Abstract
Anaplasma marginale is an ehrlichial pathogen of cattle that establishes lifelong persistent infection. Persistence is characterized by rickettsemic cycles in which new A. marginale variant types, defined by the sequence of the expressed msp2 transcripts, emerge. The polymorphic msp2 transcripts encode structurally distinct MSP2 proteins and result in an antigenically diverse and continually changing A. marginale population within the blood. In this manuscript, we used sequence analysis of msp2 transcripts to show that a restricted repertoire of variant types, designated SGV1 and SGV2, is expressed within the tick salivary gland. The same SGV1 and SGV2 variant types were expressed in ticks regardless of the variant types expressed in the blood of infected cattle at the time of acquisition feeding by the ticks. Importantly, subsequent tick transmission to susceptible cattle resulted in acute rickettsemia composed of organisms expressing only the same SGV1 and SGV2 variant types. This indicates that the msp2 expressed by organisms within the tick salivary gland predicts the variant type responsible for acute rickettsemia and disease. This restriction of transmitted A. marginale variant types, in contrast to the marked diversity within persistently infected cattle, supports development of MSP2 vaccines to prevent acute rickettsemia in tick-transmitted infections.
Anaplasma marginale is a pathogen of cattle that is closely related, both genetically and in mechanisms of transmission, to other tick-borne ehrlichiae, including Ehrlichia equi, the apparent agent of human granulocytic ehrlichiosis (HGE), and Ehrlichia chaffeensis (1, 2). The hallmarks of A. marginale infection are severe anemia during acute, cell-associated rickettsemia (>109 rickettsiae/ml) and subsequent persistent infection characterized by low (<107 per ml), microscopically undetectable rickettsemia (1, 3, 4). Persistent infection results regardless of the infecting A. marginale strain and has been demonstrated to persist, without reinfection, for at least 7 years (3, 4). This lifelong persistence is fundamental to continued transmission because transovarial passage of A. marginale within the tick vector does not occur (1, 5, 6). Consequently, A. marginale transmission, similar to other tick-borne ehrlichiae, is dependent on the infected animal reservoir (1, 2, 6–8).
Persistent A. marginale infection is characterized by sequential cycles of rickettsemia, each composed of a progressive, logarithmic increase in rickettsemia followed by a precipitous decrease (3, 4, 9). Each cycle reflects the emergence of A. marginale expressing unique transcripts encoding structurally variant major surface protein 2 (MSP2), typified by deletions, substitutions, and insertions within a central hypervariable region (9–11). Importantly, this hypervariable region encodes the hydrophilic domain of MSP2, and variation can alter the epitopes exposed on the organism surface (9–13). Consequently, the structural and antigenic phenotype of the A. marginale population during persistent rickettsemia is diverse and continually changing.
Ixodid ticks feeding on persistently rickettsemic calves acquire A. marginale in the bloodmeal and, after development of infective stages in the salivary glands, efficiently transmit the infection to susceptible cattle (6, 14–17). Do the A. marginale that develop within the tick express the same MSP2 variant types as in the acquisition-feed bloodmeal? If so, acquisition feeding by ticks at different cycles during persistent rickettsemia, with each cycle being composed of organisms expressing unique variant types, would result in an antigenically diverse set of challenges to cattle upon tick transmission. Alternatively, development within the tick may result in organisms expressing one or more specific MSP2 variant types. This would provide a more antigenically homogeneous challenge and support the feasibility of vaccine development based on the MSP2 present on A. marginale within the tick salivary gland. In this manuscript we address this question and show that the tick-transmitted A. marginale do not reflect the heterogeneity of MSP2 variant types expressed in infected cattle but, rather, specific variant types that arise within the tick. Furthermore, we show that this restricted set of tick-transmitted MSP2 variant types are also expressed by A. marginale during acute rickettsemia in cattle, but that the establishment of persistent infection is associated with expression of new MSP2 variant types.
MATERIALS AND METHODS
Infection of Ticks The six male Holstein calves used (755, 767, 769, 778, 786, 788) were confirmed free of A.
marginale by using MSP5 competitive inhibition–ELISA and msp5 PCR (18, 19) before initiation of the experiments. Calf 755 was infected by using a stabilate of the South Idaho strain of A. marginale (12, 14, 17, 20). This stabilate, derived from an acutely rickettsemic calf, contained 109 infected erythrocytes. Approximately 700 uninfected laboratory-reared Dermacentor andersoni adult males were placed in an orthopedic stockinette and allowed to attach and feed on calf 755 when the rickettsemia reached 109 per ml (14). The South Idaho strain of A. marginale is naturally transmitted by D. andersoni, and both the South Idaho strain and adult males of this D. andersoni stock have been used previously for experimental transmission to cattle (14, 17). For identification of the expressed A. marginale MSP2 variant types throughout the 7-day period of tick attachment and feeding, whole blood was collected in acid citrate dextrose and total RNA was extracted using TRIzol reagent (BRL) for subsequent use in reverse transcriptase–PCR (RT-PCR) (9). On day 7, ticks were removed from the calf and placed in a 26°C incubator with 90–98% relative humidity and a 14-hr photoperiod and held for a week (14). As a negative control, an equal number of uninfected D. andersoni adult males of the same stock were allowed to attach and feed for 7 days on an uninfected calf (no. 778). These negative-control ticks were incubated under identical conditions in a separate humidity chamber.
MSP2 expression in ticks infected by acquisition feeding on calf 755 was analyzed either immediately after incubation or after transmission feeding on calf 767. This second attachment and feeding of A. marginale-infected ticks stimulates development of infective stages within the salivary gland and results in transmission (6, 14–17). Salivary glands were dissected from infected ticks (14), or an equal number of uninfected control ticks, and collected in pools of 100 salivary glands in either proteinase inhibitor buffer (19) for immunoblot analysis or in TRIzol for total RNA isolation. DNA was extracted from an aliquot of each salivary gland pool (19) using Puregene (Gentra Systems) and used to determine the number of A. marginale based on the msp5 quantitative competitive PCR (9). The tick infection and transmission were repeated in a second experiment of identical design in which ticks acquisition-fed on calf 786 and, after incubation, transmission-fed on calf 788. Finally, a third tick acquisition feeding and analysis of expressed salivary gland variant types was done using calf 769, 7 months after its infection by tick transmission of the South Idaho strain.
Transmission to Cattle.
After the transmission feeding of 100 A. marginale-infected ticks on calves 767 and 788, blood was collected, as described above, beginning on the first day of microscopically detectable acute rickettsemia for identification of MSP2 variants. Rickettsemia exceeds 109 organisms/ml in the acute phase but then decreases dramatically to below 107 organisms per ml, the levels that define persistent infection (3, 4, 9). One and 4 weeks after the decrease to <107 A. marginale per ml in calves 767 and 788, blood was collected and used to identify MSP2 variants in persistent rickettsemia.
Immunoblot Analysis.
Equal numbers of A. marginale (104 organisms) were lysed, separated using a 7.5–17.5% polyacrylamide gel gradient, and transferred to nitrocellulose membranes (19), and MSP2 expression was detected by using a rabbit antiserum induced by immunization with purified native MSP2 (13). This antiserum binds A. marginale MSP2 proteins regardless of variant type-specific changes in B cell epitopes (11, 12) or differences in the hypervariable regions (amino acids 189–322, based on pCKR11.2 sequence; ref. 11).
Cloning and Sequencing of msp-2 cDNA.
Total RNA was reverse-transcribed by using random hexamers and msp2 cDNA was amplified using PCR (9). The full-length msp2 transcript was amplified by using primers based on sequences conserved in the msp2 homologues among ehrlichial species (21–24). The forward and reverse primers were derived from nucleotides −9 to 15 and 1,219 to 1,242, respectively (numbering based on pCKR11.2; ref. 11). The msp2 hypervariable region was amplified specifically by using primers derived from conserved regions that flank the central, hypervariable, 595-bp region of msp2 (9–11). These forward and reverse primers represented, respectively, nucleotides 375–401 and 945–965 (11). The RT-PCR, ligation of amplicons into pCR 2.1 (Invitrogen), and transformation of Escherichia coli were done as described previously (9). Plasmid DNA was extracted from individual clones and insert DNA was sequenced in both directions. Sequence analysis was performed on a Vax11/785 computer.
RESULTS
Enhanced Expression of MSP2 in Tick Salivary Glands During Transmission Feeding.
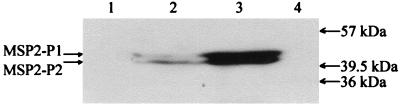
Adult male D. andersoni were acquisition-fed on calf 755 for 7 days, transferred to a humidity chamber, and held at 26°C (14). Salivary glands then were dissected from ticks before or after 3 days of transmission feeding on calf 767. Using a polyclonal antiserum against MSP2 (13), the expression levels of MSP2 were analyzed in equal numbers of A. marginale from D. andersoni salivary glands obtained either before (Fig. 1, lane 2) or after 3 days of transmission feeding (Fig. 1, lane 3). Transmission feeding clearly enhanced the level of MSP2 expressed per organism in the salivary glands. Incubation of ticks at 37°C for 60 hr, which also enhances infectivity (16), increased expression of salivary gland MSP2 to levels similar to transmission feeding (data not shown).
Figure 1.
Enhanced expression of MSP2 in tick salivary glands after transmission feeding. Equal numbers of A. marginale (104 per lane) from D. andersoni salivary glands obtained either before (lane 2) or 3 days after (lane 3) transmission feeding were reacted with rabbit antisera against MSP2. Salivary glands from uninfected ticks before (lane 1) or after (lane 4) 3 days of feeding were included as negative controls. Molecular size markers (kDa) are shown in the right margin. MSP2–P1 and MSP2–P2 indicate the positions of polypeptides specifically detected by the antibody to MSP2.
Detection of New MSP2 Variant Types Within Infected Ticks.
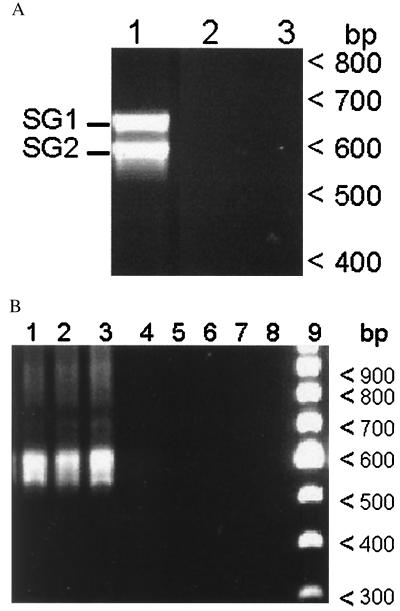
Immunoblots revealed two MSP2 polypeptides expressed within the salivary glands with approximate molecular sizes of 42 and 43 kDa (Fig. 1, lane 3). In contrast, four MSP2 polypeptides, ranging in size from <40 to 42 kDa, were expressed by organisms in the blood of calf 755 at the time when the ticks were acquisition feeding (data not shown). This size difference of several kilodaltons is characteristic of the polymorphism among MSP2 variant types, which arise during persistent rickettsemia within cattle (9), and suggested that MSP2 variants expressed within the salivary gland may not reflect the variants expressed in the acquisition bloodmeal. Definitive identification of variant types required sequence comparison of the expressed msp2 in the blood of calf 755 throughout the period of acquisition feeding to the expressed msp2 in the infective salivary glands. The cDNA clones were derived by RT-PCR using msp2-specific primers flanking the hypervariable region (Fig. 2). Two expressed salivary gland variant types were identified from sequencing a total of 20 hypervariable region clones and verified by sequencing of the complete expressed msp2 in an additional 10 independently derived clones. These two expressed variant types, designated salivary gland variant (SGV)1 and SGV2, had identical nucleotide sequences with the sole exception of a 48-bp deletion in SGV2. SGV1 and SGV2 have complete ORFs and are predicted to encode proteins with molecular sizes of 43.5 and 42.1 kDa, respectively, which are similar to the MSP2 polypeptides identified by immunoblotting (Fig. 1). These identical SGV1 and SGV2 types were also expressed in the salivary glands of a cohort of the ticks fed on calf 755 but held at 26°C for an additional 30 days before transmission feeding (on calf 769).
Figure 2.
(A) RT-PCR of total RNA from tick salivary glands obtained 3 days after transmission feeding on calf 767 (lane 1). PCR without RT was used to control for amplification of contaminating DNA (lane 2). RT-PCR without RNA was included as a template control (lane 3). The two bands in the amplicon are designated as SG1 and SG2 (left margin). SG1 and SG2 were shown by sequencing to represent SGV1 and SGV2, respectively. Molecular size markers (bp) are shown in the right margin. (B) RT-PCR of total RNA from rickettsemic blood (calf 755) during acquisition feeding (lane 1, at time of initial tick attachment; lane 2, day 3 after attachment; lane 3, day 7 after attachment). Lanes 5–7 represent the identical reactions without RT, and lanes 4 and 8 represent RT-PCR without template. Lane 9 contains a 100-bp DNA ladder, and the molecular sizes (bp) are shown in the right margin.
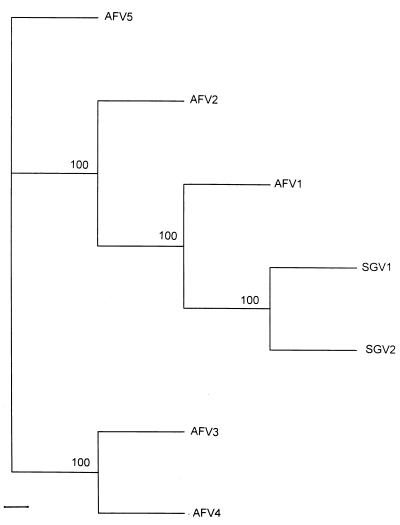
Five expressed msp2 variant types were identified similarly in the rickettsemic blood of calf 755 during the acquisition-feeding period, including the first day of tick attachment and the last day of tick feeding. All the variant types, designated acquisition-feed variants (AFV) 1–5, encoded MSP2 proteins smaller than SGV1 and SGV2 and were each distinctly different because of polymorphism in the central hypervariable region. A comparison of the predicted complete MSP2 amino acid sequences for SGV1, SGV2, and the most similar acquisition feed variant type, AFV1, reveals deletions, insertions, and substitutions within the hypervariable regions (Fig. 3). The difference is even greater for AFV2–5 as shown by the phylogram of the predicted amino acid sequences of all acquisition feed and salivary gland variant types (Fig. 4). Within the hypervariable regions, amino acid identity ranged from 63% between AFV5 and SGV1 to 79% between AFV1 and SGV2.
Figure 3.
Alignment of predicted MSP2 amino acid sequences from salivary gland variant types, SGV1 and SGV2, with the most similar AFV type, AFV1, in the rickettsemic blood of calf 755 at the time of tick acquisition feeding. Areas of amino acid substitutions, insertions, and deletions are indicated by a white background, areas of amino acid identity have a black background, and gray shading indicates conservative amino acid changes. The dashes designate deletions.
Figure 4.
A phylogram of AFV types from calf 755 and subsequent SGV types based on predicted amino acid sequences. seqboot, protdist, and protpars consense programs in the phylip phylogenetic-inference package (version 3.5) were used for the derivation of the data used in the phylogram (28). Bar indicates 1% divergence in amino acid sequences. Bootstrap values from 100 analyses are shown at the branch points of the tree.
The experiment was repeated by using a second calf (786) infected with the South Idaho strain of A. marginale. The acquisition feeding, incubation in the humidity chamber, and second attachment and transmission feeding on a new host (calf 788) used the same experimental design. As expected because of the variation in msp2 transcripts expressed within rickettsemic cattle (9, 10), the sequences of the AFV types in the blood of calf 786 all differed in the hypervariable region as compared with AFV1–5 from calf 755 (data not shown). However, analysis of the variant types expressed within the salivary gland of the ticks that had acquisition-fed on calf 786 revealed only the same SGV1 and SGV2 identified previously. The nucleotide sequences of SGV1 expressed within ticks acquisition-fed on the two different calves were identical, as were the SGV2 sequences. Finally, a third acquisition feeding was done using calf 769, after seven months of persistent infection (data not shown). The identical SGV1 and SGV2 variant types again were expressed within the tick salivary gland. Thus, SGV1 and SGV2 expression recurs in D. andersoni adult male ticks even though the ticks initially acquire A. marginale expressing different msp2 variant types, from different animals, and at different time points in infection.
Expression of MSP2 Variant Types During Acute and Persistent Rickettsemia.
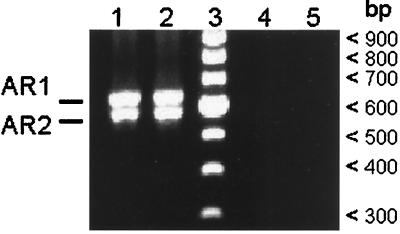
To determine whether the variant types expressed in the tick salivary gland, SGV1 and SGV2, were expressed by the A. marginale that were transmitted and replicate in acute infection, msp2 transcripts were amplified by using RT-PCR on total RNA obtained during acute rickettsemia in calf 767 (Fig. 5). Sequencing of 28 cDNA clones derived independently from the two amplicons revealed two expressed variant types, acute rickettsemia variant type 1 (ARV1) (15 clones) and ARV2 (13 clones). The nucleotide sequence of ARV1 was identical to SGV1, and ARV2 was identical to SGV2. Immunoblot analysis of A. marginale isolated from acute rickettsemia revealed MSP2 polypeptides of approximately 43 and 42 kDa, consistent with expression of the ARV1 and ARV2 transcripts (data not shown).
Figure 5.
RT-PCR of total RNA from blood collected during acute rickettsemia in calf 767, respectively, 21 and 22 days after tick attachment (lanes 1 and 2). PCR without RT was used to control for amplification of contaminating DNA (lanes 4 and 5). Lane 3 contains a 100-bp DNA ladder, and molecular sizes are indicated in the right margin. The two bands in the amplicon are designated as AR1 and AR2 (left margin). AR1 and AR2 were shown by sequencing to represent ARV1 and ARV2, respectively.
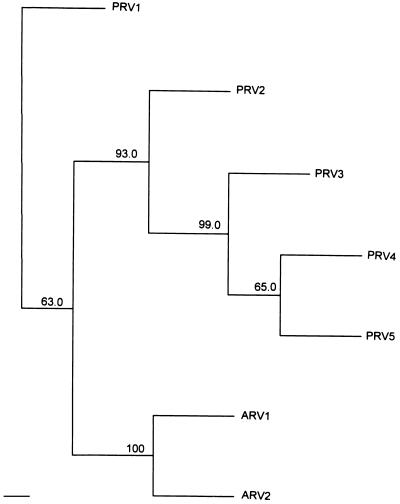
Acute rickettsemia peaks at ≥109 per ml and, concomitant with development of antibody to surface epitopes, A. marginale levels rapidly decrease to <107 per ml, indicating onset of persistent infection (3, 4, 19). Persistent rickettsemia variant types, designated PRV1–5, were identified in the blood of calf 767 1 week after the rickettsemia dropped below 107 per ml. None of the persistent rickettsemia variant types corresponded to either of the acute rickettsemia variant types, ARV1 or 2, indicating that organisms expressing these two variant types during the acute rickettsemia had been completely cleared. A comparison of the predicted hypervariable region amino acid sequences for ARV1 and ARV2 with the most similar persistent rickettsemia variant type, PRV1, reveals this polymorphism (Fig. 6). The relationships among the variant types from calf 767 including those from acute rickettsemia and the first week of persistent rickettsemia are shown in Fig. 7. The amino acid identity within the hypervariable regions ranged from 64% between PRV2 and ARV1 to 82% between PRV1 and ARV2. At the 4th week of persistent infection, a second set of expressed variant types was identified in the blood of calf 767 (data not shown). Notably, none of these variant types, designated PRV6–13, represented either of the acute rickettsemia variants or any of the variant types identified in the 1st week of persistent infection. The most similar to the acute rickettsemia variant types was PRV8, which was 72% identical on an amino acid basis to ARV2 within the hypervariable region. The amino acid identity between variant types from the two time points of persistent rickettsemia ranged from 59 to 90%, similar to the previously reported variation between sequential cycles of persistent rickettsemia (9).
Figure 6.
Alignment of the predicted hypervariable amino acid sequences of the acute rickettsemia variant types, ARV1 and ARV2, with the most similar, persistent rickettsemia variant type, PRV1. All three variant types were derived from blood obtained from calf 767 after tick transmission of A. marginale. ARV1 and ARV2 are identical to, respectively, SGV1 and SGV2.
Figure 7.
A phylogram of ARV types and PRV types based on predicted amino acid sequences. All variant types were derived from blood obtained from calf 767. The distance value determination, tree construction, scale, and calculation of bootstrap values were done as for Fig. 4.
The analysis of expressed variant types in acute and persistent rickettsemia was repeated in calf 788 using the same protocols. The same ARV1 and ARV2 transcripts were expressed in acute rickettsemia and were 100% identical on the nucleotide level to the SGV1 and SGV2 transcripts from the tick. As predicted, these variant types were not detected in the 1st week of persistent infection after control of the acute rickettsemia.
DISCUSSION
The expression of a new set of MSP2 variant types by A. marginale in sequential cycles of persistent rickettsemia appears to reflect the selective pressure of the immune response against the surface epitopes encoded by the msp2 hypervariable regions (9–13). In contrast, development of A. marginale within the tick was characterized by expression of two very closely related variant types, SGV1 and SGV2. Only SGV1 and SGV2 were identified from a total of 60 msp2 cDNA clones derived from infected tick salivary glands. SGV1 and SGV2 were present in approximately equal proportions; 55% of the cDNA clones were SGV1. Although it is possible that other msp2 transcripts were expressed in the tick salivary gland but not detected, the analysis of 60 clones provides a 95% probability that all variant types were detected, assuming that any additional variant types would compose at least 5% of the total transcript population (25). Notably, SGV1 and SGV2 were identically expressed in ticks that had acquired A. marginale by feeding on three different calves, feeding during both acute and persistent rickettsemia, and, most importantly, feeding on rickettsemic blood containing the distinctly different variant types.
Interestingly, the 48-bp deletion in SGV2 relative to SGV1 is not typical of the hypervariable region polymorphism among msp2 transcripts from acute and persistent rickettsemia. The substitutions, insertions, and deletions that characterize polymorphic msp2 variant types expressed in the blood appear to result from gene conversion among duplicated gene copies (9–11). In contrast, it appears more likely that SGV2 arose from SGV1 by gene duplication with later excision of a 48-bp loop.
SGV1 and SGV2 clearly emerge during development within the tick, reflecting either strong selection for these variants or regulated, de novo expression. Although we cannot exclude the possibility that a minor population of organisms expressing the SGV are acquired in the bloodmeal and then predominate in the salivary gland because of a selective advantage in replication within the tick, we hypothesize that expression of the msp2 transcripts is specifically regulated in the tick. Thus, signals within the tick would induce the expression of the SGV transcripts and either actively repress or fail to induce other msp2 transcripts. Notably, the expression of SGV MSP2 per organism is enhanced dramatically by transmission feeding on a new bovine host, an effect that can be mimicked by an increase in temperature to 37°C. Because MSP2 has been shown to function as an adhesin for A. marginale binding to bovine erythrocytes (13), the increased infectivity at the time of transmission feeding may reflect specifically enhanced expression of adhesins as well as an increase in total organism numbers per salivary gland (15). This enhanced MSP2 expression is consistent with our hypothesis that msp2 transcription is a specifically regulated event within the tick. If this hypothesis is correct, identification of the signaling pathways that link transmission feeding to enhanced MSP2 expression may provide new opportunities for disrupting tick-borne transmission of A. marginale.
This apparent switch to expression of preferred MSP2 variant types within the tick is similar to the regulation of the variable major proteins (Vmps) in tick transmission of the spirochete Borrelia hermsii (26). Like msp2, vmps encode variant specific B cell epitopes and unique Vmps arise in sequential cycles of spirochetemia, followed by control of organisms expressing the specific Vmp (26). Once within the tick vector, a new variant, Vmp33, is expressed singularly in the salivary glands regardless of whether tick infection was initiated with spirochetes expressing Vmp 7 or Vmp 8 (26). However, upon tick transmission to a mammalian host, B. hermsii reverts to express the initial variant acquired by the tick, either Vmp 7 or Vmp 8 (26). In contrast, the MSP2 variant types, SGV1 and SGV2, that emerge within the tick are expressed after transmission and continue to be expressed throughout acute rickettsemia. Consequently, the diversity of tick-transmitted A. marginale appears to be restricted as compared with the MSP2 variant-type heterogeneity in persistent infection (9). This raises the possibility that immunization against SGV1 and SGV2 could prevent the acute rickettsemia and associated disease induced by tick transmission. MSP2 has been shown to induce protection (20), and the clearance of organisms expressing the salivary gland variant types concomitant with resolution of acute rickettsemia supports testing of this immunization strategy. The use of Cowdria ruminantium-infected Amblyomma hebraeum as a vaccine to prevent African heartwater in ruminants provides a precedent for developing immunogens based on tick stages of ehrlichial pathogens (27).
In contrast to the limited MSP2 heterogeneity represented by organisms expressing SGV1 and SGV2 within the tick and during the subsequent acute rickettsemia, persistent infection is characterized by continuous emergence of MSP2 variant types (9). The variation encoded by the different msp2 transcripts is clustered within the hydrophilic domain and can result in B cell epitope variation (9–13). Thus, the complete clearance of acute rickettsemia variant types and emergence of new MSP2 variant types within 1 week after control of acute infection support the hypothesis that antigenic variation, rather than a poor immune response, is responsible for persistent infection. As predicted by this hypothesis (4, 9), the examination of a second time point in persistent rickettsemia, 4 weeks after control of acute rickettsemia, again showed expression of a completely different set of MSP2 variants types.
MSP2 homologues have been identified in other closely related ehrlichial pathogens, including human granulocytic ehrlichiosis (HGE) (21), E. canis (22), E. chaffeensis (23), and Cowdria ruminantium (24). Like A. marginale MSP2, these outer membrane proteins are encoded by polymorphic multigene families and include one or more hypervariable regions located in surface exposed domains. The overall similarity in MSP2 homologues among these ehrlichiae, combined with the common features of persistence within mammalian reservoir hosts and transmission by ixodid ticks, suggests that the importance of A. marginale MSP2 variants in persistence and transmission will be broadly applicable among ehrlichial pathogens.
Acknowledgments
We thank Donald P. Knowles, Jr., and Travis C. McGuire for helpful discussion and review of the manuscript. The technical help of Yvonne McGhee and Ralph Horn is appreciated. This work was supported by U.S. Department of Agriculture Grant 96-37204-3610, National Institutes of Health Grant R01 AI44005, and National Institutes of Health Fellowship K08 AI01371.
ABBREVIATIONS
- MSP
major surface protein
- SGV
salivary gland variant
- AFV
acquisition feed variant
- ARV
acute rickettsemia variant
- PRV
persistent rickettsemia variant
- VMP
variable major protein
- RT-PCR
reverse transcriptase–PCR
Footnotes
References
- 1.Losos G J. In: Infectious Tropical Diseases of Domestic Animals. Losos G J, editor. New York: Longman; 1986. pp. pp.742–795. [Google Scholar]
- 2.Walker D H, Dumler J S. Emerg Inf Dis. 1996;2:18–29. doi: 10.3201/eid0201.960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriks I S, Palmer G H, McGuire T C, Allred D R, Barbet A F. J Clin Microbiol. 1989;27:279–284. doi: 10.1128/jcm.27.2.279-284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kieser S T, Eriks I S, Palmer G H. Infect Immun. 1990;58:1117–1119. doi: 10.1128/iai.58.4.1117-1119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stich R W, Kocan K M, Palmer G H, Ewing S A, Hair J A, Barron S J. Am J Vet Res. 1989;50:1377–1380. [PubMed] [Google Scholar]
- 6.Kocan K M. In: Morphology, Physiology and Behavioral Biology of Ticks. Sauer J R, Hair J A, editors. Chichester, U.K.: Horwood; 1986. pp. 472–505. [Google Scholar]
- 7.Telford S R, Dawson J E, Katavolos P, Warner C K, Kolbert C P, Persing D H. Proc Natl Acad Sci USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLeod J, Gordon W S. Parasitology. 1933;25:273–283. [Google Scholar]
- 9.French D M, McElwain T F, McGuire T C, Palmer G H. Infect Immun. 1998;66:1200–1207. doi: 10.1128/iai.66.3.1200-1207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eid G, French D M, Lundgren A M, Barbet A F, McElwain T F, Palmer G H. Infect Immun. 1996;64:836–841. doi: 10.1128/iai.64.3.836-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer G H, Eid G, Barbet A F, McGuire T C, McElwain T F. Infect Immun. 1994;62:3808–3816. doi: 10.1128/iai.62.9.3808-3816.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuire T C, Palmer G H, Goff W L, Johnson M I, Davis W C. Infect Immun. 1984;45:697–700. doi: 10.1128/iai.45.3.697-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGarey D J, Allred D R. Infect Immun. 1994;62:4587–4593. doi: 10.1128/iai.62.10.4587-4593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stiller D, Kocan K M, Edwards W, Ewing S A, Hair J A, Barron S J. Am J Vet Res. 1989;50:1281–1386. [PubMed] [Google Scholar]
- 15.Kocan K M, Stiller D, Goff W L, Claypool P L, Edwards W, Ewing S A, McGuire T C, Hair J A, Barron S J. Am J Vet Res. 1992;53:499–507. [PubMed] [Google Scholar]
- 16.Kocan K M, Goff W L, Stiller D, Edwards W, Ewing S A, Claypool P L, McGuire T C, Hair J A, Barron S J. Am J Vet Res. 1993;54:107–112. [PubMed] [Google Scholar]
- 17.Eriks I S, Stiller D, Palmer G H. J Clin Microbiol. 1993;31:2091–2096. doi: 10.1128/jcm.31.8.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torioni de Echaide S, Knowles D P, McGuire T C, Palmer G H, Suarez C E, McElwain T F. J Clin Microbiol. 1998;36:777–782. doi: 10.1128/jcm.36.3.777-782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowles D P, Torioni de Echaide S, Palmer G H, McGuire T C, Stiller D, McElwain T F. J Clin Microbiol. 1996;34:2225–2230. doi: 10.1128/jcm.34.9.2225-2230.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer G H, Oberle S M, Barbet A F, Davis W C, Goff W L, McGuire T C. Infect Immun. 1988;56:1526–1531. doi: 10.1128/iai.56.6.1526-1531.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhi N, Ohashi N, Rikihisa Y, Horowitz H W, Wormser G P, Hechemy K. J Clin Microbiol. 1998;36:1666–1673. doi: 10.1128/jcm.36.6.1666-1673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy G R, Sulsona C R, Barbet A F, Mahan S M, Burridge M J, Alleman A R. Biochem Biophys Res Commun. 1998;247:636–643. doi: 10.1006/bbrc.1998.8844. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi N, Zhi N, Zhang Y, Rikihisa Y. Infect Immun. 1998;66:132–139. doi: 10.1128/iai.66.1.132-139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Vliet A H, Jongejan F, van Kleef M, van der Zeijst B A. Infect Immun. 1994;62:1451–1456. doi: 10.1128/iai.62.4.1451-1456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott L. An Introduction to Statistical Methods and Data Analysis. 3rd Ed. Boston: PWS-KENT Publishing Company; 1984. pp. 467–518. [Google Scholar]
- 26.Schwan T G, Hinnebusch B J. Science. 1998;280:1938–1940. doi: 10.1126/science.280.5371.1938. [DOI] [PubMed] [Google Scholar]
- 27.Oberem P T, Bezuidenhout J D. Onderstepoort J Vet Res. 1987;54:485–488. [PubMed] [Google Scholar]
- 28.Felsenstein J. Clad. 1989;5:164–166. [Google Scholar]