Abstract
Background
Previous studies have shown that infection with human immunodeficiency virus type 1 (HIV-1) causes acceleration of the synthesis of glutamine tRNA (tRNAGln) in infected cells. To investigate whether this might influence HIV-1 to utilize tRNAGln as a primer for initiation of reverse transcription, we have constructed HIV-1 proviral genomes in which the PBS and the A-loop region upstream of the PBS have been made complementary to either the anticodon region of tRNAGln,1 or tRNAGln,3 and 3' terminal 18 nucleotides of each isoacceptor of tRNAGln.
Results
Viruses in which the PBS was altered to be complementary to tRNAGln,1 or tRNAGln,3 with or without the A-loop all exhibited a lower infectivity than the wild type virus. Viruses with only the PBS complementary to tRNAGln,1 or tRNAGln,3 reverted to wild type following culture in SupT1 cells. Surprisingly, viruses in which the PBS and A-loop were complementary to tRNAGln,1 did not grow in SupT1 cells, while viruses in which the PBS and A-loop were made complementary to tRNAGln,3 grew slowly in SupT1 cells. Analysis of the PBS of this virus revealed that it had reverted to select tRNAThr as the primer, which shares complementarity in 15 of 18 nucleotides with the PBS complementary to tRNAGln,3.
Conclusion
The results of these studies support the concept that the HIV-1 has preferred tRNAs that can be selected as primers for replication.
Background
HIV-1 reverse transcription is initiated with the extension of the cellular tRNA that is bound to a specific sequence on the viral RNA genome known as the primer-binding site (PBS) [1-3]. The PBS is an 18-nucleotide sequence located near the 5' end of viral RNA that is complementary to the 3' terminal nucleotides of the primer tRNA used for initiation [3]. HIV-1 specifically selects tRNALys,3 from the intracellular milieu to be used as the primer for initiation of reverse transcription [4,5]. The mechanism of how HIV-1 specifically selects tRNALys,3 from the intracellular milieu is not completely understood. Previous studies have established that tRNALys,3 as well as tRNALys1,2 are enriched in HIV-1 virions [6-8]. The Gag-Pol polyprotein of HIV-1 is responsible, in part, for this enrichment of tRNALys1,2,3 into the virions [4,6,8]. Studies have also demonstrated that lysl tRNA synthetase can specifically interact with HIV-1 Gag to facilitate incorporation of tRNALys1,2,3 into HIV-1 virions [9-11]. Once this complex is incorporated into virions though, it is not clear how and why tRNALys,3 is specifically utilized as the primer for initiation of reverse transcription.
Previous studies from our lab and others have taken a genetic approach to understanding elements of HIV-1 primer selection [12-14]. For these studies, we have mutated the PBS to be complementary to tRNAs other than tRNALys,3. In general, mutation of the PBS to be complementary to other tRNAs, including tRNALys1,2, results in a virus that can transiently utilize the specific tRNA but most of the time reverts back to rapidly utilize tRNALys,3 following in vitro culture [12-14]. Stabilization of alternative tRNAs use has been accomplished through additional mutations upstream in the U5 region designated as the A-loop, which is complementary to the anticodon region of tRNALys,3 [15-19]. For some, but not all tRNAs, mutation of the A-loop region as well as the PBS to be complementary to the anticodon and 3' terminal nucleotides, respectively, of the tRNA allows this tRNA to be stably utilized by HIV-1 as a primer for reverse transcription. Using this strategy, we have generated viruses which stably utilized tRNALys1,2, tRNAMet, tRNAHis, and tRNAGlu [15-19]. A recent study has also found that HIV-1 can be forced to use tRNALys1,2 if mutations are made complementary to nucleotides in the TϕC loop of tRNALys1,2 in a second region upstream of the PBS, called the primer activation site [20].
All viruses that utilize alternative tRNAs do not replicate as efficiently as the wild type virus that utilizes tRNALys,3. This result has lead to the speculation that the availability of tRNA for primer selection might not be the same for all tRNAs. To test this it will be necessary to alter the levels of individual tRNA isoacceptors in cells. However, it is difficult to modulate the levels of tRNA in mammalian cells without leading to toxicity. Previous studies by Kuchino et al. though have found that the levels of a natural glutamine suppressor tRNA which exists as a minor species of glutamine tRNA (tRNAGln,3) in normal cells is increased in murine leukemia virus (MuLV) infected cells [21,22]. In follow up studies, Muller et al. found that although the amount of the suppressor tRNAGln,3 was only 6% of the major glutamine tRNAGln,1 levels the amount of suppressor increased almost 20 fold while the levels of non-suppressor tRNAGln,1 remained the same in cells infected with MuLV or HIV-1 [23,24]. Since the levels of a particular tRNA (tRNAGln,3) increase following infection with HIV-1, it might be possible to force HIV-1 to use this isoacceptor of tRNAGln as a primer for replication. To test this, we created viruses in which the PBS is complementary to the minor and major species of tRNAGln. We also constructed viruses which contain additional mutations in the A-loop regions to determine if this will affect the stable use of these tRNAs as primers for HIV-1 reverse transcription. Results of our study show that these viruses with the PBS complementary to either tRNAGln species were unstable and rapidly reverted back to utilize tRNALys,3. Inclusion of the A-loop complementary to the anticodon of tRNAGln,3 resulted in a virus that did not revert to utilize tRNALys,3 but selected an unexpected tRNA, tRNAThr. The results of these studies suggest that certain tRNAs are favored by HIV-1 for the selection as a primer for initiation of reverse transcription.
Results
Construction of HIV-1 proviral genomes with PBS and A-loop complementary to tRNAGln
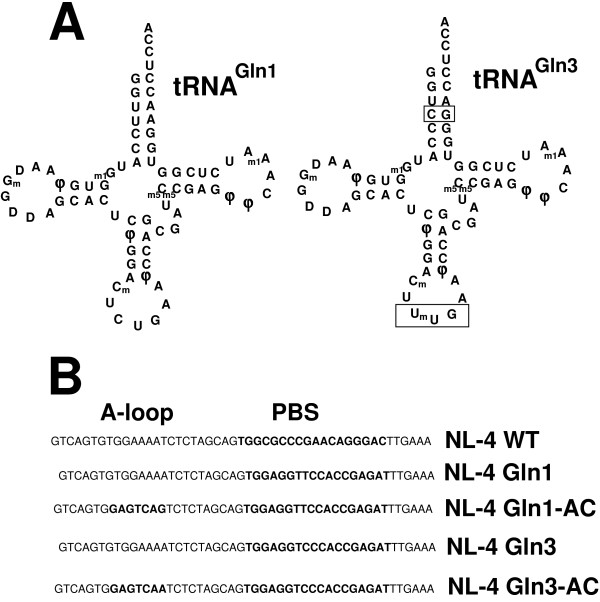
To determine if HIV-1 can utilize tRNAGln as a primer for reverse transcription, we mutated the PBS to be complementary to a 3' terminal nucleotide of tRNAGln. The major isoacceptor for tRNAGln (tRNAGln,1) has an anticodon CUG. A second tRNAGln has an anticodon UUG and is referred to as the minor tRNAGln or tRNAGln,3 [21,22] (Figure 1A). Previous studies have shown that in HIV-1 infected cells, the levels of tRNAGln,3 are increased 20 fold over that of uninfected cells [23]. The 3' terminal nucleotides of tRNAGln,1 and tRNAGln,3 differ only by a single nucleotide (Figure 1B). We have also constructed two additional proviruses in which the A-loop region of HIV-1 was mutated to correspond to the anticodon sequences of tRNAGln,1 and tRNAGln,3, respectively (Figure 1B).
Figure 1.
tRNAGln and mutated proviral genomes. Panel A. Cloverleaf structure of tRNAGln,1 and tRNAGln,3. tRNAGln,1 (major) and tRNAGln,3 (minor) are depicted. The tRNAs differ in the nucleotides within the PBS (boxed) as well as nucleotides in the anticodon region (boxed). The modified nucleotides are noted. The structures are taken from Kuchino et al. [22]. Panel B. Modifications in the NL-4 proviral genome. NL-4 WT refers to the wild type NL-4 genome with the PBS and A-loop complementary to tRNALys,3. NL-4-Gln1 refers to a modified proviral genome in which the PBS was modified to be complementary to the 3' terminal nucleotides of tRNAGln,1. NL-4 Gln1-AC also contains a PBS complementary to the 3' terminal nucleotides of tRNAGln,1 with additional modifications of the A-loop region (GAGTCAG) noted in bold. NL-4-Gln3 is an HIV-1 with the PBS modified to be complementary to the 3' terminal 18-nucleotides of tRNAGln,3. Note that the PBS is nearly identical with the exception of the T to C change in the PBS. NL-4 Gln3-AC refers to an HIV-1 in which the PBS was modified to be complementary to tRNAGln,3 with additional modification in the A-loop region consisting of GAGTCAA which is complementary to the anticodon region of tRNAGln3.
Characterization of mutant HIV-1
The first step in the characterization of HIV-1 with the PBSs alone or PBSs in combination with A-loop modifications to be complementary to tRNAGln was to determine the effects on the infectivity of viruses following transfection. For these studies, we transfected the proviral genomes into 293T cells and assayed the supernatants for infectious virus using the JC53βL assay. We also determined the amounts of virus in the supernatants by using a p24 antigen capture ELISA. The infectivity of the viruses is represented as the amount of infectious units divided by the p24 levels. Previous studies from our laboratory have shown that for the most part, mutations within the PBS of HIV genome results in viruses that exhibit infectivity approximately 20% (or lower) of the wild type virus [25]. Similar results were found for viruses in which the PBS was made complementary to tRNAGln,1 or tRNALys,3. No significant differences were observed between viruses with the PBS alone complementary to tRNAGln and viruses with the PBS and A-loop complementary to tRNAGln. The virus with a PBS and A-loop complementary to tRNAGln,1 though had the lowest infectivity, approximately 10% of the wild type virus and half as much as the other viruses in which the PBS was altered to be complementary to tRNAGln,3 (data not shown).
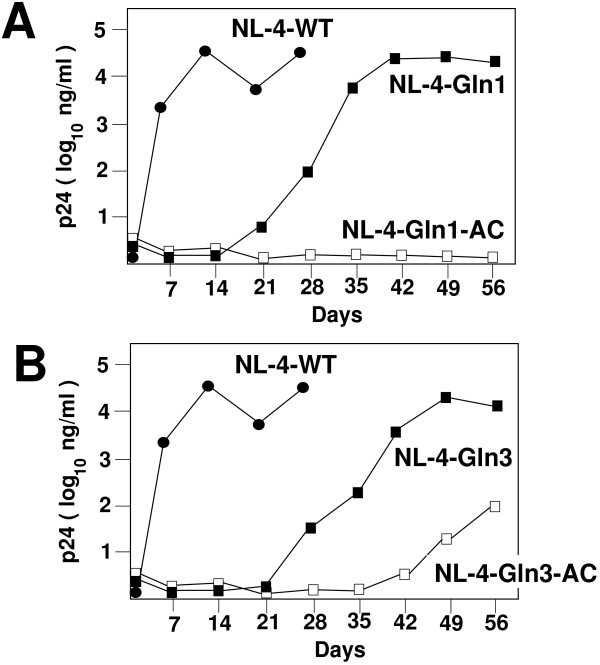
We next analyzed the replication of these viruses in SupT1 cells. Infections were established with equal amounts of infectious virus and replication was monitored by analysis of p24 in the culture supernatant. The wild type virus demonstrated a rapid increase in p24 antigen in the culture supernatant, peaking at approximately 14 days following initiation of the infection; the cultures for the wild type virus were halted at day 28 post initiation of culture. In contrast, viruses in which just the PBS alone was made complementary to tRNAGln,1 or tRNAGln,3 exhibited slower infection compared to the wild type. The p24 levels in the culture supernatants increased slowly, reaching a maximum at days 35 to 49 post initiation of culture. The final levels of p24 antigen detected in the culture supernatants from these viruses were similar to those of the wild type virus (Figure 2A). Viruses in which the PBS and A-loop were made complementary to tRNAGln,1 or tRNAGln,3 had considerably different replication profiles compared to the viruses with mutations in the PBS alone. Viruses with the PBS and A-loop complementary to tRNAGln,1 showed no increase in p24 antigen culture over the period examined (56 days of in vitro culture), indicating that the virus with this mutation in the PBS and A-loop did not undergo detectable replication and re-infection. In contrast, viruses with the PBS and A-loop complementary to tRNAGln,3 did replicate and eventually demonstrated an increase in p24 antigen during the 56 day culture period (approximately 100 fold over the starting amount of virus (p24 antigen) (Figure 2B).
Figure 2.
Replication of HIV with PBS and A-loop complementary to tRNAGln. Panel A. Replication of wild type and viruses with PBS complementary to tRNAGln,1. Infections were established in SupT1 cells with equal amounts of virus as determined by infectious units. p24 antigen was then assayed in the culture supernatants at weekly intervals following initiation of the experiment. Values for the wild type virus increased to greater than 104 nanograms/ml by approximately 14 days following initiation of the infection. The cultures were terminated at day 28. Viruses derived from NL-4-Gln1 and NL-4-Gln1-AC were carried out to approximately 56 days post initiation of culture. Note that viruses derived from NL-4-Gln1-AC did not grow, as evidenced by p24 antigen that were near the levels of mock infected cells. Cultures were terminated at day 56. Data is representative from three independent experiments. Panel B. Replication of viruses with the PBS complementary to tRNAGln,3. The replication of the wild type virus is depicted. Cultures initiated with viruses derived from NL-4-Gln3 and NL-4-Gln3-AC were monitored over 56 days of culture. The viruses derived from NL-4-Gln3 eventually reached levels approximating that of the wild type virus by day 42 through 56. Viruses derived from NL-4-Gln3-AC demonstrated a slow and gradual increase reaching levels approximately 1/100 of that of the wild type virus at the time of termination of the culture (day 56). Data is representative of three independent experiments.
We utilized PCR to amplify the U5-PBS region from integrated proviruses found in cellular genomic DNA to identify the PBS of viruses following in vitro culture. We analyzed cellular DNA obtained at day 42 from cultures infected with viruses in which the PBS alone was mutated to be complementary to tRNAGln,1 or tRNAGln,3 (Table I). In both instances, we found that analysis of U5-PBS obtained from viruses at 42 days post initiation of culture, which corresponded to the time at which there was a rise in p24 antigen, resulted in some of the viruses containing PBS complementary to the starting tRNAGln. Surprisingly, the major PBS recovered from analysis of both viruses was complementary to tRNAThr, indicating both viruses had switched their preference from tRNAGln to tRNAThr. By day 56, though, when both cultures had plateaued with the p24 antigen and the cultured supernatant, we recovered PBS that were complementary to tRNALys,3. Most probably, the process of reversion for this virus occurred through the formation of the PBS complementary to tRNAThr followed by the subsequent conversion to a PBS complementary to tRNALys,3 which resulted in the high level replication observed for both of these viruses. In contrast, analysis of viruses in which the U5-PBS was complementary to tRNAGln,3 gave a different pattern. In this case, all of the PBS recovered were complementary to tRNAThr, suggesting that the virus had selected tRNAThr from the intracellular milieu rather than the starting tRNA(tRNAGln,3) and was now stably using tRNAThr as the primer for reverse transcription.
Discussion
The original intent of the experiments was to determine whether HIV-1 would accept tRNAGln as a primer for initiation of reverse transcription. Our experiments were based on a previous study in which we found that MuLV with a PBS mutated to be complementary to tRNAGln,1 grew well in tissue culture, even though MuLV prefers to use tRNAPro as the primer for initiation of reverse transcription [26]. In addition to the viruses with the PBS complementary to tRNAGln,1, we also constructed viruses in which the PBS was complementary to the minor species, tRNAGln,3. Since previous studies have shown that this tRNA is induced in MuLV and HIV-1 infected cells at levels approximately 20 fold over the basal level found in cells [23]. Thus, we expected that HIV-1 might tolerate the selection of tRNAGln as the primer for reverse transcription. However, it was clear from our studies that viruses with a PBS alone complementary to tRNAGln,1 or tRNAGln,3 were unstable and reverted back to use tRNALys,3. Thus, even though expression of tRNAGln,3 might be enhanced in HIV-1 infected cells, this tRNA is not a preferred tRNA for selection.
Previous studies from our laboratory and others have found that regions within U5 can be altered in such a way as to facilitate the selection of alternative primers by HIV-1 for reverse transcription [15-20]. A mutation of the region upstream of the PBS (designated the A-loop) so as to be complementary to the anticodon region of certain tRNAs allows these tRNAs to be selected by HIV-1 as the primer for reverse transcription. However, the inclusion of regions within the A-loop that were complementary to tRNAGln in combination with a PBS complementary to tRNAGln had substantial effects on the stability and replication of these viruses. Viruses with a PBS and A-loop complementary to tRNAGln,1 were essentially non-infectious. While viruses in which only the PBS was altered to be complementary to tRNAGln,1 (the major tRNAGln) were infectious, they reverted back to utilize tRNALys,3 following short-term in vitro culture. Interestingly, viruses in which the PBS and A-loop were complementary to the minor species of tRNAGln,3 were infectious albeit at a greatly reduced level compared to the wild type virus. Thus, forcing HIV-1 to use tRNAGln,1 or tRNAGln,3 severely reduced the capacity for replication, indicating that this particular tRNA was not available to the virus for primer selection, even for low level of virus replication.
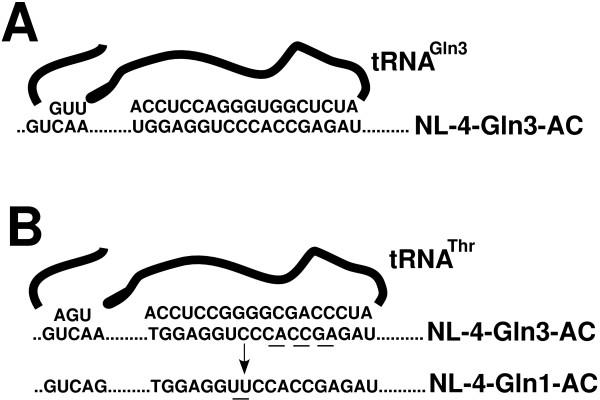
The surprising result of this study was the reversion of viruses with the PBS complementary to tRNAGln to utilize tRNAThr. How this selection occurred is not clear at this time. Comparison of the PBS sequences between those complementary to tRNAGln and tRNAThr revealed considerable homology between the first nine nucleotides as well as the last three nucleotides (Figure 3). Previous studies from our laboratory have shown that the first nine and last three to five nucleotides can facilitate the reverse transcription of HIV-1 in which the PBS was made complementary to alternative tRNAs [27]. It is clear that following selection of tRNAThr the virus could, through the process of reverse transcription, convert the PBS to be complementary to this tRNA and allow limited growth. Why the virus with a PBS and A-loop complementary to tRNAGln,1 did not convert to use tRNAThr is unknown. It is possible that the selection of tRNAThr is passive, rather than active. Thus, if the virus happens to capture tRNAThr, it will grow, albeit more slowly than the wild type virus. The fact that the process of conversion goes through an intermediate with a PBS complementary to tRNAThr suggests this tRNA has a greater availability for capture than tRNAGln. Additional studies will be needed to address this possibility.
Figure 3.
Sequence complementarity of tRNAGln and tRNAThr with mutant proviral genomes. Panel A. Sequence complementary of tRNAGln,3 with NL-4-Gln3-AC. Depicted is the predicted complementarity between the 3' terminal nucleotides and the PBS and the anticodon of tRNAGln,3 with the modified A-loop region of NL-4-Gln3-AC. Panel B. Complementarity between 3' terminal nucleotides of tRNAThr with the PBS of NL4-Gln3-AC. Nucleotide differences within the PBS and tRNAThr are underlined. The anticodon region of tRNAThr has complementarity with the modified A-loop region of NL-4-Gln3. Additional complementarity between tRNAThr and the PBS of NL-4-Gln1-AC is also shown. The single nucleotide difference between the PBS is underlined. The resulting GC pair of tRNAThr and the PBS of NL-4-Gln3-AC should be compensated for by a GU base pair. Note also the predicted complementarity between the anticodon region of tRNAThr with the modified A-loop region of NL-4-Gln1-AC.
Conclusion
In the current study, we have characterized the replication of HIV-1 in which the PBS has been altered to be complementary to tRNAGln. Viruses were constructed in which the PBS or PBS and A-loop were modified to be complementary to either tRNAGln,1 or tRNAGln,3. All viruses were found to have poor replicative capacity and the PBS was unstable following in vitro culture. However, analysis of the PBS from integrated proviruses revealed that a new tRNA, tRNAThr was preferred by HIV-1 for replication indicating that HIV-1 prefers tRNAThr as a primer for replication.
The results of our study re-enforces the idea that HIV-1 has preferences for the selection of certain tRNAs for replication. Obviously, the most preferred primer for selection is tRNALys,3. However, the results from our current and previous studies indicate HIV-1 can tolerate other tRNAs as primers. For example, in a previous study, we found that viruses in which the PBS was mutated to be complementary to tRNATrp reverted to select tRNAMet as the primer for reverse transcription [28]. Viruses such as those with a PBS and A-loop complementary to tRNAHis and tRNALys1,2 and tRNAGlu have been generated in our laboratory, suggesting the these tRNAs also are acceptable for selection as primers [15-19,29]. Since HIV-1 can select other tRNAs as the primer for reverse transcription, why HIV-1 does not use these other tRNAs for replication is unknown. It is possible that HIV-1 could have access to several different tRNAs during primer selection. However, under certain circumstances where tRNALys,3 is not favored, such as that with proviral genomes with certain A-loop modifications, the virus can select other tRNAs, such as tRNAMet and tRNAThr as the primer for reverse transcription if sufficient complementarity with the PBS exists. Further understanding of the process and what influences the preference for certain tRNAs will be important to resolve the mechanism of primer selection.
Materials and methods
Tissue culture
293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), and SupT1 cells were grown in RPMI 1640 medium supplemented with 15% FBS.
Construction of mutant proviral genomes
Mutagenesis was performed by using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. The PBS sequence in the shuttle vector pUC119PBS [29] was changed to be complementary to the 18 3'-terminal nucleotides of tRNAGln3 using the primers 5'-TGGAAAATCTCTAGCAGTGGAGGTCCCACCGAGATCTGAAAGCGAAAGGGAAACC-3' and 5'-GGTTTCCCTTTCGCTTTCAGATCTCGGTGGGACCTCCACTGCTAGAGATTTT CCA-3', creating the plasmid pUC-Gln3. pUC-Gln3 was then used as a template to mutate the PBS to be complementary to tRNAGln1, with the primers 5'-CTCTAGCAGTGGAGGTTCCAC CGAGATCTGAAAG-3' and 5'-CTTTCAGATCTCGGTGGAACCTCCACTGCTAGAG-3', resulting in plasmid pUC-Gln1. To create the plasmid pUC-Gln3AC, which contains further mutations in the U5 region complementing the anti-codon loop of tRNAGln3, PUC-Gln3 was used as a template, along with the primers 5'-ACCTCCACTGCTAGAGATTGACTCCACTGACTA AAAGGGTCTGAGG-3' and 5'-CCTCAGACCCTTTTAGTCAGTGGAGTCAATCTCTAGC AGTGGAGGT-3'. Likewise, pUC-Gln1AC with U5 sequence complementary to the anti-codon loop of tRNAGln1 was made by using PUC-Gln1 as a template, with the primers 5'-CCTCAGACCCTTTTAGTCAGTGGAGTCAGTCTCTAGCAGTGGAGGT-3' and 5'-ACCTCCACTGCTAGAGACTGACTCCACTGACTAAAAGGGTCTGAGG-3'. Subsequently, the HpaI-BssHII fragments of pUC-Gln3, pUC-Gln3AC, pUC-Gln1 and pUC-Gln1AC containing the U5-PBS region were sub-cloned between the SmaI and BssHII sites of pNL4-3 to form the complete pro-viral clones of pNL4-3-Gln3, pNL4-3-Gln3AC, pNL4-3-Gln1 and pNL4-3-Gln1AC. Sequences of pro-viral clones were verified by DNA sequencing.
Transfection and analysis of viral infectivity
Plasmids were transfected into 293T cells using the Fugene 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, IN) according to the protocol. Briefly, 2 μg of pro-viral plasmid DNA and 3 μl of Fugene 6 reagent were combined in 100 ul serum free DMEM, and incubated at room temperature for 30 min. The mixture was then added to one well of 6-well plate containing 60% confluent 293T cells in 2 ml fresh medium. The transfections was incubated at 37°C overnight, before replaced with fresh medium, and supernatants were collected after 48 hours and stocked at -80°C in aliquots. Levels of infectious virus (IU/μL) in 293T supernatants were determined using the JC53βL assay as previously described [25,30].
Infection and maintaining of viral cultures
Virus supernatant containing 250 infectious units were added to 106 SupT1 cells in 125 μl RPMI supplemented with 2% FBS in a 15 ml Falcon conical tube (BD Bioscience) with caps loosened, and incubated at 37°C for 2 hrs to allow absorption, then transferred to a tissue culture flask containing 10 ml RPMI supplemented with 15% FBS to further culture the infected cells. Every 3–4 days, 8 ml of culture were replaced with 8 ml fresh medium, and supernatants and cell pellets were collected every 7 days and stocked at -80°C. Once the infected SupT1 cultures were found to be cleared of cells, 106 new SupT1 cells were added to continue the culture.
DNA sequence analysis of pro-viral U5 and PBS region
High-molecular-weight DNA was isolated from SupT1 cell pellets using the Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. A fragment containing the U5 and PBS regions of the integrated provirus was PCR amplified from the high-molecular weight DNA using primers 5'-CGGAATTCTCTCCTTCTAGCCTCCGCTAGTC-3' and 5'-CCTTGAGCAT GCGATCTACCACACACAAGGC-3'. The PCR products were run on a 1% agarose gel and DNA running approximately 750 bp size were extracted using the Qiagen Gel Purification Kit (Qiagen, Valencia, CA) and sub-cloned into pGEM-T-Easy vector (Promega Madison, WI) according to the protocol. White colonies were picked and grown to produce DNA, which were screened for inserts by EcoRI enzyme digestion. The U5-PBS sequence of TA clones containing the approximately 750 bp inserts were analyzed by automated DNA sequencing, using the primer corresponding to the T7 promoter sequence flanking the multiple cloning site of the vector.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
ML, PGE, NN and CDM conceived the studies and ML, PGE and NN performed the experiments. CDM and ML wrote the manuscript.
Acknowledgments
Acknowledgements
We thank members of the Morrow laboratory for helpful comments and Adrienne Ellis for preparation of the manuscript. CDM acknowledges helpful comments from MAR. DNA sequencing was carried out by the UAB CFAR DNA Sequencing Core (AI 27727). The research was supported by a grant from the NIH to CDM (AI 34749).
Contributor Information
Meng Li, Email: prisslimli@yahoo.com.
Peter G Eipers, Email: peipers@uab.edu.
Na Ni, Email: niinaa1@uab.edu.
Casey D Morrow, Email: caseym@uab.edu.
References
- Panet A, Berliner H. Binding of tRNA to reverse transcriptase of RNA tumor viruses. J Virol. 1978;26:214–220. doi: 10.1128/jvi.26.2.214-220.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G, Dahlberg JE. RNA-directed DNA synthesis in Moloney murine leukemia virus: Interaction between the primer tRNA and the genome RNA. J Virol. 1979;31:398–407. doi: 10.1128/jvi.31.2.398-407.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin HM. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981;27:1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak MA, Prasad VR, Wainberg MA, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys,3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- Mak J, Jiang M, Wainberg MA, Hammarskjold ML, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorchid A, Javannbakht H, Wise S, Halwani R, Parniak MA, Wainberg MA, Kleiman L. Sequences within Pr160gag-pol affecting the selective packaging of primer tRNALys,3 into HIV-1. J Mol Biol. 2000;299:17–26. doi: 10.1006/jmbi.2000.3709. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, Halwani R, Cen S, Saadatmand J, Musier-Forsyth K, Gottlinger H, Kleiman L. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J Biol Chem. 2003;278:27644–27651. doi: 10.1074/jbc.M301840200. [DOI] [PubMed] [Google Scholar]
- Cen S, Javanbakht H, Kim S, Shiba K, Craven RC, Rein A, Ewalt KL, Schimmel P, Musier-Forsyth K, Kleiman L. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J Virol. 2002;76:13111–13115. doi: 10.1128/JVI.76.24.13111-13115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Mak J, Huang Y, Kleiman L. Reverse transcriptase is an important factor for the primer tRNA selection in HIV-1. Leukemia. 1994;8:S149–S151. [PubMed] [Google Scholar]
- Wakefield JK, Wolf AG, Morrow CD. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNALys,3. J Virol. 1995;69:6021–6029. doi: 10.1128/jvi.69.10.6021-6029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mak J, Arts EJ, Gu Z, Kleiman L, Wainberg MA, Parniak MA. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J Virol. 1994;68:6198–6206. doi: 10.1128/jvi.68.10.6198-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AT, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNALys,3. J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield JK, Kang SM, Morrow CD. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J Virol. 1996;70:966–975. doi: 10.1128/jvi.70.2.966-975.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kang SM, Li Y, Morrow CD. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA. 1998;4:394–406. doi: 10.1017/S1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Zhang Z, Morrow CD. Identification of a human immunodeficiency virus type 1 that stably uses tRNALys1,2 rather than tRNALys,3 for initiation of reverse transcription. Virology. 1999;257:95–105. doi: 10.1006/viro.1999.9615. [DOI] [PubMed] [Google Scholar]
- Kang SM, Morrow CD. Genetic analysis of a unique human immunodeficiency virus type 1 (HIV-1) with a primer binding site complementary to tRNAMet supports a role for U5-PBS stem-loop RNA structures in initiation of HIV-1 reverse transcription. J Virol. 1999;73:1818–1827. doi: 10.1128/jvi.73.3.1818-1827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Zhang Z, Morrow CD. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J Virol. 1997;71:207–217. doi: 10.1128/jvi.71.1.207-217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbink TEM, Beerens N, Berkhout B. Forced selection of a human immunodeficiency virus type 1 that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme. J Virol. 2004;78:10706–10714. doi: 10.1128/JVI.78.19.10706-10714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Nishimura S, Schroder HC, Rottmann M, Muller WE. Selective inhibition of formation of suppressor glutamine tRNA in Moloney murine leukemia virus-infected NIH-3T3 cells by Avarol. Virology. 1988;165:518–526. doi: 10.1016/0042-6822(88)90596-X. [DOI] [PubMed] [Google Scholar]
- Kuchino Y, Beiere H, Akita N, Nishimura S. Natural UAG suppressor glutamine tRNA is elevated in mouse cells infected with Moloney murine leukemia virus. Proc Natl Aca Sci (USA) 1987;84:2668–2672. doi: 10.1073/pnas.84.9.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WE, Schroder HC, Reuter P, Sarin PS, Hess G, Meyer zum Buschenfelde KH, Kuchino Y, Nishimura S. Inhibition of expression of natural UAG suppressor glutamine tRNA in HIV-infected human H9 cells in vitro by Avarol. AIDS Res and Human Retro. 1988;4:279–286. doi: 10.1089/aid.1988.4.279. [DOI] [PubMed] [Google Scholar]
- Muller WE, Schroder HC. Cell biological aspects of HIV-1 infection: effect of the anti-HIV-1 agent Avarol. Int J Sports Med. 1991;12:S43–49. doi: 10.1055/s-2007-1024749. [DOI] [PubMed] [Google Scholar]
- Moore-Rigdon KL, Kosloff BR, Kirkman RL, Morrow CD. Preferences for the selection of unique tRNA primers revealed from analysis of HIV-1 replication in peripheral blood mononuclear cells. Retrovirology. 2005;2 doi: 10.1186/1742-4690-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MT, Morrow CD. Analysis of murine leukemia virus replication complemented by yeast tRNAPhe reveals inherent preferences for the tRNA primer selected for reverse transcription. Virology. 2004;324:430–438. doi: 10.1016/j.virol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Rhim H, Park J, Morrow CD. Deletions in the tRNALys primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J Virol. 1991;65:4555–4564. doi: 10.1128/jvi.65.9.4555-4564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SM, Wakefield JK, Morrow CD. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology. 1996;222:401–414. doi: 10.1006/viro.1996.0437. [DOI] [PubMed] [Google Scholar]
- Dupuy LC, Kelly NJ, Elgavish TE, Harvey SC, Morrow CD. Probing the importance of tRNA anticodon: human immunodeficiency virus type 1 (HIV-1) RNA genome complementarity with an HIV-1 that selects tRNAGlu for replication. J Virol. 2003;77:8756–8764. doi: 10.1128/JVI.77.16.8756-8764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]