Abstract
The expression of several virulence factors of Vibrio cholerae is coordinately regulated by the ToxT molecule and the membrane proteins TcpP/H and ToxR/S, which are required for toxT transcription. To identify proteins that negatively affect toxT transcription, we screened transposon mutants of V. cholerae carrying a chromosomally integrated toxT∷lacZ reporter construct for darker blue colonies on media containing 5-bromo-4-chlor-3-indolyl β-d galactoside (X-gal). Two mutants had transposon insertions in a region homologous to the nqr gene cluster of Vibrio alginolyticus, encoding a sodium-translocating NADH–ubiquinone oxidoreductase (NQR). In V. alginolyticus, NQR is a respiration-linked Na+ extrusion pump generating a sodium motive force that can be used for solute import, ATP synthesis, and flagella rotation. Inhibition of NQR enzyme function in V. cholerae by the specific inhibitor 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO) resulted in elevated toxT∷lacZ activity. Increased toxT∷lacZ expression in an nqr mutant strain compared with the parental strain was observed when the TcpP/H molecules alone were strongly expressed, suggesting that the negative effect of the NQR complex on toxT transcription is mediated through TcpP/H. However, the ability of the TcpP/H proteins to activate the toxT∷lacZ reporter construct was greatly diminished in the presence of high NaCl concentrations in the growth medium. The flagellar motor of V. cholerae appears to be driven by a sodium motive force, and modulation of flagella rotation by inhibitory drugs, high media viscosity, or specific mutations resulted in increases of toxT∷lacZ expression. Thus, the regulation of the main virulence factors of V. cholerae appears to be modulated by endogenous and exogenous sodium levels in a complex way.
Vibrio cholerae is a Gram-negative bacterium that causes the diarrheal disease cholera. To establish infection and cause disease, V. cholerae must express a variety of virulence factors, including cholera toxin (CT), and colonization factors such as the toxin coregulated pilus (TCP). Expression of CT and TCP is coordinately regulated and strongly influenced by environmental stimuli (1). Transcription of the genes encoding these virulence factors is controlled by a regulatory cascade in which ToxR and TcpP control expression of ToxT, a transcriptional activator that directly controls expression of several virulence genes (2–4). ToxR and TcpP are inner membrane proteins that contain cytoplasmic DNA-binding domains. The periplasmic domains of ToxR and TcpP are thought to interact with other transmembrane regulatory proteins, ToxS and TcpH, respectively, that stimulate their activities (4–7). ToxT is a cytosine arabinonucleoside-like transcriptional activator that activates transcription of several genes, including the ctx and tcp operons, encoding CT and TCP, respectively (3).
In the present study, we isolated several V. cholerae transposon mutants that showed increased expression of a chromosomal toxT∷lacZ reporter construct. Two of the isolated mutants had transposon insertions in a region with high homology to the Vibrio alginolyticus nqr gene cluster, which encodes a sodium-translocating NADH–ubiquinone oxidoreductase (NQR) (8, 9). We had previously isolated a mutant strain of V. cholerae with a transposon insertion in a nqr gene homolog by selecting for V. cholerae cells that produce TCP even when grown under noninducing growth conditions (4). Because overexpression of toxT can result in cells constitutively expressing TCP (10), this prompted further investigation of the role of the nqr gene homologs in the expression of virulence genes in V. cholerae.
MATERIALS AND METHODS
Strains, Plasmids, and Culture Conditions.
V. cholerae strain O395N1 toxT∷lacZ (4) was host for the tansposon mutagenesis. The transposon TnMar was introduced into V. cholerae on a suicide plasmid by conjugation with the Escherichia coli strain β2155 carrying pFD1 (11). Transposon-mutagenized V. cholerae cells were selected by plating dilutions of the conjugation mixture on streptomycin (100 μg/ml) and kanamycin (Kan; 50 μg/ml) containing media with 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-gal) at 20 μg/ml. Darker blue colonies were obtained by visually scoring and were tested for increased β-galactosidase activity compared with the parent strain after overnight growth in LB at 30°C. For noninducing culture conditions, the bacteria were grown at 30°C either in LB where the pH was increased to 8.5 by adding NaOH or in LB with various amounts of NaCl added. To increase media viscosity, a 10% polyvinylpyrrolidone (PVP-360) or 15% Ficoll solution was prepared in LB and dialyzed against LB. X-gal, monensin, carbonyl cyanide m-chlorophenylhydrazone (CCCP), and 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO) were purchased from Sigma. Phenamil was purchased from Research Biochemicals (Natick, MA).
Genetic Manipulations.
Chromosomal DNA was extracted from V. cholerae cells by using the Easy-DNA kit (Invitrogen). The genomic regions surrounding the transposon insertions were cloned into the suicide vector pCVD442 carrying the kan-resistance gene (pCVD442π) as described (12). Briefly, the plasmid pCVD442π was introduced into the transposon carrying V. cholerae strain by conjugation, selecting for ampicillin-resistant and Kan-resistant cells. This should result in the integration of the plasmid into the transposon insertion site via homologous recombination between the kan-resistance genes present on the plasmid and the transposon. Chromosomal DNA from these strains was prepared and digested with the restriction enzyme BglII, which does not cut in pCVD442π or in the transposon. The chromosomal digest was then diluted, ligated, and transformed into DH5αλpir E. coli cells. Plasmid DNA was prepared by using the Qiagen (Chatsworth, CA) Miniprep extraction kit and sequenced with primers specific to the transposon ends (11). The nonmotile mutants were generated by homologous recombination. The plasmid pKEK93 (13) was used to generate the flaA∷cat mutation. The motY and fliG genes were amplified in PCR reactions by using specific primers and cloned into the plasmid vector pCR2.1 (Invitrogen). Internal deletions were generated by using convenient restriction sites present in the genes, and the DNA was then subcloned into pCVD442. The mutated alleles of these genes were introduced into the chromosome of the O395N1 toxT∷lacZ strain following sucrose selection as described (14).
Motility Assays.
Motility phenotypes were assessed for swarm diameter following inoculation into 0.3% soft agar. Bacterial cells also were assayed for swimming speed under a dark-field microscope after the addition of various drugs.
Biochemical Assays.
β-Galactosidase activities were assayed as described (4, 15).
RESULTS
Identification of a Gene Cluster in V. cholerae with High Homology to Genes Encoding a Sodium-Translocating NQR from V. alginolyticus.
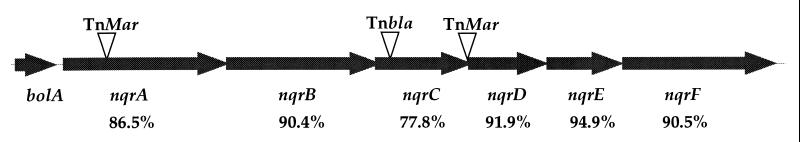
We recently reported the construction of a V. cholerae strain carrying a chromosomal toxT∷lacZ reporter construct (4). To identify genes involved in the negative regulation of toxT transcription, transposon mutagenesis of this strain was performed followed by screening for darker blue colonies on medium containing X-gal. Several mutants were isolated that exhibited increased β-galactosidase activity when compared with the parent strain. Sequencing of the DNA adjacent to the transposon insertion in these strains revealed regions with homology to both genes of known function and genes of unknown function (data not shown). Two mutant strains had transposon insertions in a chromosomal region with high homology to the nqr gene cluster from V. alginolyticus (Fig. 1). We had previously isolated a V. cholerae mutant with a Tnbla insertion in an nqr homolog, and this mutant displayed constitutive TCP expression (4). In V. alginolyticus, the nqr genes are part of a cluster of six genes encoding various subunits of a sodium-translocating NQR (8, 9), a respiration-driven Na+ pump that establishes an electrochemical gradient of sodium ions across the membrane (16). By using the partial genome sequences deposited by The Institute for Genomic Research (TIGR) and various primers, we confirmed that the nqr gene homologs are also linked in the V. cholerae chromosome (Fig. 1). The nqr gene regions from V. cholerae and V. alginolyticus showed high sequence homology; for example, the deduced amino acid sequences of the two NqrA proteins are 86.5% identical (Fig. 1).
Figure 1.
Chromosomal region of the V. cholerae nqr gene cluster. The positions of different transposon insertions are indicated. The amino acid sequence homologies of the various Nqr proteins from V. cholerae and V. alginolyticus are shown below.
Effects of Loss of NQR Activity on toxT∷lacZ expression in V. cholerae.
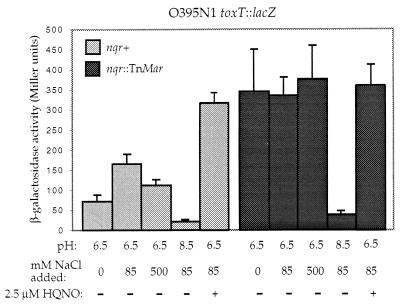
The nqr mutant strains were isolated as colonies that appeared darker blue on X-gal-containing medium compared with the parental V. cholerae strain, and the mutant strains produced ≈2-fold more β-galactosidase activity than the parent strain when assayed in liquid media (Fig. 2). The NQR enzyme is thought to be involved in pH and ion homeostasis in V. alginolyticus (16). In V. cholerae, high pH and low or high NaCl are known to negatively affect CT and TCP production, which is believed to occur via modulation of toxT transcription (1, 10). We assayed the toxT∷lacZ expression in both the parental and nqr mutant strains after growth in LB with a starting pH of 6.5 or 8.5 (Fig. 2). Consistent with previous findings that the transcription of toxT is strongly affected by the external pH (10), β-galactosidase activity in the parental strain is much reduced when the cells are grown in media with a high starting pH. In the nqr mutant strains, β-galactosidase activity also was strongly reduced after growth at elevated pH (Fig. 2). In contrast, very low or high concentrations of NaCl in the growth medium resulted in reduced toxT∷lacZ expression in the parental but not the nqr mutant strain (Fig. 2). It is interesting to note that the nqr mutant strain showed a slight growth defect compared with the parental strain and showed poorest growth at the low and high NaCl concentrations, suggesting an important role of this enzyme in ion homeostasis in V. cholerae.
Figure 2.
Effects of growth conditions on β-galactosidase activities in wild-type and nqr mutant V. cholerae strains carrying a toxT∷lacZ reporter construct. Cells were grown in LB with a starting pH of 6.5 or 8.5, in LB (pH 6.5) with low (0 mM), normal (85 mM), or high (500 mM) concentrations of NaCl, or in LB (pH 6.5) with 2.5 μM HQNO.
HQNO was reported to be a specific inhibitor of the NQR enzyme complex from V. alginolyticus and blocks its activity at micromolar concentrations (17). Addition of 2.5 μM HQNO to the growth medium resulted in markedly increased β-galactosidase activities only in the parental V. cholerae strain (Fig. 2). These results suggest that it is the activity of the NQR enzyme rather than the absence of the protein complex that is responsible for the observed effect on toxT transcription.
Enhanced toxT∷lacZ activity in a nqr mutant strain occurs even when only TcpP/H are expressed.
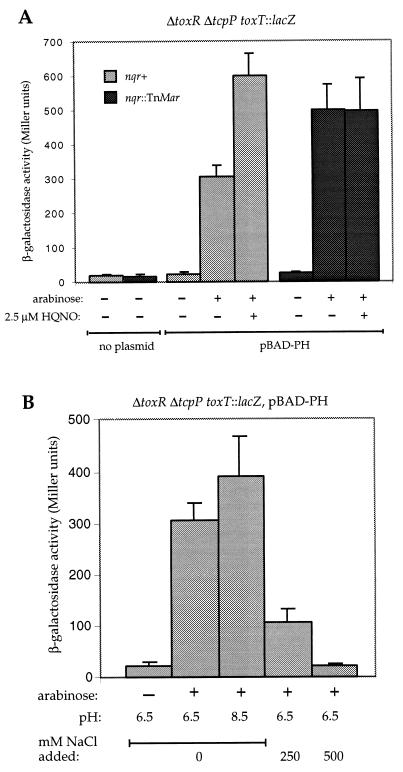
To investigate whether the effects of this sodium pump on the toxT promoter are mediated via the ToxR/S or TcpP/H proteins, we introduced the nqr∷TnMar transposon insertion into a ΔtoxR ΔtcpP toxT∷lacZ V. cholerae strain (4). As reported (4), the ΔtoxR ΔtcpP toxT∷lacZ parent strain showed very low β-galactosidase activity, and the derivative strain containing the nqr mutation showed similarly low β-galactosidase activity (Fig. 3A). Overexpression of the tcpP and tcpH genes from an arabinose-dependent promoter can partially complement the toxR deletion for activation of the toxT∷lacZ reporter construct (4) (Fig. 3A). If tcpPH are induced with the same arabinose concentrations in the strain carrying the nqr mutation, significantly higher β-galactosidase activities were observed compared with the parental strain (Fig. 3A). Furthermore, the addition of 2.5 μM HQNO to the growth media results in increased β-galactosidase activites in the parental but not in the nqr mutant strain (Fig. 3A). Together, these results indicate that the TcpP/H molecules are required for the increased toxT transcription in a nqr mutant background.
Figure 3.
Comparison of β-galactosidase activities in ΔtoxR ΔtcpP toxT∷lacZ V. cholerae strains with or without the nqr∷TnMar mutation carrying a plasmid expressing the tcpPH genes from an arabinose-inducible promoter (pBAD-PH). Arabinose (0.02%) and HQNO (2.5 μM) (A) were added as indicated. Effects of different media pH or various concentrations of NaCl are shown (B).
The Activity of the TcpP/H Proteins Is Sensitive to NaCl but Not to pH.
Carroll et al. recently reported (7) that the transcription of tcpPH is strongly reduced at high pH and temperature. Consistent with this, expression of tcpPH from an independent promoter resulted in toxT∷lacZ expression even under alkaline conditions (Fig. 3B) that result in strong repression of toxT transcription in a wild-type strain (Fig. 2) (10). In contrast, the β-galactosidase activity levels from the toxT∷lacZ reporter achieved by overexpression of the tcpPH genes are dramatically reduced if the cultures are grown in the presence of elevated concentrations of NaCl (Fig. 3B). Together, these results indicate that the activities of the TcpP/H proteins are sensitive to high salt concentrations but not to elevated pH. Of interest, the negative effects of high NaCl concentrations on toxT transcription appear to be more pronounced when TcpP/H are expressed in the absence of ToxR/S than in a wild-type background. Induction of TcpP/H also leads to elevated β-galactosidase activities in an E. coli strain carrying a chromosomally integrated copy of the toxT∷lacZ reporter construct (data not shown). Of interest, these expression levels are not negatively affected by the addition of NaCl to the growth medium (data not shown), suggesting that the TcpP/H molecules are sensitive to elevated salt concentrations only in a V. cholerae background.
The Flagellum of V. cholerae Is Energized by Sodium Ions.
In several marine and halophilic bacterial species, the respiration-driven Na+ pump, NQR, produces an electrochemical gradient of sodium ions across the membrane, which can be utilized for solute import, ATP synthesis, and flagella rotation (16). The single polar flagella of V. alginolyticus and Vibrio parahaemolyticus are energized by the translocation of sodium ions (18, 19). It was previously observed that, like in other Vibrio species, motility of V. cholerae increases with increased NaCl concentrations, indicating that Na+ plays an important role in the motility in this organism (20). Furthermore, the partial genomic sequences of V. cholerae recently released by TIGR contain several homologs of genes encoding subunits specific for a Na+-driven flagellar motor (21)—including motX, motY, pomA, and pomB (data not shown)—that when mutagenized result in nonmotile phenotypes (see below) (22). To determine whether the flagellum of V. cholerae is driven by sodium ions, we analyzed motility behavior of the V. cholerae wild-type strain O395 after the addition of various inhibitors. Swimming speed of V. cholerae was dramatically reduced after the addition of the protonophore CCCP at pH 6.5 but not at pH 8.5 (data not shown), analogous to the results obtained for V. parahaemolyticus (19). Furthermore, the addition of micromolar concentrations of phenamil, an amiloride compound believed to specifically block the sodium ion-conducting portion of flagellar motors (23, 24), resulted in dramatically reduced motility (data not shown). Together, these results and data recently obtained by Kojma et al. (25) indicate that the V. cholerae flagellum is energized by the translocation of sodium ions.
Effects of Modulation of Flagella Rotation on toxT∷lacZ Expression.
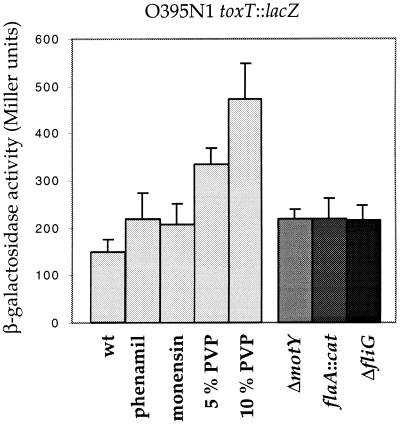
It was previously found that motility and virulence factor expression are inversely correlated in V. cholerae (22). As loss of NQR activity, an enzyme complex involved in generating a sodium motive force (smf) that can be utilized by flagella rotation, resulted in increased toxT∷lacZ expression, we wished to analyze whether changes in Na+ flux through the flagellum also result in altered toxT transcription. The addition of phenamil, an inhibitor of sodium-driven flagella, as well as addition of monensin, an ionophore that changes the level of Na+ chemical potential (25), resulted in a moderate increases in β-galactosidase activity in the O395N1 toxT∷lacZ reporter strain (Fig. 4). Similarly, introduction of various specific mutations that produced nonmotile phenotypes resulted in slightly increased β-galactosidase activities compared with the parental strain (Fig. 4). Furthermore, increasing the media viscosity by adding 5% or 10% polyvinylpyrrolidone (Fig. 4) or 15% Ficoll (data not shown) more dramatically induced toxT∷lacZ expression, reminiscent of the laf gene induction observed by increased media viscosity in V. parahaemolyticus (26).
Figure 4.
Effects of modulation of flagella rotation and specific mutations affecting motility on toxT∷lacZ expression. Cells were grown in LB or in LB containing 20 μM phenamil, 20 μM monensin, 5% polyvinylpyrrolidone (PVP), or 10% polyvinylpyrrolidone as indicated.
DISCUSSION
The main virulence factors of V. cholerae, CT and TCP, are coordinately regulated by a cascade of regulatory proteins in response to environmental conditions (1). The ToxT protein directly activates the ctx and tcpA promoters, and transcription of the toxT gene depends on the ToxR/S and TcpP/H proteins. Thus far, only the Cya and Crp proteins are known to negatively affect toxT transcription (27). To identify additional negative regulators of toxT transcription, we performed transposon mutagenesis of a V. cholerae toxT∷lacZ reporter strain followed by a screen for darker blue colonies. Several mutants were isolated that showed increased β-galactosidase activities compared with the parental strain. Analysis of the DNA sequences adjacent to the transposon insertions revealed homologies to several genomic regions of known as well as unknown functions. Interestingly, two mutant strains had transposon insertions in genes homologous to the nqr gene cluster from V. alginolyticus (8, 9). We had previously isolated an nqr∷Tnbla mutant of V. cholerae that showed increased TCP expression in media of elevated pH (4). Thus, we have isolated three independent mutants in the nqr gene cluster that resulted in elevated toxT transcription and/or TCP production in V. cholerae.
The NQR enzyme has been extensively studied in V. alginolyticus and is a respiration-linked Na+ pump (a Na+-dependent NQR) (16) establishing an electrochemical gradient of sodium ions across the membrane, resulting in a smf. Several bacterial species can use a smf for solute transport, ATP synthesis, and flagella rotation. This alternative energy coupling of sodium ions rather than protons enables the bacteria to maintain a cytoplasmic pH near neutrality in an alkaline environment. At alkaline pH, a strong reduction in toxT transcription is observed in both the wild-type and to a somewhat lesser extent in the nqr mutant strain, suggesting that the NQR enzyme is not the primary factor involved in the transcriptional repression of toxT and ToxT-regulated virulence genes in V. cholerae in response to alkaline conditions. In contrast, very low or high NaCl concentrations resulted in decreased toxT∷lacZ expression in the parental but not in the nqr mutant strain, suggesting that the NQR enzyme may play a role in the response of virulence factor expression to changes in NaCl concentration.
The ability of TcpP/H to activate the toxT∷lacZ fusion is dramatically reduced at high NaCl levels in V. cholerae. This suggests that TcpP/H may directly sense elevated Na+ ion concentrations or some other signal associated with high osmotic stress (e.g., turgor pressure or perhaps the conformation of other membrane proteins that undergo osmotically triggered structural changes). If so, it would not be surprising that loss of the NQR activity (by mutation or HQNO intoxication) causes elevated toxT∷lacZ activity, because the effect of the NQR complex is to pump out Na+ ions. However, TcpP/H mediated activation of toxT∷lacZ does not respond to elevated Na+ ion concentrations in the E. coli heterologous background. Thus, the negative signal that TcpP/H sense as a result of high Na+ concentrations may depend on another V. cholerae-specific product or physiological state. For example, another protein that negatively modulates TcpP/H activity may be induced by growth under elevated levels of NaCl. Because TcpP/H are putative membrane proteins, they may sense the activation state of the NQR complex directly through protein–protein interactions in the membrane. Alternatively, TcpP/H may sense the level of sodium gradient rather than high Na+ concentrations per se.
Motility is an important virulence factor in a variety of pathogenic bacteria and, in some cases, is inversely regulated with other virulence factors (28). Motility in V. cholerae is known to be negatively regulated by the ToxR regulon. At least two ToxR-regulated genes on the TCP–ACF island, tcpI and acfB, encode proteins with high homology to methyl-accepting chemotaxis proteins, suggesting they are chemoreceptors, and mutations in these two genes negatively affect motility of V. cholerae as assayed by swarm plate assays (29, 30). Furthermore, toxR mutant strains display a hypermotile phenotype (22). Conversely, some nonmotile mutants showed constitutive expression of CT and TCP at alkaline conditions, whereas some hypermotile mutants expressed no CT and TCP under normally inducing conditions (22). It may be that the effects of the nqr mutation on toxT transcription are mediated indirectly via motility. Unlike E. coli, the single polar flagella of several Vibrio species are energized by sodium ions rather than protons. Inhibition of motility by phenamil or monensin as shown here strongly suggests that the flagellar motor of V. cholerae is also energized by the smf via translocation of sodium ions. Because the activity of the NQR enzyme complex is believed to generate a smf that can energize flagella rotation, perhaps a lack of the NQR activity reduces smf, which in turn slows flagella. TcpP/H or another regulatory factor might sense flagellar rotation rates directly via a mechanosensory mechanism or by sensing sodium flux through the flagellar motor. Consistent with this idea, we found that inhibiton of flagellar rotation by the addition of phenamil (a known inhibitor of sodium-driven flagellar motors) or monensin (an ionophore that changes the level of Na+ chemical potential) or introduction of mutations resulting in nonmotile phenotypes lead to moderate increases in toxT∷lacZ expression and CT production under in vitro expression conditions. Furthermore, increasing the media viscosity resulted in an even more dramatic induction of virulence factor expression. Viscosity is thought to increase laf gene expression in V. parahaemolyticus by a signaling process that involves the sensing of flagellar rotation speed (26), and perhaps a similar mechanism explains the relationship between virulence gene expression and flagellar function in V. cholerae. During infection, V. cholerae encounters a high-viscosity environment in the mucus lining of the gut. Sensing of the changes in viscosity may be one of the signals that converts this organism from its environmental to its pathogenic phase.
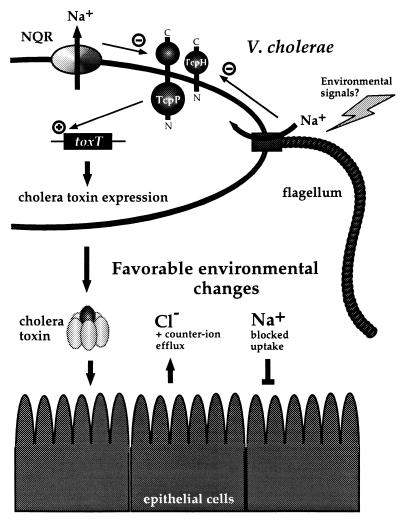
Further experiments will be necessary to elucidate how changes in membrane sodium flux and motility affect virulence gene expression in V. cholerae. It is tempting to speculate that many signals affecting the ToxR regulon may do so by altering motility, smf, or Na+ flux (Fig. 5). In agreement with this model are the observations that some of the conditions that negatively affect the ToxR regulon, such as high pH and bile, result in hypermotility (20, 31). Alternatively, it is possible that the effects of the nqr mutation on toxT transcription are mediated indirectly by affecting the ATP levels and hence cAMP levels in the cell. This could lead to altered states of the CRP protein, which is known to negatively affect toxT transcription (27). However, although we obtained three independent nqr mutants and mutations in several other genes, including hns, fumA, and glmS, that resulted in significantly raised toxT transcription, none of these were in either crp or cya, suggesting that the effects of these mutations may be more prominent than the latter two.
Figure 5.
Model of the interactions of some of the molecules affected by changes in membrane Na+ flux. See text for detailed explanation.
The data presented here strongly suggest that the expression of the main virulence factors of V. cholerae appears to be intimately connected to the sodium energetics in this halophilic organism. Sodium regulation probably plays a role in both of the major environments of V. cholerae, the intestine and water sources. It has been argued that one of cholera toxin’s functions is to generate a high-Na+ environment for V. cholerae in the lumen of the intestine (32). It is clear that the toxin causes electrolyte levels in the intestinal lumen to increase, and perhaps this milieu is a more favorable environment for the intraintestinal growth of V. cholerae (Fig. 5). This might lead to a negative-feedback mechanism, because elevated extracellular NaCl concentrations result in reduced cholera toxin production. Furthermore, it has been hypothesized that the sodium cycle of energy plays a role in the persistence of V. cholerae in the environment, as induction of this type of energy coupling may increase the resistance of bacteria to various environmental factors (33). It is conceivable that changes in the sodium cycle of energy are the primary signals that this bacterial species uses to sense whether it is in the extrahost environment or the human gut.
Acknowledgments
We thank Eric J. Rubin, Su Chiang, and Ann Hochschild for many helpful discussions and critical reading of this manuscript and Karl Klose for the plasmid pKEK93. This work was supported by National Institutes of Health Grant AI-18045.
ABBREVIATIONS
- CT
cholera toxin
- TCP
toxin coregulated pilus
- smf
sodium motive force
- NQR
NADH–ubiquinone oxidoreductase
- X-gal
5-bromo-4-chlor-3-indolyl β-d galactoside
- Kan
kanamycin
References
- 1.Skorupski K, Taylor R K. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 2.Higgins D E, DiRita V J. Mol Microbiol. 1994;14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 3.DiRita V J. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 351–365. [Google Scholar]
- 4.Häse C C, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller V L, DiRita V J, Mekalanos J J. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiRita V J, Mekalanos J J. Cell. 1991;64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 7.Carroll P A, Tashima K T, Rogers M B, DiRita V J, Calderwood S B. Mol Microbiol. 1997;25:1099–1111. doi: 10.1046/j.1365-2958.1997.5371901.x. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi M, Hirai K, Unemoto T. FEBS Lett. 1995;363:75–77. doi: 10.1016/0014-5793(95)00283-f. [DOI] [PubMed] [Google Scholar]
- 9.Beattie P, Tan K, Bourne R M, Leach D, Rich P R, Ward F B. FEBS Lett. 1994;356:333–338. doi: 10.1016/0014-5793(94)01275-x. [DOI] [PubMed] [Google Scholar]
- 10.DiRita V J, Parsot C, Jander G, Mekalanos J J. Proc Natl Acad SciUSA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin E J, Akerley B J, Husson R N, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skorupski K, Taylor R K. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 13.Klose K E, Mekalanos J J. J Bacteriol. 1998;180:303–316. doi: 10.1128/jb.180.2.303-316.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 16.Unemoto T, Hayashi M. J Bioenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- 17.Tokuda H, Unemoto T. J Biol Chem. 1982;257:10007–10014. [PubMed] [Google Scholar]
- 18.Dibrov P A, Kostryko V A, Lazarova R L, Skulachev V P, Smirnova I A. Biochim Biophys Acta. 1986;850:449–457. doi: 10.1016/0005-2728(86)90113-1. [DOI] [PubMed] [Google Scholar]
- 19.Atsumi T, McCarter L, Imae Y. Nature (London) 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 20.Kiiyukia C, Kawakami H, Hashimoto H. Microbios. 1993;73:249–255. [PubMed] [Google Scholar]
- 21.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardel C, Mekalanos J J. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atsumi T, Sugiyama S, Cragoe E J, Jr, Imae Y. J Bacteriol. 1990;172:1634–1639. doi: 10.1128/jb.172.3.1634-1639.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima S, Atsumi T, Muramoto K, Kudo S, Kawagishi I, Homma M. J Mol Biol. 1997;265:310–318. doi: 10.1006/jmbi.1996.0732. [DOI] [PubMed] [Google Scholar]
- 25.Kojima, S., Yamamoto, K., Kawagishi, I. & Homma, M. (1999) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 26.Nachliel E, Finkelstein Y, Gutman M. Biochim Biophys Acta. 1996;1285:131–145. doi: 10.1016/s0005-2736(96)00149-6. [DOI] [PubMed] [Google Scholar]
- 27.Belas R, Simon M, Silverman M. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skorupski K, Taylor R K. Proc Natl Acad SciUSA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottemann K M, Miller J F. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 30.Everiss K D, Hughes K J, Kovach M E, Peterson K M. Infect Immun. 1994;62:3289–3298. doi: 10.1128/iai.62.8.3289-3298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harkey C W, Everiss K D, Peterson K M. Infect Immun. 1994;62:2669–2678. doi: 10.1128/iai.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S, Chowdhury R. Infect Immun. 1997;65:1131–1134. doi: 10.1128/iai.65.3.1131-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakeeva L E, Chumakov K M, Drachev A L, Metlina A L, Skulachev V P. Biochim Biophys Acta. 1986;850:466–472. doi: 10.1016/0005-2728(86)90115-5. [DOI] [PubMed] [Google Scholar]
- 34.Brown I I, Sirenko L A. Biochemistry. 1997;62:225–230. [PubMed] [Google Scholar]