat ELISA plates, we isolated five stable hybridomas that produced mAbs of the IgG1/κ type or in one case IgG2a/κ (mAb 14). The specificity of the antibodies was tested on a series of TTR mutants by a direct ELISA, and the results are summarized in Table 1. The TTR antigens tested were produced in two ways. Most of them were expressed with the pET3a vector (19) introducing an N-terminal methionine. To exclude that this methionine contributed to the epitopes recognized by the antibodies, some of the TTR mutants were expressed in secreted form with a leader peptide, which was cleaved off, resulting in a TTR molecule lacking an N-terminal methionine (21).
Table 1.
Primary screening of monoclonal antibodies for reactivity with different mutant TTR molecules
| TTR | mAb 47 | mAb 11 | mAb 14 | mAb 17 | mAb 15 |
|---|---|---|---|---|---|
| Wild type | + | − | + | + | + |
| V30M | + | − | ND | ND | + |
| s53-55 | + | + | + | + | + |
| del53-55 | + | − | + | + | + |
| pINTR-RM (wild type) | − | − | − | − | + |
| pINTR-10 (C10S) | + | − | − | − | + |
| pINTR-30 (V30M) | + | − | − | − | + |
| pINTR-55 (L55P) | + | − | − | − | + |
| pINTR-119 (T119M) | + | − | − | − | + |
The antibodies were tested with a direct ELISA. ND, not determined, plasmid designation according to Materials and Methods.
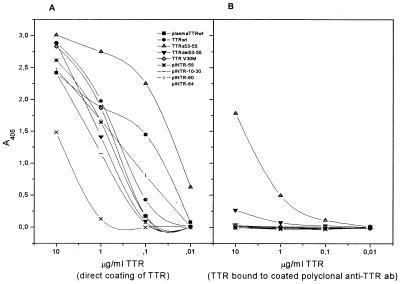
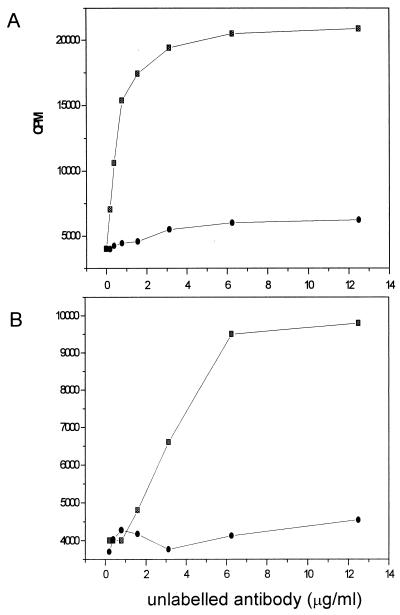
mAbs 11, 14, and 17 were discarded, because they had specificity for either the three introduced amino acids at positions 53–55 or the N-terminal methionine. Two antibodies, mAb 15 and mAb 47, hereafter designated mAb 39–44 and mAb 56–61, respectively (see below), reacted in the direct ELISA with all tested TTR mutants (Fig. 1A). However, when the specificity was further tested in a sandwich ELISA with a rabbit polyclonal antibody for coating, a different pattern appeared with mAb 56–61 (Fig. 1B). In these less denaturing conditions, only TTRdel53–55 and TTRs53–55 were detected, whereas all other tested TTR mutants, as well as the wild-type TTR, were not detectable. The same results were obtained with mAb 39–44 (data not shown).
Figure 1.
Reactivity of mAb 56–61 with recombinant wild-type TTR and a panel of mutant TTR molecules. (A) Direct ELISA, with which the plates were coated with the TTR variants indicated. (B) Sandwich ELISA, with which the plates were coated with a rabbit polyclonal anti-TTR antibody (Dako). Binding of mAb 56–61 was demonstrated by using a horseradish peroxidase-conjugated rabbit anti-mouse antibody.
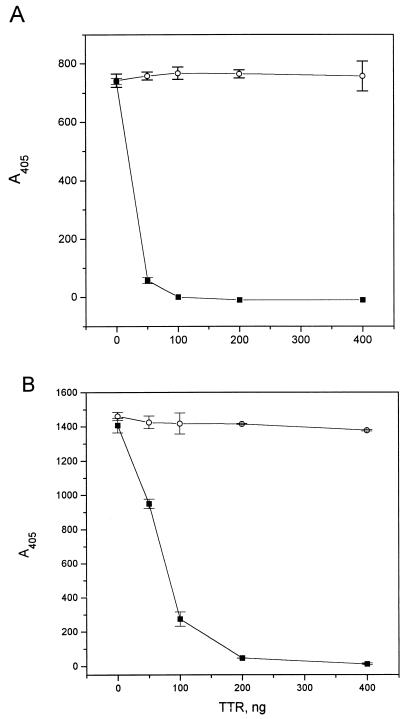
The possibility that the polyclonal rabbit anti-TTR antibody reacted with an immunodominant epitope on the nondenatured TTR molecule and blocked the binding of the monoclonal antibodies seemed less probable because TTR is a tetramer. In a control experiment, it was shown (Fig. 2) that the monoclonal antibodies bound to an epitope expressed on TTRs53–55 and not to wild-type TTR, because the substitution mutant and not the soluble, wild-type protein displaced binding of mAb 39–44 and mAb 56–61 to wild-type TTR coated to a plastic surface. Furthermore, both D strand mutants could be immunoprecipitated by both antibodies, in contrast to wild-type TTR (data not shown). Thus, both antibodies display specificity for a new epitope on TTR not exposed on soluble TTR in a native configuration.
Figure 2.
The amyloidogenic mutant TTRs53–55 but not wild-type TTR inhibits interaction between mAb 56–61 (A) or mAb 39–44 (B) and TTR coated on a plastic surface. ○, Wild-type TTR; ■, TTRs53–55. Microtiter plates were coated with 5 mg/ml wild-type TTR in PBS overnight at 4°C and then blocked with 5% skimmed milk. The mAbs and dilutions of TTR were preincubated at 37°C for 1 h in plates blocked with 5% milk, then transferred to plates coated with wild-type TTR, and incubated for 1 h. Bound mAb was visualized with horseradish peroxidase-conjugated anti-mouse IgG antibodies.
A similar pattern of reactivity was seen when the TTR mutants were separated either by native PAGE or by SDS/PAGE, followed by immunoblotting with mAb 39–44 or mAb 56–61 to detect TTR. As detailed below, only TTRdel53–55 and TTRs53–55 were detected with nondenaturing PAGE, whereas all TTR variants were detected after boiling and separation by SDS/PAGE (Fig. 3 and data not shown).
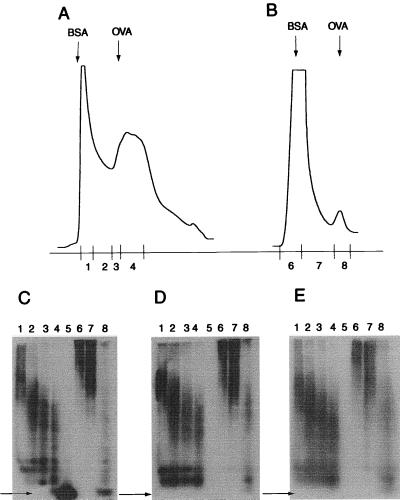
Figure 3.
Immunoreactivity of size variants of the TTRs53–55 mutant separated by size-exclusion HPLC. Size-exclusion HPLC of TTRs53–55 mutant freshly prepared (A) or aggregated in 37°C for 24 h (B). Immunoblot from native PAGE of size-exclusion HPLC fractions with polyclonal antibody (C), mAb 56–61 (D), or mAb 39–44 (E). Lanes 1–4 and 6–8 correspond to HPLC fractions as indicated; lane 5 was loaded with wild-type TTR. Lanes: 1–4, fresh material; 6–8, aggregated TTR. Arrows indicate the position of TTR tetramer; OVA (ovalbumin) and BSA indicate size markers.
The results indicate that mAb 39–44 and mAb 56–61 detected cryptic epitopes that were not exposed on the normally folded protein. This epitope was exposed in all TTR species after partial denaturing caused by coating on a plastic surface or after boiling in sample buffer with SDS and 2-mercaptoethanol. However, this epitope was permanently expressed on the D strand mutants in nondenaturing conditions.
The reactivity of the two antibodies was further analyzed by immunoblotting after an analytical separation by size-exclusion HPLC chromatography and compared with that of a polyclonal rabbit antibody. As has been demonstrated (18), the substitution mutant resolves as two peaks. Immediately after extraction, the smaller fraction (fraction 4) was contaminated by low Mr material (Fig. 3A) that did not show up on Coomassie or silver staining of PAGE gels (not shown). It disappeared after an incubation of 24 h and obviously obscured a sharp peak made up by the TTR tetramer. A separate TTR peak (Fig. 3A, fractions 1 and 2) increased in size with time and reached the size-exclusion limit of the column (Fig. 3B, fractions 6 and 7). Later it disappeared concomitantly with formation of Congo Red staining precipitates (18). Fig. 3 C–E is an immunoblot of a native PAGE separation of the HPLC fractions. Fig. 3C shows that the polyclonal antibody detects both the TTR tetramer and large Mr aggregates, increasing in size with time. In contrast, the mAbs detect only the aggregates and no TTR tetramer.
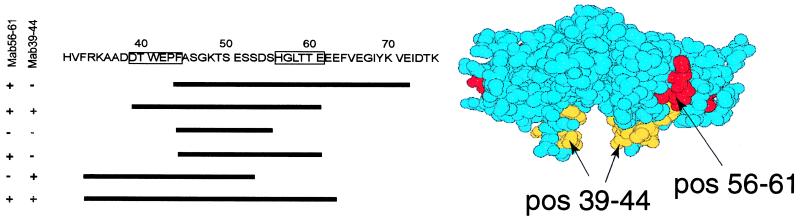
The epitopes for the two mAbs were defined by screening a TTRs53–55 cDNA fragment library fused to the T7 gene 10 protein, and several diagnostic clones were identified, as summarized in Fig. 4. The antibodies detected two distinct regions comprising amino acid residues 39–44 and 56–61, respectively. Because both antibodies also bind to the D strand deletion mutant (TTRdel53–55), residues 53–55 do not seem to be a significant part of either epitope (cf. Fig. 1B). The epitope including positions 39–44 seems to be embedded in the structure, as depicted in the space-filling model (Fig. 4), whereas the other epitope points to the cleft formed on the surface between the two monomers. The epitopes were distinct as shown by a cross-competition experiment, in which labeled mAb 56–61 was not blocked by competition by mAb 39–44 or an irrelevant antibody, whereas nonlabeled mAb 56–61 did compete for binding (Fig. 5).
Figure 4.
Epitope mapping of mAb 39–44 and mAb 56–61. (Left) Part of the sequence of TTRs53–55 with the defined epitopes boxed. The sequenced fragments obtained by screening the DNA fusion library as described in Materials and Methods are depicted as lines. (Right) A space-filling model of a dimer of TTR, by using the Molscript program, with the minimal epitopes defined by mAb 39–44 and mAb 56–61 depicted. The view is along the β-strand direction. The epitopes seem to be embedded in the TTR monomer.
Figure 5.
Distinct binding of mAb 39–44 and mAb 56–61 to the TTRs53–55 mutant. In an ELISA experiment, TTRs53–55 was coated in the wells, to which was added horseradish peroxidase-labeled mAb 56–61 together with unlabeled mAb 56–61 (■), mAb 39–44 (●), or an irrelevant antibody (▴).
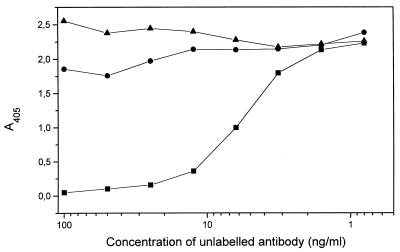
Finally, we wanted to test whether the mAbs could bind to amyloid obtained ex vivo. Amyloid from the vitreous body of the eye can be extracted under mild conditions without manipulations, which might affect conformation (23). Fig. 6 shows that both antibodies were able to bind to ex vivo amyloid in a specific way, because unlabeled antibody competed with the labeled antibody, whereas an irrelevant antibody did not compete.
Figure 6.
Competition of anti-TTR mAbs with iodinated antibodies for binding to TTR met30 fibrils isolated from vitreous deposits. Consumption of labeled antibodies was measured by estimating radioactivity in the supernatants. (A) mAb 39–44 (■), irrelevant antibody (●); (B) mAb 56–61 (■), irrelevant antibody (●).
DISCUSSION
It has long been known that TTR is a remarkably stable tetrameric molecule, retaining quaternary structure in denaturating conditions, such as high concentrations of SDS, urea, or guanidine⋅HCl (24). The tetramer is destabilized in amyloidogenic mutants of TTR (7) and it is hypothesized that the formation of monomers is the first step in amyloid formation (13). Computer modeling data (8, 9) suggest that TTR subunits incorporated in the amyloid fibril have a fold different from native one. Hydrogen exchange data suggest that the amyloidogenic intermediate might be a partially unfolded TTR monomer (25). Further indirect evidence for partial unfolding is provided by the demonstration of a hidden protease cleavage site in highly amyloidogenic TTR mutants and amyloid fibrils (18, 23).
The approach used in this study to disclose structural changes important for generation of amyloid from TTR built on the availability of mutants, which represent precursors for amyloid fibrils, with a partially misfolded conformation (11, 18). Through the generation of mAbs toward such conformational intermediates, we can provide new information concerning structural requirements for generation of amyloid from the plasma protein TTR. Two cryptic epitopes were detected in ex vivo amyloid, obtained by mild extraction procedures from the vitreous body of the eye in affected individuals. Most importantly, the epitopes are not detectable on wild-type TTR or on two well known mutants, namely, TTR V30M and TTR L55P, the latter giving rise to a clinically aggressive form of amyloidosis. However, after denaturation, all TTR species expose these cryptic epitopes.
In a separate study, we analyzed plasma from individuals with the TTR V30M mutation and found that these epitopes were detectable under mild denaturing conditions (26). This mutation displays only minor changes in the three-dimensional structure (14, 15), and it does not form amyloid in vitro in physiological buffers. However, amyloid is formed after rather harsh denaturing conditions, such as reduction of pH (7, 17) or refolding after SDS treatment (G.G., unpublished observations). A kinetic trap for amyloid formation can be envisaged, and we propose that the cryptic epitope is a marker for an amyloidogenic fold. In addition to their expression on mutants that spontaneously form amyloid and on amyloid fibrils, we have also shown (A.O., unpublished observations) that their expression correlates with increased aggregation rate at low pH, a well established experimental model for in vitro generation of amyloid. Generation of amyloid through the activity of so-called pathological chaperones (27) has not been demonstrated in the case of TTR-associated amyloidosis. The availability of the antibodies described here might be useful both for affinity selection of rare intermediates of TTR V30M in body fluids and for detection of factors that might generate their formation.
We have not been able to block in vitro generation of amyloid by using the mAbs. Therefore, we propose that the cryptic epitopes represent a structural determinant not present in the core of the fibrils but rather exposed on their surface. As pointed out (22), the method used for epitope mapping gives only a minimal estimate of an epitope. mAb 56–61 has specificity for a region at the N-terminal part of the loop connecting the small D strand in the inner sheet with the E strand on the outer loop. In the native configuration, this epitope is buried by the N-terminal end of the peptide (cf. Fig. 4). mAb 39–44 recognizes an epitope, which includes the N-terminal part of the C strand, and the C-terminal part of the loop connecting the B and C strands. This epitope is more exposed on the external surface but projects toward a valley formed by the D–E loops on both monomers, which might cause a steric hindrance toward antibody binding to this epitope in the native molecule.
Partial misfolding of TTR has been proposed to be a starting point for amyloid formation (7). Our data provide evidence that partial denaturation of the C and D strands with loss of both the β-structure and their integration in the two β-sheets is a prerequisite for self-aggregation. This has been proposed by other authors based on modeling studies (8) or biophysical studies of amyloid formed by acid denaturation (17). The model predicts that this region should be accessible on the fibril surface, which is in agreement with our finding that TTR in amyloid could bind both mAbs in contrast to the native soluble molecule. The finding of N-terminal truncations (28) and an accessible trypsin cleavage site in amyloid from the vitreous body at position 48/49 in the loop between the C and D strands (18, 23) supports the notion that the edge structures, including the C and D strands, project out from the fibril. Furthermore, recent high resolution studies of TTR L55P by Sebastião et al. (16) demonstrate disrupture of hydrogen bonds between the D and A strands in the edge region. This implies that the barrel structure is broken and new surfaces are exposed, which can now be involved in aggregation, e.g., the A and B strands. The definition of surfaces important for the interaction of TTR molecules in fibrils might be important for the development of new treatment strategies.
Acknowledgments
During the final phase of the study, Håkan Persson died; the study is dedicated to his memory. This study has been supported by a European Commision Biomed 2 grant, the Swedish Medical Research Council, the patient′s association FAMY, the Medical Faculty, Umeå University, the County of Västerbotten (E.L.), and Program Praxis XXI, Portugal (M.J.S.).
ABBREVIATIONS
- TTR
transthyretin
- TTRdel53–55
TTR, where positions 53–55 were deleted
- TTRs53–55
TTR with substitutions G53S, E54D, and L55S
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cornwell G G, III, Sletten K, Johansson B, Westermark P. Biochem Biophys Res Commun. 1988;154:648–653. doi: 10.1016/0006-291x(88)90188-x. [DOI] [PubMed] [Google Scholar]
- 2.Saraiva M J M. Hum Mutat. 1995;5:191–196. doi: 10.1002/humu.1380050302. [DOI] [PubMed] [Google Scholar]
- 3.Cohen A S, Jones L A. Curr Opin Rheumatol. 1993;5:62–76. doi: 10.1097/00002281-199305010-00010. [DOI] [PubMed] [Google Scholar]
- 4.Burke M J, Rougvie M A. Biochemistry. 1972;11:2435–2439. doi: 10.1021/bi00763a008. [DOI] [PubMed] [Google Scholar]
- 5.Glenner G G, Eanes E D, Bladen H A, Linke R P, Termine J D. J Histochem Cytochem. 1974;22:1141–1158. doi: 10.1177/22.12.1141. [DOI] [PubMed] [Google Scholar]
- 6.Blake C C F, Geisow M J, Oatley S J, Rérat B, Rérat C. J Mol Biol. 1978;121:339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 7.Colon W, Kelly J W. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 8.Blake C, Serpell L. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 9.Inouye H, Domingues F S, Damas A M, Saraiva M J, Lundgren E, Sandgren O, Kirschner D A. Amyloid. 1998;5:163–174. doi: 10.3109/13506129809003842. [DOI] [PubMed] [Google Scholar]
- 10.Schormann N, Murrell J R, Benson M D. Amyloid. 1998;5:175–187. doi: 10.3109/13506129809003843. [DOI] [PubMed] [Google Scholar]
- 11.Serpell L C, Goldsteins G, Dacklin I, Lundgren E, Blake C C F. Amyloid. 1996;3:75–86. [Google Scholar]
- 12.Blake C C F, Oatley S J. In: Conformation in Biology, the Festschrift Celebrating the Sixtieth Birthday of G. N. Ramachandran F. R. S. Srinivasan R, Sarma R H, editors. New York: Academic; 1982. pp. 29–38. [Google Scholar]
- 13.Kelly J W, Lansbury P T. Amyloid. 1994;1:186–205. [Google Scholar]
- 14.Hamilton J A, Steinrauf L K, Liepnieks J, Benson M D, Holmgren G, Sandgren O, Steen L. Biochim Biophys Acta. 1992;1139:9–16. doi: 10.1016/0925-4439(92)90075-x. [DOI] [PubMed] [Google Scholar]
- 15.Terry C J, Damas A M, Oliveira P, Saraiva M J M, Alves I L, Costa P P, Matias P M, Sakaki Y, Blake C C F. EMBO J. 1993;12:735–741. doi: 10.1002/j.1460-2075.1993.tb05707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastião M P, Saraiva M J, Damas A M. J Biol Chem. 1998;273:24715–24722. doi: 10.1074/jbc.273.38.24715. [DOI] [PubMed] [Google Scholar]
- 17.Lai Z, Colon W, Kelly J W. Biochemistry. 1996;35:6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 18.Goldsteins G, Andersson K, Olofsson A, Dacklin I, Edvinsson Å, Baranov V, Sandgren O, Thylén C, Hammarström S, Lundgren E. Biochemistry. 1997;36:5346–5352. doi: 10.1021/bi961649c. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg A, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Gene. 1987;56:5463–5467. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–383. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 21.Furuya H, Saraiva M J M, Gawinowicz M A, Alves I L, Costa P P, Sasaki H, Goto I, Sakaki Y. Biochemistry. 1991;30:2415–2421. doi: 10.1021/bi00223a017. [DOI] [PubMed] [Google Scholar]
- 22.Stanley K K, Herz J. EMBO J. 1987;6:1951–1957. doi: 10.1002/j.1460-2075.1987.tb02457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thylén C, Wahlqvist J, Haettner E, Sandgren O, Holmgren G, Lundgren E. EMBO J. 1993;12:743–748. doi: 10.1002/j.1460-2075.1993.tb05708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branch W T, Robbins J, Edelhoch H. J Biol Chem. 1971;246:6011–6018. [Google Scholar]
- 25.Nettleton E J, Sunde M, Lai Z, Kelly J W, Dobson C M, Robinson C V. J Mol Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- 26.Saraiva M J, Almeida M R, Alves I L, Bonifacio M J, Damas A M, Palha J A, Goldsteins G, Lundgren E. Ciba Found Symp. 1996;199:47–52. doi: 10.1002/9780470514924.ch4. [DOI] [PubMed] [Google Scholar]
- 27.Wisniewski T, Frangione B. Neurosci Lett. 1992;135:235–238. doi: 10.1016/0304-3940(92)90444-c. [DOI] [PubMed] [Google Scholar]
- 28.Westermark P, Sletten K, Johansson B, Cornwell I G G. Proc Natl Acad Sci USA. 1990;87:2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]