Abstract
Characterizing the biological effects of metabolic transformations (or biotransformation) is one of the key steps in developing safe and effective pharmaceuticals. Sulfate conjugation, one of the major phase II biotransformations, is the focus of this study. While this biotransformation typically facilitates excretion of metabolites by making the compounds more water soluble, sulfation may also lead to bioactivation, producing carcinogenic products. The end result, excretion or bioactivation, depends on the structural features of the sulfation sites, so obtaining the structure of the sulfated metabolites is critically important. We describe herein a very simple, high-throughput procedure for using mass spectrometry to identify the structure—and thus the biological fate—of sulfated metabolites. We have chemically synthesized and analyzed libraries of compounds representing all the biologically relevant types of sulfation products, and using the mass spectral data, the structural features present in these analytes can be reliably determined, with a 97% success rate. This work represents the first example of a high-throughput analysis that can identify the structure of sulfated metabolites and predict their biological effects.
Keywords: Mass spectrometry, ICP-MS, Bioanalytical methods, Pharmaceuticals, Biological samples, Metabolites, Sulfation
Introduction
The study of drug metabolism is one of the critical steps in drug development [1–4]. At physiological conditions, drugs undergo a variety of biotransformations, which produce metabolites with different chemical structures. The resulting metabolites differ in both pharmacological and toxicological properties compared to their parent drugs [1]. Therefore, to identify and characterize the structures and properties of drug metabolites formed in vivo, an extensive investigation of the structures generated by biotransformation is required.
Sulfate conjugation (sulfation) is one of the most important phase II reactions that occurs during the biotransformation of a variety of structurally diverse endogenous compounds, xenobiotics, and drugs [5–9]. During this process, a sulfonate moiety (SO3) is transferred from the donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) to the substrates through the catalysis of sulfotransferase enzymes, resulting in sulfated products [10]. Sulfation is generally considered as a detoxification pathway, because the sulfated products are more water-soluble; this facilitates their elimination from the body [11–13]. However, sulfation can also lead to the bioactivation of certain type of compounds which include benzylic, allylic alcohols and aromatic hydroxylamines. Sulfated products of these compounds can undergo loss of 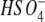 , resulting in reactive electrophilic carbocation or nitrenium ion intermediates, which covalently bind to cellular macromolecules, leading to mutagenicity and carcinogenicity [6, 14, 15]. The production of the electrophilic intermediates relies on the structural feature of sulfation sites, as shown in Fig. 1. Under physiological conditions, the sulfate anions in substrates a–c in Fig. 1 act as good leaving groups to form resonance-stabilized intermediates [6, 14]. For the other types of sulfated products in Fig. 1, this process is not favorable due to instability of cation products.
, resulting in reactive electrophilic carbocation or nitrenium ion intermediates, which covalently bind to cellular macromolecules, leading to mutagenicity and carcinogenicity [6, 14, 15]. The production of the electrophilic intermediates relies on the structural feature of sulfation sites, as shown in Fig. 1. Under physiological conditions, the sulfate anions in substrates a–c in Fig. 1 act as good leaving groups to form resonance-stabilized intermediates [6, 14]. For the other types of sulfated products in Fig. 1, this process is not favorable due to instability of cation products.
Fig. 1.
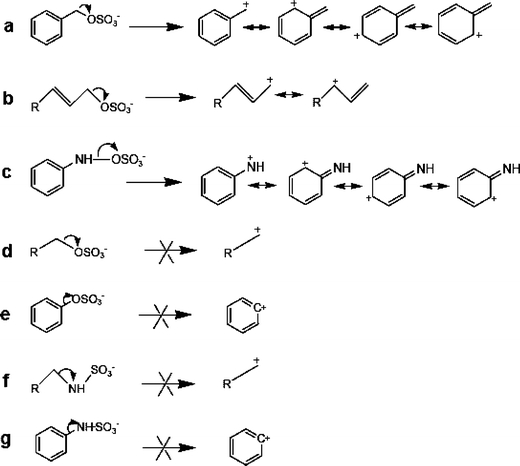
Mechanism of forming bioactive carbocation or nitrenium ion intermediates from sulfated products. Substrates a, b and c undergo the bioactivation process while d–g do not. a sulfated benzylic alcohol; b sulfated allylic alcohol; c sulfated aromatic hydroxylamine; d) sulfated aliphatic alcohol; e sulfated phenol; f sulfated aliphatic amine; g sulfated aromatic amine
The biological sulfation of tamoxifen is one example that demonstrates how the sulfation site dictates the biological fate of the molecule. Tamoxifen is a pharmaceutical that is widely used in the treatment and prevention of breast cancer. It is hydroxylated during phase I metabolism to generate two products: 4-hydroxytamoxifen (4-OH-TAM) and alpha-hydroxytamoxifen (α-OH-TAM). The sulfation of 4-OH-TAM and that of α-OH-TAM have completely different biological effects. The sulfation of 4-OH-TAM, which occurs at a phenol site (as in Fig. 1), leads to detoxification, whereas the sulfation of α-OH-TAM, which involves sulfation of a benzylic alcohol (as in Fig. 1a), leads to bioactivation [16, 17]. This example (along with many others) demonstrates the fact that the biological effects of sulfation can be determined by characterizing the sulfation site of the substrate.
It is widely known that aromatic or aliphatic hydroxyl and amine groups are the major sites in molecules that can be sulfated during metabolism [18]. However, identifying the sulfation sites can be difficult because many potential sulfation sites can be produced or modified during phase-I biotransformations [5]. For example, hydroxyl groups can be added to a benzene ring or carbon chain through aromatic or aliphatic hydroxylation, generating new potential sulfation sites as in the tamoxifen example. An amine group can be changed to a hydroxylamine group through N-oxidation [1], which modifies the structural feature of the potential sulfation site. Since these new sites are formed in vivo, characterizing the biological effect of their sulfation can be problematic. It is thus essential to develop a method that can characterize the sulfation sites of unknown sulfated metabolites. With this information, the biological effect of sulfation can be estimated, and the properties of sulfated metabolites can be characterized.
Analytical techniques such as NMR spectroscopy and mass spectrometry have been utilized to identify sulfated metabolites [19–31]. NMR is a powerful technique that can not only detect sulfated metabolites [20, 21, 24], but also help to determine the position of sulfation in the substrate molecules [19, 22, 23]. However, due to the relatively high sample requirements, the complexity of data analysis, and the need to characterize purified compounds, the application of NMR in identification and characterization of drug metabolites is limited in high-throughput metabolite profiling. Compared to NMR, mass spectrometry is more widely used in drug metabolite identification, due to its high selectivity and sensitivity, low detection limit, and ability to analyze mixtures [32]. It is known that mass spectrometry can identify sulfation by detecting the 80 Da mass increase in MS mode, or the characteristic ions in MS/MS mode [21, 25–31]. However, limited information about structural features of sulfation sites currently can be obtained from MS/MS analysis, with the well-known exception that the product ion m/z 97 (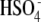 ) can be used to differentiate alicyclic sulfates from aromatic sulfates [25, 26].
) can be used to differentiate alicyclic sulfates from aromatic sulfates [25, 26].
In order to facilitate the identification of sulfation sites in pharmaceuticals, we developed a mass spectrometry-based protocol that differentiates between the biologically relevant sulfation sites. After completing the studies on several classes of sulfated products, a set of rules was developed to predict the sulfation sites. With these prediction rules, structural feature of sulfation sites can be determined by detecting MS/MS fragmentation pathways of their corresponding sulfated products. With the structural information of sulfation sites, the two key different biological effects of sulfation, “detoxification” and “bioactivation”, can be differentiated, based on literature precedence that links the type of sulfation to its biological fate. Based on that information, relevant properties of sulfated products can be estimated.
Experimental
Reagents Sulfates and sulfamates which include 4-nitrocatechol sulfate dipotassium salt, L-ascorbic acid 2-sulfate dipotassium salt, indoxyl sulfate potassium salt, β-estradiol 3-sulfate sodium salt, potassium 4-nitrophenyl sulfate, β-Estradiol 3-sulfate sodium salt, 5-Br-4-Cl-3-indolyl sulfate potassium salt, 4-methylumbelliferyl sulfate potassium salt, 2-aminoethyl hydrogen sulfate, poly (vinyl sulfate) potassium salt, D-glucose 6-sulfate potassium salt, chondroitin disaccharide Δdi-6S sodium salt, N-acetylglucosamine 6-sulfate sodium salt, N-cyclohexylsulfamic acid, 3-hydroxypropyl-sulfamic acid monopotassium salt, D-glucosamine 2-sulfate sodium salt, and butyl-sulfamic acid were purchased from Sigma-Aldrich (St. Louis, MO). One sulfamate, 4-methylphenyl- sulfamic acid, was purchased from Scientific Exchange, Inc. (Center Ossipee, NH). (R)(+)-α-phenethylsulfamic acid was purchased from Norse Laboratories (Newbury Park, CA).The benzylic alcohols, allylic alcohols, hydroxylamines which include (R)-(+)-α-methyl-2-naphthalenemethanol, benzyl alcohol, (R)-1-phenyl-2-propen-1-ol, 4-chloro-2-methylbenzyl alcohol, 3-ethoxybenzyl alcohol, 2-ethoxybenzyl alcohol, 4-ethoxybenzyl alcohol, crotyl alcohol, furfuryl alcohol, N-methylhydroxylamine hydrochloride, N-isopropylhydroxylamine hydrochloride, N-cyclohexylhydroxylamine hydrochloride, N, N-diethylhydroxylamine and N-benzoyl-N-phenylhydroxylamine were purchased from Sigma-Aldrich (St. Louis, MO). These benzylic alcohols, allylic alcohols, and hydroxylamines were utilized to synthesize their corresponding sulfated products. The other reagents used in the sulfation of alcohols and hydroxylamines were also purchased from Sigma-Aldrich (St. Louis, MO).
Sulfation of benzylic, allylic alcohols and tertiary hydroxylamines Sulfated products were prepared based on slight modifications to a previous protocol [33] by dissolving 1.0 equivalent (5.0 mmol) of the substrate in 5.0 ml dimethylformamide (DMF) and adding sulfur trioxide-dimethylformamide complex (DMF-SO3 5.5 mmol) and pyridine (5.5 mmol). The reaction mixture was stirred at 40 °C for 1 h. H2O (35 ml) was added to the product solution, and the product was extracted with 3×15 mL ethyl acetate (EtOAc). Evaporation of the solvent afforded the corresponding sulfated product.
Sulfation of secondary hydroxylamines To a dry THF (10 ml) solution of NaH (10.0 mmol) was added secondary hydroxylamines (5 mmol) at 0 °C. Then the solution was stirred at room temperature for 2 h, followed by reaction with DMF-SO3 (5.0 mmol) overnight. The corresponding sulfated products were obtained by evaporation of THF.
Sample pretreatment Sulfated compounds that were purchased were dissolved to a final concentration of 1.0×10−4 M with 50% MeOH/ 50% H2O, and directly injected into the mass spectrometer. Sulfated benzylic and allylic alcohols and tertiary hydroxylamines were prepared by diluting 10 μl of the liquid product by 100 fold with 50% MeOH/50% H2O, and then injected to the mass spectrometer. Sulfated secondary hydroxylamines were prepared by dissolving 10 mg solid product in 1.0 ml 50% MeOH/ 50% H2O, followed by injection into the mass spectrometer.
Mass spectrometry The mass spectra were recorded using a Quattro Ultima (Waters Corp., Milford, MA) triple quadrupole mass spectrometer equipped with an electrospray ionization source. Data was acquired in the negative ion mode using a capillary voltage of 2.80 kV and a cone voltage was 45 V. The source temperature and desolvation gas temperature were 80 and 150 °C, respectively. Argon is used as the collision gas, and the pressure in collision cell is 1.7E(−3) mbar. The collision energy applied in MS/MS for all the samples was 35 eV.
Results and discussion
The structures of the sulfated products are in Fig. 2. These compounds are representative of different types of sulfated products that are produced by sulfation at different sites. Mass spectrometric studies were conducted in the negative ion mode since sulfated products are deprotonated at physiological pH [11, 15]. The common product ions from the MS/MS experiments are summarized in Table 1. As the table indicates, common product ions include m/z [M-H-80], m/z 80, m/z 96 and m/z 97. The presence of m/z [M-H-80], m/z 96 or m/z 97 ions are quite dependent on the site of sulfation. The presence of the negative ion with m/z 80, however, is common in the fragmentation of almost every type of sulfated products, so it is less helpful in identification of sulfation sites. Thus this ion, m/z 80, is used to identify sulfation sites when no other characteristic ions can be detected. Based on the MS/MS data, each type of sulfated product possesses a characteristic fragmentation pathway, which is described below. An example of MS/MS data for each type of sulfated product is provided in Fig. 3 with the characteristic ion labeled by an asterisk.
Fig. 2.
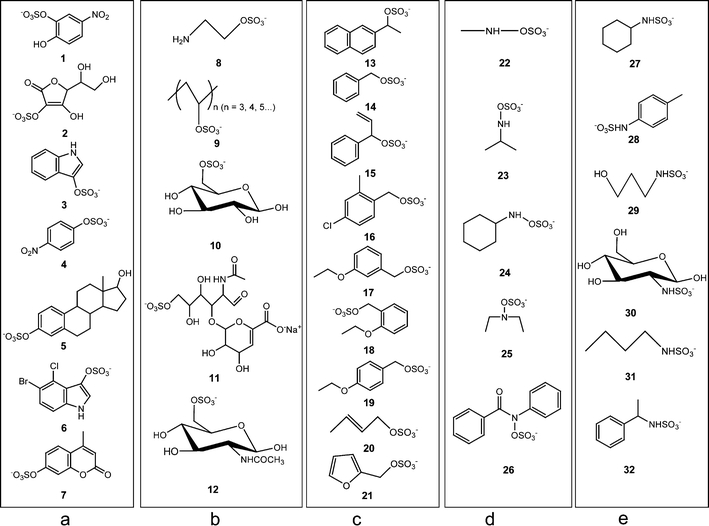
Structures of selected sulfated compounds a sulfated aromatic alcohols or enols; b sulfated aliphatic alcohols with β hydrogens on sp3carbons; c sulfated benzylic or allylic alcohols; d sulfated hydroxylamines; e sulfated amines
Table 1.
Characteristic fragmentation of selected sulfated products
| Group | No. | Chemical name | Abundance of characteristic ions in MS/MS (%) | |||
|---|---|---|---|---|---|---|
| m/z [M-80]- | m/z 80 | m/z 96 | m/z 97 | |||
| a | 1 | 4-nitrocatechol sulfate | 100a | – | – | – |
| 2 | L-ascorbic acid 2-sulfate | 31a | – | – | – | |
| 3 | Indoxyl sulfate | 24a | 52 | – | – | |
| 4 | 4-nitrophenyl sulfate | 100a | – | – | – | |
| 5 | β-estradiol 3-sulfate | 100a | – | – | – | |
| 6 | 5-Br-4-Cl-3-indolyl sulfate | 47a | 59 | – | – | |
| 7 | 4-methylumbelliferyl sulfate | 100a | – | – | – | |
| b | 8 | 2-aminoethyl hydrogen sulfate | – | 100 | 18 | 67a |
| 9 | Poly (vinyl sulfate) | – | – | – | 97a | |
| 10 | D-glucose 6-sulfate | – | – | – | 100a | |
| 11 | Chondroitin disaccharide Δdi-6S | – | – | – | 31a | |
| 12 | N-acetylglucosamine 6-sulfate | – | – | – | 93a | |
| c | 13 | 2-naphthalenemethanol, α-methyl- sulfate | – | 6.7 | 7.3a | 100 |
| 14 | Benzyl sulfate | – | 16 | 100a | – | |
| 15 | (R)-1-phenyl-2-propen-1-sulfate | – | 26 | 100a | 12 | |
| 16 | 4-Cl-2-methylbenzyl sulfate | – | – | 100a | 95 | |
| 17 | 3-ethoxybenzyl sulfate | – | 100 | 13a | 6.4 | |
| 18 | 2-ethoxybenzyl sulfate | – | 100 | 17a | 8.5 | |
| 19 | 4-ethoxybenzyl sulfate | – | 100 | 20a | 8.7 | |
| 20 | crotyl sulfate | – | 55 | 91a | 50 | |
| 21 | furfuryl sulfate | – | – | 53a | 39 | |
| d | 22 | N-methyl-hydroxylamine-O-sulfonic acid | – | 100 | 73a | 9.2 |
| 23 | N-isopropyl- hydroxylamine-O-sulfonic acid | – | 68 | 100a | 5.1 | |
| 24 | N-cyclohexyl-hydroxylamine-O-Sulfonic acid | – | 27 | 100a | 11 | |
| 25 | N,N-diethyl-hydroxylamine-O-sulfonic acid | – | 92 | 100a | 19 | |
| 26 | N-benzoyl-N-phenyl-hydroxylamine-O-sulfonic acid | – | – | 100a | – | |
| e | 27 | N-cyclohexylsulfamic acid | – | 100a | – | – |
| 28 | 4-methylphenyl-Sulfamic acid | – | 100a | – | – | |
| 29 | 3-hydroxypropyl-sulfamic acid | – | 100a | – | – | |
| 30 | D-glucosamine 2-sulfate | – | 100a | – | – | |
| 31 | butyl-sulfamic acid | – | 100a | – | – | |
| 32 | (R)(+)-α-phenethylsulfamic acid | – | 100a | – | – | |
*A threshold of 5.0% is used for the relative abundance of characteristic ions. The long dash (–) means that the characteristic ion can not be detected or the relative abundance is below 5.0%
aRepresents the specific characteristic ion of each group that can help to identify the sulfation site
Fig. 3.
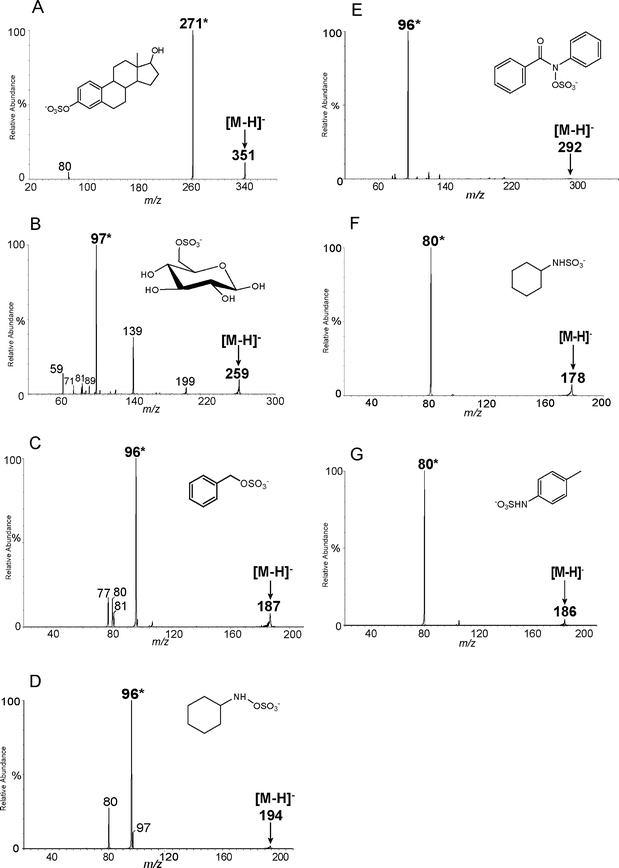
Characteristic (-) ESI-MS/MS data for compounds in this study: a sulfated aromatic alcohol, from group a; b sulfated aliphatic alcohol with β hydrogen on sp3carbon, from group b; c sulfated benzylic alcohol, from group c; d sulfated aliphatic hydroxylamine, from group d; e sulfated aromatic hydroxylamine, from group d; f sulfated aliphatic amine, from group e; g sulfated aromatic amine, from group e
Group a: sulfated aromatic alcohols or enols
Compounds 1–7 are sulfated aromatic alcohols or enols. For these compounds, the sulfate group is attached to an sp2 carbon (benzene ring or double bond). These sulfated products share the same fragmentation pathway by undergoing the neutral loss of 80 Da, and forming a characteristic ion at m/z [M-H-80], as seen in Fig. 3a. After undergoing the neutral loss of SO3, phenoxide or enolic anions are formed, and the negative charge is resonance stabilized, as shown in Scheme 1 [33].
Scheme 1.
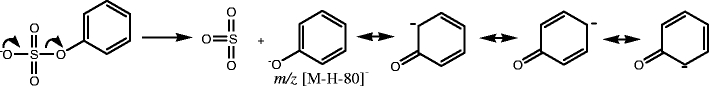
Sulfated aromatic alcohols or enols (group a) dissociate to produce a characteristic ion with neutral loss of 80 Da. Group b: sulfated aliphatic alcohols with β hydrogens on sp3carbons
The second group of sulfated products originates from the sulfation of aliphatic alcohols: compounds 8–12. For these species, the sulfate group is attached to an sp3 carbon. All sulfated products of this type can produce the characteristic ion m/z 97 in MS/MS, and one example is shown in Fig. 3b. The ion m/z 97 is generated when the proton from the β carbon is transferred to the sulfate moiety, and the C-O bond is broken to form a bisulfate anion [33]. See Scheme 2.
Scheme 2.
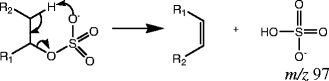
Sulfated aliphatic alcohols (group b) produce the characteristic ion m/z 97
This rearrangement occurs via an energetically accessible six-membered ring transition state, and a stable neutral product is formed. As a result, this fragmentation is predominant in the MS/MS data of this group of sulfated products. The mechanism shows that the availability of a β hydrogen on an sp3 carbon is required for this fragmentation. Unlike compounds in group a, neutral loss of 80 Da is not observed from this group of compounds. This is likely due to the fact that the product ion that would be generated from loss of 80 Da (SO3) is not resonance stabilized, so the loss is not favorable. Compound 8 is a special case because it gives a characteristic ion with m/z 96, in addition to m/z 97. The relevant mechanism for this loss is addressed in the discussion of compounds in group d.
Group c: sulfated benzylic or allylic alcohols
In the third group of sulfated compounds (13–21), each contains a sulfate group attached to benzylic or allylic carbon. All the compounds in this group produce a characteristic ion with m/z 96. An example of an MS/MS data for this type is shown in Fig. 3c. The characteristic ion is produced by homolytic cleavage, producing benzylic or allylic radicals. The radical is resonance stabilized, which is demonstrated in Scheme 3 [33].
Scheme 3.
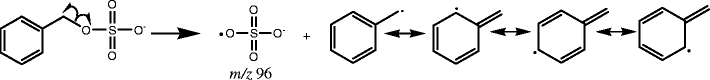
Sulfated benzylic and allylic alcohols (group c) produce the characteristic ion m/z 96
In addition to m/z 96, the negative ion m/z 97 can also be obtained for some of the compounds in this category. For example, compounds 13 and 16 both have an abundant peak at m/z 97, due to the availability of a hydrogen on the sp3 carbon in close proximity to the sulfate group. Even though compound 13 is a benzylic sulfate, a hydrogen attached to the β sp3 carbon is present. Therefore, just like compounds in group b, it undergoes a rearrangement to form the ion, m/z 97. Compound 16 possesses a methyl group in the ortho position, and the availability of hydrogen attached to the sp3 carbon in methyl group enables a similar rearrangement (eight-membered ring) to form the product ion, m/z 97, as illustrated in Scheme 4. Even though β hydrogens are available in other benzylic and allylic sulfates in this group, the fragmentation to produce m/z 97 is not as favorable, since the hydrogens are on the sp2 hybridized carbons. The neutral loss of 80 Da is not favorable either, because the product ion that would be generated from such a loss is not resonance stabilized.
Scheme 4.
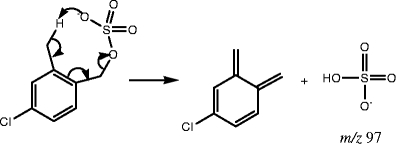
Fragmentation mechanism producing m/z 97 from compound 16
Group d: sulfated hydroxylamines
The fourth group of sulfated products originates from sulfation of hydroxylamines, and as a result, the sulfate group is directly attached to the amine: See compounds 22–26. All the sulfated products of this group can produce the characteristic ion m/z 96, and examples of MS/MS data are shown in Fig. 3. A characteristic ion with m/z 96 can be obtained for sulfated products originating from both aliphatic (Fig. 3d) and aromatic (Fig. 3e) hydroxylamines. For these sulfated compounds, homolytic cleavage is favorable. This is due to the small electronegativity difference (about 0.5) between N and O, compared to electronegativity difference of around 1.0 for C –N and O-S [34]. The small electronegativity difference causes the electrons to be split equally (homolytic cleavage) between the oxygen and nitrogen when the bond breaks, instead of both electrons moving onto the oxygen or nitrogen, which would be the case during heterolytic cleavage. In Scheme 5, the mechanism of this homolytic cleavage is depicted. An amine radical is formed as the product.
Scheme 5.
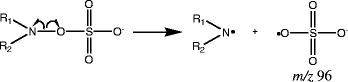
Sulfated hydroxylamines (group d) produce the characteristic ion m/z 96
This mechanism can also be used to explain the presence of the ion with m/z 96 in the MS/MS spectrum of compound 8. Scheme 6 shows how this ion is produced from compound 8. In this case, homolytic cleavage can be used to distribute a single electron onto the nitrogen. Compounds that contain a nitrogen two carbons away from the sulfation site, like this one, could also produce m/z 96 as a product ion.
Scheme 6.
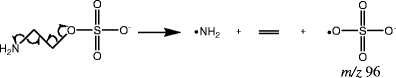
Fragmentation mechanism producing m/z 96 from compound 8
Group e: sulfated amines
The last group of sulfated products, sulfamates, originates from the sulfation of amines. Compounds 27–32 belong to this type. The only characteristic ion for this type of sulfated products is the negative ion, m/z 80. This characteristic ion can be obtained from fragmentation of sulfated products originating from both aliphatic amines (Fig. 3f) and aromatic amines (Fig. 3g). This group of sulfated products has another type of bond, a N-S bond, that has a small electronegativity difference between the two atoms (0.5) [34], making the homolytic cleavage between the nitrogen and sulfur favorable. The mechanism that shows production of m/z 80 is illustrated in Scheme 7. No other fragmentation pathway is as favorable as this homolytic cleavage pathway, making m/z 80 the predominant ion in all of the MS/MS data for these compounds.
Scheme 7.
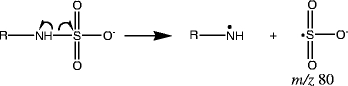
Sulfated amines (group e) produce the characteristic ion m/z 80
Prediction rules
For each type of sulfated product, there is a predominant fragmentation pathway that results in a characteristic product ion during MS/MS fragmentation. The specific characteristic ion for each group is obtained from every sulfated product in that group, as shown in Table 1: the ion with the neutral loss of 80 Da can be seen in MS/MS spectrum of every sulfated product in group a; an ion with m/z 97 can be detected in all the MS/MS data of group b compounds; an ion with m/z 96 can be obtained from MS/MS data of all the sulfated products of both group c and group d. A negative ion with m/z 80 can always be found in MS/MS data of sulfated products in group e.
We have shown that characteristic fragmentation pathways of sulfated products are dependent upon the structural features of the sulfation sites. Conversely, relevant structural information of sulfation sites can be obtained by determining characteristic fragmentation pathways or characteristic ions. To facilitate this information transformation, a set of prediction rules is described as follows:
If the characteristic ion with a neutral loss of 80 Da can be detected, this indicates that the sulfate group is attached to an sp2 carbon, the sulfated product is therefore produced by the sulfation of a phenol or an enol.
If the characteristic ion with m/z 96 can be detected, the sulfated product is produced by sulfation of a benzylic or allylic alcohol, or a hydroxylamine.
If the characteristic ion with m/z 97 can be obtained, and it is more abundant than the ion, m/z 96 (if it is detected), the sulfate group is attached to an sp3 carbon. This is the sulfated product produced by sulfation of aliphatic alcohols with an available β hydrogen attached to an sp3 carbon;
If none of the ions, m/z [M-H-80]−, m/z 96, or m/z 97 can be detected, and only the ion m/z 80 is observed, then the sulfated product is produced by sulfation of an amine.
There are occasions when the conditions in rule 2 and 3 might both be observed. For example, both of these apply to compound 13, which suggests that the sulfation site is not only an alcohol with available β hydrogen (on an sp3 carbon), but also a benzylic or allylic alcohol; and this estimation is consistent with the actual structure of compound 13. Among the selected 32 sulfated products, 31 of them follow the proposed prediction rules. The exception, compound 8, possesses an amine group in the β position. The amine at this position enables the compound undergo the fragmentation pathway to produce ion with m/z 96 (see Scheme 6). In this case, rule 2 incorrectly predicts the sulfation site for compound 8. As a result, the obtained data in Table 1 demonstrates a 97% (31/32=0.97) success rate of these prediction rules.
Potential application of the prediction rules in characterizing unknown sulfated metabolites
As one of the major phase II biotransformations, sulfate conjugation is involved in the metabolism of an enormous range of substrates [5]. In general, sulfation is a detoxification or deactivation pathway, but it can also bioactivate the substrate molecules when sulfation sites are benzylic, allylic alcohols (group c) or aromatic hydroxylamines (group d) [6, 8, 14]. According to the prediction rules developed herein, sulfation at any of these sites will generate products that produce the ion m/z 96 in MS/MS, so the structural feature of the sulfation sites can be determined by rule 2. Sulfation of other sites, like phenols, aliphatic alcohols or amines, will cause other characteristic ions to appear in MS/MS data. Therefore, the sulfation sites can be determined by rule 1, 3 or 4. In these cases, sulfation facilitates detoxification.
With the proposed prediction rules, biological effects of sulfation can be determined, and the properties of sulfated metabolites can be estimated. Figure 4 demonstrates how the proposed method works to characterize unknown sulfated metabolites. Since high-quality MS/MS data is a necessary prerequisite of using this method, if the metabolite is present in a complex biological matrix, some mass spectral optimization may be necessary. After obtaining reliable (-)ESI-MS/MS data on the unknown sulfated metabolite, the prediction rules are used to determine sulfation site by identifying the characteristic ions present in the MS/MS data. If the MS/MS data matches rule 1, 3 or 4 (instead of rule 2), the sulfation sites can be phenols (or enols), aliphatic alcohols (with β hydrogen on sp3 carbon) or amines. In these cases, the sulfation undergoes a detoxification pathway and the sulfated metabolite is more easily eliminated, compared to the parent drug. If the MS/MS data is consistent with rule 2, the sulfation site should be benzylic, allylic alcohols or hydroxylamines. This would indicate that the sulfation might follow a bioactivation pathway, and lead to carcinogenic sulfated products. One potential caveat to this second condition is that a few nontoxic sulfated metabolites that have structures similar to compound 8, might also produce the characteristic ion with m/z 96, following the mechanism in Scheme 6. Another possible drawback of this characterization is that the diagnostic ion m/z 96 does not discriminate between sulfated products of aliphatic and aromatic hydroxylamines; and thus far only aromatic hydroxylamines have been proven to be toxic. However, distinguishing between aromatic and aliphatic hydroxylamines is not critically important because sulfated aliphatic hydroxylamines are rather uncommon metabolites. While it is possible that a few compounds could be misassigned as “toxic”, none of the sulfated metabolites that are known to be bioactivated would be misdiagnosed as “nontoxic”, as demonstrated herein.
Fig. 4.
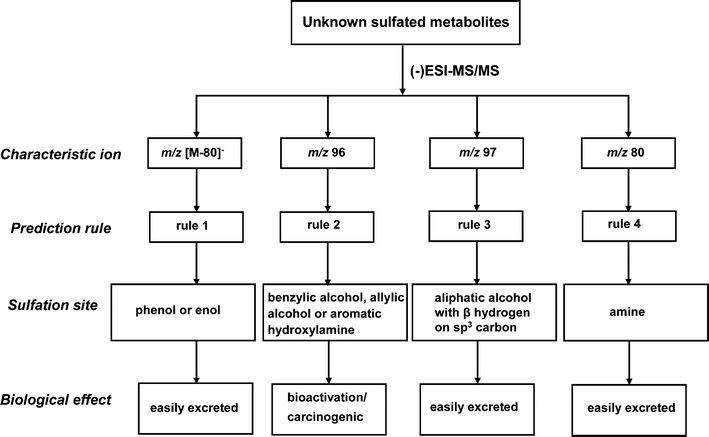
The method for characterizing unknown sulfated metabolites
Conclusion
A method was developed to determine the structural features of sulfation sites, by detecting the characteristic fragmentation pathway of the corresponding sulfated products in (-) ESI-MS/MS. By summarizing MS/MS data from five different types of sulfated products originating from different sulfation sites, their characteristic fragmentation pathways and characteristic ions were determined. Based on this information, a set of prediction rules was developed to transfer information about the fragmentation pathway of sulfated products to the structural features of the sulfation site. As a result, the proposed prediction rules can be applied in drug metabolite profiling to characterize sulfation sites, to further estimate the biological effect of sulfation, and to evaluate relevant properties of sulfated metabolites.
Acknowledgements
The authors gratefully acknowledge the University of Kansas New Faculty Graduate Research Fund for financial support, and Dr. Todd Williams and the KU Mass Spectrometry facility for the use of the triple quadruple mass spectrometer.
References
- 1.Gibson GG, Skett P (1986) Introduction to drug metabolism. Chapman and Hall, New York
- 2.Welling PG (1996) Changes in pharmacokinetics and drug metabolism responsibility in drug discovery and development. In: Welling PG, Lasagna L, Banakar UV (eds) The drug development process: increasing efficiency and cost-effectiveness; drugs and the pharmaceutical sciences, vol 76, Chapter 9. Marcel Dekker, New York
- 3.Sneader W (1986) Production and formulation. In: Drug development: from laboratory to clinic, chapter 2. Wiley, Chichester
- 4.Lin JH, Lu AYH (1997) Pharmacol Rev 49:403–449 [PubMed]
- 5.Falany CN, Wilborn TW (1994) Biochemistry of cytosolic sulfotransferases involved in bioactivation. In: Anders MW, Dekant W (eds) Advances in pharmacology, vol 27. Academic Press, New York, pp 301–329 [DOI] [PubMed]
- 6.Banoglu E (2000) Curr Drug Metab 1:1–30 [DOI] [PubMed]
- 7.Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB (1997) FASEB J 11:3–14 [PubMed]
- 8.Coughtrie MWH, Sharp S, Maxwell K, Innes NP (1998) Chem Biol Interact 109:3–27 [DOI] [PubMed]
- 9.Coughtrie MWH, Bamforth KJ, Sharp S, Jones AL, Borthwick EB, Barker EV, Roberts RC, Hume R, Burchell A (1994) Chem Biol Interact 92:247–256 [DOI] [PubMed]
- 10.Klaassen CD, Boles JW (1997) FASEB J 11:404–418 [DOI] [PubMed]
- 11.Pang KS, Schwab AJ, Goresky CA, Chiba M (1994) Chem Biol Interact 92:179–207 [DOI] [PubMed]
- 12.McFadden SA (1996) Toxicology 111:43–65 [DOI] [PubMed]
- 13.Cerniglia CE, Freeman JP, Mitchum RK (1982) Appl Environ Microbiol 43:1070–1075 [DOI] [PMC free article] [PubMed]
- 14.Glatt H (1997) FASEB J 11:314–321 [DOI] [PubMed]
- 15.Glatt H (2000) Chem Biol Interact 129:141–170 [DOI] [PubMed]
- 16.Chen G, Yin S, Maiti S, Shao X (2002) J Biochem Molecular Toxicology 16:279–285 [DOI] [PubMed]
- 17.Shibutani S, Ravindernath A, Terashima I, Suzuki N, Laxmi YRS, Kanno Y, Suzuki M, Apak TI, Sheng JJ, Duffel MW (2001) Cancer Res 61:3925–3931 [PubMed]
- 18.Hemmerich S, Verdugo D, Rath VL (2004) Drug Discov Today 9:967–975 [DOI] [PubMed]
- 19.Lerch ML, Harper MK, Faulkner DJ (2003) J Nat Prod 66:667–670 [DOI] [PubMed]
- 20.He X, Li J, Gao H, Qiu F, Cui X, Yao X (2003) Chem Pharm Bull 51:586–589 [DOI] [PubMed]
- 21.Mutlib AE, Shockcor J, Chen S, Espina RJ, Pinto DJ, Orwat MJ, Prakash SR, Gan L (2002) Chem Res Toxicol 15:48–62 [DOI] [PubMed]
- 22.Ibrahim AS, Galal AM, Ahmed MS, Mossa GS (2003) Chem Pharm Bull 51:203–206 [DOI] [PubMed]
- 23.Schaber G, Wiatr G, Wachsmuth H, Dachtler M, Albert K, Gaertner I, Breyer-pfaff U (2001) Drug Metab Dispos 29:923–931 [PubMed]
- 24.Spraul M, Hofmann M, Lindon JC, Farrant RD, Seddon MJ, Nicholson JK, Wilson ID (1994) NMR Biomed 7:295–303 [DOI] [PubMed]
- 25.Lafaye A, Junot C, Gall BR, Fritsch P, Ezan E, Tabet J (2004) J Mass Spectrom 39:655–664 [DOI] [PubMed]
- 26.Zhang H, Henion J (1999) Anal Chem, 71:3955–3964 [DOI] [PubMed]
- 27.Metzger K, Rehberger PA, Erben G, Lehmann WD (1995) Anal Chem 67:4178–4183 [DOI]
- 28.Sun L, Huang H, Liu L, Zhong D (2004) Appl Environ Microbiol 70:2722–2727 [DOI] [PMC free article] [PubMed]
- 29.Keski-Hynnilä H, Kurkela M, Elovaara E, Antonio L, Magdalou J, Luukkanen L, Taskinen J, Kostiainen R (2002) Anal Chem 74:3449–3457 [DOI] [PubMed]
- 30.Hong Y, Mitchell AE (2004) J Agric Food Chem 52:6794–6801 [DOI] [PubMed]
- 31.Frederiksen H, Frandsen H (2004) Drug Metab Dispos 32:661–665 [DOI] [PubMed]
- 32.Siuzdak G (2003) Pharmacokinetics in the expanding role of mass spectrometry in biotechnology, Chapter 9. MCC Press, San Diego
- 33.Attygalle AB, García-Rubio S, Ta J, Meinwald J (2001) J Chem Soc, Perkin Trans 2:498–506
- 34.Chang R, Cruickshank B (2005) Chemistry bonding I: Basic concepts. In: Chemistry, 8th edn, Chapter 9. McGraw-Hill, New York


