Abstract
The recent isolation of a novel DNA virus from the serum of a Japanese patient (T.T.) has provided the latest possible candidate virus associated with cryptogenic hepatitis. In the present study, we report the complete nucleotide sequence of this virus (TTV) isolated from the serum of a West African. Based on PCR studies designed to amplify overlapping regions of the viral genome and sensitivity to digestion with mung bean nuclease, the viral genome is circular and negative stranded, and comprises 3,852 nt, which is 113 nt longer than the prototype isolate from Japan. Cesium chloride density gradient centrifugation demonstrated banding of the virus at 1.31–1.34 g/ml; filtration studies indicated that TTV had a particle size of 30–50 nm. These results suggest that the virus is similar to the Circoviridae, viruses known to infect plants and vertebrates (e.g., birds and swine); however, sequence similarity searches of available databases did not reveal identity between TTV and other viruses. Phylogenetic analyses of a 260-nt region from 151 globally distributed isolates demonstrated the existence of three major TTV genotypes. Several individuals at high risk for infection with parenterally transmitted viruses were infected with more than one genotype. There was no correlation between genotype and geographic origin. Finally, intravenous inoculation of TTV-positive human serum into chimpanzees demonstrated that TTV can be transmitted to primates; no biochemical or histological evidence for hepatitis was obtained. The distinct biophysical and molecular characteristics of TTV suggest that it is a member of a new family of viruses, which we have tentatively named the Circinoviridae.
Recently, a novel human DNA virus was isolated from the serum of a Japanese patient (initials T.T.) with cryptogenic hepatitis (1). TT virus (TTV) was detected by PCR in sera from three of five patients with non-A to GBV-C hepatitis. Subsequently, the nearly complete nucleotide sequence of the TTV genome, encompassing 3,739 bases, and a more sensitive PCR assay for the detection of virus in serum were reported (2). In addition, based on sensitivity to single-strand- but not double-strand-specific endonucleases, the virus appeared to possess a single-stranded DNA genome. Data presented regarding the size of the genome, its single-strandedness, and resistance to detergents suggested that TTV was similar to the parvoviruses (2). However, the buoyant density in CsCl (1.31–1.32 g/ml) was lighter than that reported for the parvoviruses. Further studies (3) demonstrated that TTV was excreted in the feces of infected humans and the buoyant density of virus present in fecal extracts was similar to virus in serum. The presence of TTV in feces could explain the rather high prevalence of TTV infection, as determined by PCR, in healthy blood donors in Japan and the United States (3, 4). Based on limited prevalence studies (5–7), the association between TTV infection and human hepatitis is questionable.
In an effort to further characterize TTV, we repeated and extended the studies of Nishizawa et al. (1) and Okamoto et al. (2) through biophysical characterization of the virus, as well as cloning and sequencing of the complete viral genome. Our data confirm the single-strandedness of the DNA genome and strongly suggest that it is circular and not linear, as previously believed. In addition, nuclease protection assays with strand-specific probes suggest that TTV possesses a negative-stranded genome. Sequence analysis of a 260-nt region within the largest ORF revealed up to 44% nucleotide sequence divergence among 151 globally distributed TTV isolates. Phylogenetic analysis of this region revealed three major TTV genotypes with no geographic correlation. Comparison of the genomic sequence, its encoded proteins, and the biophysical characteristics of the virus suggests that TTV is most closely related to the Circoviridae.
MATERIALS AND METHODS
Human Serum.
Human sera known to be positive for TTV DNA (4) were used as the virus source for further study. Other TTV isolates were obtained from patients from Argentina (8) and children from New Zealand (9). Serum from a TTV-positive West African individual (10) was used as the source of virus for cloning and sequencing of the genome and for filtration experiments.
Filtration.
TTV-positive serum (50 μl) combined with parvovirus B19-containing human serum (106 virus particles) was used for filtration studies to determine the approximate size of the putative virion. The sample was diluted to 1.0 ml with PBS and spun for 10 min at 12,000 × g at 4°C. The supernatant was passed sequentially through 13-mm polycarbonate filters (Costar) with decreasing pore sizes of 200, 100, 50, 30, and 15 nm. PBS (100 μl) containing 0.1 mg/ml BSA was passed through all filters before use. Aliquots (100 μl) of unfiltered serum and the resulting filtrates were extracted for total nucleic acid with a DNA/RNA isolation kit (United States Biochemical) as directed by the manufacturer. TTV and B19 sequences were detected by PCR (20 μl final reaction volume) with AmpliTaq Gold DNA polymerase (Perkin–Elmer) and 2 μl (20%) of each extracted sample. TTV primers were as described (set B, see ref. 6 and below), Parvovirus B19 primers were as follows: forward primer, 5′-GATGGTGCAAAC(T/C)TTTGCCTCC-3′; reverse primer, 5′-GCATGACTTCAGTTAATTCTGCA-3′. Reactions were heated for 8 min at 94°C, followed by 40 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 30 s, and then a 3-min final extension at 72°C. The second round of amplification was performed as described above with an aliquot of the first-round products and either the same primers (B19) or nested primers (TTV). Products were analyzed by agarose gel electrophoresis with visualization by ethidium bromide fluorescence.
CsCl Density Gradient Centrifugation.
Human sera containing TTV (200 μl) or parvovirus B19 (20 μl, approximately 2 ng of B19 DNA) were mixed and centrifuged at 14,000 × g for 15 min at 4°C. Supernatants were combined and mixed with 11.5 ml of CsCl (1.302 g/ml). Isopycnic gradients were formed by centrifugation in a Beckman SW41Ti rotor at 35,000 rpm (150,000 × g) for 65 h at 6°C. Fractions (≈800 μl) were collected from the bottom of each gradient, refractive indices were measured to determine the density, and 200 μl of each fraction was extracted for total nucleic acids with the High Pure Viral RNA kit (Boehringer Mannheim). One-tenth of the isolated nucleic acids was tested for TTV or B19 by PCR with AmpliTaq Gold (Perkin–Elmer) as directed by the manufacturer. For the PCR reactions (20 μl), 1 μM primers were used (TTV, 5′-AGACAGAGGAGAAGGCAACA-3′ and 5′-GACCAAAACATACACATGAA-3′; B19, 5′-GTAAGCGGGAACACTACAAC-3′ and 5′-CGGAGGAAACTGGGCTTCCG-3′). Reactions were thermocycled (94°C, 9 min; 40 cycles of 94°C for 20 s, 55°C for 30 s, 72°C for 30 s; final extension at 72°C for 10 min), and 10 μl from each reaction was separated by agarose gel electrophoresis, capillary transferred to Hybond-N+ (Amersham), and visualized via Southern hybridization with an amplicon-specific 32P-labeled DNA probe.
Animal Transmission Studies.
Nonhuman primate transmission studies were conducted at the Southwest Foundation for Biomedical Research in San Antonio, TX. All animals were maintained and monitored according to protocols that met all relevant requirements for the humane care and ethical use of primates in an approved facility. Baseline serum levels were established for the liver-specific enzymes alanine aminotransferase, γ-glutamyltransferase, and aspartate aminotransferase. Animals were inoculated i.v. with TTV-containing human serum or plasma from individuals with the diagnosis of chronic non-A–E hepatitis and then monitored twice weekly for serum levels of the above-mentioned liver-specific enzymes. Chimpanzee 314 (CH314) was inoculated with 20 ml of human plasma from patient A and chimpanzee 306 (CH306) was inoculated with 2.0 ml of human serum from patient B.
TTV viremia was determined by nested PCR as described (see ref. 6, primer set B), except that a modified A5427 primer was used in the first-round amplification (A5427m, 5′-TACCAYTTAGCTCTCATTCTWA-3′). Total nucleic acids extracted from 100 μl of chimpanzee serum were used for PCR with the QIAamp blood kit (Qiagen, Chatsworth, CA) as directed by the manufacturer. Nucleic acids were ethanol precipitated and suspended in 25 μl of water. For first-round PCR, 4 μl of the extracted nucleic acids was used in a 20-μl reaction volume. Cycling conditions for first-round (45 cycles) and second-round (40 cycles) amplification were 94°C for 1 min followed by 94°C for 20 s, 55°C for 30 s, and 72°C for 30 s; final extension was at 72°C for 5 min. Products were analyzed by agarose gel electrophoresis with visualization by ethidium bromide fluorescence. The approximate titers of TTV in the human inocula were determined by making serial 2-fold dilutions of extracted nucleic acids and performing PCR as described above.
Fecal material was suspended in PBS (15%, wt/vol), vortexed, and centrifuged at 3,000 × g at 4°C for 10 min. The supernatant was transferred to a clean tube and centrifuged at 8,000 × g at 4°C for 5 min. Nucleic acids were extracted from 200 μl of the resulting supernatant with the QIAamp blood kit. Nucleic acids were ethanol precipitated and resuspended in 15 μl of water. TTV PCR was performed as described above.
Genomic Extension.
To obtain the genomic sequence, total nucleic acids were extracted from a West African individual (GH1) by using a DNA/RNA isolation kit (United States Biochemical) as recommended by the manufacturer. Initial anchored PCR extension products were generated up- and downstream of the N22 clone region (1) by anchored PCR (11, 12). The TTV-specific primers used to obtain sequences upstream of the N22 region were N22-A1 [2,132–2,109; numbering system of prototype isolate reported by Nishizawa et al. (1); 5′-GGGTCTGTGTGTACTAAGAGTTGG-3′] and N22-A2 (2,083–2,061; 5′-AAAGTCTGGCATTCATGTGTATG-3′). The anchored primers used to obtain sequences downstream of the N22 region were N22-S1 (2,181–2,203; 5′-GCCAGGAGGTAGCAGCAATGTGC-3′) and N22-S2 (2,204–2,230; 5′-CTATTAGAATGAGAGCTAAGTGGTACC-3′). To test the possibility that the TTV genome is circular, inverted PCR with nested primers derived from the anchored PCR products was performed with Takara LA TAQ (Panvera, Madison, WI) as described by the manufacturer with the following primers: UFGH1-A1 (418–394; 5′-GCTAAGTACACTTGAGTACCATTGC-3′), UFGH1-A2 (386–362; 5′-GGTACTGTTGGTCATTGCGAGGTGG-3′), DFGH1-S1 (2,955–2,978; 5′-CTGGAAGGAAGAGTATGAGGCCTG-3′), and DFGH1-S2 (2,987–3,011; 5′-CCCTAGAGGCAATCTAAGAGACACC-3′.
The circular nature of the virus was confirmed by nested genome-length PCR by using primers derived from the N22 region and Takara LA TAQ (first round: UFTTV1, 5′-AGCCTTTTGTGGGGTCTGTGTGTACTA-3′, and DFTTV1, 5′-TGGAAATGGTAAAATGCCAGGAGGTAG-3′; second round: UFTTV2, 5′-GTCTGGCATTCATGTGTATGTTTTGGTC-3′, and DFTTV2, 5′-GCAGCAATGTGCCTATTAGAATGAGAGC-3′). All PCR products were cloned into pGEM-T EASY vector (Promega), and two to four clones were sequenced. Sequencing reactions were performed with ABI Big Dye or Prism dGTP Big Dye (Applied Biosystems-Perkin-Elmer). Reactions were electrophoresed under denaturing conditions, and sequence data were collected on the Applied Biosystems 377 DNA automated sequencer as directed by the manufacturer. Sequences were compiled and edited with sequencher, version 3.0 (GeneCodes), and analyzed with the programs of the Wisconsin Sequence Analysis Package, version 9.0. The genomic sequence TTV-GH1 was submitted to GenBank (accession no. AF122913).
Genome Polarity.
To establish the polarity of the TTV genome, a hybridization/nuclease protection assay (13) was performed with serum total nucleic acids containing TTV DNA and strand-specific RNA run-off transcripts made from plasmids containing identical TTV sequences but in opposite orientations. Reduction of template plasmid DNA concentration to below detectable limits was achieved by repeated digestion with DNase I and organic extraction with TRIzol reagent (GIBCO/BRL). Plus- or minus-strand, single-strand phagemid DNAs made from the same plasmids were used for control experiments.
Total nucleic acid was extracted as described above from TTV-positive human serum (100 μl) with a sequence identical to that of the cloned TTV sequence over the region to be analyzed, and it was resuspended in 40 μl of water. Plus- or minus-strand RNA transcripts (2 ng, 1010 copies) were mixed, in separate reactions, with (i) plus- or minus-strand phagemid DNA (300 copies), (ii) 10 μl of the extracted nucleic acids, or (iii) water without DNA. The samples were dried under vacuum, dissolved in 8 μl of 30 mM EPPS, pH 8.1 [N-(2-hydroxyethyl)-piperazine-N′-(3-propanesulfonic acid)], containing 3 mM EDTA, overlaid with mineral oil, and heated for 3 min at 99°C. After 2 μl of NaCl (5 M) was added, the samples were hybridized at 67°C for 21 h. One-half (5 μl) of each hybridization was added to 45 μl of buffer (33.3 mM sodium acetate, pH 5.2/1.44 mM ZnSO4/5.5% glycerol), with or without mung bean nuclease (6 units per reaction), and incubated for 30 min at 30°C. The nuclease was inactivated by adding 6 μl of 467 mM Tris⋅HCl, pH 8.9, and 14 mM EDTA and heating for 5 min at 99°C. Nucleic acid was ethanol precipitated and resuspended in 20 μl of water. Each sample (4 μl) was tested for the presence of TTV sequences by nested PCR (20 μl), followed by agarose gel electrophoresis as described above. The first-round primers were A8761 (6) and A1 (5′-CCTGGCATCTTTCCATTTCCAAAG-3′); the second-round primers were S2 (5′-GACTGGCTAACTAAAGATACCTCAG-3′) and A2 (5′-TCCAAAGTTAAAACTGTAGGGTACG-3′). These primers are specific for, and contained within, the cloned TTV region from which the run-off transcripts and phagemid were made.
Phylogenetic Analysis.
PCR products from 151 TTV DNA-positive individuals (4) were chosen for sequence analysis. PCR products were generated by using primers described earlier (6) by a heminested PCR method: primers A5430 and A5427m (see above) were used for first-round amplification, followed by A8761 and A5427m for second-round amplification, applying the cycling method (4). Products were separated by agarose gel electrophoresis and gel purified with the Qiaex II gel extraction kit (Qiagen). Purified products were sequenced directly (see above). Those products that yielded uninterpretable sequences (significant degree of ambiguities) were cloned into pGEM-T EASY (Promega), and at least six clones of each PCR product were sequenced (see above).
The TTV sequences determined in this study and those deposited in GenBank that overlapped the amplified region were aligned by using the program pileup (Wisconsin Package, version 9.0). PCR primer sequences were not included in the sequence alignment. The final alignment of 163 sequences (260 nt in length) was used to determine the evolutionary relationship between isolates by using the programs of the phylip package, version 3.5c (1993, distributed by J. Felsenstein, Department of Genetics, Univ. of Washington, Seattle). Nucleotide sequence distances were determined with dnadist. Amino acid sequence distances were determined with protdist; calculated distances were then used by neighbor to generate unrooted trees. The program retree with the midpoint rooting option was used to plot the trees. Bootstrap values were determined on 100 resamplings of amino acid sequences and 1,000 resamplings of nucleotide sequences applying seqboot, dnadist for nucleotide sequences or protdist for amino acid sequences, neighbor, and finally consense to generate the majority rule consensus tree. Bootstrap values greater than 70% were considered supportive of the observed groupings. The final trees were visualized with treeview (14).
RESULTS
Estimation of TTV Particle Size.
To estimate the particle size of TTV, virus-containing serum was successively passed through filters of decreasing pore size, and then TTV was detected by PCR. As controls for filterability, parvovirus B19-containing serum was included. As expected, parvovirus B19 particles (a nonenveloped, single-stranded DNA virus with a reported diameter of 18–22 nm) were detected in the 200-, 100-, 50-, and 30-nm filtrates but not in the 15-nm filtrate. TTV was detected in the 200-, 100-, and 50-nm filtrates but not in the 30- or 15-nm filtrates. Thus, TTV virions appear to exist in serum with a particle diameter between 30 and 50 nm.
Determination of TTV Buoyant Density.
Isopycnic CsCl gradients of two TTV-positive sera were examined to determine the buoyant density of virus particles. Parvovirus B19 was included as an internal control. PCR analysis of the gradient fractions located TTV in fractions with a density of 1.31–1.34 g/ml (data not shown). This value is similar to the CsCl buoyant density reported by Okamoto et al. (2). In contrast, parvovirus B19 was found in fractions with a density of 1.38–1.51 g/ml (data not shown). Thus, TTV possesses a buoyant density significantly lighter than parvovirus B19.
Transmission of TTV to Nonhuman Primates.
Several studies have observed a high prevalence of TTV in individuals at risk for infection with parenterally transmitted viruses, suggesting that TTV can be transmitted by blood and/or blood products. However, there have been no cases of TTV transmission reported in the literature to date. To investigate whether TTV is a transmissible agent, and whether it can be transmitted parenterally, serum or plasma from two patients with chronic non-A-GBV-C hepatitis known to be infected with TTV were inoculated i.v. into chimpanzees. Plasma (20 ml) from one patient (approximate TTV titer 2 × 103 genome copies per ml) was inoculated into chimpanzee 314 (CH314). Serum (2 ml) from a second patient (approximate TTV titer 1 × 103 genome copies per ml) was inoculated into chimpanzee 306 (CH306). TTV DNA was detected in CH314 serum for 28 days starting 93 days postinoculation (PI). The duration of the viremia is unclear because samples between 121 and 226 days PI were not available. However, sera collected after 226 days were negative. TTV DNA was detected in CH306 serum starting at 149 days PI and remained positive until 219 days PI, at which time viral DNA became undetectable. The later appearance of TTV viremia in CH306, compared with CH314 (149 vs. 93 days PI), may be because of the lower volume and titer of the inoculum used. Nucleic acids extracted from CH306 fecal samples spanning 133–175 days PI were tested for TTV via nested PCR; however, TTV DNA was not detected. Thus, TTV is (i) not shed in the feces of CH306, (ii) not present in the feces during the time tested, or (iii) below the limit of detection. TTV sequences present in the human inocula and in the corresponding chimpanzee recipients were found to be 100% identical, whereas the sequences of the TTV PCR products from the two human inocula were only 91% identical (data not shown). The complete conservation of TTV sequences between source and recipient indicates that TTV infection was derived from its corresponding inoculum, thus demonstrating the infectious nature of the inoculum and the parenteral transmissibility of the virus. Neither chimpanzee exhibited any biochemical or histological evidence of hepatitis.
Determination of TTV Genome Polarity.
The single-stranded nature of the TTV genome was determined by sensitivity to mung bean nuclease digestion (data not shown) as reported (2). Polarity of the genome was determined by a hybridization/nuclease protection assay. TTV DNA extracted from human serum was hybridized with excess strand-specific RNA probes, and the hybridization products were digested with single-strand-specific mung bean nuclease. The formation of nuclease-resistant DNA:RNA hybrids was determined by PCR analysis. In the absence of nuclease, TTV was always detected except in the RNA-only hybridizations (data not shown). In the presence of nuclease, however, TTV viral DNA was detected only in hybridizations containing plus-strand RNA. The TTV-containing phagemid DNA controls were detected only when the hybridizations contained the opposite-strand RNA. These results strongly suggest that TTV is a negative-stranded DNA virus.
Genomic Extension and Organization.
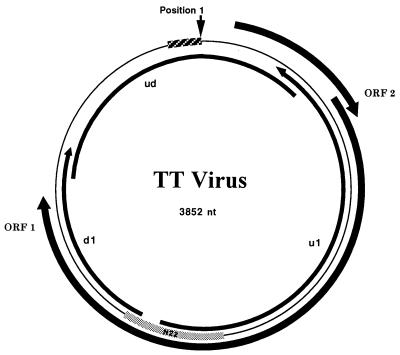
To obtain the genome length sequence of TTV, anchored PCR was performed on nucleic acids extracted from the serum of the West African individual (GH1). Amplified genome fragments were generated upstream (1,766 bp) and downstream (882 bp) from the N22 region of TTV-TA278 (1). Assuming that the TTV genome was circular, inverted PCR with upstream antisense primers and downstream sense primers was performed, generating a 1,300-bp product representing the remainder of the genome. The circular structure of the viral genome was reproducibly confirmed by nested genome-length PCR originating from the N22 region that produced the expected product of approximately 3,700 bp. The genomic sequence of this isolate (designated GH1) comprises 3852 nt (Fig. 1), 113 nt longer than that of TTV-TA278 (2). This additional sequence, located at the extreme 3′ end of the linear TA278 sequence (positions 3,740–3,852), consists of 89% G or C residues and possesses multiple inverted repeats. Alignment of TTV-GH1 and TTV-TA278 reveals 93% identity across the entire genome. The region with the lowest degree of conservation lies between bases 1,440 and 1,827 and exhibits only 73.6% identity. The region of highest identity lies between bases 2,240 and 2,911, exhibiting 99.5% identity. Both TTV-TA278 (2) and TTV-GH1 encode two large ORFs of 203 and 770 aa (Fig. 1) exhibiting 95% and 96% identity between the isolates, respectively. The ORF1 protein of both isolates possesses an arginine-rich region at its amino terminus (44 of the first 82 aa). The ORF1 regions from amino acids 1–274 exhibit essentially 100% identity, but positions 275–405 exhibit only 69% identity. The remainder of the ORF1 protein is essentially 100% conserved between the two isolates. No significant identity with non-TTV sequences were obtained when TTV-GH1 was analyzed by blast against the GenBank (Release 110.0) or SwisProt (Release 36.0) databases.
Figure 1.
TTV circular genome (thin line) with ORFs 1 and 2 indicated. Anchored PCR extension clones extending upstream and downstream from the N22 clone sequence (gray box) described by Nishizawa et al. (1) are indicated by the arrows inside the circular genome. The inverse PCR product (ud) that overlaps the anchored PCR products is also shown. The 113-nt sequence identified in the GH1 isolate is indicated by the hatched box. Position 1 is indicated by the arrow.
Phylogenetic Analysis.
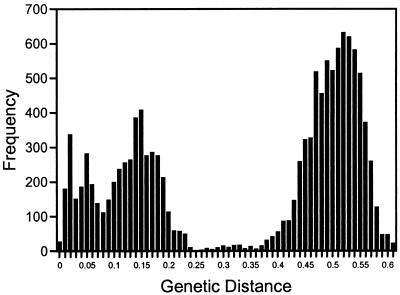
To determine the degree of sequence variability of TTV, we examined a 260-bp region of the genome (excluding primer sequences) amplified from 151 globally distributed individuals. The region analyzed (1,939–2,198) overlaps the N22 clone sequence (1), which is not within the hypervariable region of the genome described above (1,440–1,827). Of the 151 PCR products directly sequenced, 54 yielded more than 10% nucleotide sequence ambiguities, suggesting mixed infections. This was confirmed by cloning and sequencing of PCR products from 12 of the 54 individuals, revealing up to 36% DNA sequence variability among multiple sequences cloned from a single individual. The sequences we determined were aligned with 36 TTV sequences deposited in GenBank, including representatives of the putative genotypes 1a, 1b, 2a, and 2b (2, 6). The pairwise genetic distances calculated for all 163 aligned sequences clearly show two tiers of sequence diversity (Fig. 2), suggesting the existence of distinct genetic groups or genotypes. There was a high degree of variability, up to 0.62 substitutions per position (or an uncorrected distance of 44%), within this putative coding region of the virus. In general, pairwise distances of less than 0.26 represent intragroup distances and values greater than 0.34 represent intergroup distances.
Figure 2.
Distribution of the pairwise genetic distances observed between 157 TTV nucleotide sequences. Genetic distances are on the x axis; the frequency of occurrence of discrete distances is on the y axis.
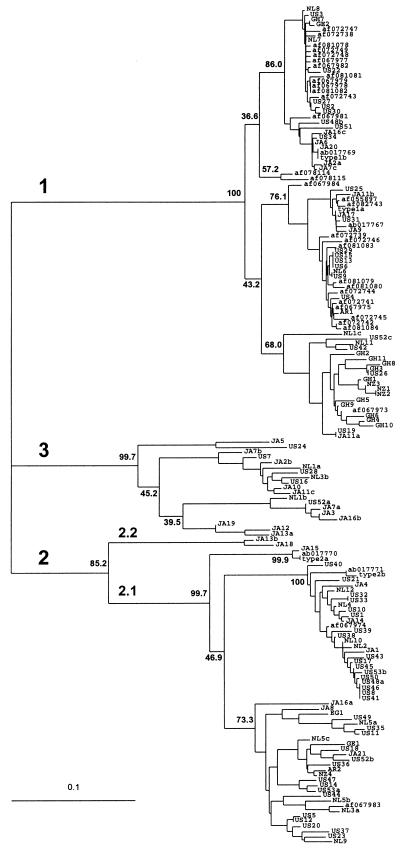
These pairwise distances were used to generate an unrooted phylogenetic tree (Fig. 3). Three major groups were observed with approximately equal divergence from each other. These major groups were strongly supported by bootstrap analysis: group 1 or 3 sequences associated in nearly 100% of the trees, whereas group 2 sequences grouped in 85% of the trees. Although subgroups 1a and 1b have been described (2, 6), and the branching order of group 1 sequences suggests the presence of at least two subgroups, the associated bootstrap values are less than 44%. Thus, we found no support for group 1 subtypes. Based on pairwise distances, the two Japanese sequences constituting subtype 2.2 (Fig. 3) are more closely related to subtype 2.1 isolates than those in group 1 or 3 (Table 1) and, thus, have been segregated into a subtype of group 2 rather into their own major group. There is strong bootstrap support for the existence of these subgroups within group 2, each with at least 85% support. Subtypes 2.1 and 2.2 do not correspond to the previously reported 2a and 2b subgroups (2, 6), the latter of which, based on bootstrap analysis, did not segregate (46.4%). Support for subgroups among genotype 3 sequences was not obtained. Finally, each of the three groups contained isolates from around the world, demonstrating no clear correlation between genotype and country of origin.
Figure 3.
Consensus phylogenetic tree (unrooted) of 260 nt (amplification primers removed) from 163 TTV isolates. Branch lengths are proportional to the genetic distance (scale shown). Isolates representing the subtypes (1a, 1b, 2a, and 2b) proposed by Okamoto et al. (2) are labeled. GenBank accession numbers are given where appropriate. Geographical designations: AR, Argentina; EG, Egypt; GE, Greece; GH, Ghana; JA, Japan; NL, The Netherlands; NZ, New Zealand; US, United States. Sequences isolated from a single individual are designated with the isolate number followed by the lower case letters a, b, or c. Bootstrap values are shown at the nodes for 1,000 resamplings of the data. Genetic groups are indicated as genotypes 1, 2, and 3 and subtypes 2.1 and 2.2.
Table 1.
The range of genetic distances observed between and within groups for which significant bootstrap support was obtained
| 1 | 2.1 | 2.2 | 3 | |||
|---|---|---|---|---|---|---|
| 1 | 0–0.18 | 0–0.18 | 0.41–0.62 | 0.42–0.52 | 0.39–0.55 | |
| 2.1 | 0.46–0.61 | 0–0.17 | 0–0.23 | 0.26–0.36 | 0.34–0.54 | |
| 2.2 | 0.35–0.48 | 0.16–0.25 | 0.06 | 0.03 | 0.36–0.47 | |
| 3 | 0.43–0.63 | 0.33–0.52 | 0.34–0.41 | 0–0.22 | 0.02–0.26 | |
| 1 | 2.1 | 2.2 | 3 | |||
DNA sequence distances are shown at the upper right and amino acid sequence distances at the lower left.
Given the high degree of nucleotide sequence variability within the TTV genome region analyzed and our inability to obtain bootstrap support for subtypes, it is possible that the true phylogenetic relationships are obscured by a high rate of synonymous substitutions. To examine this possibility, phylogenetic analysis was performed on the deduced amino acid sequences of the TTV isolates. The segregation of amino acid sequences into three major groups was supported in 96% of the trees by bootstrap analysis (data not shown). As with nucleotide sequence analysis, there was no significant bootstrap support for group 1 or 3 subtypes, although support was obtained for two group 2 subtypes in 89% of the trees. As shown for the nucleotide sequences, pairwise amino acid sequences indicate that the two subtype 2.2 sequences are more closely related to subtype 2.1 isolates than those in groups 1 or 3 (Table 1).
DISCUSSION
Previous interpretation of biophysical and molecular data suggested that TTV resembled members of the Parvoviridae (2, 3). In the present study, we have confirmed that TTV possesses a single-stranded DNA genome, consistent with the Parvoviridae. However, the buoyant density in CsCl of TTV (1.31–1.34 g/cm3) and its particle size determined by filtration (30–50 nm) are not like those of other parvoviruses (1.39–1.42 g/cm3 and 18–22 nm). Most notably, the TTV genome was found to be circular, not linear as previously reported (2). This was demonstrated by inverse PCR and primers located at the termini of anchored PCR products located up- and downstream of the original N22 sequence to generate an amplicon of about 1,300 bp (Fig. 1). Had the genome been linear, no amplicon would have been produced. Furthermore, inverse PCR with primers derived from the N22 region were able to produce a 3,700-bp product encompassing nearly the entire genome, including those sequences originally believed to be at the 5′ and 3′ termini. Similar products have been generated from several other TTV-positive samples (data not shown). The genome sequence of this TTV isolate, designated GH1, was found to be 3,852 nt long, which is 113 nt longer than previously reported (2). The newly discovered region is GC-rich (89%) and contains several potential stem-loop structures. Amplification of this region was possible only when contained within PCR products greater than 700 bp. These findings may explain the failure of previous attempts to demonstrate the circular nature of the genome that used inverse PCR with primers located near the presumed termini (2).
Other than its single-stranded genome and lack of an envelope, TTV does not share any other characteristics of the Parvoviridae. TTV does share some attributes of the Circoviridae, the members of which include chicken anemia virus (CAV), psittacine beak and feather disease virus, and porcine circovirus (15). Circoviruses are nonenveloped, 15–22 nm in diameter, and band in CsCl at 1.33–1.37 g/ml. Their genomes make up a single molecule of circular, single-stranded DNA, 1.7–2.3 kb long and with either positive or ambisense polarity (15, 16). The TTV genome is approximately 4 kbp long and, based on nuclease/hybridization protection assays, appears to encapsidate the negative strand with respect to the ORF1 gene that is encoded on the complementary, or positive strand (Fig. 1). Although the particle size and circular DNA genome of TTV are larger than that reported for the Circoviridae, TTV and circoviruses possess similar densities in CsCl, suggesting a similar protein to DNA ratio.
Nucleotide and amino acid sequence database searches failed to identify significant sequence similarity between TTV-GH1 and other viruses, as has been reported (2). The similarities in genome structure and composition between TTV and the Circoviridae prompted a more detailed comparison. Circoviruses contain stem-loop structures essential for DNA replication in which the loop possesses a nonanucleotide motif conserved among plant and animal circoviruses. However, CAV is an exception in that the nonanucleotide motif is semiconserved but is not associated with a stem-loop structure (16). This motif was not identified in TTV. In TTV, the three largest stem-loop structures identified lie outside the ORF1- and ORF2-coding regions, and two of these stem-loops are located within the 113-nt region cloned from TTV-GH1.
TTV encodes two large (202 and 770 aa) and several small ORFs (33–105 aa). The circoviruses encode up to seven ORFs (17), including the Rep protein, involved in rolling circle replication (16). The Rep protein possesses up to four amino acid sequence motifs conserved among many plant and animal circoviruses and bacteriophage ΦX-174 (16). Conserved motifs 1 (FTL) and 3 (YXXK) were identified in ORF1 of TTV-GH1. The active site tyrosine in motif 3 was conserved in the ORF1 proteins of TTV-GH1 and TTV-TA278. Motif 4, or the P-loop (putative ATP/GTP-binding motif), was not found. This motif is also absent in the putative Rep protein (encoded by ORF1) of CAV. The capsid or coat proteins of most circoviruses are encoded by separate genes and are highly basic (rich in arginine or lysine) (16). CAV is the exception, however, in that its ORF1 protein contains a highly basic amino terminus and also possesses three of the four conserved Rep protein motifs closer to the carboxyl end. ORF1 of TTV encodes 44 arginine residues of the first 100 aa and, toward the carboxyl end of ORF1, possesses two of the four conserved Rep protein motifs. Thus, TTV ORF1 appears to resemble the CAV ORF1 protein (16), and the presence of these conserved features in TTV ORF1 suggests that TTV may replicate by a rolling circle mechanism. However, until viral transcripts and their encoded gene products are identified, the actual coding regions of TTV and their function will be difficult to determine with certainty.
Phylogenetic analysis with bootstrapping provided strong support for the existence of three major groups of TTV sequences exhibiting approximately equal divergence. Others have suggested the presence of TTV subtypes (2, 6), but we did not obtain bootstrap support for segregation of the previously reported 1a and 1b sequences, or any of the other group 1 sequences we determined, into subgroups. This was also true for the purported 2a and 2b genotypes. Attempts to eliminate the potential obfuscation of subtypes because of the high rate of synonymous substitutions through analysis of only first- and second-codon positions or deduced amino acid sequences were not successful in obtaining support for subtypes. It remains possible, however, that the region analyzed is insufficient for subtype identification because of its short length (260 nt) and/or high variability. Verification of our results will require the analysis of longer genomic segment, or ideally, full-length genome sequences.
Sequence analysis performed on 30 cloned TTV sequences from 12 individuals demonstrated that 10 individuals were infected with two different TTV genotypes and two individuals were infected with representatives of all three genotypes (Fig. 3). These mixed infections occurred in individuals at high risk for infection with parenterally transmitted viruses, such as i.v. drug users, hemophiliacs, and patients with non-A–E hepatitis. In contrast, only one of 36 TTV-positive U.S. donors was coinfected. It remains to be determined whether this observation is the result of repeated infection with variant genotypes, rapid mutation of the virus within the individual, or some other mechanism.
TTV-GH1 and TA278 are 93% identical across the entire genome, but local regions of lower or higher identity exist. These two isolates exhibit 92% identity within the 260-base region analyzed for phylogenetic relationships. Genetic divergence in this region among the globally distributed isolates examined was up to 44% at the nucleotide level and 36% at the amino acid level. This degree of variability among geographically remote isolates has not been previously observed for circoviruses. Comparison of the four CAV genome sequences present in GenBank revealed 4% maximum diversity. Analysis of 11 CAV VP1 sequences from GenBank revealed only 1.5% sequence divergence (data not shown). Similar results are obtained when porcine circovirus sequences are compared (18). A recent report described a hypervariable region within ORF1 of CAV spanning 13 aa (up to 38% divergence) based on comparison of eight isolates (19). In contrast, the ORF1 proteins of TTV-GH1 and TA278 exhibit 5% divergence but also contain a hypervariable region spanning 126 aa with 31% divergence. Thus, TTV exhibits much greater variability than CAV or porcine circovirus.
From the data presented here, it is clear that TTV cannot be classified within an existing virus family. The circular nature of the genomic DNA, in addition to the virion size, buoyant density, and lack of sequence identity, precludes its membership among the Parvoviridae. However, by virtue of its negative-stranded, circular DNA genome, TTV is most closely related to the Circoviridae, although TTV possesses a larger genome and viral particle relative to members of this family. Furthermore, the absence of significant sequence similarities between TTV and circoviruses, beyond the possible conservation of motifs involved in rolling circle replication, does not support inclusion of TTV in the Circoviridae. Therefore, we propose that TTV is a member of a new virus family that infects humans, tentatively named the Circinoviridae, derived from the Latin circinatio meaning “the describing of a circle.”
ABBREVIATIONS
- TTV
virus isolated from Japanese patient T.T
- PI
postinoculation
- CAV
chicken anemia virus
Footnotes
References
- 1.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 3.Okamoto H, Akahane Y, Ukita M, Fukada M, Tsuda F, Miyakawa Y, Mayumi M. J Med Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- 4.Desai, S. M., Muerhoff, A. S., Leary, T. P., Erker, J. C., Simons, J. N., Chalmers, M. L., Birkenmeyer, L. G., Pilot-Matias, T. & Mushahwar, I. K. (1999) J. Infect. Dis., in press. [DOI] [PubMed]
- 5.Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Weisner R. Hepatology. 1998;28:839–842. doi: 10.1002/hep.510280335. [DOI] [PubMed] [Google Scholar]
- 6.Simmonds P, Davidson F, Lycett C, Prescott L E, MacDonald D M, Ellender J, Yap P L, Ludlam C A, Haydon G H, Gillon J, Jarvis L M. Lancet. 1998;352:191–194. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 7.Naoumov N V, Pertova E P, Thomas M G, Williams R. Lancet. 1998;352:195–197. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 8.Frider B, Sookoian S, Castano G, Gonzalez J, Flichman D, Viudez P, Dawson G J, Schlauder G G, Mushahwar I K. J Viral Hepatitis. 1998;5:161–164. doi: 10.1046/j.1365-2893.1998.00094.x. [DOI] [PubMed] [Google Scholar]
- 9.Tobias M I, Miller J, Mushahwar I K. N Z Med J. 1986;99:488–490. [PubMed] [Google Scholar]
- 10.Martinson F E A, Weigle K A, Mushahwar I K, Weber D J, Royce R, Lemon S M. J Med Virol. 1996;48:278–283. doi: 10.1002/(SICI)1096-9071(199603)48:3<278::AID-JMV11>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 11.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M C, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 12.Sorensen A B, Duch M, Jorgensen P, Pedersen F S. J Virol. 1993;67:7118–7124. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Page R D M. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 15.Lukert P D, Boer G F d, Dale J L, Keese P, McNulty M S, Randles J W, Tisher I. The Circoviridae. Vienna: Springer; 1995. [Google Scholar]
- 16.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 17.Bassami M R, Berryman D, Wilcox G E, Raidal S R. Virology. 1998;249:435–459. doi: 10.1006/viro.1998.9324. [DOI] [PubMed] [Google Scholar]
- 18.Meehan B M, McNeilly F, Todd D, Kennedy S, Jewhurst V A, Ellis J A, Hassard L E, Clark E G, Haines D M, Allan G M. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 19.Renshaw R W, Soine C, Weinkle T, O’Connell P H, Ohashi K, Watson S, Lucio B, Harrington S, Schat K A. J Virol. 1996;70:8872–8878. doi: 10.1128/jvi.70.12.8872-8878.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]