Abstract
The modulation of utrophin gene expression in muscle by the nerve-derived factor agrin plausibly involves the trophic factor ARIA/heregulin. Here we show that heregulin treatment of mouse and human cultured myotubes caused a ≈2.5-fold increase in utrophin mRNA levels. Transient transfection experiments with utrophin promoter-reporter gene constructs showed that this increase resulted from an enhanced transcription of the utrophin gene. In the case of the nicotinic acetylcholine receptor δ and ɛ subunit genes, heregulin was previously reported to stimulate transcription via a conserved promoter element, the N-box, which binds the multimeric Ets-related transcription factor GA binding protein (GABP). Accordingly, site-directed mutagenesis of a single N-box motif in the utrophin gene promoter abolished the transcriptional response to heregulin. In addition, overexpression of heregulin, or of the two GABP subunits in cultured myotubes, caused an N-box-dependent increase of the utrophin promoter activity. In vivo, direct gene transfer into muscle confirmed that heregulin regulates utrophin gene expression. Finally, electrophoretic mobility shift assays and supershift experiments performed with muscle extracts revealed that the N-box of the utrophin promoter binds GABP. These findings suggest that the subsynaptic activation of transcription by heregulin via the N-box motif and GABP are conserved among genes expressed at the neuromuscular junction. Because utrophin can functionally compensate for the lack of dystrophin, the elucidation of the molecular mechanisms regulating utrophin gene transcription may ultimately lead to therapies based on utrophin expression throughout the muscle fibers of Duchenne muscular dystrophy patients.
Duchenne muscular dystrophy (DMD) is the most prevalent primary myopathy because it affects ≈1 out of every 3,500 male births (1). The disease is characterized by repeated cycles of muscle degeneration/regeneration with an eventual failure to regenerate leading to the replacement of muscle fibers by fat and connective tissues. DMD progresses rapidly because patients are functionally impaired before their teen years and death usually occurs in the second or third decade of life, most often as a result of respiratory or cardiac failure. The genetic defect underlying DMD was simultaneously mapped to chromosome Xp21 in different laboratories (for reviews, see refs. 2 and 3). The gene responsible for this disease was termed dystrophin, and it codes for a large cytoskeletal protein known to accumulate at the sarcolemma of muscle fibers. Mutations and/or deletions of this gene, as seen in DMD, lead to an absence of full-length dystrophin, thereby making muscle fibers extremely fragile to the effects of repetitive cycles of muscle contraction and relaxation. Although several therapeutic strategies have been envisaged, including dystrophin gene replacement and pharmacological interventions (see refs. 4–6), there is currently no cure available for DMD.
Several years ago, an autosomal homologue to dystrophin was identified (7). This gene, now referred to as utrophin, also codes for a large cytoskeletal protein (8). In contrast to the homogeneous distribution of dystrophin along muscle fibers, utrophin preferentially accumulates within the postsynaptic domain of the neuromuscular junction in both normal and DMD muscle fibers (9–12). Because of the high degree of sequence identity between dystrophin and utrophin as well as their ability to bind a group of proteins referred to as the dystrophin-associated proteins (13), it has been suggested that increased expression of utrophin into extrasynaptic regions of dystrophic muscle fibers may represent an alternate therapeutic strategy for DMD (14, 15). Recent studies using transgenic mouse model systems have clearly demonstrated that expression of utrophin throughout muscle fibers can indeed functionally compensate for the lack of dystrophin and hence, prevent the muscle pathology (16–18). It thus becomes important to elucidate the molecular and cellular mechanisms presiding over utrophin expression at the neuromuscular junction to ultimately modulate specific regulatory events that could lead to expression of the endogenous gene product along the length of dystrophic muscle fibers.
In a recent series of studies, we began to examine the role of the nerve in regulating utrophin expression at the neuromuscular junction. Initially, we showed that local transcriptional activation of the utrophin gene in myonuclei located within the postsynaptic sarcoplasm contributes to the synaptic localization of utrophin (19). We have next examined the contribution of nerve-derived trophic factors in the regulation of utrophin in muscle cells and showed that agrin induced the expression of utrophin in cultured muscle cells via a transcriptional regulatory mechanism (20). ARIA-heregulin, another nerve-derived trophic factor, is known to regulate acetylcholine receptor (AChR) subunit gene expression (for reviews, see refs. 21 and 22). In addition, agrin is thought to stimulate transcription of the AChR genes via heregulin. In the present study, we demonstrate that heregulin modulates utrophin gene expression and analyze the molecular mechanisms of this regulation in cultured myotubes and in muscle fibers in vivo.
METHODS
Tissue Culture.
Mouse C2 muscle cells were cultured as described (20). Normal human skeletal muscle cells were obtained from Clonetics-BioWhittaker (San Diego), and they were grown and maintained according to the supplier’s recommendations. Three- to five-day-old myotubes were treated with 3 or 30 nM heregulin (kindly supplied by M. Sliwkowski, Genentech) for 48 hr.
RNA Extraction, Reverse Transcription, and PCR.
Total RNA was extracted from samples by using Tripure as recommended by the manufacturer (Boehringer Mannheim). The RNA concentration was determined by using a GeneQuant II RNA/DNA spectrophotometer (Amersham Pharmacia), and the samples were rediluted to a final concentration of 50 ng/μl. Only 2 μl (100 ng of total RNA) of this dilution was used for reverse-transcription and amplification with the PCR.
Reverse transcription–PCR analysis was performed to strictly determine the relative abundance of transcripts under different experimental conditions. Utrophin cDNAs were specifically amplified by using primers synthesized on the basis of available sequences for human (10) and mouse (20) cDNAs. PCR experiments were performed as described elsewhere (20). Typically, 30–36 cycles of amplification were performed because control experiments showed that these cycle numbers were within the linear range of amplification. After amplification, PCR products were separated on ethidium bromide-stained agarose gels, and the size of the products was estimated by using the 100-bp molecular mass marker (GIBCO/BRL). For quantitative assays, the PCR products were separated on Vistragreen (Amersham)-containing gels and the fluorescent intensity of the products, which is linearly related to the amount of DNA, was quantitated by using a Storm PhosphorImager (Molecular Dynamics) and analyzed by using the accompanying imagequant software (Molecular Dynamics). In these assays, we verified that equal amounts of total RNA were indeed used for each sample by monitoring from the same reverse transcription mixtures, the abundance of either S12 rRNA or glyceraldehyde-3-phosphate dehydrogenase (23).
Expression of Utrophin Promoter-Reporter Gene Constructs.
In these experiments, we used the same human utrophin promoter-reporter gene constructs that we recently described (see refs. 19 and 20). Specifically, we used the 1.3-kb utrophin promoter fragment and the N5 N-box mutant. These promoter fragments were inserted upstream of the reporter gene LacZ and a nuclear localization signal. In addition, we used plasmids containing the heregulin β cDNA driven by the cytomegalovirus promoter, and the GA binding proteins α and β (GABP α and β) cDNAs placed downstream of the murine sarcoma virus promoter (24). Plasmid DNA was prepared by using the Qiagen (Chatsworth, CA) Mega-Prep procedure.
C2 myoblasts were transfected with 3 μg of the appropriate utrophin promoter-reporter gene construct by using the Mammalian Transfection System—Calcium Phosphate kit (Promega). Once the cultures became confluent, the medium was switched to the differentiation medium and treated with heregulin as described above. Forty-eight hours later, cells were scraped into 300 μl of 1× Reporter Lysis buffer (Promega) and freeze thawed twice. After centrifugation, the supernatants were collected and the level of β-galactosidase (β-gal) activity was determined by using a luminescent assay (Luminescent β-gal Enzyme Kit; CLONTECH) and normalized to a cotransfected chloramphenicol acetyltransferase (CAT) plasmid (Promega) and protein content. CAT activity was determined by using a CAT Enzyme Assay system (Promega), and protein content was determined by the bicinchoninic acid method (Pierce).
For direct gene transfer into mouse tibialis anterior (TA) muscles, experiments were performed as described (19, 20, 25–27). Briefly, 25 μl of DNA solution was injected directly into TA muscles of 4-week-old mice. Muscles were excised 2 weeks later, frozen in liquid nitrogen, and homogenized in 500 μl of 1× Reporter Lysis buffer (Promega) by using a Polytron (Kinematica, Lucerne, Switzerland). After centrifugation, the supernatants were collected and the activities of β-gal and CAT were determined as described above.
Muscle Extracts and Electrophoretic Mobility Shift Assays.
Muscle extracts were prepared as described (26). Electrophoretic mobility shift assays were performed by using 32P-labeled probes encompassing the utrophin N-box region (sense: 5′-GGCTGATCTTCCGGAACAAAGT-3′ and antisense: 5′-ACTTTGTTCCGGAAGATCAGCC-3′). The binding reaction mixture included 0.2 ng of labeled probes, 1.0 μg of poly(dI-dC), and 20 μg of muscle extract and was incubated for 30 min on ice before electrophoresis in a 5% polyacrylamide gel. The specificity of the binding reaction was assessed by adding a 50- or 500-fold molar excess of unlabeled probe in the reaction mixture. For the supershift assays, antibodies to GABP α and β were kindly provided by Steve McNight (Tularik, San Francisco). These antibodies were added to the reaction mixture for 20 min on ice after the initial 30-min incubation and before electrophoresis.
RESULTS
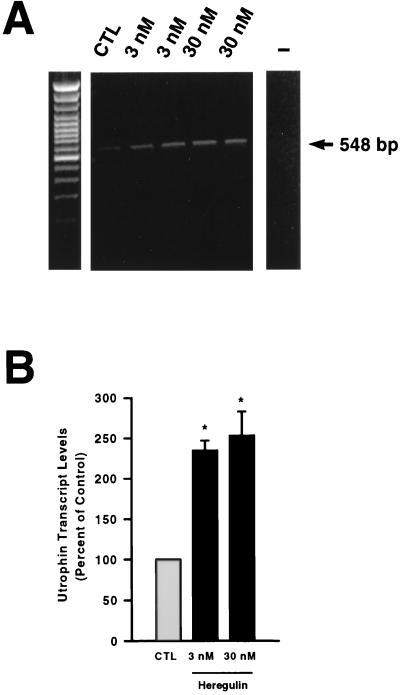
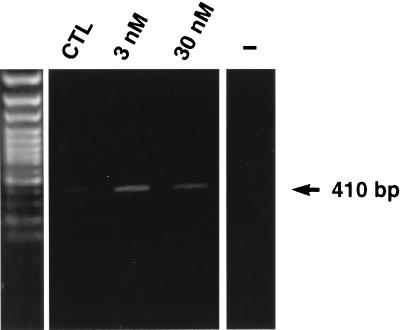
In a first set of experiments, we examined whether heregulin increased utrophin gene expression in cultured myotubes. In comparison with untreated cultures, we found that the levels of utrophin transcripts were increased by heregulin treatment (Fig. 1A). The abundance of utrophin mRNA was ≈2.5-fold higher (P < 0.05) after treatment with either 3 or 30 nM heregulin (Fig. 1B). By contrast, treatment of myotubes with epidermal growth factor, platelet-derived growth factor, and insulin-like growth factors I and II, which are known to influence expression of muscle proteins in tissue culture (28), failed to modify expression of utrophin transcripts (data not shown). Consistent with the results obtained with mouse muscle cells, we also noted that treatment of human myotubes with either 3 or 30 nM heregulin also led to a ≈2-fold increase in utrophin mRNA levels (Fig. 2).
Figure 1.
(A) A representative example of an ethidium bromide-stained gel of utrophin PCR products (548 bp) obtained from nontreated (control; CTL) versus heregulin-treated (3 or 30 nM) mouse myotubes. Note the increase in utrophin mRNA levels after heregulin treatment. The negative control lane is marked with a minus sign. The left panel is the 100-bp marker (GIBCO/BRL). (B) Quantitative analysis of utrophin mRNA levels in control and heregulin-treated myotubes. Utrophin transcript levels are expressed as a percent of control. Asterisks denote significant differences from control (CTL) levels (Student’s t test, P < 0.05).
Figure 2.
Representative example of an ethidium bromide-stained gel of utrophin PCR products (410 bp) obtained from nontreated (control; CTL) versus heregulin-treated (3 and 30 nM) human myotubes. Note the increase in utrophin mRNA levels after heregulin treatment. The negative control lane is marked with a minus sign. The left panel is the 100-bp marker (GIBCO/BRL).
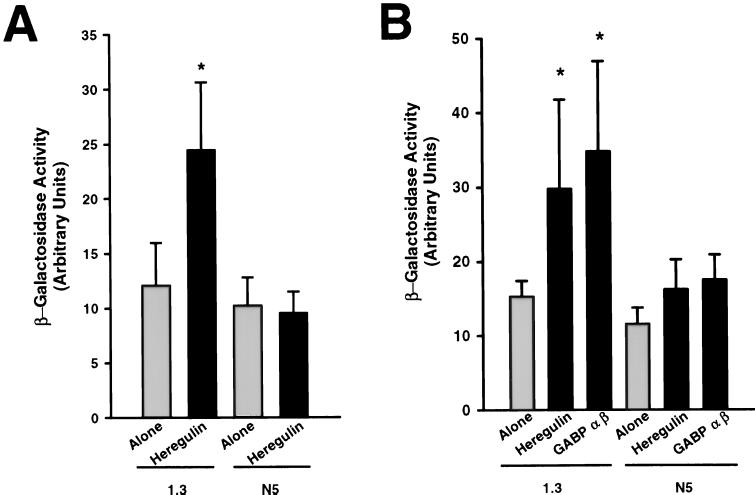
To determine whether the increase in utrophin transcripts after heregulin treatment resulted from enhanced transcriptional activation of the utrophin gene, we next transfected C2 myoblasts with plasmids containing the reporter gene LacZ driven by the 1.3-kb wild-type utrophin promoter or its N-box mutated counterpart (see N5 mutant construct in ref. 20) and treated myotubes for 48 hr with heregulin. As illustrated in Fig. 3A, we observed a significant increase (P < 0.05) in the expression of β-gal in cells transfected with the construct containing the wild-type promoter fragment and treated with heregulin. Moreover, expression of the reporter gene was not affected after heregulin treatment in cultures transfected with constructs containing the N5-mutated utrophin promoter fragment. Similarly, cotransfection of C2 cultures with plasmids containing the cDNAs of heregulin or the two subunits of GABP driven by constitutive promoters and the wild-type utrophin promoter fragment caused an increase in the expression of the reporter gene (Fig. 3B). In parallel, cultures transfected with plasmids containing the N-box mutant promoter construct, and overexpressing either heregulin or GABP α and β, failed to affect expression of β-gal.
Figure 3.
(A) Mouse myotubes transfected with plasmids containing human utrophin promoter fragments (either the 1.3-kb wild-type or the N5 mutant; see ref. 20) and the reporter gene LacZ were treated with heregulin. Note the increase in activity in cultures transfected with the wild-type utrophin promoter fragment. (B) Cotransfection of the utrophin wild type or N5 mutant promoter fragments with cDNAs encoding heregulin or GABP α and β. Note the increase in activity of the reporter gene driven by the 1.3-kb wild-type promoter after overexpression of heregulin or GABP α and β. For all these experiments, the levels of β-gal activity were determined and normalized to CAT and protein content. Asterisks denote significant differences from control levels (Student’s t test, P < 0.05).
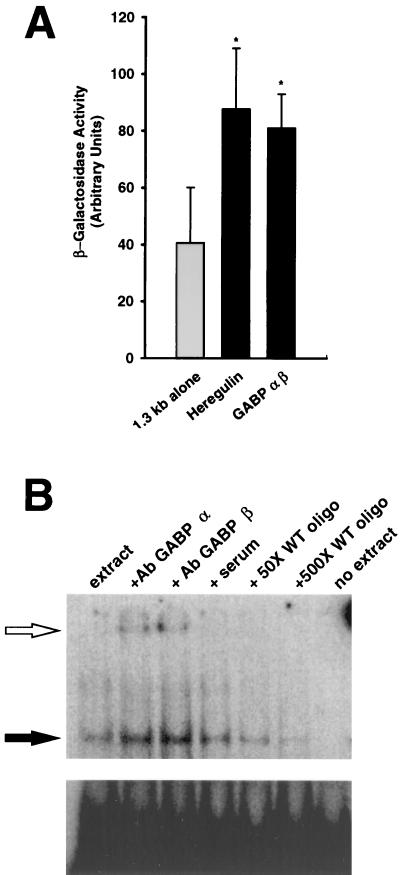
To verify that similar regulatory mechanisms could contribute to the regulation of the utrophin gene in vivo, we performed a series of experiments in which plasmid DNA was directly injected into mouse TA muscles. In comparison with injection of the 1.3-kb utrophin promoter-reporter gene constructs, coinjection with a plasmid containing the heregulin cDNA constitutively expressed led to a ≈2-fold increase (P < 0.05) in the expression of β-gal (Fig. 4A). In these experiments, we also examined the contribution of GABP α and β in the control of utrophin gene expression in vivo. As shown in Fig. 4A, coinjection of the wild-type utrophin promoter construct with plasmids constitutively expressing GABP α and β also caused a significant increase (P < 0.05) in the activity of the reporter gene.
Figure 4.
(A) Ectopic overexpression of heregulin or GABP α and β increases expression of the wild-type utrophin promoter-reporter gene construct in vivo. Mouse TA muscles were coinjected with plasmids containing the wild-type human utrophin promoter fragment along with plasmids encoding heregulin or GABP α and β, and the level of β-gal activity was determined 2 weeks later and normalized to CAT and protein content. Asterisks denote significant differences from control levels (Student’s t test, P < 0.05). (B) The Ets-related transcription factor GABP binds to the N-box motif contained within the utrophin promoter. DNA-binding activity to the N-box motif (solid arrow) was detected by using electrophoretic mobility shift assays and extracts from TA muscles. This band was competed by incubation with either 50 or 500 M excess of wild-type (WT) unlabeled probe. In addition, this band was supershifted (open arrow) by an additional incubation with antibodies against either GABP α and β, but not by incubation with the preimmune serum. (Lower) Unbound radioactivity.
Finally, to confirm the binding of GABP to the N-box present in the utrophin promoter, we performed a series of electrophoretic mobility shift assays using muscle extracts and probes encompassing the utrophin N-box region. In these experiments, we observed specific protein binding activity that was competed by an excess of unlabeled oligonucleotides (Fig. 4B). Furthermore, this binding activity was supershifted by incubating the reaction mixtures with antibodies against GABP α or β, thereby confirming the involvement of these subunits from an Ets-related protein in the transcriptional regulation of the utrophin gene.
DISCUSSION
Recently, we showed that maintenance of high levels of utrophin at the neuromuscular junction involved the local transcriptional regulation of the utrophin gene in myonuclei of the postsynaptic sarcoplasm (19, 20). In addition, we demonstrated the important contribution of the nerve in maintaining utrophin expression because induction of ectopic synapses at sites distant from the original neuromuscular junctions resulted in the appearance of utrophin at these newly formed synaptic contacts (19). Because expression of utrophin is largely insensitive to nerve-evoked electrical activity (29, 30), we postulated that expression of utrophin in muscle is strictly positively regulated in the subsynaptic myonuclei by trophic factors (15). In the present study, we show that the trophic factor heregulin, which is concentrated at the neuromuscular junction (for a review, see ref. 21), can increase utrophin gene expression in cultured muscle cells as well as in muscle fibers in vivo.
It is well established that ARIA/heregulin exerts a profound influence on expression of the AChR subunit genes in myogenic cells maintained in culture (for reviews, see refs. 21 and 22). Such observation has led to the notion that the localization of this molecule under nerve terminals and its subsequent interaction with ErbB receptors located on the postsynaptic membrane of the neuromuscular junction trigger a signaling cascade that culminates in the local activation of specific AChR subunit genes within myonuclei of the postsynaptic sarcoplasm (31–33). Until recently, the nature of the signaling pathway involved in this trans-synaptic control of gene expression was unknown. However, promoter analysis has led to the identification of a DNA element termed N-box that is critical for the synapse-specific expression of the AChR δ and ɛ subunit genes (26, 27). Additional studies have shown that the N-box plays a central role in the transcriptional activation of AChR genes by heregulin. These studies demonstrated that the response to heregulin involved binding of Ets transcription factors to the N-box (24, 34). The candidate factor implicated in this regulation was shown to be the multimeric Ets-related factor GABP (24), a finding recently confirmed by Fromm and Burden (35). Interestingly, it was also shown that the Ras/mitogen-activating protein kinase pathway, through which heregulin stimulates AChR genes transcription (36, 37), controls the N-box-dependent response to heregulin and modulates the phosphorylation of GABP (24). Taken together, these data are consistent with a model whereby selective expression of AChR subunit genes at the neuromuscular junction is achieved via interaction of heregulin with ErbB receptors, which in turn, results in the transactivation of AChR subunit promoters through Ets-related transcription factors binding to the N-box motif. Our current results showing that heregulin and GABP α and β increase utrophin gene expression in muscle cells via the N-box, are consistent with this model. A conserved mechanism involving the N-box and GABP may thus regulate the expression of multiple synapse-specific genes at the level of the fundamental nuclei.
In a recent study, we showed that treatment of myogenic cells in culture with agrin increased the expression of utrophin via a transcriptional regulatory mechanism involving the N-box (20). However, the exact nature of the regulatory events underlying this increase in utrophin expression remained unclear. In this context, it is noteworthy that Brenner and colleagues have shown that agrin treatment enhances the transcription of the AChR ɛ subunit gene in cultured muscle cells (38, 39), by first inducing the local accumulation of muscle-derived heregulin and its ErbB tyrosine kinase receptors, subsequently leading to the transcriptional activation of the AChR ɛ subunit gene via an autocrine/paracrine pathway (40). Based on our current findings showing that overexpression of heregulin in muscle cells increases utrophin gene expression, it is likely that a similar mechanism accounts for the increase in utrophin expression after agrin treatment.
Recent studies performed with transgenic mouse model systems have revealed that an increase in the expression of utrophin in extrasynaptic compartments of dystrophic muscle fibers prevents the occurrence of the muscle pathology (16–18), thereby indicating that up-regulation of utrophin is indeed a viable approach for treating DMD. Therefore, the results demonstrating that both agrin and heregulin can modulate expression of the utrophin gene in myogenic cells in culture (this study and ref. 20) as well as in muscle fibers in vivo (this study and refs. 41 and 42) have definite implications for the treatment of DMD because they offer the possibility to pharmacologically stimulate the signaling cascade that links events taking place at the membrane to alterations in utrophin gene expression. In this context, our results showing that heregulin treatment increased expression of utrophin transcripts not only in mouse muscle cells but also in human myotubes is particularly relevant because they provide the necessary basis to begin designing specific pharmacological interventions in a clinically relevant system.
Acknowledgments
This work was supported by grants from the Muscular Dystrophy Association of America, the Muscular Dystrophy Association of Canada, the Association Française contre les Myopathies, the Medical Research Council of Canada and Great Britain, the Collège de France, and the Centre National de la Recherche Scientifique. During the course of this work, A.O.G. was an Arthur Minden Predoctoral Fellow from the Muscular Dystrophy Association of Canada and is now supported by a Ministry of Ontario Graduate Scholarship. L.M.A. holds a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship. E.A.B. is an Action Research Training Fellow. B.J.J. is a Scholar of the Medical Research Council of Canada.
ABBREVIATIONS
- AChR
acetylcholine receptor
- DMD
Duchenne muscular dystrophy
- GABP
GA-binding protein
- CAT
chloramphenicol acetyltransferase
- β-gal
β-galactosidase
- TA
tibialis anterior
References
- 1.Emery A. Neuromusc Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Sadoulet-Puccio H M, Kunkel L M. Brain Pathol. 1996;6:25–35. doi: 10.1111/j.1750-3639.1996.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 3.Worton R. Science. 1995;270:755–760. doi: 10.1126/science.270.5237.755. [DOI] [PubMed] [Google Scholar]
- 4.Ahn A H, Kunkel L M. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 5.Khan M A. J Neurol Sci. 1993;120:8–14. doi: 10.1016/0022-510x(93)90017-s. [DOI] [PubMed] [Google Scholar]
- 6.Matsumura K, Campbell K P. Muscle Nerve. 1994;17:2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- 7.Love D R, Hill D F, Dickson G, Spurr N K, Byth B C, Marsden R F, Walsh F S, Edwards Y H, Davies K E. Nature (London) 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 8.Tinsley J M, Blake D J, Roche A, Fairbrother U, Riss J, Byth B C, Knight A E, Kendrick-Jones J, Suthers G K, Love D R, Edwards Y H, Davies K E. Nature (London) 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 9.Fardeau M, Tome F M, Collin H, Augier N, Pons F, Leger J, Leger J. C R Acad Sci. 1990;311:197–294. [PubMed] [Google Scholar]
- 10.Khurana T S, Watkins S C, Chafey P, Chelly J, Tome F M, Fardeau M, Kaplan J, Kunkel L M. Neuromusc Disord. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- 11.thi Man N, Ellis J M, Love D R, Davies K E, Gatter K C, Dickson G, Morris G E. J Cell Biol. 1991;115:1695–1700. doi: 10.1083/jcb.115.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohlendieck K, Ervasti J M, Matsumura K, Kahl S D, Leveille C J, Campbell K P. Neuron. 1991;7:499–508. doi: 10.1016/0896-6273(91)90301-f. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura K, Ervasti J M, Ohlendieck K, Kahl S D, Campbell K P. Nature (London) 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- 14.Blake D J, Tinsley J M, Davies K E. Brain Pathol. 1996;6:37–47. doi: 10.1111/j.1750-3639.1996.tb00781.x. [DOI] [PubMed] [Google Scholar]
- 15.Gramolini A O, Jasmin B J. Neuromusc Disord. 1998;8:351–361. doi: 10.1016/s0960-8966(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 16.Tinsley J M, Potter A C, Phelps S R, Fisher R, Trickett J I, Davies K E. Nature (London) 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- 17.Deconinck N, Tinsley J M, DeBacker F, Fisher R, Kahn D, Phelps S, Davies K, Gillis J M. Nat Med. 1997;3:1216–1221. doi: 10.1038/nm1197-1216. [DOI] [PubMed] [Google Scholar]
- 18.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis J M, Davies K E. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 19.Gramolini A O, Dennis C L, Tinsley J M, Roberston G S, Cartaud J, Davies K E, Jasmin B J. J Biol Chem. 1997;272:8117–8120. doi: 10.1074/jbc.272.13.8117. [DOI] [PubMed] [Google Scholar]
- 20.Gramolini A O, Burton E A, Tinsley J M, Ferns M J, Cartaud A, Cartaud J, Davies K E, Lunde J A, Jasmin B J. J Biol Chem. 1998;273:736–743. doi: 10.1074/jbc.273.2.736. [DOI] [PubMed] [Google Scholar]
- 21.Duclert A, Changeux J P. Physiol Rev. 1995;75:339–368. doi: 10.1152/physrev.1995.75.2.339. [DOI] [PubMed] [Google Scholar]
- 22.Burden S J. Genes Dev. 1998;12:133–148. doi: 10.1101/gad.12.2.133. [DOI] [PubMed] [Google Scholar]
- 23.Gramolini, A. O., Karpati, G. & Jasmin, B. J. (1999) J. Neuropathol. Exp. Neurol., in press. [DOI] [PubMed]
- 24.Schaeffer L, Duclert N, Huchet M, Changeux J P. EMBO J. 1998;17:3078–3090. doi: 10.1093/emboj/17.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duclert A, Savatier N, Changeux J P. Proc Natl Acad Sci USA. 1993;90:3043–3047. doi: 10.1073/pnas.90.7.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike S, Schaeffer L, Changeux J P. Proc Natl Acad Sci USA. 1995;92:10624–10628. doi: 10.1073/pnas.92.23.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duclert A, Savatier N, Schaeffer L, Changeux J P. J Biol Chem. 1996;271:17433–17438. doi: 10.1074/jbc.271.29.17433. [DOI] [PubMed] [Google Scholar]
- 28.Husmann I, Soulet L, Gautron J, Martelly I, Barritault D. Cytokine Growth Factor Rev. 1996;7:249–258. doi: 10.1016/s1359-6101(96)00029-9. [DOI] [PubMed] [Google Scholar]
- 29.Jasmin B J, Alameddine H, Lunde J A, Stetzkowksi-Marden F, Collin H, Tinsley J M, Davies K E, Tome F M S, Parry D J, Cartaud J. FEBS Lett. 1995;374:393–398. doi: 10.1016/0014-5793(95)01131-w. [DOI] [PubMed] [Google Scholar]
- 30.Biral D, Senter L, Salviati G. J Muscle Res Cell Motil. 1996;17:523–532. doi: 10.1007/BF00124352. [DOI] [PubMed] [Google Scholar]
- 31.Altiok N, Bessereau J, Changeux J P. EMBO J. 1995;14:4258–4266. doi: 10.1002/j.1460-2075.1995.tb00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn Jo S, Zhu X, Marchionni M A, Burden S J. Nature (London) 1995;373:158–161. doi: 10.1038/373158a0. [DOI] [PubMed] [Google Scholar]
- 33.Moscoso L M, Chu G C, Gautam M, Noakes P G, Merlie J P, Sanes J R. Dev Biol. 1995;172:158–169. doi: 10.1006/dbio.1995.0012. [DOI] [PubMed] [Google Scholar]
- 34.Sapru M, Florence S K, Kirk C, Goldman D. Proc Natl Acad Sci USA. 1998;95:1289–1294. doi: 10.1073/pnas.95.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fromm L, Burden S J. Genes Dev. 1998;12:3074–3083. doi: 10.1101/gad.12.19.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tansey M G, Chu G C, Merlie J P. J Cell Biol. 1996;134:465–476. doi: 10.1083/jcb.134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altiok N, Altiok S, Changeux J P. EMBO J. 1997;16:717–725. doi: 10.1093/emboj/16.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones G, Herczeg A, Ruegg M A, Lichtsteiner M, Kröger S, Brenner H R. Proc Natl Acad Sci USA. 1996;93:5985–5990. doi: 10.1073/pnas.93.12.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones G, Lichtsteiner M, Witzemann V, Sakmann B, Brenner H R. Proc Natl Acad Sci USA. 1997;94:2654–2659. doi: 10.1073/pnas.94.6.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier T, Masciulli F, Moore C, Schoumacher F, Eppenberger U, Denzer A J, Jones G, Brenner H R. J Cell Biol. 1998;141:715–726. doi: 10.1083/jcb.141.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier T, Hauser D M, Chiquet M, Landman L, Ruegg M A, Brenner H R. J Neurosci. 1997;17:6534–6544. doi: 10.1523/JNEUROSCI.17-17-06534.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen I, Rimer M, Lomo T, McMahan U J. Mol Cell Neurosci. 1997;9:237–253. doi: 10.1006/mcne.1997.0623. [DOI] [PubMed] [Google Scholar]