Abstract
A rare set of hematopoietic stem cells (HSC) must undergo a massive expansion to produce mature blood cells. The phenotypic isolation of HSC from mice offers the opportunity to determine directly their proliferation kinetics. We analyzed the proliferation and cell cycle kinetics of long-term self-renewing HSC (LT-HSC) in normal adult mice. At any one time, ≈5% of LT-HSC were in S/G2/M phases of the cell cycle and another 20% were in G1 phase. BrdUrd incorporation was used to determine the rate at which different cohorts of HSC entered the cell cycle over time. About 50% of LT-HSC incorporated BrdUrd by 6 days and >90% incorporated BrdUrd by 30 days. By 6 months, 99% of LT-HSC had incorporated BrdUrd. We calculated that approximately 8% of LT-HSC asynchronously entered the cell cycle per day. Nested reverse transcription–PCR analysis revealed cyclin D2 expression in a high proportion of LT-HSC. Although ≈75% of LT-HSC are quiescent in G0 at any one time, all HSC are recruited into cycle regularly such that 99% of LT-HSC divide on average every 57 days.
Hematopoiesis is a dynamic process resulting in the continuous production of at least eight separate lineages of cells. The hematopoietic tissues of an adult mouse must replace approximately 2.4 × 108 red blood cells and 4 × 106 nonlymphoid peripheral blood cells each day (1) and compensate for hematological stresses such as blood loss, infection, and ingestion of cytotoxic chemicals. Such demands necessitate strict control over hematopoietic progenitor proliferation. All cells produced by the hematopoietic system are derived from hematopoietic stem cells (HSC) that reside mainly in the bone marrow (BM). HSC are defined at the single-cell level by their ability to self-renew and to give rise to all lineages of blood cells (2). The multipotent progenitor pool is heterogeneous and can be divided into colonogenic long-term self-renewing HSC (LT-HSC), transiently self-renewing HSC (short-term HSC), and non-self-renewing multipotent progenitors (MPP) (3).
The clonal-succession hypothesis (4) proposed a model for how LT-HSC coordinate cell cycle events to meet the demands of hematopoiesis. The model postulated that there is a large supply of HSC representing the only source of mature blood cells for the lifetime of the animal, but that only one or a few HSC clones give rise to mature blood cells at any time. Other HSC were postulated to remain quiescent and not contribute to hematopoiesis until the proliferative capacity of the active LT-HSC clone is exhausted. The reconstitution patterns from several experiments in which retrovirally marked hematopoietic progenitors were used to reconstitute irradiated mice were interpreted to support the clonal-succession model (5–7). The contributions of individual clones of retrovirus-infected HSC to hematopoiesis were followed by monitoring the different retroviral integration sites in mature blood cells. In these experiments, only one or a few LT-HSC clones appeared to contribute to blood cell production at any one time (5–7). Subsequent retroviral marking studies observed that many clones contributed to hematopoiesis, with extensive clonal fluctuation for a long post-transplant period, followed by equilibration to oligoclonal reconstitution (8, 9); part of this post-transplant fluctuation could reflect the fact that different classes of HSC have different self-renewal potentials (3, 10). Analogous experiments in a large animal model, the cat, also revealed large clonal fluctuations post-transplant that have been interpreted to support clonal succession (11); however, some of the cats failed to engraft and had to be retransplanted. It was later calculated that the bone marrow used to reconstitute these cats contained only 6–12 HSC per kg body mass (12). This raises the question of whether enough HSC engraft in transplantation models to test whether many HSC normally contribute to hematopoiesis. Indeed, it has been argued that reconstitution assays may not be an appropriate test of the clonal succession hypothesis because they do not represent steady-state hematopoiesis and because only limited numbers of HSC typically engraft (9). Therefore, it may not be surprising that in reconstituted animals, only a few transplanted clones contribute to hematopoiesis. To avoid reconstitution effects, Harrison et al. (13) prepared embryonic chimeric mice in which HSC were derived from embryonic stem (ES) cells of two different genotypes. Because there was little variability in the relative contributions of the two genotypes over time, Harrison et al. concluded that many HSC simultaneously contribute to hematopoiesis in mice.
Although experiments by Harrison and colleagues demonstrated that many HSC contribute simultaneously to hematopoiesis, it remains possible that only certain subsets of HSC are active during each period in the life history of an animal. To investigate this possibility, the proliferation history of HSC has been analyzed directly. The BrdUrd incorporation rates of primitive hematopoietic progenitors have been analyzed by exposing mice to subtoxic doses of BrdUrd to label the nuclei of dividing cells. In an early and important study, Pietrzyk et al. (14) analyzed the BrdUrd incorporation rates of day-14 colony-forming units spleen (CFU-S), a population that includes HSC as well as other MPP and myeloerythroid progenitors. They demonstrated that ≈75% of progenitors capable of forming day-14 spleen colonies had incorporated BrdUrd over a 32-day period of normal hematopoiesis. Bradford et al. (15) analyzed the proliferation kinetics of highly enriched HSC defined by Hoechst (Ho) and Rhodamine 123 (Rho) exclusion. They demonstrated that up to 89% of HoloRholo progenitors incorporated BrdUrd over a period of 12 weeks. Thus, both the Pietrzyk et al. and Bradford et al. studies were consistent with the idea that a majority of HSC divide regularly.
Despite these important advances, a more complete view of LT-HSC cell cycle kinetics has not been presented. The relative distribution between the G1 and G0 phases of the cell cycle, as well as the average cell cycle times have not been determined for HSC in vivo. Furthermore, it remains possible that up to 10% of HSC may remain quiescent for very long periods of time. If true, this could have fundamental implications for heterogeneity in and regulation of the HSC pool. In this study, we determined the in vivo proliferative history and cell cycle status of mouse HSC purified by fluorescence-activated cell sorting. All HSC activity in C57BL/Ka-Thy 1.1 mice is contained within c-kitbright, Thy 1.1lo, Sca-1+, Lineage −/lo (KTSL−/lo) fraction of BM (3, 16–18). KTSL−/lo cells are heterogeneous, containing all LT-HSC as well as short-term HSC and MPP (3, 10). The LT-HSC subset can be isolated by purifying the lineage-negative subset of these cells (KTSL−) (3). We examined the proliferation history of the KTSL− and KTSL−/lo HSC populations by studying their in vivo BrdUrd incorporation kinetics. Cells from either population (99% total) incorporated BrdUrd over a period of 3–6 months, demonstrating that all HSC cycle regularly. At any one time, ≈25% of LT-HSC were either in G1 or S/G2/M phases of the cell cycle. Consistent with this, a high proportion of LT-HSC expressed cyclin D2 mRNA. We present a model describing the LT-HSC cell cycle kinetics.
MATERIALS AND METHODS
Mouse Strains.
C57BL/Ka-Ly 5.1, Thy 1.1 and C57BL/Ka-Ly 5.2, Thy 1.1 mice were bred and maintained in the Stanford University School of Medicine animal care facility. Mice were maintained on acidified water (pH 2.5) until BrdUrd treatment. All mice were 6–12 weeks old at the initiation of treatment.
BrdUrd Treatment of Mice.
Aged-matched mice were allowed to freely imbibe sterile deionized water containing BrdUrd (1 mg/ml; Sigma) for 3, 6, 10, 15, 20, 30, or 180 days. Containers of BrdUrd water were wrapped in aluminum foil to prevent light exposure and were changed every 3–6 days. For the zero time point, mice were given an intraperitoneal (i.p.) injection of BrdUrd in 0.9% saline (1 mg of BrdUrd per 6 g of body weight). After 90 min, the mice were sacrificed for isolation of HSC.
Control Experiments for BrdUrd Toxicity.
To ensure there was no alteration of hematopoiesis by BrdUrd, complete blood counts were performed by using tail-vein blood from age-matched mice that were fed BrdUrd for 6 days (n = 5) and from mice not fed BrdUrd (n = 9). BM cells from the left leg of mice exposed to BrdUrd for 10 days (n = 6) and age-matched controls (n = 6) were counted by using trypan blue exclusion. BM cells were also stained with antibodies against myeloid [M1/70 (anti-Mac-1) and 8C5 (anti-Gr-1)] and lymphoid [KT31.1 (anti-CD3) and 6B2 (anti-B220)] markers and analyzed by dual-laser fluorescence-activated cell sorting (FACS, Vantage, Becton Dickinson). The Hoechst DNA profiles of BM and HSC from BrdUrd-exposed mice were compared with controls as described (3). To test whether BrdUrd administration had an effect on HSC function, cells from treated donors were examined by a limit dilution competitive reconstitution assay as described (3).
Antibodies.
The antibodies used in immunofluorescence staining included: 19XE5 (anti-Thy 1.1), E13 (anti-Sca-1, Ly6 A/E), 2B8 (anti-c-kit), and the Lineage markers GK1.5 (anti-CD4), 53–56.7 (anti-CD8), KT31.1 (anti-CD3), 53.73 (anti-CD5), 6B2 (anti-B220), Ter 119 (anti-erythroid specific antigen), 8C5 (anti-Gr-1), M1/70 (anti-Mac-1). Monoclonal antibody BU-1 (Caltag, South San Francisco, CA) was used for BrdUrd analysis.
HSC Isolation.
All procedures were done in UV-free light to prevent the degradation of BrdUrd in samples and the photoactivation of BrdUrd in DNA. Marrow was flushed and pooled from the femurs and tibias of groups of five aged-matched mice that had been allowed to imbibe water containing BrdUrd for equal amounts of time. BM was stained for HSC isolation as described (3). HSC were isolated with a dual-laser FACS. BM preparations were sorted on the basis of high expression of c-kit, low expression of Thy 1.1, positive expression of Sca-1, and a negative-to-low level expression of lineage markers (KTSL−/lo). This isolated the entire HSC pool, including LT-HSC, short-term HSC, and MPP (3, 10). Nearly pure populations of LT-HSC were isolated by sorting cells with no detectable levels of lineage marker staining and were thus Lineage-negative (KTSL−) (3, 10). To ensure purity, all HSC populations were double sorted to 95% purity. HSC were directly sorted onto a chambered glass microscope slide pretreated with the histological adhesive Cell-Tak (Collaborative Biomedical Products, Bedford, MA), according to manufacturer’s instructions.
BrdUrd Staining.
HSC sorted on to chambered slides were spun down and fixed with 70% ethanol at minus 20°C for 30 minutes. DNA was denatured with 2 M HCL + 0.8% Triton X-100. Between each procedure, the slides were washed 3 times for 5 min in PBS. The acid was neutralized with 0.1 M sodium borate. The slide was then incubated with staining medium (PBS + 0.1% Nonidet P-40 + 4% goat serum) for 1 hour. The HSC were stained with anti-BrdUrd mAb for 1 hour at room temperature, washed, and stained with fluorescein isothiocyanate-conjugated anti-mouse antibodies (Caltag, South San Francisco, CA) for 1 hour at room temperature. All antibodies were diluted in the staining medium described above. Finally, cells were mounted in 1:1 glycerol/PBS containing 1 μg/ml Hoechst 33342 (Sigma).
Microscopic Analysis of BrdUrd Staining.
BrdUrd-positive HSC were counted by using a Nikon FXA fluorescent microscope at ×400 magnification. Cell nuclei were first counted by using the Hoechst filter, then the fluorescein isothiocyanate/Texas Red filter was used to count the number of BrdUrd-positive (green) HSC in the same field. Thymocytes or double-sorted Thy1.1lo BM cells from mice not exposed to BrdUrd were used as negative controls for each experiment. The fluorescein isothiocyanate stain from the Thy 1.1 antibody was not visible on cells after BrdUrd staining. For most time points, HSC were obtained from at least three groups with five mice per group, and 1,500 HSC were scored. For three time points (day 0, day 20-KTSL−, and day 30 KTSL−/lo), HSC were obtained from two groups of five mice, but in each case at least 500 cells were counted.
Cyclin D2 Reverse Transcription–PCR (RT-PCR).
Single KTSL− LT-HSC were directly clone sorted into U-bottom 96-well plates filled with 19 μl of RT lysis buffer containing cyclin D2-specific RT primer and salts as described (19) by using nested cyclin D2-specific PCR primers. After 70 (35 × 2) amplification cycles, 10 μl of PCR products were electrophoresed through a 1.5% agarose gel, and the PCR products were visualized in the ethidium bromide-stained gels. The sequences of all RT and PCR primers can be obtained from S.H.C. (e-mail: cheshier@leland.stanford.edu). The PCR primers were selected to be specific only for cyclin D2 and gave no PCR product when used in PCR reactions containing cyclin D1 or D3 cDNAs or in reactions with mouse genomic DNA or splenic T cells that do not express cyclin D2 (data not shown). PCR with cyclin D2 primers always resulted in a single band of the appropriate size (331 bp) that hybridized to an internal cyclin D2-specific probe on Southern blot analysis (data not shown).
Pyronin Y (PY)/Hoechst Staining.
KTSL− LT-HSC were isolated as described above, double-sorted into a V-bottom 96-well plate containing 100 μl of Hanks Balanced Salt Sownen (GIBCO) + 2% bovine calf serum, and stained for Hoechst and PY as described (20). Samples were then placed on ice for 15 min, and propidium iodide was added to a final concentration of 0.5 μg/ml for exclusion of dead cells. Fluorescence-activated cell sorting analysis of these cells was performed on a FACSTAR Plus (Becton Dickinson) equipped with UV-laser.
RESULTS
BrdUrd Does Not Alter Normal Hematopoiesis or HSC Function.
HSC were labeled in vivo by allowing mice to drink BrdUrd in their water. Some studies have demonstrated that low levels of BrdUrd are not toxic to cells (14, 21), but others have documented alterations of gene expression and cellular toxicity because of the presence of this molecule (22, 23). To ascertain whether the BrdUrd doses used in this study altered normal hematopoiesis, we measured several BM and HSC functional parameters in mice exposed to BrdUrd and compared them to normal controls. We saw no differences in peripheral blood counts (white blood cells, red blood cells, neutrophils, band cells, and lymphocytes), or in the proportion of lymphoid cells in the BM (Table 1). There was, however, a slight increase in both Gr-1 and Mac-1-positive cells in the BM of mice that received BrdUrd for 10 days (Table 1). Although statistically significant, the increases were very small. If BrdUrd exposure altered HSC proliferation kinetics, changes in the normal proportions of HSC in S/G2/M phases of the cell cycle should have been apparent. However, the percentage of BM and HSC (KTSL− and KTSL−/lo) with >2n DNA was not significantly altered even after a 10 day exposure to BrdUrd (Table 2). KTSL− HSC from mice fed BrdUrd for 10 days were just as effective as normal KTSL− HSC in the limit−dilution long-term competitive reconstitution assay, with limit−dilution dose of around 1 in 12 cells (Table 2). We conclude that the doses of BrdUrd used in this study did not significantly alter normal HSC biology.
Table 1.
Control experiments for the effects of BrdUrd on peripheral blood and BM populations
| +BrdU | −BrdU | ||
|---|---|---|---|
| Peripheral Blood Counts | White blood cells | 6.2 ± 1.6 | 8.0 ± 2.2 |
| Platelet counts | 1,488 ± 531 | 1,404 ± 374 | |
| Neutrophils | 10.0 ± 2.9 | 9.9 ± 3.8 | |
| Lymphocytes | 85.8 ± 3.6 | 87.8 ± 4.3 | |
| Reticulocytes | 1.96 ± 0.58 | 2.1 ± 0.42 | |
| WBM cell populations | Total cell # Leg Bone | 3.64 ± 0.43 | 3.86 ± 0.51 |
| Gr-1‡ | 35.7 ± 3.3 | 29.4 ± 0.4 | |
| Mac-1‡ | 38.9 ± 2.75 | 34.9 ± 1.5 | |
| B220 | 30.3 ± 5.1 | 34.8 ± 0.45 | |
| CD3 | 2.37 ± 0.12 | 2.04 ± 0.06 |
All data are given as mean ± SD. Peripheral blood counts from the tail vein of mice exposed to BrdU for 6 days (n = 5) compared to aged matched controls not receiving BrdU (n = 9). No statistical differences (P > 0.05) were seen in any parameters using Student’s t test statistics. BM Cells from the left leg bones of mice exposed to BrdUrd for 10 days (n = 6) and age-matched controls (n = 6) were counted by using Trypan blue exclusion. BM cells were also stained with antibodies against myeloid and lymphoid lineage markers and analyzed by using fluorescence-activated all sorting. No statistical differences (P > 0.05) were seen in BM cell numbers or % B220 and CD3 in BM using Student’s t test statistics.
The differences in % Gr-1- and Mac-1-positive BM were statistically significant (P = 0.001 and P = 0.01 respectively) using Student’s t test statistics.
Table 2.
Control experiments for effects of BrdU on KTSL−/lo and KTSL− HSC
| BrdUrd | Hoechst DNA Staining
|
CLDCR | ||
|---|---|---|---|---|
| S/G2/M WBM, % | S/G2/M KTSL−/lo, % | S/G2/M KTSL−, % | ||
| Present | 11.1 ± 2.6 | 10.9 ± 3.7 | 6.2 ± 2.6 | 9/17 |
| Absent | 12.85 ± 2.0 | 11.2 ± 2.6 | 5.7 ± 2.2 | 6/10 |
All values are given as mean ± SD. BM from mice exposed to BrdUrd for 10 days (n = 5) and age-matched controls (n = 5) was stained with Hoechst 33342 and then stained with antibodies against c-Kit, Thy 1.1, Sca-1, and Lineage markers as described in Materials and Methods. DNA analysis was performed on dual-laser FACS Vantage equipped with UV laser. No statistical differences (P > 0.05) were seen using Student’s t test. To test whether BrdU administration had an effect on HSC function, cells from treated donors were examined by a clonal limit dilution competitive reconstitution assay (CLDCR). Twelve KTSL− HSC from C57BL/Ka-Ly 5.1, Thy 1.1 mice exposed to BrdUrd for 10 days and 2 × 105 Ly 5.2 BM cells were injected into lethally irradiated (4.75 Gy × 2 = 9.5 Gy) Ly 5.2 congenic hosts (n = 17). 12 KTSL− HSC from non-Brd Urd-exposed mice and 2 × 105 Ly 5.2 BM cells injected into Ly 5.2 congenic hosts were used as controls (n = 10). Recipient mice were considered Ly 5.1-reconstituted if greater than background (0.1%) levels of Ly 5.1 myeloid and lymphoid cells, as determined by fluorescence-activated cell sorting, were present in the peripheral blood for at least 12 weeks post-transplantation. MBT = Myeloid, B cell, and T cell. Yates-corrected χ2 analysis did not reveal any statistical difference (P > 0.05) between BrdUrd KTSL− and control KTSL−. CLDCR values are donor MBT-reconstituted mice/total no. of host mice.
In Vivo Proliferation Kinetics of HSC.
To determine the proliferative history of HSC, we isolated KTSL−/lo and KTSL− HSC (Fig. 1) from the BM of mice that were fed BrdUrd. Consistent with our observation that ≈11% of KTSL−/lo cells were in S/G2/M phases of the cell cycle (ref. 30; Table 2), an average of 13.2% of these cells labeled immediately after a single pulse of BrdUrd. KTSL−/lo cells entered S phase at a rapid rate, with an average of 78.4% BrdUrd-labeled cells by day 6, 87.8% by day 10, and virtually all cells by day 30 (99.4%; Fig. 2A). The fact that >99% of KTSL−/lo cells incorporate BrdUrd over a 30-day period suggests that no subset of HSC remains quiescent for more than 30 days. We wished to test further whether there was a significant lag in BrdUrd incorporation in LT-HSC (KTSL−). Again consistent with the steady-state level of ≈4–6% of these cells in S/G2/M (ref. 3; Table 2), an average of 4.7% of these cells were labeled immediately after BrdUrd administration; an average of 52.2% were labeled by 6 days, and after 30 days, >90% were BrdUrd-positive (Fig. 2A). In mice fed for 180 days with BrdUrd, >99% of KTSL− incorporated BrdUrd (Fig. 2A). Thus, all LT-HSC divide regularly during steady-state hematopoiesis, and no subset remains dormant.
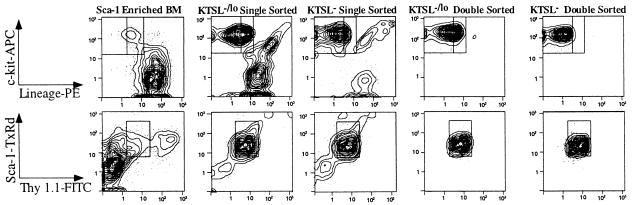
Figure 1.
Double-sorting is necessary to obtain pure KTSL−/lo and KTSL− cells. (Upper) c-kit (y-axis) and lineage (x-axis) density plots with outliers. Boxes indicate sort gates for KTSL−/lo (inner and outer boxes) and KTSL− (inner boxes only). (Lower) Sca-1 (y-axis) and Thy 1.1 (x-axis) density plots with outliers. Boxes indicate sort gates for KTSL−/lo and KTSL− cells. As seen in the second and third columns, significant numbers of cells outside the sort gates are present in a reanalysis of a single sorted sample. After the samples are double-sorted, the great majority of cells (≥95%) are within the sort gates that define KTSL−/lo (fourth column) and KTSL− (fifth column) HSC.
Figure 2.
In vivo BrdUrd incorporation kinetics of KTSL−/lo and KTSL− cells. (A) BrdUrd incorporation rate of KTSL−/lo cells (□) and KTSL− cells (●). Each data point represents mean percentages of BrdUrd positive HSC from at least two separate experiments with a combined analysis of at least 500 cells. Some error bars are smaller than their associated symbol. (B) BrdUrd incorporation means plotted in semilogarithmic fashion as the proportion of BrdUrd-negative KTSL−/lo cells (□) and KTSL− cells (●) against time in days. The lines were generated by using least squares fit linear regression.
When the percentage of BrdUrd negative cells was plotted against time in a log-linear fashion as in Fig. 2B, the data closely approximated a straight line. This indicates that a constant fraction of HSC, about 7.8% per day, entered the cell cycle in an apparently random fashion (24). From the log-linear plot we interpolated the 50% BrdUrd incorporation point for all MPP and for LT-HSC: 2.8 and 7.4 days, respectively. The potential doubling time of asynchronously dividing cells is the amount of time necessary for 77% of cells to incorporate BrdUrd (25). Thus, the potential doubling time of all MPP and LT-HSC was 7.7 days and 17.8 days, respectively. Finally, the 99% BrdUrd-incorporation point was extrapolated from the log-linear plot as about 57 days for LT-HSC.
LT-HSC Express Cyclin D2 mRNA.
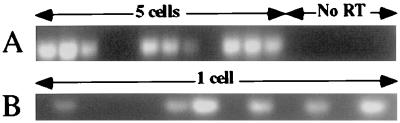
The small percentage of LT-HSC with >2n DNA, and their relative resistance to cytoreductive agents such as 5′-fluorourocil led to the hypothesis that LT-HSC are mostly quiescent (26–29). However, their surprisingly rapid uptake of BrdUrd demonstrates that LT-HSC are more active that previously thought. D cyclins are a family of highly homologous proteins with three known members expressed in a tissue-specific manner (30). D cyclins integrate extracellular proliferative signals with the cell cycle machinery to control progress through G1 (31). D cyclin mRNA expression begins in early G1, and both protein and mRNA are rapidly turned over (32, 33). Thus, only actively proliferating cells should express significant amounts of cyclin D mRNA. We performed RT-PCR on double-sorted KTSL− LT-HSC to determine whether a significant fraction of these cells express cyclin D. RT-PCR revealed that LT-HSC express cyclin D2 (Fig. 3) and D3 but not D1 (data not shown). Cyclin D2-positive results were obtained for 22 of 26 RT-PCR reactions (85%) with 5 LT-HSC (Fig. 3A), and 27 of 63 RT-PCR reactions (43%) with one cell were positive (Fig. 3B). Thus, nearly half of LT-HSC express cyclin D2 mRNA, suggestive of cell cycle activity in a significant fraction of LT-HSC.
Figure 3.
Cyclin D2 expression in five (A) and one (B) KTSL− cells as determined by using nested RT-PCR. No RT indicates lanes with RT-PCR reactions performed on KTSL− without the initial addition of reverse transcriptase to the RT reactions as a control.
Determination of the Proportion of LT-HSC in Cycle Using PY and Hoechst.
During cell cycle progression, there is a constant increase of cellular RNA, mainly because of the increased production of ribosomal RNA (34, 35). Quiescent cells have, on average, 10–20% the RNA as their cycling counterparts (36). Simultaneous staining of viable cells with the RNA-specific flourochrome PY and the DNA-specific fluorochrome Hoechst has been widely used to determine the cell cycle status of many different cell types (20). By using this technique, the fraction of KTSL− in G1/S/G2/M vs. G0 can be determined, revealing the fraction of actively proliferating cells (growth fraction) in vivo. As seen in Fig. 4, a significant fraction of KTSL− cells with a 2n amount of DNA had the same or greater PY staining levels as S/G2/M KTSL−. According to this analysis, these PY-high, 2n cells were in G1 phase of the cell cycle. In three separate experiments analyzing 2,000–5,000 cells, 23.5 ± 3.4% of KTSL− contained RNA levels consistent with G1/S/G2/M cells. Thus, during steady-state hematopoiesis, about one-fourth of LT-HSC were in the cell cycle, whereas the remaining were quiescent G0 cells.
Figure 4.
Growth Fraction of KTSL− LT-HSC. FACS analysis of double-sorted KTSL− cells stained with PY (y axis) and Hoechst 33342 (x axis). The growth fraction of a population of cells is the fraction of actively dividing cells (G1/S/G2/M). On average, 23.5 ± 3.4% of KTSL− LT-HSC were in G1/S/G2/M at any one time.
Throughout the age cohort used in this study, LT-HSC maintain constant numbers, balancing cell production with cell loss (3, 37). Because LT-HSC are a population of asynchronously dividing cells undergoing steady-state kinetics, BrdUrd incorporation rates can be related to the fraction of proliferating cells and average cell cycle time by the following equation: RB = GF/TC, where RB is the BrdUrd incorporation rate (0.078/day in this study), GF is the fraction of proliferating cells or growth fraction (0.235 in this study), and TC is the apparent cell cycle time (equation adapted from ref. 24). The TC in this equation averages the cell cycle time of cells in cycle and cells not yet in cycle. Thus, the actual cell cycling time of a LT-HSC once in cycle is actually shorter (24). By using the equation above, the estimated apparent in vivo cell cycle time of KTSL− LT-HSC under steady-state conditions was 3.0 days.
DISCUSSION.
We analyzed the in vivo proliferation and cell cycle kinetics of C57BL/Ka-Thy 1.1 mouse KTSL−/lo and KTSL− HSC. Over 90% of LT-HSC entered the cell cycle at least once every 30 days, and by 6 months >99% of LT-HSC divided. The subsets of cells analyzed in this experiment included all HSC in this mouse strain (18). Thus, there was no subset of LT-HSC that remained dormant for long periods of time. As determined by the slope of the log-linear plot of BrdUrd incorporation versus time (Fig. 2B), the probability of a LT-HSC entering the cell cycle was about 0.08 each day. The apparent duration of the LT-HSC cell cycle was 3.0 days. RT-PCR analysis of LT-HSC revealed that >40% expressed significant amounts of G1 cyclin D2. Consistent with this, simultaneous PY and Hoechst staining of LT-HSC revealed that about one-fourth of LT-HSC were in G1/S/G2/M during steady -state hematopoiesis. Assuming that 8% of LT-HSC go into cycle each day and that 24% of HSC are in G1/S/G2/M phases of the cell cycle, we calculated an apparent cell cycle time of 3.0 days. Although this number is helpful in considering the potential doubling time of the population, it represents an average across an asynchronous population of cycling and quiescent cells. Thus, the actual cell cycle time of the dividing cells may be different. It should also be noted that we used a conservative gate to determine the percentage of LT-HSC in G1 phase by PY staining. It is possible that some early G1 cells were excluded by this gate. Our own experiments suggest that ≈40% of LT-HSC express the G1 cyclin D2. If 40% of LT-HSC were in G1/S/G2/M phases of the cell cycle, then the apparent cell cycle time would be closer to 6 days.
The results of this study are consistent with those of Pietrzyk et al. (14) and Bradford et al. (15), although we observed somewhat faster incorporation of BrdUrd in the LT-HSC fraction than was observed by Bradford et al. This could be caused by the differences in mouse strains used, because the cell cycle dynamics of HSC are likely to be mouse strain-specific (38, 39). Also, Bradford et al. used the RhloHolo method to isolate HSC, whereas we used KTSL as described (3). Although both populations are highly enriched for long-term reconstituting cells, they may not overlap completely in terms of their composition.
In addition to varying between strains, cell cycle parameters vary during ontogeny. For example, the number of fetal liver HSC doubles daily, and around one-fourth of LT-HSC in the fetal liver are in S/G2/M phases of the cell cycle at any one time (40). HSC in old (>20 months) but otherwise normal C57BL mice exhibit increased proportions of HSC in cycle (37). Thus, whereas HSC in normal steady-state bone marrow cycle slowly, at other times during ontogeny increased proportions of HSC can be recruited into cycle, and cell cycle times can be much faster.
Because the size of the LT-HSC pool remains constant and does not increase under steady-state conditions in mice of this age (37), dividing LT-HSC must give rise to equal numbers of LT-HSC and other cell types, presumably short-term HSC. This could be accomplished by each LT-HSC undergoing an asymmetric division or by undergoing symmetric divisions in which half yield two LT-HSC and half yield two cells that differentiate or die.
The in vivo proliferation kinetics of HSC does not support the clonal-succession model. Our data demonstrate that LT-HSC enter the cell cycle in an asynchronous manner, that proliferation was not dominated by a few clones, and that on average 99% of LT-HSC divide every 57 days. Harrison et al. (13, 41) showed that in normal chimeric mice, many HSC contribute simultaneously to hematopoiesis. Recent experiments in our laboratory show that if several hundred retrovirus-infected LT-HSC were used to rescue lethally irradiated mice, then at least seven clones contributed to mature blood cells by Southern analysis at 25 weeks post-transplantation (C. Klug and I.L.W., unpublished observations). These results were consistent with the idea that if larger numbers of retrovirus-marked HSC are transplanted, larger numbers of clones durably contribute to hematopoiesis.
Model of LT-HSC Cell Cycle Regulation.
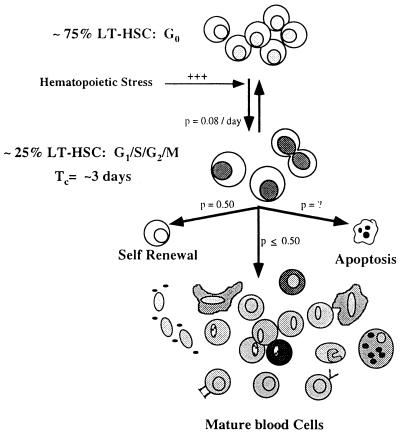
In light of our data on LT-HSC cell cycle kinetics, a more complete model of how these cells proliferate and contribute to blood cell production under normal hematopoietic situations can be considered (Fig. 5). At any one time, a fraction of LT-HSC would be in the cell cycle (at least 23.5 ± 3.4% in this study), and the remaining LT-HSC would be in G0. Because most LT-HSC are quiescent, the only explanation for the rapid incorporation of BrdUrd into LT-HSC is that these cells are continuously moving into and out of the cell cycle, as indicated by arrows in Fig. 5. The fraction of cells in each compartment remains relatively constant unless hematopoietic demands are altered significantly. For example, we have observed that most or all LT-HSC are driven to self-renew with a cell cycle time of <12 hours in mice that are stressed by administration of cyclophosphamide and granulocyte colony stimulating factor (42). All LT-HSC in cytoxan/G-CSF-treated mice label with BrdUrd after only 4 days of continuous feeding (D. Wright and I.L.W., unpublished observation). Also, blocking KTSL−/lo in the S phase by the administration of hydroxyurea to mice leads to a rapid increase in the percentage of these cells in-cycle (43). Thus, although the cell cycle time of HSC was relatively slow under steady-state conditions, it is highly responsive to changes in the hematopoietic environment. The proposed model provides a highly adaptable system for hematopoiesis in which blood cell production could be controlled in part by regulating the fraction of LT-HSC in cycle and/or their average cell cycle time.
Figure 5.
Model of LT-HSC cell cycle regulation. LT-HSC are asynchronously dividing with a constant fraction of cells in the cell cycle and a constant fraction in G0. LT-HSC are continuously moving into and out of the cell cycle at rates indicated next to the arrows (p denotes probability of labeled event occurring) as determined from the BrdUrd incorporation kinetics of KTSL− cells. Tc = apparent cell cycle time.
Acknowledgments
We thank Annete Schlageter, Jos Domen, Douglas Wright, Andrew BitMansour, and Dennise Dalma-Weiszhausz for critical review and help in preparing this manuscript. We also thank Libuse Jerabek, Veronica Braunstein, and Joe Verdi for antibodies and experimental expertise and Lucindo Hidalgo for animal care. This work was supported by USDH8 National Cancer Institute Grant CA-42551 to I.W.; S.H.C. was supported by the Medical Scientist Training Program Grant 5T32GM07365 at Stanford University School of Medicine.
ABBREVIATIONS
- HSC
hematopoietic stem cells
- BM
bone marrow
- LT-HSC
long-term hematopoietic stem cells
- MPP
multipotent progenitors
- BrdUrd
bromodeoxyuridine
- KTSL− or KTSL−1/lo
c-kit−bright, Thy 1.1lo, Sca-1+, Lineage negative or negative/lo HSC
- PY
pyronin Y
- RT-PCR
reverse transcription–PCR
References
- 1.Metcalf D. The Molecular Control of Blood Cells. Cambridge, MA: Harvard Univ. Press; 1988. [Google Scholar]
- 2.McCulloch E A, Till J E. Radiat Res. 1960;13:115. [PubMed] [Google Scholar]
- 3.Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 4.Kay H E M. Lancet. 1965;ii:418–419. doi: 10.1016/s0140-6736(65)90763-4. [DOI] [PubMed] [Google Scholar]
- 5.Lemischka I R, Raulet D H, Mulligan R C. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 6.Snodgrass R, Keller G. EMBO J. 1987;6:3955–3960. doi: 10.1002/j.1460-2075.1987.tb02737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capel B, Hawley R, Covarrubias L, Hawley T, Mintz B. Proc Natl Acad Sci USA. 1989;86:4564–4568. doi: 10.1073/pnas.86.12.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan C T, Lemischka I R. Genes Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- 9.Lemischka I R. Curr Top Microbiol Immunol. 1992;177:59–71. doi: 10.1007/978-3-642-76912-2_5. [DOI] [PubMed] [Google Scholar]
- 10.Morrison S J, Wandycz A M, Hemmati H D, Wright D E, Weissman I L. Development. 1997;124:1929–1939. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- 11.Abkowitz J, Linenberger M, Newton M, Shelton G, Ott R, Guttorp P. Proc Natl Acad Sci USA. 1990;87:9062–9066. doi: 10.1073/pnas.87.22.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abkowitz J L, Catlin S N, Guttorp P. Nat Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- 13.Harrison D E, Lerner C, Hoppe P C, Carlson G A, Alling D. Blood. 1987;69:773–777. [PubMed] [Google Scholar]
- 14.Pietrzyk M E, Priestley G V, Wolf N S. Blood. 1985;66:1460–1462. [PubMed] [Google Scholar]
- 15.Bradford G B, Williams B, Rossi R, Bertoncello I. Exp Hematol. 1997;25:445–453. [PubMed] [Google Scholar]
- 16.Ikuta K, Weissman I L. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida N, Aguila H L, Fleming W H, Jerabek L, Weissman I L. Blood. 1994;83:3758–3779. [PubMed] [Google Scholar]
- 18.Uchida N, Weissman I L. J Exp Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klug C A, Morrison S J, Masek M, Hahm K, Smale S T, Weissman I L. Proc Natl Acad Sci USA. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro H M. Cytometry. 1981;2:143–150. doi: 10.1002/cyto.990020302. [DOI] [PubMed] [Google Scholar]
- 21.Schneider E L, Sternberg H, Tice R R. Proc Natl Acad Sci USA. 1977;74:2041–2044. doi: 10.1073/pnas.74.5.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stockdale F, Okazake K, Nameroff M, Holtzer H. Science. 1964;186:533–535. doi: 10.1126/science.146.3643.533. [DOI] [PubMed] [Google Scholar]
- 23.Yen A, Forbes M E. Cancer Res. 1990;50:1411–1420. [PubMed] [Google Scholar]
- 24.Aherne W A, Camplejohn R S, Wright N A. An Introduction to Cell Population Kinetics. Baltimore: Univ. Park Press; 1977. [Google Scholar]
- 25.Colquhoun D. Lectures on Biostatistics. London: Oxford Univ. Press; 1971. pp. 81–85. [Google Scholar]
- 26.Lerner C, Harrison D E. Exp Hematol. 1990;18:114–118. [PubMed] [Google Scholar]
- 27.Randall T, Weissman I. Blood. 1997;89:3596–3606. [PubMed] [Google Scholar]
- 28.Van Zant G. J Exp Med. 1984;159:679–690. doi: 10.1084/jem.159.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming W H, Alpern E J, Uchida N, Ikuta K, Spangrude G J, Weissman I L. J Cell Biol. 1993;122:897–902. doi: 10.1083/jcb.122.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherr C J. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C J, Kato J, Quelle D E, Matsuoka M, Roussel M F. Cold Spring Harb Symp Quant Biol. 1994;59:11–19. doi: 10.1101/sqb.1994.059.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Matsushime H, Roussel M F, Sherr C J. Cold Spring Harb Symp Quant Biol. 1991;56:69–74. doi: 10.1101/sqb.1991.056.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Matsushime H, Roussel M F, Ashmun R A, Sherr C J. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 34.Darzynkiewicz Z, Evenson D P, Staiano-Coico L, Sharpless T K, Melamed M L. J Cell Physiol. 1979;100:425–438. doi: 10.1002/jcp.1041000306. [DOI] [PubMed] [Google Scholar]
- 35.Darzynkiewicz Z. Leukemia. 1988;2:777–787. [PubMed] [Google Scholar]
- 36.Johnson L F, Williams J G, Abelson H T, Green H, Penman S. Cell. 1975;4:69–75. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
- 37.Morrison S J, Wandycz A M, Akashi K, Globerson A, Weissman I L. Nat Med. 1996;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- 38.de Haan G, Van Zant G. J Exp Med. 1997;186:529–536. doi: 10.1084/jem.186.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Haan G, Nijhof W, Van Zant G. Blood. 1997;89:1543–1550. [PubMed] [Google Scholar]
- 40.Morrison S J, Hemmati H D, Wandycz A M, Weissman I L. Proc Natl Acad Sci USA. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison D E, Astle C M, Lerner C. Proc Natl Acad Sci USA. 1988;85:822–826. doi: 10.1073/pnas.85.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison S J, Wright D E, Weissman I L. Proc Natl Acad Sci USA. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida N, Friera A M, He D, Reitsma M J, Tsukamoto A S, Weissman I L. Blood. 1997;90:4354–4362. [PubMed] [Google Scholar]