Abstract
We characterized the molecular genetic consequences of a balanced chromosome translocation t(8;22)(p21;q12) which occurred as the sole cytogenetic aberration in short-term cultured cells from an intrathoracic mature teratoma in a 15-year-old girl. Fluorescence in situ hybridization and reverse transcription-polymerase chain reaction disclosed that t(8;22) resulted in the fusion of the genes PPP2R2A and CHEK2, with an inserted fragment belonging to class I endogenous retrovirus-related sequences at the junction. Sequencing of the two genes did not reveal any additional mutation. None of the three detected PPP2R2A/CHEK2 fusion transcripts resulted in an in-frame PPP2R2A/CHEK2 chimerical open reading frame; however, in all of them, the known open reading frame of CHEK2 was preserved. Thus, promoter swapping leading to deregulated CHEK2 expression would be the most likely oncogenic mechanism. Whereas inactivating mutations of CHEK2 previously have been described in a variety of sporadic tumors and in inherited cancer-predisposing syndromes, PPP2R2A, encoding a regulatory subunit of the multimeric enzyme phosphatase 2, has not been directly implicated in tumorigenesis. Our findings suggest that deregulation of CHEK2 and/or PPP2R2A is of pathogenetic importance in at least a subset of germ cell tumors.
Keywords: Mature teratoma, translocation, PPP2R2A, CHEK2, fusion gene
Introduction
Mature teratomas are germ cell neoplasms composed of adult-type tissues derived from at least two embryonic layers [1,2]. Most mature teratomas present congenitally in the sacrococcygeal region or within the ovaries or testes of adolescents, but they may occur throughout the body at any age. As the development of teratomas resembles embryogenesis in various aspects, they have been used as a model system to study both embryonic and tumorigenic processes.
Little is known about the genetic mechanisms involved in the pathogenesis of mature teratomas. In contrast to more aggressive subtypes of germ cell tumors, which typically have aneuploid karyotypes with a gain of the short arm of chromosome 12 as the most common feature, mature teratomas often display normal karyotype on cytogenetic analysis, and no consistent pattern of chromosomal gains or losses has been disclosed by comparative genomic hybridization [3,4]. Furthermore, neither in mature teratomas nor in germ cell tumors, in general, has any recurrent, acquired, tumor-specific, balanced chromosome rearrangement been detected. Recently, however, a constitutional t(12;15)(q13;q25), resulting in the fusion of the genes SENP1 and MESDC2, was identified in a patient with infantile teratoma [5].
We have previously reported cytogenetic findings in two teratomas, one of which displayed a balanced translocation t(8; 22)(p21 ;q12) [6]. In the present study, precise mapping of breakpoints was carried out by fluorescence in situ hybridization (FISH) on metaphase spreads from this tumor. Based on FISH results implicating a fusion between the PPP2R2A and CHEK2 genes, further molecular investigations were undertaken.
Materials and Methods
Patient
A 15-year-old girl was admitted to a hospital because of viral meningoencephalitis. Due to increasing respiratory difficulty, X-ray and computed tomography scan of the thorax were performed, revealing a large mediastinal tumor. On cytologic analysis of cells obtained from a fine-needle aspiration biopsy of the tumor, no malignant cells were identified, and the tumor was tentatively diagnosed as cystic mature teratoma. After the patient had recovered from meningoencephalitis, the mediastinal tumor was radically excised. Histologic examination of the excised tumor demonstrated a mature teratoma without malignant features. Postoperatively, the patient had a slow recovery but remained disease-free for 6 years, after which she was lost to follow-up.
G-banding
Cytogenetic analysis was performed on two occasions [6]. On diagnosis, both a bone marrow sample and tumor cells derived from the fine-needle aspiration biopsy of the mediastinal mass were analyzed. All 25 metaphase cells from the bone marrow sample showed normal female karyotype, whereas short-term cultured cells from the fineneedle aspiration biopsy displayed a balanced translocation t(8;22)(p21;q12) as the sole change in 9 of 14 metaphases; the remaining five mitoses were normal. Cytogenetic investigation of the radically excised tumor showed t(8;22) as the sole change in all 25 metaphase spreads analyzed.
FISH
Vital frozen cells stored in liquid nitrogen were thawed and plated in culture flasks with RPMI 1640 medium supplemented with 17% serum and antibiotics. Chromosome preparations for FISH were harvested at passages 2 and 4, as described [7]. To confirm whether the cells still carried t(8;22), they were also subjected to conventional G-banding analysis.
The breakpoint on chromosome 22 was first investigated using commercially available probes for the EWSR1 (22q12.2) and BCR (22q11.23) genes (Vysis, Downers Grove, IL). To further characterize the two chromosomal breakpoints, 32 BAC clones (16 spanning 8p12-8p21 and 16 spanning 22q11.2-22q12.2) were selected based on their location on the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/cgi-bin/Entreez/map-search, 2005) and the UCSC Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway). Clones were propagated, and DNA was extracted by standard methods [8]. BAC DNA was labeled by random hexamer priming (megaprime DNA labeling system; Amersham, Buckinghamshire, UK) with a single fluorochrome or ligand (i.e., biotin-16-dUTP, Cy3-dCTP, digoxigenin-11-dUTP, or FluorX-dCTP). Labeled probes were purified, precipitated, and dissolved in a standard hybridization solution. Slides and probes were denatured simultaneously by incubation on a hot plate at 72°C. After overnight hybridization at 37°C in a humidified chamber, the slides were washed at 74°C in 0.4x SSC for 2 minutes, DAPI-stained, and analyzed.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Sequence Analyses
Total RNA was extracted from cultured cells using Trizol reagent, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). cDNA synthesis was conducted using 5 µg of total RNA in a 20-µl reaction mixture containing 1x first-strand buffer, 10 mM DTT, 1 mM of each deoxynucleoside triphosphate (dNTP), 20 U of RNAse inhibitor (RNA guard; Amersham Biosciences, Piscataway, NJ), 500 pmol of random hexamers, and 200 U of M-MLV reverse transcriptase (Invitrogen). The reaction was carried out at 37°C for 60 minutes, heated at 65°C for 5 minutes, and then kept at 4°C until analysis. PCR amplifications were performed using 1 µl of cDNA as template in a final volume of 50 µl containing 1x PCR buffer, 0.2 mM of each dNTP, 1.25 mM MgCl2, 0.5 µM of each forward primer and reverse primer, and 1 U of Platinum Taq DNA polymerase (Invitrogen), and run on a PCT-200 DNA Engine (MJ Research, Waltham, MA).
Primers specific for CHEK2 and PPP2R2A were designed to detect possible fusion transcripts (Table 1). Transcripts were amplified using initial denaturation for 5 minutes at 94°C; 30 cycles of 1 minute at 94°C, 1 minute at 60°C, and 1 minute at 72°C; and final extension for 10 minutes at 72°C. Amplified fragments were run on a 1.6% agarose gel stained with ethidium bromide, purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany), and directly sequenced with the dideoxy procedure using an ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) on the Applied Biosystems Model 3100-Avant DNA sequencing system. BLAST software (http://www.ncbi.nlm.nih.gov/blast) was used for the analysis of CHEK2 and PPP2R2A sequence data.
Table 1.
Primers for RT-PCR and Direct Sequencing.
| Primer | Sequence |
| PPP2R2A-242F | GTCTCCGCTTCCTGAACTCACCC |
| PPP2R2A-286F | CCATGTTGCGCTGCAAATGGT |
| CHEK2-294R | TCCTCAGGTTCTTGGTCCTCAGGA |
| CHEK2-473R | GGTGCCTCACACCTCTTATCCCAG |
| CHEK2-686R | ACAAAGGTTCCATTGCCACTGTGAT |
| CHEK2-982R | AGAGCTGGGTCTGCCTCTCTTGC |
| CHEK2-906R | CATGTTTTCCTCTCGAAAGCCAGC |
Screening for Additional CHEK2 and PPP2R2A Mutations
To search for additional CHEK2 mutations, PCR amplifications of exons 4 to 9 were performed using 160 ng of genomic DNA as template in a volume of 50 µl containing 1x PCR buffer, 0.8 mM dNTP, 1.25 mM MgCl2, 0.5 µM of each forward primer and reverse primer, and 1 U of Platinum Taq DNA polymerase (Invitrogen). The annealing temperature for exons 4 to 9 was 58°C. PCR reactions were carried out on a PCT-200 DNA Engine (MJ Research) with initial denaturation for 5 minutes at 94°C; 30 cycles of 1 minute at 94°C, 1 minute at 58°C, and 1 minute at 72°C; and final extension for 5 minutes at 72°C and for 2 minutes at 26°C. Amplified fragments were run on a 1.6% agarose gel stained with ethidium bromide, purified using the QIAquick gel extraction kit (Qiagen), and sequenced as described above.
For the analysis of exons 10 to 14, the methods described by Sodha et al. [9] were used. Long-range PCR across exons 10 to 14 of CHEK2 was performed, and the product was used to individually amplify exons 10 to 14.
To search for PPP2R2A mutations, exons 1 to 10 were amplified by PCR and sequenced as described above. Primers for the sequencing of CHEK2 and PPP2R2A are available at request.
The BLAST software (http://www.ncbi.nlm.nih.gov/blast) was used for the mutational analysis of CHEK2 and PPP2R2A.
Results
FISH Characterization of Breakpoints in 8p21 and 22q12
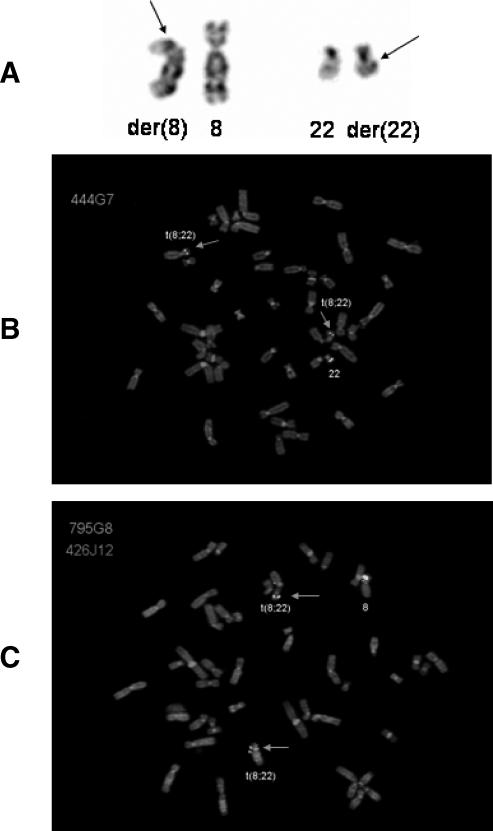
All metaphase cells harvested from passages 2 and 4 carried t(8;22) (Figure 1A). With the use of probes for the EWSR1 (22q12.2) and BCR genes (22q11.23), the chromosomal breakpoint in chromosome 22 was narrowed down to the 6-Mb region between these two loci. Subsequently, 16 BAC clones within this region were selected for further FISH, and the hybridization of one of these clones (i.e., RP11-444G7, covering the CHEK2 locus) showed split signals (Figure 1B). The breakpoint in chromosome 8 was then investigated by the use of 16 BAC clones spanning 8p12-p21. The hybridization of probe RP11-795G8 (containing the PPP2R2A locus) resulted in split signals (Figure 1C).
Figure 1.
(A) Partialkaryogram showing a reciprocal translocation t(8;22)(p21; q12). Arrows indicate breakpoints. (B) Metaphase cell hybridized with the BAC probe RP11-444G7 (covering the CHEK2 locus; green signals). Arrows indicate split signals on derivative chromosomes 8 and 22. (C) Metaphase cell hybridized with BAC probes RP11-795G8 (covering the PPP2R2A locus; green signals) and RP11-426J12 (mapping to the centromeric side of the breakpoint on chromosome 8; red signal). Arrows indicate split signals for RP11-795G8 on the derivative chromosomes 8 and 22.
Detection and Characterization of PPP2R2A/CHEK2 Fusion Transcripts
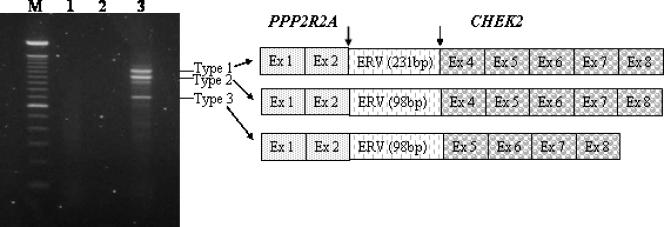
RT-PCR with various PPP2R2A and CHEK2 primer combinations was carried out to detect fusion transcripts. The use of PPP2R2A forward primer PPP2R2A-242F and CHEK2 reverse primer CHEK2-982R gave rise to three amplified fragments, suggesting the presence of a PPP2R2A/CHEK2 fusion gene (Figure 2). The three fusion transcripts were then analyzed by direct sequencing with the use of several different PPP2R2A forward primers and CHEK2 reverse primers for further characterization of the fusion breakpoint. Furthermore, a fourth fainter band was seen, presumably representing another splice variant of the fusion product. This band, however, was not sequenced.
Figure 2.
Analysis of the PPP2R2A/CHEK2 fusion transcripts. M: 100-bp DNA ladder. Lane 1: No fusion transcript was found while using the forward primer CHEK2-82F and the reverse primer PPP2R2A-956R. Lane 2: Blank. Lane 3: Three fusion transcripts were detected with the forward primer PPP2R2A-242F and the reverse primer CHEK2-982R.
In fusion transcripts 1 and 2, exon 2 of PPP2R2A (NM_002717) was fused with exon 4 of CHEK2 (NM_007194.3) (Figures 2 and 3), whereas in transcript 3, exon 2 of PPP2R2A was fused with exon 5 of CHEK2. In all three transcripts, inserted fragments were found at the junctions (Figure 3). In transcript 1, a 231-bp DNA fragment was inserted between exon 2 of PPP2R2A and exon 4 of CHEK2. In transcripts 2 and 3, a 98-bp fragment was inserted between the two genes. BLAST analysis revealed that the inserted fragment corresponded to nucleotide positions 22187 to 21953 (231-bp insert) and 22187 to 22086 (98-bp insert) of AL023494 (RP3-366L4), which flanks the telomeric 5′ part of CHEK2. Using a repeat masker engine (http://www.repeatmasker.org), the inserts were found to belong to class I endogenous retrovirus (ERV)-related sequences [10].
Figure 3.
Complete nucleotide sequence of PPP2R2A/CHEK2 chimeric transcript 1 detected by RT-PCR. The primers PPP2R2A-242F and CHEK2-982R are underlined. Arrowheads indicate breakpoints, and vertical lines indicate exon boundaries. The inserted DNA between exon 2 of PPP2R2A and exon 4 of CHEK2 is a repetitive sequence from chromosome 22, which belongs to type 1 of ERV-related sequences. Sequence in bold (part of the inserted DNA) is absent in type 2 chimeric transcript. Sequence in brackets is not present in type 3 of the PPP2R2A/CHEK2 cDNA fragment (part of inserted sequence and exon 4 of CHEK2).
None of the three amplified transcripts resulted in a chimeric in-frame PPP2R2A/CHEK2 open reading frame. In all detected fusion transcripts, the first 28 translated amino acids of PPP2R2A were followed by amino acids GQ and the terminal codon TGA.
Sequencing of CHEK2 exons 4 to 14 and of PPP2R2A exons 1 to 10 did not reveal any additional mutations.
Discussion
In the present study, we used FISH and RT-PCR to characterize a balanced translocation t(8;22) in a mediastinal mature teratoma and showed that this translocation leads to the creation of a novel fusion gene PPP2R2A/CHEK2. The fact that t(8;22) was the sole chromosomal change in this tumor makes it reasonable to assume that it was important in pathogenesis. Although the t(8;22) seen in our case has not been described before, it should be emphasized that only a few teratomas, in general, and pediatric extragonadal mature teratomas, in particular, have been cytogenetically analyzed. It should also be noted that various structural rearrangements with breakpoints in 8p or 22q have been reported in teratomas [11].
The mechanism by which t(8;22) may promote tumor development is unclear. All detected PPP2R2A/CHEK2 fusion transcripts were out of frame, making it unlikely that they would be translated into proper fusion proteins. However, transcripts 1 and 2 preserve the known CHEK2 open reading frame, which begins at exon 4, suggesting that one consequence may be aberrant expression of CHEK2 from the PPP2R2A promoter. Such transcriptional deregulation through juxtapositioning with an ectopic promoter (an oncogenic mechanism known as promoter swapping) has been well documented for several other solid tumors, including pleomorphic adenoma of the salivary glands, lipoblastoma, and aneurysmal bone cyst [12–14]. Although this seems to be the most likely mechanism in the present case, it should be noted that, theoretically, the transcripts could be translated into the first 28 amino acids of PPP2R2A followed by two amino acids from the inserted ERV sequence, and it cannot be excluded that this chimera could be of pathogenetic importance.
CHEK2, which encodes a checkpoint kinase, is considered a tumor-suppressor gene based on its known role in cellular responses to DNA double-strand breaks, the finding of biallelic inactivating mutations in various sporadic tumors, and epidemiological data showing that certain constitutional mutations confer increased risk for breast cancer, for example [15–17]. In response to double-strand DNA breaks, CHEK2 becomes activated and phosphorylates various substrates, including CDC25A, CDC25C, BRCA1, E2F1, PML, PLK3, and TP53, thereby promoting a variety of cellular responses, such as cell cycle arrest, apoptosis, and DNA repair. Somatic inactivating mutations of CHEK2 have been found in small subsets of diverse types of sporadic human malignancies, including osteosarcomas, lymphomas, and carcinomas of the breast, vulva, urinary bladder, and ovary, but they seem to be rare in pediatric cancer [18]. The majority of somatic mutations are missense or truncating mutations clustered in three domains of the CHEK2 protein: the Nterminal SQ/TQ-rich regulatory domain, the protein-protein interaction FHA domain, and the C-terminal catalytic domain [16]. 1100delC mutation, which was first reported in a subset of patients with Li-Fraumeni syndrome, and other constitutional variants are overrepresented in families predisposed to breast cancer and other malignancies [19–21]. It is also well known that CHEK2 is subject to extensive alternative splicing, with close to 90 splice variants being detected in one study [22]. At present, it is not known whether this complex alternative splicing could influence tumorigenesis. However, the finding of at least three splice variants of the PPP2R2A/CHEK2 fusion product in the present teratoma is well in line with previously reported data for CHEK2.
PPP2R2A, however, has not been directly implicated in tumorigenesis. The PPP2R2A protein is a regulatory subunit of the protein phosphatase 2 (PP2A) enzyme, a highly conserved Ser/Thr phosphatase that regulates cell growth and differentiation through a variety of cellular processes, including signal transduction, DNA replication, apoptosis, and cell cycle progression [23–25]. The PP2A holoenzyme consists of a common dimeric core of a catalytic (C) subunit and a structural (A) subunit, which, in turn, may be associated with a variety of regulatory (B) subunits influencing the subcellular localization and substrate specificity of the phosphatase. Seventy-five different dimeric and trimeric PP2A holoenzymes can thus be generated [23,25]. The PPP2R2A gene encodes an α isoform (aka B55α or PR55α) of the regulatory B55 subunit family. Drosophila melanogaster mutants carrying an inactivated B55α/PR55α gene show an abnormal anaphase distribution of chromosomes; in mammalian cells, the B55α/PR55α subunit seems to be essential for the dephosphorylation of cytoplasmic intermediate filaments [26,27]. Furthermore, PP2A is thought to prevent premature entry into mitosis by negatively regulating CDC2 by keeping CDC25C in a dephosphorylated low-activity state [28]; seemingly, the dephosphorylation/inhibition of CDC25C is exerted by a form of PP2A containing the regulatory subunit B55a [29]. Other support for a suppressor function of PP2A in tumorigenesis stems from the observation that the tumor-promoting agent okadaic acid is a potential inhibitor of PP2A [30]. Additionally, mutations in PPP2R1A and PPP2A1B genes, encoding the PP2A subunits RP65α and RP65α, respectively, have been detected in melanomas and carcinomas of the lung, breast, and colon [25,31,32]. Of particular interest, it was recently shown that PP2A can dephosphorylate and inactivate CHEK2 and, vice versa, that CHEK2 can phosphorylate the B subunit of PP2A, thereby increasing its activity [33].
The present study showed that the outcome of t(8;22), found as the sole chromosome aberration in a mature teratoma, is the fusion of PPP2R2A and CHEK2—the first acquired fusion gene to be detected in germ cell tumors. In the majority of cases, previous identification of gene fusions in human malignancies has been guided by the finding of balanced chromosome rearrangements in short-term cultured tumor cells, and it has been hypothesized that the present relative lack of known fusion genes in certain tumor types, such as epithelial malignancies and germ cell tumors, might be partly explained by the small number of such tumors that have been properly analyzed by chromosome banding techniques [34]. In line with this reasoning, it was recently shown that a large fraction of prostate carcinomas carries subtle chromosome rearrangements, leading to highly recurrent fusion genes [35]. Whether the presently detected PPP2R2A/CHEK2 fusion is a common and tumor-specific event needs to be analyzed in a larger series of teratomas. In addition, if the essential molecular outcome in the present case were deregulation of CHEK2 and/or PPP2R2A rather than creation of a specific fusion transcript, it might be worthwhile to study the expression patterns of these genes in germ cell neoplasms.
Footnotes
This study was supported by grants from the Swedish Children's Cancer Foundation and the Gunnar Nilsson's Cancer Foundation.
References
- 1.Nogales F, Talerman A, Kubik-Huch RA, Tavassoli FA, Devouassoux-Shisheboran M. Germ cell tumors. In: Tavassoli FA, Devile P, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Lyon: IARC Press; 2003. pp. 163–175. [Google Scholar]
- 2.Gatcombe HG, Assikis V, Kooby D, Johnstone PA. Primary retroperitoneal teratomas: a review of the literature. J Surg Oncol. 2004;86:107–113. doi: 10.1002/jso.20043. [DOI] [PubMed] [Google Scholar]
- 3.Surti U, Hoffner L, Chakravarti A, Ferrell RE. Genetics and biology of human ovarian teratomas: I. Cytogenetic analysis and mechanism of origin. Am J Hum Genet. 1990;47:635–643. [PMC free article] [PubMed] [Google Scholar]
- 4.Veltman I, Veltman J, Janssen I, Hulsbergen-vande Kaa C, Oosterhuis W, Schneider D, Stoop H, Gillis A, Zahn S, Looijenga L, et al. Identification of recurrent chromosomal aberrations in germ cell tumors of neonates and infants using genomewide array-based comparative genomic hybridization. Genes Chromosomes Cancer. 2005;43:367–376. doi: 10.1002/gcc.20208. [DOI] [PubMed] [Google Scholar]
- 5.Veltman IM, Vreede LA, Cheng J, Looijenga LH, Janssen B, Schoenmakers EF, Yeh ET, Geurts van Kessel A. Fusion of the SUMO/Sentrin-specific protease 1 gene SENP1 and the embryonic polarity-related mesoderm development gene MESDC2 in a patient with an infantile teratoma and a constitutional t(12;15)(q13;25) Hum Mol Genet. 2005;4:1955–1963. doi: 10.1093/hmg/ddi200. [DOI] [PubMed] [Google Scholar]
- 6.Mertens F, Mandahl N, Hägerstrand I, Kullendorff C-M, Heim S. Cytogenetic findings in pediatric germ cell tumors. Int J Oncol. 1995;6:401–404. doi: 10.3892/ijo.6.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Mandahl N. Methods in solid tumor cytogenetics. In: Rooney DE, editor. Human Cytogenetics: Malignancy and Acquired Abnormalities. 3rd ed. New York: Oxford University Press; 2001. pp. 165–203. [Google Scholar]
- 8.Sambrook J, Fritsch EF, Maniatis T. In: Molecular Cloning: A Laboratory Manual. 2nd ed. Nolan C, editor. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Sodha N, Houlston RS, Williams R, Yuille MA, Mangion J, Eeles RA. A robust method for detecting CHK2/RAD53 mutations in genomic DNA. Hum Mutat. 2002;19:173–177. doi: 10.1002/humu.10031. [DOI] [PubMed] [Google Scholar]
- 10.Anderssen S, Sjottem E, Svineng G, Johansen T. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology. 1997;234:14–30. doi: 10.1006/viro.1997.8590. [DOI] [PubMed] [Google Scholar]
- 11.Mitelman F, Johansson B, Mertens F, editors. Mitelman Database of Chromosome Aberrations in Cancer. 2005 ( http://cgap.nci.nih.gov/Chromosomes/Mitelman)
- 12.Kas K, Voz ML, Roijer E, Åström AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3; 8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- 13.Hibbard MK, Kozakewich HP, Dal Cin P, Sciot R, Tan X, Xiao S, Fletcher JA. PLAG1 fusion oncogenes in lipoblastoma. Cancer Res. 2000;60:4869–4872. [PubMed] [Google Scholar]
- 14.Oliveira AM, Perez-Atayde AR, Dal Cin P, Gebhardt MC, Chen CJ, Neff JR, Demetri GD, Rosenberg AE, Bridge JA, Fletcher JA. Aneurysmal bone cyst variant translocations upregulate USP6 transcription by promoter swapping with the ZNF9, COL1A1, TRAP150, and OMD genes. Oncogene. 2005;24:3419–3426. doi: 10.1038/sj.onc.1208506. [DOI] [PubMed] [Google Scholar]
- 15.McGowan CH. A checkpoint kinase and tumor suppressor. Bioessays. 2002;24:502–511. doi: 10.1002/bies.10101. [DOI] [PubMed] [Google Scholar]
- 16.Bartek J, Lukas Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN, Xia F. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YY, Takita J, Tanaka K, Ida K, Koh K, Igarashi T, Hanada R, Kikuchi A, Tanaka Y, Toyoda Y, et al. Aberrations of the CHK2 gene are rare in pediatric solid tumors. Int J Mol Med. 2005;16:85–91. [PubMed] [Google Scholar]
- 19.Schutte M, Seal S, Barfoot R, Meijers-Heijboer H, Wasielewski M, Evans DG, Eccles D, Meijers C, Lohman F, Klijn J, et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet. 2003;72:1023–1028. doi: 10.1086/373965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, Teodorczyk U, Byrski T, Gronwald J, Matyjasik J, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson N, Fletcher O, Naceur-Lombardelli C, dos Santos Silva I, Ashworth A, Peto J. Interaction between CHEK2*1100delC and other low-penetrance breast-cancer susceptibility genes: a familial study. Lancet. 2005;366:1554–1557. doi: 10.1016/S0140-6736(05)67627-1. [DOI] [PubMed] [Google Scholar]
- 22.Staalesen V, Falck J, Geisler S, Bartkova J, Borresen-Dale A-L, Lukas J, Lillehaug JR, Bartek J, Lonning PE. Alternative splicing and mutation status of CHEK2 in stage III breast cancer. Oncogene. 2004;23:8535–8544. doi: 10.1038/sj.onc.1207928. [DOI] [PubMed] [Google Scholar]
- 23.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schönthal AH. Role of serine/threonine protein phosphatase 2A in cancer. Cancer Lett. 2001;170:1–13. doi: 10.1016/s0304-3835(01)00561-4. [DOI] [PubMed] [Google Scholar]
- 25.Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15:34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Mayer-Jaekel RE, Ohkura H, Gomes R, Sunkel CE, Baumgartner S, Hemmings BA, Glover DM. The 55 kD regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell. 1993;72:621–633. doi: 10.1016/0092-8674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- 27.Turowski P, Myles T, Hemmings BA, Fernandez A, Lamb NJ. Vimentin dephosphorylation by protein phosphatase 2A is modulated by the targeting subunit B55. Mol Biol Cell. 1999;10:1997–2015. doi: 10.1091/mbc.10.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke PR, Hoffmann I, Draetta G, Karsenti E. Dephosphorylation of cdc25-C by a type-2A protein phosphatase: specific regulation during the cell cycle in Xenopus egg extracts. Mol Biol Cell. 1993;4:397–411. doi: 10.1091/mbc.4.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TH, Turck C, Kirschner MW. Inhibition of cdc2 activation by INH/PP2A. Mol Biol Cell. 1994;5:323–338. doi: 10.1091/mbc.5.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bialojan C, Takai A. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Biochem J. 1988;256:283–290. doi: 10.1042/bj2560283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang SS, Esplin ED, Li JL, Huang L, Gazdar A, Minna J, Evans GA. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, di Iasio MG, Caprini E, Vorechovsky I, Natali PG, Sozzi G, Croce CM, Barbanti-Brodano G, Russo G, Negrini M. Low frequency of alterations of the alpha (PPP2R1A) and beta (PPP2R1B) isoforms of the subunit A of the serine-threonine phosphatase 2A in human neoplasms. Oncogene. 2000;19:1191–1195. doi: 10.1038/sj.onc.1203389. [DOI] [PubMed] [Google Scholar]
- 33.Dozier C, Bonyadi M, Baricault L, Tonasso L, Darbon JM. Regulation of Chk2 phosphorylation by interaction with protein phosphatase 2A via its B′ regulatory subunit. Biol Cell. 2004;96:509–517. doi: 10.1016/j.biolcel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 35.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]