Abstract
Rhabdomyosarcoma, the most common pediatric soft tissue sarcoma, likely results from deregulation of the skeletal myogenesis program. Although associations between PAX3, PAX7, FOXO1A, and RMS tumorigenesis are well recognized, the entire spectrum of genetic factors underlying RMS development and progression is unclear. Using a combined approach of spectral karyotyping, array-based comparative genomic hybridization (CGH), and expression analysis, we examined 10 primary RMS tumors, including embryonal, alveolar, and the rare adult pleomorphic variant, to explore the involvement of different genes and genetic pathways in RMS tumorigenesis. A complete karyotype established for each tumor revealed a high aneuploidy level, mostly tetraploidy, with double minutes and additional structural aberrations. Quantitative expression analysis detected the overexpression of the AURKA gene in all tumors tested, suggesting a role for this mitotic regulator in the aneuploidy and chromosomal instability observed in RMS. Array-based CGH analysis in primary RMS tumors detected copy number changes of genes involved in multiple genetic pathways, including transcription factors such as MYC-related gene from lung cancer and the cytoskeleton and cell adhesion-encoding genes laminin γ-2 and p21-activated kinase-1. Our data suggest the involvement of genes encoding cell adhesion, cytoskeletal signaling, and transcriptional and cell cycle components in RMS tumorigenesis.
Keywords: Rhabdomyosarcoma, chromosome aberrations, array-based comparative genomic hybridization, AURKA overexpression, cell adhesion
Introduction
Rhabdomyosarcoma (RMS), the most common pediatric soft tissue sarcoma, likely results from an imbalance in the proliferation and differentiation of precursor cells during the skeletal myogenesis program. In contrast to normal myoblasts, which fuse into myotubes to form multinucleate syncytial cells that no longer proliferate [1], RMS cells are unable to reach terminal differentiation, exhibiting interrupted myogenesis with sustained proliferation [2,3]. The tumor seems to recapitulate normal embryonal myogenesis, expressing muscle-specific markers, such as MyoD and vimentin, that are normally present during the different steps of fetal muscle development [4,5]. The exact cell type from which RMS is derived remains unclear; however, evidence of RMS in sites other than the skeletal muscle suggests that the tumor may originate from a primitive mesenchymal cell or from a committed myogenic precursor [2,3].
RMS tumors are classified into three subtypes according to histopathological description: the more prevalent embryonal rhabdomyosarcoma (ERMS), the more aggressive alveolar rhabdomyosarcoma (ARMS), and the rare adult variant pleomorphic rhabdomyosarcoma (PRMS) [6]. Karyotype descriptions of pleomorphic subtype are scarce, and only a few chromosomal and genetic characteristics have been reported. The two other RMS subtypes, ERMS and ARMS, often demonstrate a high level of aneuploidy and are associated with different cytogenetic changes [7,8]. Frequent but neither consistent nor specific chromosomal gains nor losses are detected in ERMS [9]. In contrast, the majority (about 85%) of ARMS tumors are characterized by recurrent translocation between the genes encoding for the transcription factors FOXO1A (original name, FKHR, localized at chromosome 13q14) with either PAX3 (at 2q35) or, less commonly, PAX7 (at 1p36) [10]. PAX3 and PAX7 are also overexpressed in some ARMS and ERMS tumors [11]. Chromosome instability in RMS karyotypes is also reflected as genomic amplification through double minutes (dmins) [12,13], which include various genes such as MDM2, MYCN, and GLI, and the characteristic ARMS fusion gene PAX3-FOXO1A or PAX7-FOXO1A [14–17]. Furthermore, conventional comparative genomic hybridization (CGH) studies of primary RMS tumors have confirmed quantitative genomic alterations of large chromosomal regions, including the amplification of 2q and 8p or the deletion of 17p, as seen in cytogenetic analyses and in the amplification and deletion of novel loci (15q25–26[9] and 5q32-ter[7]). Additionally, other genes and pathways, such as IGF, SHH, and Rac1, have been implicated in RMS tumorigenesis [2,18].
Materials and Methods
Primary Tumor Tissues and Cell Lines
Ten primary RMS tumors, including two ERMS, seven ARMS, and one PRMS, were analyzed. Fresh non-necrotic tissues were obtained during open biopsy or excision of the tumor. All patients underwent systemic intravenous neoadjuvant chemotherapy before excision, with the exception of ARMS5 and PRMS1, which did not receive chemotherapy before excision. ERMS2, which was a recurrent tumor, was treated with neoadjuvant chemotherapy at the time of first diagnosis, followed by radiotherapy. Routine cytogenetic and pathological analyses were performed, and the diagnosis of RMS was confirmed according to standard clinical criteria and pathological studies. The study was approved by the IRB Helsinki Committees of the Tel-Aviv Sourasky Medical Center and the Supreme National Helsinki Committee for Genetic Studies of the Israeli Ministry of Health. Table 1 lists clinical and pathological features.
Table 1.
Clinical and Pathological Features and Cytogenetic Analyses of Primary RMS Cases.
| RMS Number | Age at Biopsy (years)/Sex | Primary Site/LNM | Stage at Presentation Intergroup Rhabdomyosarcoma Study | Outcome until December 2005 | Karyotype Descriptions |
| ERMS1 | 1/F | Arm | Group 1, stage 1 | NED | 46,XX,+2,-8,der(17)t(11;17)(q13;p13),der(19)t(19;8)(q13.4;?),der(22) t(19;22)(?;q13)t(8;19)(q?11.2;?)[6]/47,idem,+7[7]/48,idem,+7,+9[1]/46,XX[2]* |
| ERMS2 | 7/M | Leg and thigh/LNM | Group 2, stage 1 | D | 46, Complex karyotype† |
| ARMS1 | 1/M | Bilateral groins/LNM | Group 3 | D | 83∼85,XXYY,+i(1)(q10),t(2;12)(p?14;q23)x2,i(3)(q10),der(3)del(p24)t(3;17) (q13;?),der(4)t(4;11)(p12;?)x2,der(5)t(5;12)(q12;?)x2,der(8)t(4;8)(?;p12), -9,-9,+der(10)t(2;10)(?;p?),-11,del(12)(q13q21)x2,del(13)(q14)x2,-15, -15,del(17)(q22),-17,-21,2 dmin (3)[cp17]/46,XY[5]* |
| ARMS2 | 2/F | Arm | Group 1, stage 1 | NED | 92,XXXX,10∼30 dmin (13)[7]/46,XX[10]* |
| ARMS3 | 3/M | Perineum | Group 2, stage 1 | D | 46,XY[15]† |
| ARMS4 | 17/F | Hand | Group 1, stage 1 | NED | 46,XX[25]† |
| ARMS5 | 18/M | Thigh/LNM | Group 1, stage 1 | D | 92,XXYY,del(13)(q14)x2,der(20)t(13;20)(q14;q13.3)x2,10†30 dmin (13)[12]/46,XY[5]* |
| ARMS6 | 24/M | Groin/LNM | Group 2, stage 1 | D | 100∼104<4n>,XXXYYY,+2,-3,+5,+6,+6,+7,+8,+8,+8,+8,-9,+10,-11, -11,+12,+12,+12,-17,-19,-19,+20,+22,+22,+22,20∼30 dmin (1)[10]/46,XY[5]* |
| ARMS7 | 31/F | Perineum/LNM | Group 2, stage 1 | D | 46,XX[15]† |
| PRMS1 | 68/M | Groin/LNM | Group 2, stage 1 | D | 74∼82,XXY,del(1)(p11)x2+idic(1)(q10),del(2)(p1?6)x2,del(3)(q13)x2,-4, der(4)t(4;16)(p11;p11),der(4)t(4;17)(q28;q11),del(5)(p13), +der(5)t(5;6)(q?35;?),der(5)t(5;10)(q3?4;q?21),del(6)(p?12),+i(6) (q10),+der(6)t(6;6)(q2?5;q?),der(7)t(6;7),der(7)del(7)(p21)del(7)(q31), +der(8)t(8;8)(p23;q22)x2,+der(8)t(4;8)(?;q11),der(9)t(9;20)(q13;q11.2), +der(9)t(9;20)(q13;q11.2),der(10)t(5;10)(?;p11)x2,+der(10)ins(5) dup(10)(q22),+del(11)(q2?1),+der(11)t(3;11)(?;q21),-13,-14,+15,+der(15) t(15;16)(p10;p10)x2,-16,der(17)t(17;22)(p12;q12)x2,+der(17)del(17) (q11)t(11;17)(?;p13),-18,+18,-19,+der(19)del(19)(p13)del(19)(q13),-20, del(20)(q11),-21,+22,+22,+22,+del(22)(q12)[cp12]* |
LNM, lymph node metastasis found; NED, no evidence of disease; D, died.
G-banding and SKY analyses.
G-banding analysis.
The RMS cell lines RD and A-204 (embryonal subtype) were obtained from the American Type Culture Collection (Manassas, VA). The RMS cell lines RH28 and RMZ-RC2 (alveolar subtype) were kindly provided by Dr. Peter J. Houghton (St. Jude's Children's Research Hospital, Memphis, TN) and Dr. Pier-Luigi Lollini (Department of Experimental Pathology, University of Bologna, Bologna, Italy), respectively.
As normal controls, we purchased four different RNA samples that were extracted from normal adult skeletal muscles (Clontech BD Biosciences, Erembodegem, Belgium; Ambion, Inc., Austin, TX; BioChain Institute, Inc., Hayward, CA).
Chromosomal Studies
Chromosomal analysis was performed on primary cultures derived from all RMS samples. Short-term cultures, chromosomal preparations, G-band staining, and spectral karyotype (SKY) analysis were performed according to standard cytogenetic techniques and manufacturer's instructions, as previously described [19]. SKY painting for ERMS1, ARMS1, ARMS2, ARMS5, ARMS6, and PRMS1 samples helped refine G-band findings and defined the origin of dmins. Karyotypes were established according to the International System for Human Cytogenetic Nomenclature [20].
DNA and RNA Extraction
DNA and total RNA were extracted from snap-frozen samples of 10 primary tumors and 4 cell line cultures, using Puregene DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, MN) and Tri Reagent (Sigma-Aldrich, St. Louis, MO), according to the manufacturer's instructions, respectively. RNA quality was determined based on OD260/280 for all RNA samples. Optical density at 260 and 280 nm was measured, and all ratios calculated were within 1.75 to 1.93.
Array-Based CGH Analysis
Detection of gene amplification by array-based CGH was performed using Vysis GenoSensor system (Abbott Vysis, Inc., Downers Grove, IL), according to the manufacturer's instructions as described previously [19]. Briefly, test DNA and normal reference DNA were labeled by random priming to incorporate fluorophores and were hybridized to either the AmpliOnc I microarray (Abbott Vysis Inc., Downers Grove, IL, USA) containing 59 probes (P1, PAC, or BAC clones) corresponding to 57 different oncogenes, or to the GenoSensor Array 300 microarray (Vysis, Inc.) containing 287 genomic clones, including tumor-suppressor genes, telomeres, and additional selected loci representing each chromosome arm. The lists of targets on the microarrays are available at www.vysis.com. Hybridization signal images in three colors [4′,6-diamidino-2-phenylindole (DAPI), Cy3, and Cy5 (or Alexa488 and Alexa594 for the AmpliOnc I microarray)] and their values were then analyzed by the Geno-Sensor Reader System.
Fluorescent In Situ Hybridization (FISH) Analysis
BAC clones were purchased from CHORI (BACPAC Resources, Oakland, CA) and were used as FISH probes. The following clones contained the corresponding genes: RPCI-1 118J21 for MYCL1 gene (clone cytogenetic locus, 1p34.1–p35.3), RPCI-11 181K3 for LAMC1/LAMC2 (1q25–q31), RPCI-11 350N15 for FGFR1 (8p11.1–p11.21), RPCI-11 635N3 for p21-activated kinase-1 (PAK1; 11q13–q14), RPCI-11 571M6 for CDK4/SAS (12q13–q14), RPCI-11 772E1 for GLI (12q13.2–q13.3), and RP11-89L15 and RP11-181D10 for FOXO1A (13q12.3–q14.12). Briefly, following extraction and purification, DNA probes were directly labeled by nick translation with SpectrumGreen dUTP (Vysis, Inc.) for the PAK1 probe and with SpectrumOrange dUTP (Vysis, Inc.) for the other probes. Chromosomal preparations on slide and fluorescent probes were denatured at 73°C before overnight hybridization at 37°C, followed by posthybridization washes and chromosome counterstaining by Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Hybridization signals were detected using corresponding filters on an Olympus B52 microscope (Olympus Life and Material Science, Hamburg, Germany). Images were captured using a charge-coupled device camera and Cytovision software (Applied Imaging, Santa Clara, CA).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
RNA was DNase-treated using the DNA-free kit (Ambion, Inc.), according to manufacturer's protocol. Complementary DNA (cDNA) was synthesized from 1 µg of total RNA using 100 U of Superscript II Reverse Transcriptase, 75 ng/µl random hexanucleotides (Invitrogen, Carlsbad, CA), and 125 µM of each dNTP (Pharmacia, Uppsala, Sweden) in a final volume of 10 Ml. RNA and primers were denatured at 70°C for 10 minutes, followed by reverse transcription at 42°C for 1 hour and termination at 90°C for 2 minutes.
Expression of the oncogenic fusion gene PAX3-FOXO1A or PAX7-FOXO1A (PAX3/7-FOXO1A) in RMS tumors was detected using RT-PCR. Amplifications of both chimeric fusion gene and FOXO1A (as control) transcripts were performed as described previously [21].
Real-Time Quantitative RT-PCR
Expression levels of PAX3 (GI: 31563350), PAX7 (GI: 4505618), FOXO1A (GI: 9257221), and AURKA (GI: 38327561) genes were determined in RNA from primary RMS tumors, cell lines, and normal controls. The primer sequences used for the amplification of each gene and the sizes of the products generated were as follows:
PAX3: 5′-CTGCGTCTCCAAGATCCTGTG-3′ (forward) and 5′-CGGCCTCCTCCTCTTCACC-3′ (reverse) generated a 269-bp fragment
PAX7: 5′-GCTCCGGGGCAGAACTACC-3′ (forward) and 5′-GCACGCGGCTAATCGAACTC-3′ (reverse) generated a 436-bp fragment
FOXOX1A: 5′-GTGTAACCTGCTCACTAACC-3′ (forward) and 5′-CCGCCTGACCCAAGTGAAG-3′ (reverse) generated a 331 -bp fragment
AURKA 5′-GGACCGATCTAAAGAAAACTGC-3′ (forward) and 5′-CTTTCCTTTACCCAGAGGGCG-3′ (reverse) generated a 428-bp fragment
for GAPDH: 5′-CCAGAACATCATCCCTGC-3′ (forward) and 5′-GGAAGGCCATGCCAGTGAGC-3′ (reverse) generated a 96-bp fragment.
All primers were exon-spanning (Sigma-Genosys Ltd., Rehovot, Israel).
Quantitative expression analysis was performed using LightCycler Technology and SYBR Green kit, according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Quantitative RT-PCR was performed in a total volume of 10 µl using LightCycler FastStart DNA Master SYBR Green I (Roche Diagnostics) with 50 nM of each primer (Sigma-Genosys Ltd.), 3 mM MgCl2, and 5% dimethyl sulfoxide only in the PAX3 and PAX7 reactions. All PCR conditions included a preincubation step of 10 minutes at 95°C, followed by 45 cycles. Each cycle consisted of denaturation at 95°C for 10 seconds, annealing at specific temperatures for 5 seconds, elongation at 72°C for 11 to 18 seconds, and fluorescence measurement at specific temperatures for 5 seconds (specific reaction conditions are available on request). The final PCR cycle was followed by a melting curve analysis to assess product specificity. Reaction efficiency was determined for all quantitative RT-PCRs. The slope of the standard curve was determined for each reaction, and only the results from RT-PCR with slopes ranging between -3.3 and -3.4 were included. Each experiment was performed in duplicate; amplified products were checked by electrophoresis on ethidium bromide-stained agarose gels and sequenced to confirm their identity using BigDye Terminator v. 1.1 on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA).
A standard curve was plotted for each gene and for the GAPDH gene that served as a reference gene, using serial dilution of cDNA. Log concentrations of the gene (X) and of GAPDH were calculated from the standard curve using LightCycler 5.1 software (Roche Applied Science, Mannheim, Germany). The quantification procedure was as follows: in each tumor, the expression level of each gene (RX) was calculated relative to GAPDH expression. The relative expression value of the specific gene (X) in each specimen ([X/GAPDH]RMS) was compared to the average expression obtained from the four normal skeletal muscle control RNA samples ([X/GAPDH]average controls). The final results, termed RX, were determined by the equation:
where RX values equal to or greater than a 1.5-fold change are overexpressions, and RX values equal to or less than a 0.5-fold change are underexpressions. For each gene, representative graphs of relative expression were constructed with Prism software (GraphPad Software, San Diego, CA).
Cytogenetic Analysis of Primary RMS Tumors
Table 1 describes patients' clinical characteristics and the cytogenetic analyses of the 10 primary RMS tumors. ERMS1 tumor cells demonstrated a modal number ranging from pseudodiploid to hyperdiploid, with trisomies, chromosome losses, and translocations that have been described previously in ERMS tumors [9,13,22]. ERMS2 was resected following both chemotherapy and radiotherapy, and displayed a complex pseudodiploid karyotype of several clones involving numerical and structural aberrations of chromosomes 1, 2, 4, 5, 8, and 11 (Table 1).
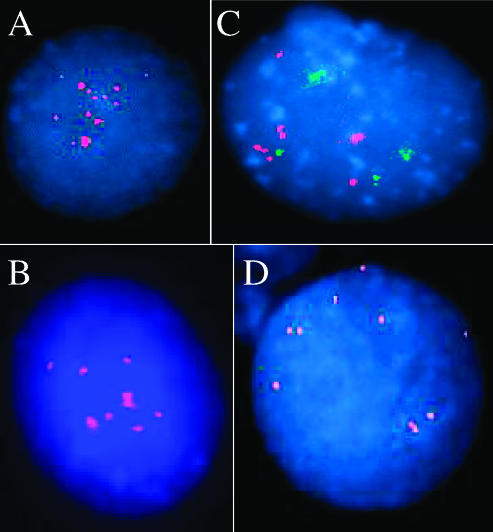
Three of 10 ARMS tumors examined (ARMS3, ARMS4, and ARMS7) demonstrated normal G-band karyotypes. It is possible that contamination by adjacent normal cells may explain these normal karyotypes or that non-neoplastic cells may have a selective advantage and thus prevail over neoplastic cells. Thus, a normal G-band karyotype does not necessarily rule out the neoplastic nature of the sample, as previously reported [12,13,22,23]. All other ARMS tumors tested displayed abnormal karyotypes. The cytogenetic aberrations detected in both ARMS2 and ARMS6 tumors were chromosomal polysomies without structural alterations apart from dmins. ARMS2 revealed a tetraploid content (Figure 1A), whereas ARMS6 demonstrated different levels of polysomies in a tetraploid karyotype (Figure 1B). In addition to numerical changes, dmins were detected in these four ARMS tumors. SKY analyses classified dmins as originating from chromosome 13 in ARMS2 and ARMS5 (Figure 1, A and C, respectively), from chromosome 1 in ARMS6 (Figure 1B), and from chromosome 3 in ARMS1 (Figure 1D). ARMS1 and ARMS5 tumor cells with tetraploidy and dmins displayed various translocations detailed in Table 1.
Figure 1.
SKY analyses of primary RMS tumors. (A) ARMS2 sample demonstrated tetraploidy and dmins classified as chromosome 13. (B) ARMS6 cells displayed dmins originating from chromosome 1 in a hypertetraploid karyotype. (C) ARMS5 presented a novel rearrangement between 13q14 and 20q13 and dmins classified as originating from chromosome 13. The trisomies of chromosomes 10 and 21 shown here are not clonal. (D) ARMS1 tumor cells display a structural rearrangement at the 13q14 region and two dmins from chromosome 3 in a complex hypotetraploid karyotype. (E) Representative SKY image of the pleomorphic RMS tumor (PRMS1) demonstrating multiple numerical and structural alterations in a hypertriploid karyotype involving all chromosomes (see also Table 1).
It is worth noting that none of the characteristic ARMS translocations [t(1;13)(p36;q14) or t(2;13)(q35;q14)] was cytogenetically detected by either G-banding or SKY analyses in the ARMS tumors examined here. However, breakpoints at 13q14 region, which harbors the FOXO1A gene, were seen in two samples ARMS1 and ARMS5 (Table 1). The ARMS5 tumor demonstrated a novel translocation of the 13q14-ter region with a chromosome 20q13.1 band in tetraploid. This translocation may be reciprocal and appears as the sole structural alteration detected in this case along with dmins (Figure 1C). The karyotype of ARMS1 tumor cells was hypotetraploid and complex, involving numerical and structural alterations (Figure 1D). The 13q14 locus was rearranged, but the reciprocal and counterpart translocant was not detectable by either G-banding or SKY painting.
To date, only three karyotypes of PRMS tumors have been published [22,24,25], whereas analysis of seven PRMS samples using conventional CGH provided additional genomic data [7]. We present here G-banding and complementary SKY karyotype descriptions of a PRMS tumor obtained at the time of diagnosis (PRMS1; Table 1 and Figure 1E). This chromosomal analysis demonstrated a hypertriploid and complex karyotype with clonal evolution. The aberrations involved all chromosomes, either numerically or structurally, including multiple translocations, deletions, and insertions (Table 1). Some of the gains and losses detected here have been published previously, particularly gains of chromosomes 5, 6q, 8, 18, and 22 and losses of 1q, 3, 13, 14, 15, and 17p [7,22,24,25].
Array-Based CGH Analysis Revealed Novel Gains and Losses of Specific Genes
To identify genetic copy number changes associated with RMS, tumor DNA was analyzed with AmpliOnc version I (Abbott Vysis, Inc.) or Array 300 microarrays (AmpliOnc version I was updated to Array 300 during the course of the experiment). DNA from ARMS1, ARMS3, ARMS4, ARMS5, ARMS7, and PRMS1 were hybridized with AmpliOnc I, whereas Array 300 was used to analyze DNA samples from ERMS1, ERMS2, ARMS2, and ARMS6 tumors.
Borderline levels of copy number gain or loss were defined for AmpliOnc I array, as previously described (0.77–1.26) [19]. Regarding Array 300, the mean G/R ratio value was 1.003, and the standard deviation (SD) was 0.107, defining a mean value of ±2 SD at the cutoff level of 0.78 to 1.22. Because this range of values is within the range used for AmpliOnc I analysis, cutoff values of 0.77 to 1.26 were designated to determine the gain and loss levels for both array versions. Values less than 0.77 were designated as losses. We further defined two classes of gain: values with 1.26- to 2-fold change represented a mild increase in copy number, and changes greater than 2-fold represented amplification. The genes that were either gained (mild increase or amplification) or lost in RMS tumors are summarized in Table 2.
Table 2.
Copy Number Changes of Specific Genes in RMS Tumors Detected by Array-Based CGH and FISH Analyses.
| Case Number | Array-Based CGH Results (Gene, Location, and Fold Change) | FISH Results | ||||||
| Mild Increase | Amplification | Loss | Gene Designation | Number of Signals per Cell | ||||
| Gene Designation (Cytogenetic Locus) | Fold Change | Gene Designation (Cytogenetic Locus) | Fold Change | Gene Designation (Cytogenetic Locus) | Fold Change | |||
| ERMS1 | MSH2* (2p22.3-p22.1) | 1.30 | AKT2* (19q13.1–q13.2) | 2.40 | 82M15* (17ptel) | 0.75 | ||
| GARP (11q13.5–q14) | 1.30 | WI-14673* (17ptel) | 0.77 | |||||
| HIC1* (17p13.3) | 0.74 | |||||||
| D17S125* (17p12-p11.2) | 0.74 | |||||||
| 22QTEL31* (22qtel) | 0.76 | |||||||
| ERMS2 | FGR (1p36.2-1) | 1.28 | Chr 3 | 0.73–0.77 | LAMC2 | 3–4 | ||
| STS* (1q21) | 1.65 | PAK1 | 2–3 | |||||
| LAMC2 (1q25–q31) | 1.33 | Two STS probes* (9p11.2) | 0.66 | |||||
| TGFB2* (1q41) | 1.37 | |||||||
| Tel probe* (1 qtel) | 1.30 | |||||||
| AIB1 (20q12) | 1.30 | |||||||
| TNFRSF6B* (20q 13) | 1.31 | |||||||
| TOM* (20qtel) | 1.33 | |||||||
| ARMS1 | PAK1 (11q13.5–q14) | 1.26 | LAMC2 | 6 | ||||
| PAK1 | 4–10 | |||||||
| ARMS2 | PDGFB (22q13.1) | 1.40 | LAMC2 | 4 | ||||
| ARMS3 | CCND2 (12p13) | 1.28 | PDGFRA (4q 12) | 0.73 | ||||
| AIB1 (20q12) | 1.33 | |||||||
| PTPN1 (20q13.1–q13.2) | 1.36 | |||||||
| ARMS4 | No change | |||||||
| ARMS5 | GLI (12q13.2–q13.3) | 2.5 | GLI | 7–8 | ||||
| SAS/CDK4 (12q13.3) | 3.2 | SAS/CDK4 | 10–12 | |||||
| ARMS6 | CSF1R* (5q33-35) | 1.27 | MYCL1 (1 p34.3) | 6.62 | Two tel probes* (4qtel) | 0.75 | MYCL1 | 8 |
| Two tel probes* (8ptel) | 1.38 | LAMC2 | 4 | |||||
| MYC (8q24.12–13) | 1.34 | |||||||
| PTK2 (8q24-ter) | 1.79 | |||||||
| Two tel probes* (1 0qtel) | 1.78 | |||||||
| WNT1 (12q12–q13) | 1.44 | |||||||
| CDK2 (12q13) | 1.30 | |||||||
| ERBB3* (12q13) | 1.30 | |||||||
| SAS/CDK4 (12q13.3) | 1.29 | |||||||
| AKT2* (19q13.1–2) | 1.30 | |||||||
| Chr 22 | 1.35–1.73 | |||||||
| ARMS7 | FGR (1p36.2-36.1) | 1.71 | HRAS (11 p15.5) | 0.74 | LAMC2 | 2 | ||
| LAMC2 (1q25–q31) | 1.41 | PAK1 | 2 | |||||
| MYCN (2p24.1) | 1.51 | |||||||
| RAF1 (3p25) | 1.42 | |||||||
| PAK1 (11q13.5–14) | 1.56 | |||||||
| CCND2 (12p13) | 1.48 | |||||||
| ERBB2 (17q21.2) | 1.43 | |||||||
| D17S1670 (17q23) | 1.29 | |||||||
| CBFA2 (21q22.3) | 1.33 | |||||||
| PRMS1 | LAMC2 (1q25–q31) | 1.27 | FGFR1 (8p11.2–p11.1) | 14.0 | GLI (12q13.2–q13.3) | 0.72 | LAMC2 | 4 |
| PAK1 (11q13.5-q14) | 1.43 | FGFR1 | 12–31 | |||||
| PAK1 | 6 | |||||||
STS, sequence-tagged site; tel, telomere; Chr, all probes on this chromosome were changed.
Probes were represented only in the Array 300 microarray.
Array-based CGH results were further correlated with cytogenetic analyses of tumor cells and were consistent with ERMS1 for gains of chromosomes 2, 11q, and 19q and for losses of 17p and 22q, and with ARMS6 for gains of chromosome regions 8ptel, 8q24-ter, 12q12—13.3, and 22q. In other cases, involvement of large chromosomal loci was depicted only from array-based CGH results: the ERMS2 tumor demonstrated a gain of chromosomes 1 and 20 with a loss of chromosomes 3 and 9p12–q21; ARMS3 displayed a gain of 20q; and DNA from ARMS7 revealed a gain of the 17q21–23 chromosomal region.
Array-based CGH analysis detected copy number changes in genes related to multiple functional classes. Some of the genes identified in our series encode or interact with transcription factors. A mild increase in copy number and amplifications of the three members of the MYC family were detected: MYCN in ARMS7, and both MYC and MYC-related gene from lung cancer (MYCL1) genes in ARMS6. The co-amplification of SAS/CDK4 and GLI genes was detected in the ARMS5 tumor. Gain of SAS/CDK4 and loss of GLI were identified in ARMS6 and PRMS1 samples, respectively. A mild increase in the copy number of the CBFA2 gene in ARMS7 and of the CDK2 gene in ARMS6 was also detected. Loss of another transcription factor, the HIC1 gene, was detected in ERMS1. Finally, a mild increase in the copy number of CCND2 in both ARMS3 and ARMS7 samples was also found.
Changes in copy number were also detected in genes that encode protein members of signaling pathways, including ligands of cell surface receptors TGFB2 (in ERMS2), PDGFB (ARMS2), and WNT1 (ARMS6), and genes encoding tyrosine kinase receptors ErbB2 (ARMS7), ErbB3 (ARMS6), TNFRSF6B (ERMS2), CSF1R (ARMS6), and FGFR1 (PRMS1). A mild increase in the copy number of genes involved in mitogen-activated protein kinase cascade and other cell signaling proteins was also identified in our series, including RAF1 (ARMS7), FGR (ERMS2 and ARMS7), AKT2 (ERMS1 and ARMS6), PTK2 (ARMS6), and PTPN1 (ARMS3). Conversely, the PDGFRA and HRAS genes were deleted in ARMS3 and ARMS7, respectively. A mild increase in copy number was also detected in two genes belonging to the steroid receptor superfamily, which are involved in cellular signaling (the AIB1 gene in both ERMS2 and ARMS3 tumors and the TOM gene in ERMS2 tumor).
A mild increase in the copy number changes of two genes, laminin γ-2 (LAMC2) and PAK1, both involved in basal membrane organization and cytoskeletal elements was detected in four RMS samples (ERMS2, ARMS1, ARMS7, and PRMS1; Table 2). FISH analysis was implemented to validate the amplification of genes with a > 2-fold increase in array-based CGH analysis and to validate the mild increase in the copy number of LAMC2 and PAK1 genes detected in three RMS samples each (Table 2). Fluorescent probes for specific genes were used; fluorescent signals were counted in 30 interphases in each analysis. The results are summarized in Table 2 and are illustrated in Figure 2, A–D. Good concordance was obtained between array-based CGH and FISH results, except for ARMS7, which displayed an increase in the copy number changes of several genes (Table 2), whereas tumor cell culture demonstrated normal karyotype (Table 1). SAS/CDK4 (Figure 2A) and GLI (Figure 2B) amplifications in ARMS5 cells were confirmed. An increased copy number of LAMC2 and PAK1 was detected by FISH in ERMS2 and PRMS1 cells (Table 2) and in ARMS1 (Figure 2C), and the amplification of MYCL1 in ARMS6 cells (Figure 2D) was also validated and confirmed.
Figure 2.
Interphase FISH analyses of primary RMS tumor cells with specific BAC and PAC clones confirmed the increased copy number change detected by array-based CGH analysis: ARMS5 tumor cells hybridized with SAS/CDK4 (A) and GLI (B) probes. LAMC2 (C; red) and PAK1 (C; green) signals were detected in ARMS1 cells. (D) Hybridization of ARMS6 cells with the MYCL1 probe.
Expression Analysis of PAX3, PAX7, FOXO1A, and AURKA Genes in Primary RMS Tumors
RT-PCR and quantitative RT-PCRwere performed in RNA extracted from the 10 primary tumors and the 4 cell lines. RT-PCR detected PAX3-FOXO1A or PAX7-FOXO1A fusion transcripts in 6 of 10 primary tumors (ERMS2, ARMS2, ARMS3, ARMS4, ARMS5, and ARMS7) and in the RMZ-RC2 and RH28 cell lines, as previously described [26,27].
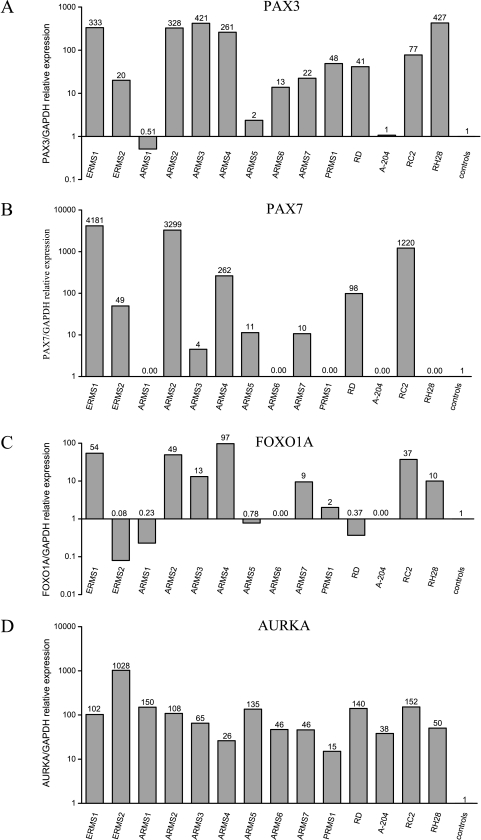
Primer pair sequences for quantitative RT-PCR were designed to amplify a part of the binding domains of PAX3 and PAX7 genes and a part of the transcriptional activation domain of FOXO1A gene. Therefore, the expression levels of each gene represented the sum of the expression detected from both the wild type and the fusion genes. The relative expression levels of these genes are presented in Figure 3, A—C, respectively, and were further correlated with RMS tumor subtypes (ERMS, ARMS, or PRMS) and with the presence of PAX3/7-FOXO1A chimeric transcripts.
Figure 3.
Quantitative RT-PCR expression analysis of primary RMS tumors and cell lines relative to average expression in normal skeletal muscle controls: (A) PAX3, (B) PAX7 (C), FOXO1A, and (D) AURKA genes. Relative expression values are calculated in log scale and are presented above each bar.
PAX3 and PAX7 genes were overexpressed in three of four embryonal RMS samples tested (ERMS1, ERMS2, and RD), with higher values in the two RMS samples ERMS1 and RD that did not express the fusion transcript. Generally, PAX7 expression values were higher than PAX3 values. The FOXO1A gene was overexpressed in ERMS1 and was underexpressed in the three other RMS samples tested.
Overexpression of PAX3 and/or PAX7 genes was detected in all of the primary alveolar subtype tumors, except for the ARMS1 sample that did not express the oncogenic fusion gene PAX3/7-FOXO1A. FOXO1A overexpression was seen in four of seven primary ARMS tumors tested, but not in the ARMS1 sample mentioned above and not in an additional sample that expressed the fusion oncogene (ARMS5). In cell line samples, overexpression of PAX7 was detected only in RMZ-RC2, which harbors the PAX7-FOXO1A translocation [26], and PAX3 overexpression was detected in the RH28 cell line, which harbors PAX3-FOXO1A translocation [27].
The PRMS tumor that did not express PAX3/7-FOXO1A transcripts demonstrated an overexpression of PAX3 and FOXO1A genes.
The AURKA gene was overexpressed in all of the RMS samples (Figure 3D). In primary RMS tumors, the highest overexpression values (103- to 1030-fold change) were detected in five samples that exhibited either complex karyotypes (ERMS1, ERMS2, and ARMS1) or tetraploid karyotypes (ARMS2 and ARMS5). All four RMS cell lines tested, which were already shown to carry aneuploidy [23,26–28], also demonstrated overexpression of the AURKA gene. It is worth noting that the lowest overexpression value of AURKA (15-fold change) was detected in the adult-type PRMS1 sample.
Discussion
Our complete spectral karyotyping, combined with array-based CGH and expression analysis of fresh tissues obtained from primary RMS tumors, revealed novel genomic aberrations and altered gene expressions. Such analyses have been reported on cell lines in the past [28,29] and on two primary RMS using G-band, RT-PCR, and conventional CGH recently [30].
Spectral analysis defined the origin of dmins from chromosomes 1 and 13, and a novel translocation between the 13q14 and 20q13.1 chromosomal regions. The multiple chromosomal aberrations identified in the PRMS sample enhance published cytogenetic data and further suggest candidate loci associated with the development and progression of this rare adult RMS subtype. Our analysis also emphasizes the chromosomal instability of RMS cells, including aneuploidy (mostly tetraploidy), multiple structural alterations, and dmins.
AURKA, a serine-threonine kinase, has been implicated in the regulation of centrosome function, spindle assembly, spindle maintenance, chromosome segregation, and cytokinesis [31]. Alterations in its activity affect genomic stability and disrupt the fidelity of centrosome duplication, resulting in tetraploidization [32]. AURKA was therefore implicated as a potent oncogene that induces cellular transformation [33]. AURKA amplification and/or overexpression was found in a series of cancer cell lines and primary tumors, such as breast, prostate, colorectal, and pancreatic tumors, and was associated with grades of tumor differentiation and invasive capability [34–37]. The high level of aneuploidy, in particular the tetraploid pattern detected in four RMS samples, led us to hypothesize that AURKA may also play a role in RMS tumorigenesis. Of note, AURKA overexpression was not accompanied here by gene amplification, as depicted by array-based CGH analysis. The demonstration of AURKA overexpression in all primary RMS tumors and cell lines tested suggests a novel association between this gene and the chromosomal instability in RMS, and may propose a therapeutic role for AURKA inhibitors in the treatment of this skeletal muscle neoplasm [38]. Other genes belonging to the mitotic spindle checkpoint complex may also be involved in RMS. For example, somatic mutations were recently detected in the BUB1B gene in familial ERMS cases [39], further suggesting an important role for regulators of mitosis in the chromosomal instability detected in RMS.
The association between PAX3, PAX7, and FOXO1A genes and RMS is well established [10] and further characterized in our RMS tumor samples. Because oncogenic PAX3/7-FOXO1A fusions were not detected by either cytogenetic or RT-PCR analyses in 4 of 10 primary tumors examined, expression analyses of these genes were performed. We confirmed previously published data and demonstrated that overexpression of PAX3 and PAX7 does not necessarily correlate with the detection of the chimeric fusion transcript PAX3-FOXO1A or PAX7-FOXO1A, suggesting that mechanisms other than oncogenic fusion cause changes in the expression patterns of these genes [11]. One example is the ARMS6 tumor that demonstrated only PAX3 overexpression (14-fold), without overexpression of PAX7 or FOXO1A, and did not express fusion transcripts. Of note, the embryonal ERMS2 tumor sample demonstrated the PAX3/7-FOXO1A fusion transcript that is characteristic of the alveolar RMS subtype. It is possible that this transcript resulted from prior chemotherapy and radiotherapy of this patient; however, previous cytogenetic analyses in both an ERMS cell line [40] and a primary ERMS tumor [41] have demonstrated translocations of the 2q35 locus that harbors PAX3, and fusions of PAX3/7-FOXO1A have been detected by either cytogenetic, FISH, or RT-PCR analyses in embryonal RMS samples [11,42]. Furthermore, we did not detect either fusion transcripts or overexpression of PAX3 or PAX7 in the primary ARMS1 and in the A-204 ERMS cell line. These findings suggest that RMS transformation is not absolutely dependent on the abnormal expression of the chimeric PAX3-FOXO1A or PAX7-FOXO1A transcripts, and might be initiated and propagated by other genetic changes. This idea is further supported by PAX3-FOXO1A knockin studies in mice, which demonstrated that oncogenic fusion is not sufficient for tumor development [43].
Array-based CGH is a powerful tool for the rapid and accurate detection of specific gene copy number changes, allowing the identification of novel candidate genes in carcinogenesis [19,44]. Using array-based CGH analysis, multiple copy number changes were detected in our RMS series. By comparison, in six Wilms tumor samples, array-based CGH analyses detected only one amplification event in the MRP1 gene [19], and no amplifications or deletions were seen in 8 synovial sarcoma and 20 Ewing sarcoma samples analyzed in our laboratory with the same arrays (A. Bar-Shira and A. Orr-Urtreger, personal communication). Our data therefore suggest that RMS tumorigenesis involves multiple amplification or deletion events in several gene families and the disruption of a number of cellular pathways.
The involvement of the MYC transcription factor family, particularly MYCN, has been reported in RMS tumorigenesis [45,46]. We detected the novel amplification of MYCL1 gene in RMS, which has already been demonstrated in other neoplasms, including small cell lung cancer [47] and hormone-resistant prostate cancer [48]. Additional changes in genes encoding for transcription factors (such as the amplification of GLI and the mild increase in the copy number of CBFA2, and the overexpression of PAX3 and PAX7) were also observed in our study and reported in RMS tumorigenesis [3,11], suggesting that the disruption of the transcriptional machinery is a major event in RMS cells, which likely leads to the abnormal expression of multiple downstream target genes.
Using array-based CGH and FISH analyses, we detected a mild increase in the copy number of genes implicated in cell-to-cell contact, cell adhesion, and motility, including WNT1, FGR, PTK2, TGFB2, and AIB1. Gain in the copy number of LAMC2 and PAK1, both related to basal membrane organization and cytoskeletal elements, was identified in three primary RMS tumors each, suggesting a novel association between these genes and RMS tumorigenesis.
The p21-activated kinase PAK1 is one of the critical effectors linking the small GTPases Rac1 and Cdc42 to cytoskeleton-dependent cell functions [49]. Overexpression of PAK1 in cancer cells increases cell migration potential and anchorage-independent growth [50]. To date, PAK1 function has not been directly related to myogenesis or RMS. However, studies on RMS cell lines have demonstrated constitutive activation of Rac1 and Cdc42, which appears to elicit the loss of cell contact inhibition and anchorage-dependent growth, possibly contributing to transformation of myoblastic cells, inhibition of myoblast differentiation, and impaired exit of myoblasts from cell cycle [51–53]. Recently, gene expression profiling of primary RMS tumors demonstrated Rac1 overexpression [18]. The increased copy number of PAK1 in primary RMS tumors may therefore be linked to these abnormal myoblast functions during tumorigenesis.
The composition of extracellular matrix components influences differentiation and myogenesis [54]. LAMC2 encodes the γ polypeptide chain specific to laminin (LN)5, an extracellular matrix protein that forms anchoring filaments, contributing to the structural and biologic relationships between the epithelium and the stroma. The γ chain is of importance in outside-in and inside-out cell signaling, which sustain cell-to-cell contact and cell adhesion [55]. LN5, particularly the γ chain, is associated with cancer aggressiveness [56]. LN5 expression enhances the tumorigenicity of a human fibrosarcoma cell line, suggesting that it may also promote tumor growth in vivo [57]. Overexpression of LAMC2 was also linked to tumor invasion and unfavorable outcomes in breast and colon carcinomas, although the mechanism(s) associating LAMC2 overexpression to cancer progression remains unclear [57–59]. Increased LAMC2 copy number in three of the RMS samples tested here may suggest its involvement in the disruption of cell adhesion and RMS progression.
Of note, although some of the alterations, such as AURKA overexpression, are described here in all primary RMS tumor samples, others (including LAMC2 and PAK1 copy number increase) were observed in only a few samples. Overall data emphasize the molecular differences underlying RMS tumors, and may also explain the variable outcome of individual patients. It is also worth noting that, although the samples studied were from primary tumors, specific genetic alterations described here, which may lead to the dysregulation of a given gene, are not sufficient to establish their causality in RMS and do not prove whether or not they are essential to RMS tumorigenesis.
In summary, cytogenetic and molecular analyses of 10 primary RMS tumors have demonstrated the complexity of the genetic mechanisms underlying the development of this neoplasm. The novel detection of AURKA overexpression suggests an association between the spindle mitotic checkpoint complex and the chromosomal instability and aneuploidy frequently seen in RMS. The copy number gain of LAMC2 and PAK1, in addition to other genes involved in cell contact and migration, may implicate the disruption of these functions and the abnormal progression of myogenesis in RMS tumorigenesis. Together, these altered pathways may interact with one another, in the cell matrix, and through intracellular signaling, influencing the transcriptional machinery and mitotic complexes and ultimately contributing to RMS malignant transformation and metastasis.
Acknowledgements
We thank Anat Bar-Shira for important technical help, Serena Rosner for editorial assistance, and Ruth Shomrat for support. We also thank Shlomo Wientroub and Yehuda Kollender.
Abbreviations
- RMS
rhabdomyosarcoma
- CGH
comparative genomic hybridization
- SKY
spectral karyotype
- dmins
double minutes
- FISH
fluorescent in situ hybridization
- LAMC2
laminin γ-2
- PAK1
p21-activated kinase-1
- MYCL1
MYC-related gene from lung cancer
Footnotes
This work was supported by the M.K. Humanitarian Fund and was performed in partial fulfillment of the requirement for the doctoral degree of Myriam Goldstein (Sackler Faculty of Medicine, Tel-Aviv University).
References
- 1.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 2.Anderson J, Gordon A, Pritchard-Jones K, Shipley J. Genes, chromosomes, and rhabdomyosarcoma. Genes Chromosomes Cancer. 1999;26:275–285. [PubMed] [Google Scholar]
- 3.Merlino G, Helman LJ. Rhabdomyosarcoma—working out the pathways. Oncogene. 1999;18:5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 4.Molenaar WM, Oosterhuis JW, Oosterhuis AM, Ramaekers FC. Mesenchymal and muscle-specific intermediate filaments (vimentin and desmin) in relation to differentiation in childhood rhabdomyosarcomas. Hum Pathol. 1985;16:838–843. doi: 10.1016/s0046-8177(85)80256-2. [DOI] [PubMed] [Google Scholar]
- 5.Tonin PN, Scrable H, Shimada H, Cavenee WK. Muscle-specific gene expression in rhabdomyosarcomas and stages of human fetal skeletal muscle development. Cancer Res. 1991;51:5100–5106. [PubMed] [Google Scholar]
- 6.Fletcher CDM, Unni KK, Mertens F, editors. World Health Organization: Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press; 2002. pp. 146–154. [Google Scholar]
- 7.Gordon A, McManus A, Anderson J, Fisher C, Abe S, Nojima T, Pritchard-Jones K, Shipley J. Chromosomal imbalances in pleomorphic rhabdomyosarcomas and identification of the alveolar rhabdomyosarcoma-associated PAX3-FOXO1A fusion gene in one case. Cancer Genet Cytogenet. 2003;140:73–77. doi: 10.1016/s0165-4608(02)00631-3. [DOI] [PubMed] [Google Scholar]
- 8.San Miguel-Fraile P, Carrillo-Gijon R, Rodriguez-Peralto JL, Badiola IA. Prognostic significance of DNA ploidy and proliferative index (MIB-1 index) in childhood rhabdomyosarcoma. Am J Clin Pathol. 2004;121:358–365. doi: 10.1309/3DD8-0UC4-DDJ0-JMT2. [DOI] [PubMed] [Google Scholar]
- 9.Bridge JA, Liu J, Qualman SJ, Suijkerbuijk R, Wenger G, Zhang J, Wan X, Baker KS, Sorensen P, Barr FG. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33:310–321. doi: 10.1002/gcc.10026. [DOI] [PubMed] [Google Scholar]
- 10.Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. 2001;20:5736–5746. doi: 10.1038/sj.onc.1204599. [DOI] [PubMed] [Google Scholar]
- 11.Tiffin N, Williams RD, Shipley J, Pritchard-Jones K. PAX7 expression in embryonal rhabdomyosarcoma suggests an origin in muscle satellite cells. Br J Cancer. 2003;89:327–332. doi: 10.1038/sj.bjc.6601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang-Wuu S, Soukup S, Ballard E, Gotwals B, Lampkin B. Chromosomal analysis of sixteen human rhabdomyosarcomas. Cancer Res. 1988;48:983–987. [PubMed] [Google Scholar]
- 13.Gordon T, McManus A, Anderson J, Min T, Swansbury J, Pritchard-Jones K, Shipley J. Cytogenetic abnormalities in 42 rhabdomyosarcoma: a United Kingdom Cancer Cytogenetics Group Study. Med Pediatr Oncol. 2001;36:259–267. doi: 10.1002/1096-911X(20010201)36:2<259::AID-MPO1063>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 14.Meddeb M, Valent A, Danglot G, Nguyen VC, Duverger A, Fouquet F, Terrier-Lacombe MJ, Oberlin O, Bernheim A. MDM2 amplification in a primary alveolar rhabdomyosarcoma displaying a t(2;13)(q35;q14) Cytogenet Cell Genet. 1996;73:325–330. doi: 10.1159/000134368. [DOI] [PubMed] [Google Scholar]
- 15.Driman D, Thorner PS, Greenberg ML, Chilton-MacNeill S, Squire J. MYCN gene amplification in rhabdomyosarcoma. Cancer. 1994;73:2231–2237. doi: 10.1002/1097-0142(19940415)73:8<2231::aid-cncr2820730832>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Roberts WM, Douglass EC, Peiper SC, Houghton PJ, Look AT. Amplification of the gli gene in childhood sarcomas. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- 17.Barr FG, Nauta LE, Davis RJ, Schafer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 18.De Pitta C, Tombolan L, Albiero G, Sartori F, Romualdi C, Jurman G, Carli M, Furlanello C, Lanfranchi G, Rosolen A. Gene expression profiling identifies potential relevant genes in alveolar rhabdomyosarcoma pathogenesis and discriminates PAX3-FKHR positive and negative tumors. Int J Cancer. 2005 doi: 10.1002/ijc.21698. (E-pub) [DOI] [PubMed] [Google Scholar]
- 19.Goldstein M, Rennert H, Bar-Shira A, Burstein Y, Yaron Y, Orr-Urtreger A. Combined cytogenetic and array-based comparative genomic hybridization analyses of Wilms tumors: amplification and overexpression of the multidrug resistance associated protein 1 gene (MRP1) in a metachronous tumor. Cancer Genet Cytogenet. 2003;141:120–127. doi: 10.1016/s0165-4608(02)00667-2. [DOI] [PubMed] [Google Scholar]
- 20.Mitelman F. Guidelines for Cancer Cytogenetics, Supplement to An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: S. Karger; 1995. [Google Scholar]
- 21.Barr FG, Xiong QB, Kelly K. A consensus polymerase chain reaction-oligonucleotide hybridization approach for the detection of chromosomal translocations in pediatric bone and soft tissue sarcomas. Am J Clin Pathol. 1995;104:627–633. doi: 10.1093/ajcp/104.6.627. [DOI] [PubMed] [Google Scholar]
- 22.Kullendorff CM, Donner M, Mertens F, Mandahl N. Chromosomal aberrations in a consecutive series of childhood rhabdomyosarcoma. Med Pediatr Oncol. 1998;30:156–159. doi: 10.1002/(sici)1096-911x(199803)30:3<156::aid-mpo5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 23.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 24.Douglass EC, Valentine M, Etcubanas E, Parham D, Webber BL, Houghton PJ, Houghton JA, Green AA. A specific chromosomal abnormality in rhabdomyosarcoma. Cytogenet Cell Genet. 1987;45:148–155. doi: 10.1159/000132446. [DOI] [PubMed] [Google Scholar]
- 25.Mertens F, Fletcher CD, Dal Cin P, De Wever I, Mandahl N, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, et al. Cytogenetic analysis of 46 pleomorphic soft tissue sarcomas and correlation with morphologic and clinical features: a report of the CHAMP Study Group. Chromosomes and morphology. Genes Chromosomes Cancer. 1998;22:16–25. doi: 10.1002/(sici)1098-2264(199805)22:1<16::aid-gcc3>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 26.Frascella E, Lenzini E, Schafer BW, Brecevic L, Dorigo E, Toffolatti L, Nanni P, De Giovanni C, Rosolen A. Concomitant amplification and expression of PAX7-FKHR and MYCN in a human rhabdomyosarcoma cell line carrying a cryptic t(1;13)(p36;q14) Cancer Genet Cytogenet. 2000;121:139–145. doi: 10.1016/s0165-4608(00)00258-2. [DOI] [PubMed] [Google Scholar]
- 27.Hazelton BJ, Houghton JA, Parham DM, Douglass EC, Torrance PM, Holt H, Houghton PJ. Characterization of cell lines derived from xenografts of childhood rhabdomyosarcoma. Cancer Res. 1987;47:4501–4507. [PubMed] [Google Scholar]
- 28.Pandita A, Zielenska M, Thorner P, Bayani J, Godbout R, Greenberg M, Squire JA. Application of comparative genomic hybridization, spectral karyotyping, and microarray analysis in the identification of subtype-specific patterns of genomic changes in rhabdomyosarcoma. Neoplasia. 1999;1:262–275. doi: 10.1038/sj.neo.7900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Perales S, Martinez-Ramirez A, de Andres SA, Valle L, Urioste M, Benitez J, Cigudosa JC. Molecular cytogenetic characterization of rhabdomyosarcoma cell lines. Cancer Genet Cytogenet. 2004;148:35–43. doi: 10.1016/s0165-4608(03)00216-4. [DOI] [PubMed] [Google Scholar]
- 30.Cerveira N, Torres L, Ribeiro FR, Henrique R, Pinto A, Bizarro S, Ferreira AM, Lopes C, Teixeira MR. Multimodal genetic diagnosis of solid variant alveolar rhabdomyosarcoma. Cancer Genet Cytogenet. 2005;163:138–143. doi: 10.1016/j.cancergencyto.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Marumoto T, Zhang D, Saya H. Aurora-A—a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 32.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutertre S, Descamps S, Prigent C. On the role of aurora-A in centrosome function. Oncogene. 2002;21:6175–6183. doi: 10.1038/sj.onc.1205775. [DOI] [PubMed] [Google Scholar]
- 34.Zhu J, Abbruzzese JL, Izzo J, Hittelman WN, Li D. AURKA amplification, chromosome instability, and centrosome abnormality in human pancreatic carcinoma cells. Cancer Genet Cytogenet. 2005;159:10–17. doi: 10.1016/j.cancergencyto.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Bar-Shira A, Pinthus JH, Rozovsky U, Goldstein M, Sellers WR, Yaron Y, Eshhar Z, Orr-Urtreger A. Multiple genes in human20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res. 2002;62:6803–6807. [PubMed] [Google Scholar]
- 36.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 38.Hata T, Furukawa T, Sunamura M, Egawa S, Motoi F, Ohmura N, Marumoto T, Saya H, Horii A. RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 2005;65:2899–2905. doi: 10.1158/0008-5472.CAN-04-3981. [DOI] [PubMed] [Google Scholar]
- 39.Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;10:10. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 40.Roberts I, Gordon A, Wang R, Pritchard-Jones K, Shipley J, Coleman N. Molecular cytogenetic analysis consistently identifies translocations involving chromosomes 1, 2 and 15 in five embryonal rhabdomyosarcoma cell lines and a PAX-FOXO1A fusion gene negative alveolar rhabdomyosarcoma cell line. Cytogenet Cell Genet. 2001;95:134–142. doi: 10.1159/000059335. [DOI] [PubMed] [Google Scholar]
- 41.Ho RH, Johnson J, Dev VG, Whitlock JA. A novel t(2;20)(q35;p12) in embryonal rhabdomyosarcoma. Cancer Genet Cytogenet. 2004;151:73–77. doi: 10.1016/j.cancergencyto.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Anderson J, Gordon T, McManus A, Mapp T, Gould S, Kelsey A, McDowell H, Pinkerton R, Shipley J, Pritchard-Jones K. Detection of the PAX3-FKHR fusion gene in paediatric rhabdomyosarcoma: a reproducible predictor of outcome? Br J Cancer. 2001;85:831–835. doi: 10.1054/bjoc.2001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagutina I, Conway SJ, Sublett J, Grosveld GC. Pax3-FKHR knock-in mice show developmental aberrations but do not develop tumors. Mol Cell Biol. 2002;22:7204–7216. doi: 10.1128/MCB.22.20.7204-7216.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holzmann K, Kohlhammer H, Schwaenen C, Wessendorf S, Kestler HA, Schwoerer A, Rau B, Radlwimmer B, Dohner H, Lichter P, et al. Genomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes. Cancer Res. 2004;64:4428–4433. doi: 10.1158/0008-5472.CAN-04-0431. [DOI] [PubMed] [Google Scholar]
- 45.Toffolatti L, Frascella E, Ninfo V, Gambini C, Forni M, Carli M, Rosolen A. MYCN expression in human rhabdomyosarcoma cell lines and tumour samples. J Pathol. 2002;196:450–458. doi: 10.1002/path.1068. [DOI] [PubMed] [Google Scholar]
- 46.Bortoluzzi S, Bisognin A, Romualdi C, Danieli GA. Novel genes, possibly relevant for molecular diagnosis or therapy of human rhabdomyosarcoma, detected by genomic expression profiling. Gene. 2005;348:65–71. doi: 10.1016/j.gene.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 47.Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, McBride OW, Bertness V, Hollis GF, Minna JD. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 48.Edwards J, Krishna NS, Witton CJ, Bartlett JM. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res. 2003;9:5271–5281. [PubMed] [Google Scholar]
- 49.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 50.Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, Chernoff J, Hung MC, Kumar R. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 51.Heller H, Gredinger E, Bengal E. Rac1 inhibits myogenic differentiation by preventing the complete withdrawal of myoblasts from the cell cycle. J Biol Chem. 2001;276:37307–37316. doi: 10.1074/jbc.M103195200. [DOI] [PubMed] [Google Scholar]
- 52.Meriane M, Charrasse S, Comunale F, Gauthier-Rouviere C. Transforming growth factor beta activates Rac1 and Cdc42Hs GTPases and the JNK pathway in skeletal muscle cells. Biol Cell. 2002;94:535–543. doi: 10.1016/s0248-4900(02)00023-0. [DOI] [PubMed] [Google Scholar]
- 53.Meriane M, Charrasse S, Comunale F, Mery A, Fort P, Roux P, Gauthier-Rouviere C. Participation of small GTPases Rac1 and Cdc42Hs in myoblast transformation. Oncogene. 2002;21:2901–2907. doi: 10.1038/sj.onc.1205396. [DOI] [PubMed] [Google Scholar]
- 54.Osses N, Brandan E. ECM is required for skeletal muscle differentiation independently of muscle regulatory factor expression. Am J Physiol Cell Physiol. 2002;282:C383–C394. doi: 10.1152/ajpcell.00322.2001. [DOI] [PubMed] [Google Scholar]
- 55.Aumailley M, El Khal A, Knoss N, Tunggal L. Laminin 5 processing and its integration into the ECM. Matrix Biol. 2003;22:49–54. doi: 10.1016/s0945-053x(03)00013-1. [DOI] [PubMed] [Google Scholar]
- 56.Giannelli G, Antonaci S. Biological and clinical relevance of Laminin-5 in cancer. Clin Exp Metastasis. 2001;18:439–443. doi: 10.1023/a:1011879900554. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima H, Hirosaki T, Miyata S, Takamura H, Miyagi Y, Miyazaki K. Expression of laminin-5 enhances tumorigenicity of human fibrosarcoma cells in nude mice. Jpn J Cancer Res. 2002;93:652–659. doi: 10.1111/j.1349-7006.2002.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schenk S, Hintermann E, Bilban M, Koshikawa N, Hojilla C, Khokha R, Quaranta V. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olsen J, Kirkeby LT, Brorsson MM, Dabelsteen S, Troelsen JT, Bordoy R, Fenger K, Larsson LI, Simon-Assmann P. Converging signals synergistically activate the LAMC2 promoter and lead to accumulation of the laminin gamma 2 chain in human colon carcinoma cells. Biochem J. 2003;371:211–221. doi: 10.1042/BJ20021454. [DOI] [PMC free article] [PubMed] [Google Scholar]