Abstract
The identification of genes with selective expression in specific organs or cell types provides an entry point for understanding biological processes that occur uniquely within a particular tissue. Using a subtraction approach designed to identify genes preferentially expressed in specific tissues, we have identified prostase, a human serine protease with prostate-restricted expression. The prostase cDNA encodes a putative 254-aa polypeptide with a conserved serine protease catalytic triad and an amino-terminal pre-propeptide sequence, indicating a potential secretory function. The genomic sequence comprises five exons and four introns and contains multiple copies of a chromosome 19q-specific minisatellite repeat. Northern analysis indicates that prostase mRNA is expressed in hormonally responsive normal and neoplastic prostate epithelial tissues, but not in prostate stromal constituents. Prostase shares 35% amino acid identity with prostate-specific antigen (PSA) and 78% identity with the porcine enamel matrix serine proteinase 1, an enzyme involved in enamel matrix degradation and with a putative role in the disruption of intercellular junctions. Radiation-hybrid-panel mapping localized prostase to chromosome 19q13, a region containing several other serine proteases, including protease M, pancreatic/renal kallikrein hK1, and the prostate-specific kallikreins hK2 and hK3 (PSA). The sequence homology between prostase and other well-characterized serine proteases suggests several potential functional roles for the prostase protein that include the degradation of extracellular matrix and the activation of PSA and other proteases.
Carcinoma of the prostate is the most commonly diagnosed noncutaneous malignancy and the second-leading cause of cancer-related death in American men. Estimates from the American Cancer Society indicate that more than 180,000 American men will be diagnosed with prostate cancer, and 39,000 will die of this malignancy in 1998 (1). Great though these numbers are, they represent but a small fraction of the histologically prevalent prostate cancers in the population. The incidence of latent, or incidental, prostate cancer approaches 35% in men over the age of 50 years, which translates into more than 10 million affected individuals (2, 3).
The treatment of prostate carcinoma has been greatly aided by the identification of genes and their cognate proteins whose expression is essentially restricted to the prostate. The most useful of these proteins to date is prostate-specific antigen (PSA), a serine protease expressed by the luminal epithelium of normal prostate tissue and also produced by malignant prostate cells (4). The positive predictive value of PSA for assessing cancer risk makes PSA the most useful tumor marker for monitoring treatment response and disease progression among patients diagnosed with prostate cancer (5). Though controversial, screening for elevated levels of PSA in asymptomatic populations recently has been suggested to decrease prostate cancer mortality (6). However, PSA is not a perfect tumor marker because levels often are elevated in men with benign prostatic hyperplasia, prostatitis, and other nonmalignant disorders (7). In addition, PSA measurements fail to discriminate accurately between indolent and aggressive cancers, PSA levels do not clearly correlate with tumor volume, PSA levels may be affected by drug treatments without affecting tumor growth, and assays for PSA may crossreact with other closely related serine proteases such as glandular kallikreins 1 and 2 (7).
In addition to diagnostic uses, tissue-specific genes such as PSA are currently under evaluation for prognostic and therapeutic applications. The hematogenous identification of PSA mRNA indicative of circulating prostate carcinoma cells may provide stage-related prognostic information. Immunotherapy strategies involving vaccine and mAbs targeting PSA and the cell-surface marker prostate-specific membrane antigen have been developed (8, 9). Prodrug activation by PSA enzymatic activity has been proposed as a means to produce tissue-specific cytotoxicity (10), and several gene therapy approaches in early clinical trials use the PSA promoter for cell type-specific gene activation (11).
In an effort to identify additional genes preferentially expressed in the human prostate that could serve as potential markers for the diagnosis and treatment of prostate cancer, we constructed a prostate cDNA library enriched by subtraction with cDNAs from four other normal tissues. Analysis of the resulting cDNA population by expressed sequence tag sequencing identified a gene, prostase, that exhibits a transcript expression pattern restricted to the normal and neoplastic prostate. Prostase exhibits features of a secreted protein and is a member of the serine protease gene family with greatest sequence homology to enamel matrix serine proteinase 1 (EMSP1), a porcine protein hypothesized to be involved in remodeling of organic matrix in tooth development (12).
MATERIALS AND METHODS
Cell Culture and General Methods.
DNA manipulations, including transformation, plasmid preparation, gel electrophoresis, and probe labeling, were performed according to standard procedures (13). Restriction and modification enzymes (Life Technologies, Rockville, MD) were used in accordance with the manufacturer’s recommendations. Prostate carcinoma cell lines LNCaP, DU145, and PC3 were cultured in RPMI medium 1640 supplemented with 10% FBS. LNCaP cells were transferred into RPMI 1640 media with 10% charcoal-stripped fetal calf serum (CS-FCS) (Life Technologies) 24 hr before androgen regulation experiments. This media was replaced with fresh CS-FCS media including 1 nM of the synthetic androgen R1881 (New England Nuclear). Cells were harvested for RNA isolation at 0-, 1-, 2-, 4-, 8-, 24-, 48-, and 72-hr time points.
cDNA Library Construction and Analysis.
Poly(A)+ RNA was isolated from flash-frozen normal prostate, brain, liver, and placental tissues by using the TRIzol reagent (Life Technologies) and oligo(dT) Sepharose columns (Amersham Pharmacia) according to the manufacturers’ instructions. Prostate cDNA synthesis was performed by using Molone murine leukemia virus reverse transcriptase (Life Technologies) and the PCR-Select cDNA subtraction kit (CLONTECH) using suppression subtractive hybridization (14). Prostate-enriched and -subtracted cDNAs were cloned into the PCRII vector (Invitrogen), transformed into Escherichia coli, and plated on Luria–Bertani/ampicillin agar. Ninety-six clones were randomly selected and subjected to PCR amplification with insert-flanking vector primers to generate template for DNA sequencing as described (15).
cDNA and Genomic DNA Cloning.
A 128-bp prostase cDNA fragment obtained from the subtractive cDNA library was labeled with 32P-dCTP by using a random priming method (Life Technologies) and used to screen 36,000 arrayed clones from a normal prostate cDNA library (15). Positive clones were isolated, propagated, and sequenced as described above. Rapid amplification of cDNA ends was used to obtain 5′ cDNA sequence by using normal prostate cDNA (CLONTECH) and oligo(dG)-tailed LNCaP cDNA as template with the gene-specific primer PRR1 5′-GGTATTTGGGGGTCAATTTCATGGG-3′. A bacterial artificial chromosome (BAC) clone containing the prostase gene was obtained by screening a human BAC library (Genome Systems, St. Louis) with prostase gene-specific oligonucleotides (PRU1 5′-CCTTGCTCGCTAACGACCTCAT-3′ and PRL1 5′-CCAGCCAGAAACGAGGCAAGAGT-3′). Cesium chloride density gradient-purified BAC DNA was sonicated, and DNA fragments 1.5–2 kb in size were separated on an agarose gel, purified, and subcloned into M13 vectors. Five hundred clones were partially sequenced as described (15) and assembled by using phred, phrap, and consed sequence assembly software (16, 17).
DNA Sequencing and Analysis.
DNA sequencing was performed by the dideoxy chain-termination method by using Taq dye primer and dye terminator kits (Applied Biosystems). The nucleotide sequences were analyzed with an ABI 377 automated sequencer (Applied Biosystems). Sequence homology searches and comparisons were performed by using blastn and blastx algorithms (18) on the National Center for Biotechnology Information web server (http://www.ncbi.nlm.nih.gov/BLAST/). The assembled genomic sequence was analyzed with the gene prediction program genescan (http://gnomic.stanford.edu/GENSCANW.html) (19) and compared with the cDNA sequence to define intron/exon boundaries and gene sequence structures. Multiple sequence alignments were performed by using the clustal algorithm (20). A neural network-based signal peptide and cleavage site prediction program was used to determine the putative signal peptide sequence (http://www.cbs.dtu.dk/services/SignalP/) (21).
Chromosomal Localization.
The G3 Gene bridge radiation hybrid panel (Research Genetics, Huntsville, AL) (22) was used to determine the chromosomal location of prostase. Prostase primers PRU1 5′-CCTTGCTCGCTAACGACCTCAT-3′ and PRL1 5′-CCAGCCAGAAACGAGGCAAGAGT-3′ were used in PCRs with DNA pools from the hybrid panel according to the manufacturer’s instructions. After 40 cycles of amplification, the reaction products were separated on a 1.2% agarose gel, and the result was analyzed through the Stanford genome center web server (http://www-shgc.stanford.edu/RH) to determine the probable chromosomal location.
Northern Analysis.
Total RNA was isolated from cell lines, normal prostate tissue, and prostate cancer xenografts by using TRIzol (Life Technologies) according to the manufacturer’s directions. The RNA samples were fractionated on a 1.2% agarose denaturing gel and transferred to nylon membrane by capillary method (13). The human multiple tissue blot was obtained from CLONTECH. Blots were probed with a cDNA fragment representing exon 4 and 5 of the prostase cDNA. Filters were imaged and quantitated by using a phosphor-capture screen and imagequant software (Molecular Dynamics).
RESULTS
Subtracted cDNA Library Analysis.
A prostate cDNA library was constructed for the enrichment of transcripts preferentially expressed in prostate tissue relative to liver, brain, and placenta. Ninety-six clones from this subtracted library were selected at random and partially sequenced. A comparison of these sequences to the GenBank and dbEST nucleotide databases using the blastn and blastx algorithms identified 69 sequences with >95% identity to known genes, seven with identity to sequences only in dbEST, and four putatively novel sequences with less than 70% nucleotide sequence homology to any database entries (Table 1). Sixteen clones contained no inserts or yielded poor sequence data. Overall, 56 of the 80 high-quality sequences (70%) were homologous to five different genes previously described to exhibit prostate and seminal vesicle-restricted or enhanced expression, prostate acid phosphatase (23), PSA (24), seminogelin (25), and prostate-specific transglutaminase (26). We previously have shown that none of these transcripts are present at a frequency greater than 3% in a nonsubtracted prostate library (15), thus indicating the subtraction procedure successfully enriched the library population for prostate-specific cDNAs.
Table 1.
Sequence analysis results of 96 randomly selected cDNA clones from the subtracted prostate cDNA library
| Identification | #Clones |
|---|---|
| Prostate acid phosphatase | 21 |
| Seminogelin I | 14 |
| Seminogelin II | 8 |
| PSA | 7 |
| Prostate-specific glutaminase | 6 |
| Myoson heavy chain | 4 |
| Other known genes* | 9 |
| Expressed sequence tag | 7 |
| Novel | 4 |
| No data | 16 |
| Total | 96 |
The Novel category indicates cDNAs without significant matches to any sequences in the public databases. The No Data category indicates the number of clones selected with no inserts or poor DNA sequence read quality.
Identification, Cloning, and Sequence Analysis of Prostase.
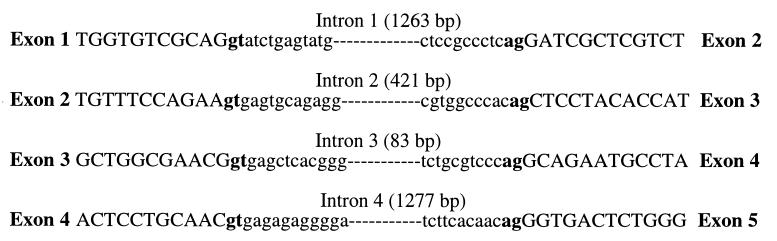
The partial ORF of one 157-bp cDNA clone, G7, exhibited significant amino acid similarity (40%–80%) to several serine proteases, including members of the kallikrein protein family and the recently described EMSP1 (GenBank accession no. U76256) (12). Additional sequence for the G7 clone was obtained by screening a normal prostate cDNA library and by performing rapid amplification of cDNA ends (RACE). However, when compared with other serine protease sequences, the final assembly of the prostase cDNA sequence failed to yield a legitimate exon 1. To obtain the first exon and to determine the genomic organization of the prostase gene, a bacterial artificial chromosome clone containing the prostase gene was partially sequenced. Analysis of the final assembled sequence with the genscan gene prediction program program (27) identified five exons with four matching to the prostase cDNA sequence obtained from RACE and library screening. The putative first exon and part of exon 2 thus were identified from genomic sequence. PCR primers were designed based on the exon sequences obtained from the exon prediction program and used to confirm the cDNA sequence (data not shown). The intron/exon splice sites (—mGT…AGm —) and their flanking sequences in the prostase exons are closely related to the consensus splicing signals (–mGTAAGT…CAGm—) (28) (Fig. 1). Although most trypsin-like serine proteases are comprised of five exons, and the gene prediction algorithm did not identify an additional exon, we cannot exclude the possibility that an additional upstream noncoding exon exists.
Figure 1.
The sequences of prostase intron/exon boundaries.
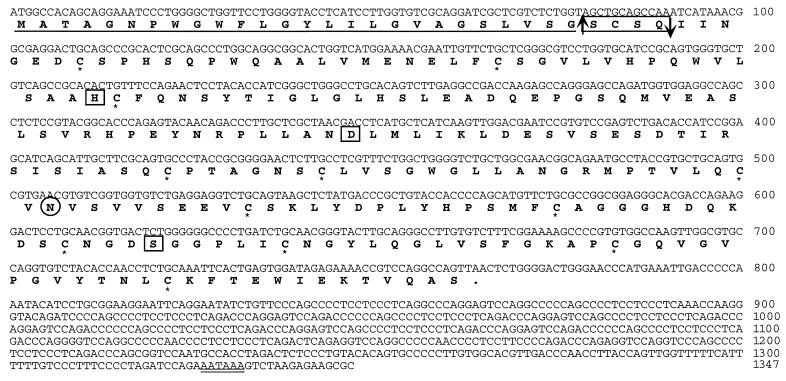
The final prostase sequence was assembled from cDNA and genomic sequence to generate a cDNA sequence of 1,347 nt with a presumptive protein-coding region of 765 nt starting at the first methionine codon preceded by an appropriate Kozak consensus sequence for the initiation of translation (29) (Fig. 2). A 582-nt 3′ untranslated region contains a polyadenylation signal, AATAAA, located 21 bases upstream of the poly(A) tail.
Figure 2.
The DNA and derived amino acid sequence of human prostase. The number of the last nucleotide in each row is indicated on the right. The putative signal peptide sequence is underlined with the predicted cleavage site indicated by an upward arrow. The propeptide sequence is boxed with the predicted cleavage site indicated by a downward arrow. Cysteine residues involved in putative disulfide bonds are marked with an ∗. The catalytic triad active-site residues (H, D, S) are boxed. One potential N-linked glycosylation site (N110) is circled. The polyadenylation signal is underlined twice.
The cDNA encodes a putative 254-aa polypeptide, with computer analysis predicting a 26-aa signal peptide with a cleavage site on the carboxyl side of Gly26 and a proprotein cleavage site on the carboxyl side of Gln30, leaving an active protein of 224 aa (http://www.cbs.dtu.dk/services/SignalP/) (Fig. 2). The catalytic triad of serine proteases His71(H), Asp116(D), and Ser207(S), (His57-Asp102-Ser195 in the bovine chymotrypsin numbering system) is conserved in prostase. The sequence alignment with EMSP1 supports, by analogy, the assignments of the translation initiation site and proprotein cleavage sites. Thus, like other serine proteases such as trypsin and PSA, prostase is likely also synthesized as a preproenzyme that contains an N-terminal signal peptide (prezymogen) followed by a short activation peptide and the enzymatic domain at the C-terminal end. The presence of a signal peptide suggests that prostase is likely to be a secreted protein. A putative N-linked glycosylation site is found at Asn110 (N)-Val111(V)-Ser112(S) (Fig. 2).
Prostase Is Highly Homologous to Members of the Serine Protease Family.
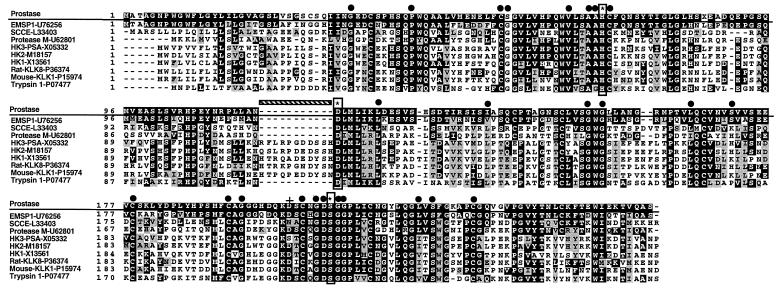
We used the nucleotide and the mature 224-aa prostase sequence to search the National Center for Biotechnology Information sequence database by using blast and beauty algorithms. Significant homology was seen with several serine protease sequences, including members of the kallikrein and trypsin families (Fig. 3). Prostase is most similar to the recently reported porcine gene EMSP1 (12) with 84% nucleotide and 72% amino acid sequence identity over the coding region. Prostase shares 37% amino acid identity and 52% amino acid similarity with PSA (Fig. 3).
Figure 3.
Alignment of the deduced amino acid sequence of prostase with the serine proteases EMSP1, SCCE, protease M, hK1, hK2, hK3, rat KLK8, mouse KLK1, and trypsin. GenBank accession numbers are listed. Amino acid sequence identity (black) or similarity (gray) between prostase and at least one other protein is shaded. Dashes represent gaps to bring the sequences to better alignment. The serine protease catalytic triad of histidine, aspartic acid, and serine are boxed vertically and denoted with an ∗. The 29 invariant serine protease residues are indicated by ●. The Asn residue conferring trypsin cleavage specificity is indicated by a cross. The hatched horizontal bar indicates the kallikrein loop sequence.
The kallikrein proteins contain a conserved 11-aa sequence immediately preceding the aspartate (D) active site residue. These residues, not found in prostase, form the kallikrein loop that is thought to determine substrate specificity (30). As previously shown for EMSP1 (12), a phylogenetic analysis separated the kallikreins from the trypsin-related proteins and grouped stratum corneum chymotryptic enzyme (SCCE, GenBank accession no. L33404), protease M (31), EMSP1, and prostase (data not shown). The presence of aspartate (D) at position 201 predicts that prostase will produce trypsin-like cleavage, unlike PSA, which has a serine at the corresponding position and produces chymotrypsin-like cleavage.
Twenty-nine “invariant” amino acids surrounding the active site of serine proteases have been described (32). Of these, 26 are conserved in prostase (Fig. 3). The three alterations (Ser57 for Gly, Ile137 for Leu, and Ser171 for Pro) are conservative changes (33). Twelve cysteine residues are present in the putative active prostase protein. Ten are conserved in all of the serine proteases aligned in Fig. 3 and would be expected to form disulfide bridges. The other two cysteines are not found in the kallikreins or human trypsin, but are found in similar positions in EMSP1, SCCE, protease M, and bovine trypsin and would be expected to form an additional disulfide bond.
Chromosomal Location of Prostase.
A chromosome 19-specific minisatellite repeat has been reported to be clustered exclusively in the q13 band of chromosome 19 (34). Eleven copies of this 37-bp repeat were identified in the 3′ untranslated region of prostase with additional copies in each intron. Three copies of this repeat also are found in the 3′ untranslated region of the porcine EMSP1 gene. The presence of this repeat suggested a 19q chromosomal location for the prostase gene. PCR analysis of a G3 radiation hybrid panel with prostase exon 5 primers mapped the prostase gene to chromosome 19q13. Several other serine proteases, including protease M, pancreatic/renal kallikrein (hK1), and the prostate-specific kallikreins glandular kallikrein 2 (hK2), and glandular kallikrein 3 (PSA, hK3) also map to this region (31, 35).
Prostase Expression Is Prostate Specific and Androgen Regulated.
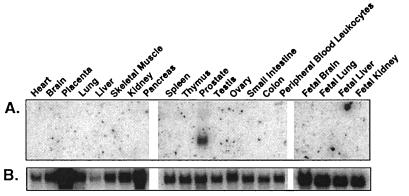
The distribution of prostase transcripts in normal human tissues was examined by Northern analysis. Of 20 adult and fetal tissues examined, prostase expression was evident almost exclusively in the prostate (Fig. 4). Prostase expression can be detected in small intestine after prolonged exposure (data not shown). Reverse transcriptase–PCR analysis of prostase expression in normal human tissues produced similar results (data not shown).
Figure 4.
Multiple tissue Northern analysis of prostase. (A) Prostase is expressed specifically in normal prostate tissue with a transcript size of approximately 1.3 kb. (B) A human glyceraldehyde-3-phosphate dehydrogenase probe was used as a control for equivalent RNA loading. Tissue types are indicated at the top of each lane.
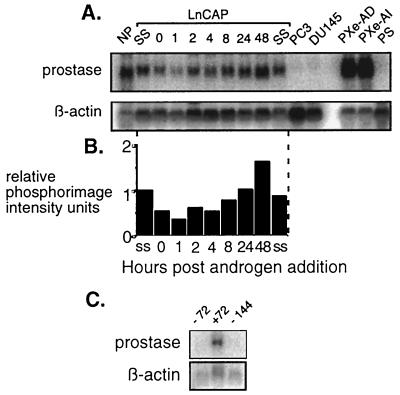
The expression of prostate-specific genes, including PSA and the homeobox gene hNKX3.1, have been shown to be regulated by androgenic hormones (36, 37). In this study, we used the androgen-responsive LNCaP prostate cancer cells as a model system to evaluate whether prostase expression is under androgenic control. After steroid depletion, the cells were supplemented with 1 nM R1881, a synthetic androgen. RNA was extracted at specific time points and examined by Northern analysis using a prostase cDNA probe. After 24 hr of androgen depletion, prostase expression decreased to 50% of steady-state levels and rose after androgen reintroduction to reach a maximum 1.7-fold increase over steady state at 48 hr (Fig. 5 A and B). Though reduced, prostase expression still could be detected in the LNCaP cells after androgen deprivation, indicating that a basal level of expression may not be androgen dependent or that residual androgens were present in the charcoal-stripped FCS media. The removal of androgens for 72 hr reduced prostase expression to undetectable levels (Fig. 5C). Prostase expression was not observed in the androgen-unresponsive prostate cancer cell lines PC-3 and DU-145 and was not seen in a short-term culture of prostate stroma consisting of fibroblasts and smooth muscle cells (Fig. 5A).
Figure 5.
Androgen regulation of prostase expression. (A) Northern analysis by using a prostase probe with RNA extracted from normal prostate (NP), LNCaP at steady state (SS), LNCaP after 24 hr of androgen deprivation (0), LNCaP at specified hours after androgen exposure (1, 2, 4, 8, 24, and 48), the PC3 (PC3) and DU145 (DU145) prostate cancer cell lines, the androgen dependent (PXe-AD) and androgen independent (PXe-AI) prostate cancer xenografts, and prostate stroma (PS). A probe for β-actin is used to estimate RNA loading. (B) Relative prostase expression levels. Expression ratios are determined by normalizing all samples for β-actin intensity relative to the LNCaP steady-state (SS) sample. Prostase expression for each sample is shown in PhosphorImager intensity units relative to the SS LNCaP sample. (C) Northern analysis of prostase expression in LNCaP cells grown either with (+) or without (−) androgen (1 nM R1881) for 72 or 144 hr. A probe for β-actin is used to estimate RNA loading.
Although normal secretory prostate epithelial cells and early-stage prostate carcinomas depend on androgens for growth, a hallmark of advanced prostate cancer is the emergence of an androgen-independent (AI) cellular phenotype. These AI neoplastic cells are also capable of producing and secreting PSA. We examined the expression of prostase in human prostate cancers propagated in a xenograft system that recapitulates the androgen-dependent (AD) and subsequent AI characteristics of human prostate cancer growth (38). Prostase was highly expressed in both the AD and AI tumors (Fig. 5A), a finding that parallels PSA expression in this system, indicating a possible dysregulation of prostase control.
DISCUSSION
In a search for genes preferentially expressed in the human prostate, we have isolated and characterized cDNA and genomic clones encoding prostase, a member of the serine protease family. Prostase expression is virtually restricted to cells of prostate epithelial origin. The predicted sequence of the encoded prostase protein is similar to trypsin and members of the kallikrein family, such as PSA, and prostase is highly homologous to the recently described EMSP1 at both the nucleic acid and amino acid level (12). Structural features essential for serine protease activity such as the His-Asp-Ser catalytic triad, the residues lining the substrate binding cleft, and the cysteine bridges, are almost perfectly conserved.
The biological function of prostase is not known. The serine protease gene family encodes protein-cleaving enzymes that play important roles in diverse physiological processes, including digestion (e.g., trypsin and chymotrypsin), tissue remodeling (e.g., SCCE), and blood clotting (e.g., plasminogen activator and thrombin). Serine proteases also act as regulators of a variety of processes by proteolytic activation of precursor proteins. The prostate-specific serine proteases PSA and hK2 both are synthesized as precursor proteins that must be activated by cleavage of the propeptide. Recombinant hK2 and a partially purified seminal fluid enzyme(s) have been shown to activate PSA in vitro (39). The activators of these enzymes have not been identified. The predicted trypsin-like cleavage specificity of prostase makes it a candidate activator of PSA and hK2. We hypothesize that the normal function of prostase may be to participate with hK2 and PSA in a “cascade” of enzymatic reactions similar to those found in the processes of fibrinolysis and blood coagulation (39, 40). This cascade would culminate in the cleavage of seminogelin and the liquefaction of seminal fluid, an essential step for maintaining sperm motility and fertility (41).
Based on similarities to serine proteases with characterized functions, prostase also may have physiologic roles involving growth and tissue remodeling with implications for cancer biology. PSA has been reported to cleave proteins involved in normal and neoplastic cellular proliferation, including insulin-like growth factor binding protein 3 (42), parathyroid hormone-related protein (43), and pro-epidermal growth factor (44). Prostase exhibits approximately 45% amino acid identity with SCCE (45), a skin-specific serine endoproteinase with a putative role catalyzing the degradation of intercellular cohesive structures in the continuous shedding of cells from the skin surface (46). It is conceivable that prostase may play a similar role in the normal turnover of prostate glandular epithelial cells and potentially also function to modulate cellular adhesive forces that are critical for neoplastic growth and metastatis. Interestingly, the cancer-associated protein designated protease M, a serine protease shown to be highly expressed in primary tumors of breast and ovarian origin, shares 38% identity with prostase (31). Investigators studying protease M expression have suggested that this protein may play a role in establishing primary breast and ovarian tumors and also may function later in the cancer progression process as a metastasis inhibitor (31).
The extensive homology between the prostase and EMSP1 proteins (72% identity and 85% similarity) suggests additional roles for prostase. EMSP1 was the first proteinase fractionated from the developing tooth enamel matrix (47) and functions to degrade enamel matrix proteins such as the amelogenins during the process of enamel maturation (48). EMSP1 also may play a role in modulating cellular junctions during ameloblast maturation. Based on the amino acid sequences of the prostase and EMSP1 proproteins, neither are predicted to be activated by trypsin or chymotrypsin cleavage. However, EMSP1 is reported to be activated by matrix metalloproteinases (MMPs) (49), a class of enzymes under intensive investigation for their role in the cancer metastatic process (50). Thus, members of the MMP family, particularly those MMPs expressed in prostate epithelial cells, should be investigated as potential activators of prostase.
The homology between prostase and EMSP1 is intriguing in view of the high propensity of prostate carcinoma to metastasize to bone, the principal metastatic site, and cause of morbidity in this disease. Bone is a metabolically active and highly organized tissue consisting of a mineral phase of hydroxyapatite and amorphous calcium phosphate crystals deposited in an organic matrix. Metastatic prostate cancer cells alter the normal bone remodeling balance involving interactions between osteoblasts, osteoclasts, and constituents of the bone matrix to favor bone formation. Both osteoblastic and osteolytic changes occur during the course of tumor progression as a consequence of significant bone resorption and bone production occurring within the metastatic sites (51, 52). Although some components of bone remodeling involving tumor metastases have been identified, including PTHrP and transforming growth factor β (52), the process remains ill-defined. If the sequence conservation of prostase and EMSP1 correlates to functional attributes, then prostase should be investigated as a potential contributor to the bone pathogenesis of metastatic prostate cancer.
The mapping of prostase to chromosome 19q13 is interesting in that two other prostate-specific genes, hKLK2 and hKLK3 (PSA), map to the same chromosomal region. The third characterized human kallikrein, hKLK1, also is clustered with hKLK2 and hKLK3 in a region spanning 60 kb on 19q13.2-q13.4 (35). In addition to prostase, several other serine proteases recently have been reported to reside on chromosome 19q13, including protease M (31), normal epithelial cell-specific gene 1 (53), and PRSSL1 (54). The localization of these genes suggests that the kallilkrein serine protease cluster is larger and more diverse than previously described. Although these genes may have originated from a single ancestral precursor gene, the low overall sequence homology between the serine proteases in this region indicates that except for the kallikreins, these proteases are likely the result of ancient gene duplication events. Subsequent somatic mutations, gene conversions, or nonreciprocal recombinations may have occurred to further adapt, diversify, and expand the genes encoding these proteases. The chromosome 19q13 syntenic region resides on mouse chromosome 7 and contains the mouse kallikrein locus comprised of at least 24 distinct kallikrein genes (55). This finding suggests that the separation between the kallikreins and other serine proteases such as prostase occurred much earlier than the expansion of the kallikrein genes, the latter arising from recent gene duplication events after the divergence of human and rodent.
We anticipate that the further characterization of the prostase gene locus coupled with functional studies of the prostase protein will advance our understanding of both normal and pathological prostate physiology. To this end, the identification of activators and substrates of prostase is required. As with the serine proteases PSA and hK2, prostase appears to be a prostate-specific secreted protein that could find utility as a target for diagnosis and therapy. Antibodies directed against peptide epitopes of the prostase protein should provide a means to evaluate prostase expression in prostate malignancies and to determine whether serum levels of the prostase protein can be used as adjuncts to currently available cancer screening methods.
Acknowledgments
We thank Steve Lasky and the sequencing group at Chiroscience for sequencing support. We are grateful to Dr. Robert Vessella for providing xenograft RNA and advice. We thank Vilaska Nguyen for tissue culture assistance. We thank Dr. Barbara Trask and Dr. Victor Ng for critical reading of the manuscript. This work was supported in part by the CaPCURE Foundation and a grant (K08 CA75173-01A1) from the National Cancer Institute (P.S.N.).
ABBREVIATIONS
- PSA
prostate-specific antigen
- EMSP1
enamel matrix serine proteinase 1
- SCCE
stratum corneum chymotryptic enzyme
- AI
androgen independent
Footnotes
References
- 1.Landis S H, Murray T, Bolden S, Wingo P A. Ca Cancer J Clin. 1998;48:6–29. doi: 10.3322/canjclin.48.1.6. [DOI] [PubMed] [Google Scholar]
- 2.Carter H B, Coffey D S. A Multidisciplinary Analysis of Controversies in the Management of Prostate Cancer. New York: Plenum; 1988. [Google Scholar]
- 3.Carter H B, Coffey D S. Prostate. 1990;16:39–48. doi: 10.1002/pros.2990160105. [DOI] [PubMed] [Google Scholar]
- 4.Oesterling J E. J Urol. 1991;145:907–923. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- 5.Catalona J J, Smith D S, Ratliff T L, Dobbs K M, Coplen D E, Yuann J J, Petros J A, Andriole G L. N Engl J Med. 1991;324:1156–1161. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- 6.Farkas A, Schneider D, Perrotti M, Cummings K B, Ward W S. Urology. 1998;52:444–448. doi: 10.1016/s0090-4295(98)00242-8. ; discussion 448–449. [DOI] [PubMed] [Google Scholar]
- 7.Oesterling J, Fuks Z, Lee C T, Scher H I. In: Cancer of the Prostate. DeVita V T, Hellman S, Rosenberg S A, editors. Philadelphia: Lippincott; 1997. pp. 1322–1386. [Google Scholar]
- 8.Tjoa B A, Simmons S J, Bowes V A, Ragde H, Rogers M, Elgamal A, Kenny G M, Cobb O E, Ireton R C, Troychak M J, et al. Prostate. 1998;36:39–44. doi: 10.1002/(sici)1097-0045(19980615)36:1<39::aid-pros6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Correale P, Walmsley K, Nieroda C, Zaremba S, Zhu M, Schlom J, Tsang K Y. J Natl Cancer Inst. 1997;89:293–300. doi: 10.1093/jnci/89.4.293. [DOI] [PubMed] [Google Scholar]
- 10.Denmeade S R, Nagy A, Gao J, Lilja H, Schally A V, Isaacs J T. Cancer Res. 1998;58:2537–2540. [PubMed] [Google Scholar]
- 11.Sikora, K. & Pandha, H. (1997) Br. J. Urol.79, Suppl. 2, 64–68. [DOI] [PubMed]
- 12.Simmer J P, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, Margolis H C, Shimizu M, DeHart B C, Hu C C, Bartlett J D. J Dent Res. 1998;77:377–386. doi: 10.1177/00220345980770020601. [DOI] [PubMed] [Google Scholar]
- 13.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson P S, Ng W L, Schummer M, True L D, Liu A Y, Bumgarner R E, Ferguson C, Dimak A, Hood L. Genomics. 1998;47:12–25. doi: 10.1006/geno.1997.5035. [DOI] [PubMed] [Google Scholar]
- 16.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 17.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari S, Ramachandran S, Bhattacharya A, Bhattacharya S, Ramaswamy R. Comput Appl Biosci. 1997;13:263–270. doi: 10.1093/bioinformatics/13.3.263. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 23.Hakalahti L, Vihko P, Henttu P, Autio-Harmainen H, Soini Y, Vihko R. Intl J Cancer. 1993;55:590–597. doi: 10.1002/ijc.2910550413. [DOI] [PubMed] [Google Scholar]
- 24.Wang M C, Valenzuela L A, Murphy G P, Chu T M. Invest Urol. 1979;17:159–163. [PubMed] [Google Scholar]
- 25.Bjartell A, Malm J, Moller C, Gunnarsson M, Lundwell A, Lilja H. J Androl. 1996;17:17–26. [PubMed] [Google Scholar]
- 26.Dubbink H J, de Waal L, van Haperen R, Verkaik N S, Trapman J, Romijn J C. Genomics. 1998;51:434–444. doi: 10.1006/geno.1998.5393. [DOI] [PubMed] [Google Scholar]
- 27.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 28.Iida Y. J Theor Biol. 1990;145:523–533. doi: 10.1016/s0022-5193(05)80486-2. [DOI] [PubMed] [Google Scholar]
- 29.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ashley P L, MacDonald R J. Biochemistry. 1985;24:4512–4520. doi: 10.1021/bi00338a005. [DOI] [PubMed] [Google Scholar]
- 31.Anisowicz A, Sotiropoulou G, Stenman G, Mok S C, Sager R. Mol Med. 1996;2:624–636. [PMC free article] [PubMed] [Google Scholar]
- 32.Dayhoff, M. O. (1978) Natl. Biomed. Res. Found.5, Suppl. 3, 79–81.
- 33.Miyata T, Miyazawa S, Yasunaga T. J Mol Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- 34.Das H K, Jackson C L, Miller D A, Leff T, Breslow J L. J Biol Chem. 1987;262:4787–4793. [PubMed] [Google Scholar]
- 35.Riegman P H, Vlietstra R J, Suurmeijer L, Cleutjens C B, Trapman J. Genomics. 1992;14:6–11. doi: 10.1016/s0888-7543(05)80275-7. [DOI] [PubMed] [Google Scholar]
- 36.Young C Y, Montgomery B T, Andrews P E, Qui S D, Bilhartz D L, Tindall D J. Cancer Res. 1991;51:3748–3752. [PubMed] [Google Scholar]
- 37.He W W, Sciavolino P J, Wing J, Augustus M, Hudson P, Meissner P S, Curtis R T, Shell B K, Bostwick D G, Tindall D J, et al. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 38.Bladou F, Vessella R L, Buhler K R, Ellis W J, True L D, Lange P H. Int J Cancer. 1996;67:785–790. doi: 10.1002/(SICI)1097-0215(19960917)67:6<785::AID-IJC6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Takayama T K, Fujikawa K, Davie E W. J Biol Chem. 1997;272:21582–21588. doi: 10.1074/jbc.272.34.21582. [DOI] [PubMed] [Google Scholar]
- 40.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 41.Lilja H, Oldbring J, Rannevik G, Laurell C B. J Clin Invest. 1987;80:281–285. doi: 10.1172/JCI113070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen P, Graves H C, Peehl D M, Kamarei M, Giudice L C, Rosenfeld R G. J Clin Endocrinol Metab. 1992;75:1046–1053. doi: 10.1210/jcem.75.4.1383255. [DOI] [PubMed] [Google Scholar]
- 43.Cramer S D, Chen Z, Peehl D M. J Urol. 1996;156:526–531. doi: 10.1097/00005392-199608000-00076. [DOI] [PubMed] [Google Scholar]
- 44.Parries G, Cohen S. J Urol. 1996;155:507. (abstr.). [Google Scholar]
- 45.Hansson L, Stromqvist M, Backman A, Wallbrandt P, Carlstein A, Egelrud T. J Biol Chem. 1994;269:19420–19426. [PubMed] [Google Scholar]
- 46.Sondell B, Thornell L E, Egelrud T. J Invest Dermatol. 1995;104:819–823. doi: 10.1111/1523-1747.ep12607007. [DOI] [PubMed] [Google Scholar]
- 47.Fukae M, Shimizu M. Arch Oral Biol. 1974;19:381–386. doi: 10.1016/0003-9969(74)90179-4. [DOI] [PubMed] [Google Scholar]
- 48.Termine J D, Belcourt A B, Christner P J, Conn K M, Nylen M U. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- 49.Tanabe T, Fukae M, Shimizu M. Adv Dent Res. 1996;10:170–172. doi: 10.1177/08959374960100020801. [DOI] [PubMed] [Google Scholar]
- 50.Denis L J, Verweij J. Invest New Drugs. 1997;15:175–185. doi: 10.1023/a:1005855905442. [DOI] [PubMed] [Google Scholar]
- 51.Revilla M, Arribas I, Sanchez-Chapado M, Villa L F, Bethencourt F, Rico H. Prostate. 1998;35:243–247. doi: 10.1002/(sici)1097-0045(19980601)35:4<243::aid-pros2>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 52.Mundy G R. Cancer. 1997;80:1546–1556. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1546::aid-cncr4>3.3.co;2-r. [DOI] [PubMed] [Google Scholar]
- 53.Luo L, Herbrick J A, Scherer S W, Beatty B, Squire J, Diamandis E P. Biochem Biophys Res Commun. 1998;247:580–586. doi: 10.1006/bbrc.1998.8793. [DOI] [PubMed] [Google Scholar]
- 54.Polikoff D, Kuo W L, Cochran J F, Wernick M, Kowbel D, Myambo K, Collins C C. Cytogenet Cell Genet. 1997;79:147–148. doi: 10.1159/000134705. [DOI] [PubMed] [Google Scholar]
- 55.Murray, S. R., Chao, J. & Chao, L. (1992) Agents Actions38, Suppl., 26–33. [DOI] [PubMed]