Abstract
APOBEC3G (A3G) and related deoxycytidine deaminases are potent intrinsic antiretroviral factors. A3G is expressed either as an enzymatically active low-molecular-mass (LMM) form or as an enzymatically inactive high-molecular-mass (HMM) ribonucleoprotein complex. Resting CD4 T cells exclusively express LMM A3G, where it functions as a powerful postentry restriction factor for HIV-1. Activation of CD4 T cells promotes the recruitment of LMM A3G into 5- to 15-MDa HMM complexes whose function is unknown. Using tandem affinity purification techniques coupled with MS, we identified Staufen-containing RNA-transporting granules and Ro ribonucleoprotein complexes as specific components of HMM A3G complexes. Analysis of RNAs in these complexes revealed Alu and small Y RNAs, two of the most prominent nonautonomous mobile genetic elements in human cells. These retroelement RNAs are recruited into Staufen-containing RNA-transporting granules in the presence of A3G. Retrotransposition of Alu and hY RNAs depends on the reverse transcriptase machinery provided by long interspersed nucleotide elements 1 (L1). We now show that A3G greatly inhibits L1-dependent retrotransposition of marked Alu retroelements not by inhibiting L1 function but by sequestering Alu RNAs in cytoplasmic HMM A3G complexes away from the nuclear L1 enzymatic machinery. These findings identify nonautonomous Alu and hY retroelements as natural cellular targets of A3G and highlight how different forms of A3G uniquely protect cells from the threats posed by exogenous retroviruses (LMM A3G) and endogenous retroelements (HMM A3G).
Keywords: RNA granules, Ro ribonucleoproteins, prespliceosomes
The intrinsic antiretroviral factor APOBEC3G (A3G) is highly active against HIV-1 and other retroviruses (1). Incorporation of A3G into budding HIV-1 virions promotes extensive mutation of nascent HIV-1 DNA formed by reverse transcription in the next round of infection (2–5). However, HIV-1 counters these effects of A3G with virion infectivity factor (Vif), which accelerates proteasome-mediated degradation of A3G (6–11) and partially impairs de novo synthesis of A3G (6, 12). These two actions in virus-producing cells effectively deplete intracellular A3G, making the enzyme unavailable for virion encapsidation. Resting CD4 T cells and monocytes, which are refractory to HIV-1 infection, express only the low-molecular-mass (LMM) form of A3G (13). siRNA-mediated knockdown of LMM A3G expression in resting CD4 T cells renders these cells permissive for HIV-1 infection, indicating that LMM A3G functions as a potent postentry restriction factor for HIV-1 (13). Conversely, resting CD4 T cells in lymphoid tissues are permissive for HIV-1 infection, and A3G is predominantly in high-molecular-mass (HMM) complexes in these cells (14) because of the lymphoid microenvironment. Locally produced cytokines, including IL-2 and IL-15, and cell–cell interactions in lymphoid tissues stimulate assembly of the HMM A3G complexes (14) and confer permissiveness for HIV-1 infection.
The genes encoding A3G and other APOBEC3 (A3) family members are clustered on human chromosome 22 (15). During mammalian evolution, this locus expanded from a single gene in mice to eight genes (A3A–H) in primates (15, 16). These genes apparently have been modulated by repeated episodes of strong positive evolutionary pressure predating the emergence of the primate lentiviruses (17, 18). The striking coincidence between the expansion of the APOBEC3 gene cluster (15, 16) and the abrupt decline in retrotransposon activity in primates (19) raises the possibility that these genes may have expanded to prevent genomic instability caused by endogenous retroelements (17).
The major classes of endogenous retroelements in mammals include autonomous long interspersed nucleotide elements (LINEs), nonautonomous short interspersed nucleotide elements, and elements with long terminal repeats, such as endogenous retroviruses. These retroelements are mobile through retrotransposition, an intracellular process involving reverse transcription. They occur in high copy number in ancestral genomes and likely played an important role in genome evolution (20).
Recent studies revealed that human A3 proteins can impair the activity of mouse endogenous retroviruses (21–24) and yeast Ty1 retrotransposons (25, 26); A3A, A3B, A3C, and A3F can inhibit human long interspersed nucleotide elements 1 (L1) (24, 27–29). A3A and A3B can also inhibit L1-mediated Alu retrotransposition (24). Because retrotransposition of L1 is not affected by A3G (21, 24, 27–30), it is unknown whether A3G helps defend human cells from the “threat within” posed by endogenous mobile genetic elements.
Results
Tandem Affinity Purification (TAP) of HMM A3G Complexes.
To characterize the protein and RNA cofactors in HMM A3G assembly, we used a TAP strategy. Sequences encoding streptavidin-binding and calmodulin-binding peptides were fused to the N terminus of A3G (NTAP–A3G) and transfected into 293T cells lacking A3G. NTAP–A3G assembled into HMM complexes (Fig. 1A) and was sensitive to Vif-mediated degradation (not shown). Molecular sieving studies using CL-2B and CL-6B matrices suggest that the HMM A3G complexes are 5–15 MDa (not shown). RNase treatment of the HMM complexes generated a LMM form of NTAP–A3G (Fig. 1A). Thus, NTAP–A3G assembles into HMM ribonucleoprotein (RNP) complexes containing putative cellular RNAs and protein cofactors. The NTAP–A3G-containing complex was isolated under native conditions from cytoplasmic extract (see Figs. 6–15, Table 1, and Supporting Text, which are published as supporting information on the PNAS web site) as described in Methods. Proteins associated with the purified HMM NTAP–A3G complexes were separated by SDS/PAGE and visualized by Coomassie staining (Fig. 1B). This complex pattern of proteins copurifying with NTAP–A3G proved highly reproducible (data not shown).
Fig. 1.
Characterization of HMM A3G complexes. (A) NTAP–A3G expressed in 293T cells principally resides in HMM complexes that are converted to LMM forms after RNase A treatment. FPLC analysis was performed as described (13). The upper band of the doublet recognized by anti-A3G corresponds to the tagged A3G, and the bottom band corresponds to nontagged A3G derived from a cleavage reaction occurring between the tags and A3G. (B) TAP of HMM NTAP–A3G complexes. Purified proteins were visualized by Coomassie staining (Left) and anti-A3G blotting (Right). Control cell lysates containing unlinked NTAP and HA–A3G were identically processed (lane 1). (C) NTAP–APO1 (lane 1) and NTAP–A3G (lane 3) bind different sets of proteins based on silver staining. ◇, NTAP-tagged proteins.
To assess whether this large ensemble of proteins reflected specific versus nonspecific interaction with NTAP–A3G, parallel purifications were performed with NTAP-tagged APOBEC1 (APO1), a well known APOBEC family member (15). APO1 functions as the catalytic subunit of an editosome complex that mediates apoB mRNA editing. The pattern of proteins copurifying with NTAP–APO1 was quite distinct from that obtained with NTAP–A3G (Fig. 1C), arguing that the proteins copurifying with NTAP–A3G are not the result of promiscuous nonspecific binding.
Identifying HMM A3G Protein Cofactors by Tandem MS.
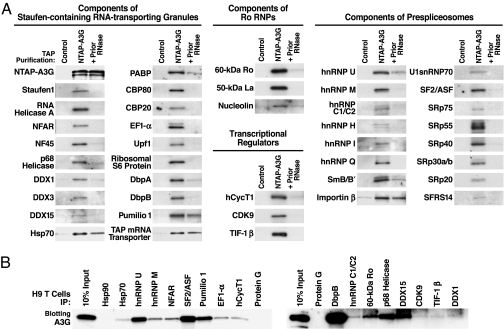
To identify candidate cofactors, the NTAP–A3G gel (Fig. 1B, lane 2) was sectioned into 40 slices. After in-gel trypsin digestion, peptides in each of the 40 slices were analyzed by tandem MS. This analysis identified ≈95 unique proteins (Table 1). Strikingly, these proteins comprise three distinct multisubunit RNPs, including Staufen-containing RNA-transporting granules (≈10 MDa in size) (31–33), Ro RNPs (34–36), and prespliceosomes (37–39). These findings raised the possibility that HMM A3G might correspond to more than a single type of RNP in human cells.
To confirm the participation of these proteins in HMM A3G complexes, we analyzed experimental and control purified complexes by immunoblotting with specific antibodies. Major components of Staufen-containing RNA-transporting granules, Ro RNPs, and prespliceosomes were readily detected by immunoblotting of the HMM A3G complex (Fig. 2A). When comparable amounts of NTAP–A3G complexes were treated with RNase A, most of these protein cofactors were no longer detected, although a subset of proteins remained bound. Thus, RNA in the HMM A3G complex likely plays an important role in cofactor recruitment. These identified cofactors also cofractionated with the endogenous HMM A3G complexes in H9 T cells and exogenously expressed HMM HA–A3G complexes formed in 293T cells (Fig. 7). The participation of the identified cofactors in HMM A3G complexes was confirmed by coimmunoprecipitation (co-IP) of endogenous A3G with these cofactors expressed in H9 T cells (Figs. 2B and 8) and co-IP of these cofactors with HMM HA–A3G expressed in 293T cells (Fig. 9). Prior RNase A treatment again disrupted the co-IP of most of these cofactors.
Fig. 2.
Verifying the tandem MS-identified protein cofactors. (A) Purified complexes (NTAP–A3G) or control purifications (unlinked NTAP plus HA–A3G) were immunoblotted with antibodies specific for the indicated proteins. To test RNase A sensitivity of cofactor binding, lysates were pretreated with 50 μg/ml RNase A (+Prior RNase) for 2 h at 4°C before purification. Shown are 42 representative components of Staufen-containing RNA-transporting granules, Ro RNPs, transcriptional regulators, and prespliceosomes. Some cofactors (e.g., Hsp70, Pumilio 1, TAP mRNA transporter, and importin-β) were detected after RNase treatment, suggesting RNA-independent interactions with A3G. Several multifunctional proteins, including nucleolin, DbpB, RNA helicase A, NFAR, and E1B–Ap5, participate in more than one RNP. (B) Endogenous A3G in H9 T cells assembles into the same RNPs. IP analyses were performed with antibodies reacting with select components of the various RNPs identified in HMM A3G complexes followed by rabbit (Left) or mouse (Right) anti-A3G blotting. IP with protein G-agarose or antibodies reacting with Hsp90, an abundant protein not copurifying with NTAP–A3G, were included as negative controls. Fig. 8 provides data on the IP efficiency of each antibody.
Specific Assembly of A3G with Select RNP Complexes.
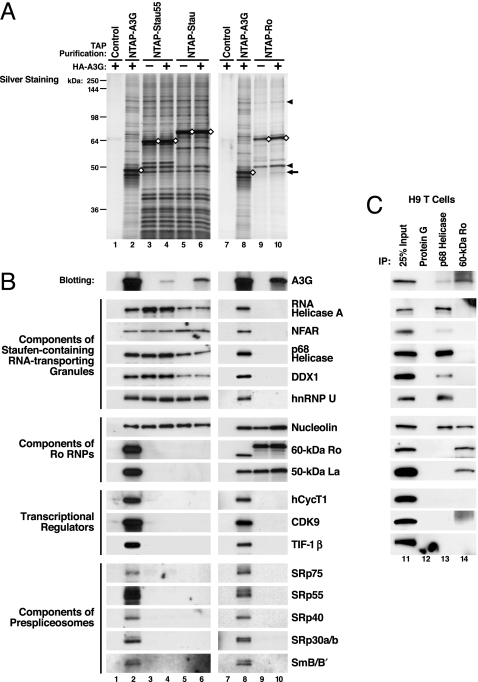
We next determined whether HMM A3G corresponds to at least three different types of cellular RNP complexes, including Staufen-containing RNA-transporting granules, Ro RNPs, and reservoirs of transcriptional regulators and components of prespliceosomes. Sequences encoding full-length Staufen1 (Stau), a 55-kDa splicing variant of Staufen1 (Stau55), and 60-kDa Ro were tagged with NTAP, expressed in 293T cells, and subjected to TAP with or without coexpressed HA–A3G. Purified NTAP–Stau (Stau55) complexes contained many but not all of the protein bands detected with NTAP–A3G (Fig. 3A Left). NTAP–Ro complexes contained a different and less complex set of proteins. Nucleolin and 50-kDa La were the two major proteins copurifying with NTAP–Ro (Fig. 3A, arrowheads). When HA–A3G was coexpressed, HA–A3G copurified with NTAP–Stau (Stau55) and NTAP–Ro (Fig. 3 A, arrow, and B, top panels). Previously identified components of RNA-transporting granules were copurified with NTAP–Stau (Stau55) and NTAP–A3G, but not with NTAP–Ro. Components of Ro RNPs were copurified with NTAP–A3G and NTAP–Ro but not NTAP–Stau (Stau55). Nucleolin, a protein participating in both RNA granules and Ro RNPs, was commonly detected with NTAP–A3G, NTAP–Stau (Stau55), and NTAP–Ro fusion proteins. Transcriptional regulators and components of prespliceosomes were copurified with NTAP–A3G but not with NTAP–Stau (Stau55) or NTAP–Ro (Fig. 3B). Endogenous A3G in human H9 T cells also assembled with the same type of RNP complexes as NTAP–A3G expressed in 293T cells (Fig. 3C). These findings highlight the specificity of the NTAPs and strongly support the conclusion that exogenous NTAP–A3G and endogenous A3G specifically enrich in at least three distinct HMM RNP complexes.
Fig. 3.
HMM A3G complexes correspond to at least three RNP complexes. (A) NTAP-tagged Staufen1 (Stau), a 55-kDa splicing variant of Staufen1 (Stau55), and 60-kDa Ro were subjected to TAP after expression in the absence or presence of HA–A3G coexpression. As controls, cell lysates containing unlinked NTAP and HA–A3G were identically processed (lanes 1 and 7). Arrow, HA–A3G copurified with NTAP–Ro; arrowheads, nucleolin and 50-kDa La. (B) Purified proteins were immunoblotted with antibodies reacting with the indicated proteins. Association of HA–A3G with NTAP–Stau (Stau55) and NTAP–Ro RNPs was detected by anti-HA (top sets of panels). (C) Endogenous A3G associates with the same RNP complexes. Lysates from A3G-expressing H9 T cells were subjected to IP with antibodies reacting with p68 helicase, a major component of RNA-transporting granules, and 60-kDa Ro, a major component of Ro RNPs, followed by immunoblotting. IP with protein G-agarose was included as a negative control.
Of note, almost all of the protein factors that participate in the formation of these three distinct cellular RNPs (described in Supporting Discussion in Supporting Text) were detected in the purified HMM A3G complexes but not in NTAP–APO1 complexes (Fig. 10). The lysis procedure used for TAP and FPLC analysis selectively solubilizes cytoplasmic proteins (Fig. 6). Thus, the identification of some factors in HMM A3G complexes that are predominantly nuclear likely reflects their nucleocytoplasmic shuttling properties rather than their recruitment after cellular lysis. We conclude that the identified factors reflect a specific association of A3G with distinct RNP complexes containing these factors.
Identification of Human Alu and Small hY Retroelement RNAs in HMM A3G Complexes.
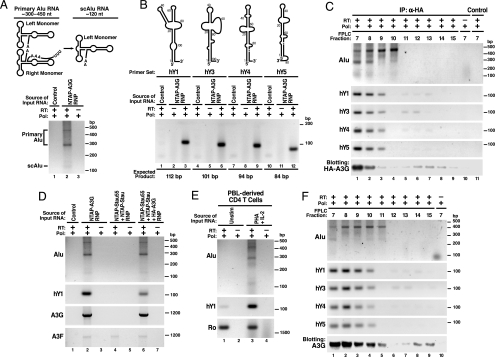
To identify the RNA species participating in the HMM A3G complexes, we treated purified NTAP–A3G complexes with DNase and extracted RNA. Purified RNAs were reverse-transcribed by using oligo(dT)20 or random hexamers as primers, and the resultant cDNAs were cloned and sequenced. Intriguingly, various Alu RNA sequences, including the Sx, Sp, Ya5, Ya8, and Yb8 subfamily members (40–43), were identified in two independent experiments (data not shown). DNA sequencing revealed both primary Alu (300–450 nt) and processed small cytoplasmic Alu (scAlu) RNAs in the HMM A3G complexes after Alu-specific RT-PCR (Fig. 4A). Consistent with the identification of SRP14 in purified HMM A3G complexes (Table 1), this protein binds to both primary Alu and scAlu RNAs (44).
Fig. 4.
Detection of nonautonomous mobile genetic elements in the HMM A3G complexes. (A Upper) Schematic of primary Alu and processed scAlu RNAs. (A Lower) RT-PCR detection of primary Alu and scAlu RNAs in HMM A3G complexes. Control purifications: unlinked NTAP+HA–A3G. (B Upper) Schematic of hY RNAs. (B Lower) RT-PCR detection of hY RNAs in HMM A3G complexes. (C) Co-IP of Alu and hY RNAs with HA–A3G from HMM fractions of 293T lysates resolved by FPLC. Protein G-agarose in the absence of anti-HA served as a control. (D) A3G-dependent recruitment of Alu and hY RNAs into the Staufen-containing RNA granules. A3G and endogenous A3F mRNAs are also present in purified NTAP–A3G and NTAP–Stau (Stau55) complexes. Of note, 293T cells expressed extremely low levels of A3F. Alu RNA was not present in the RNA granules unless A3G was coexpressed. (E) PHA–IL-2-induced expression of Alu and hY RNAs in primary CD4 T cells. Cells were either untreated or treated with PHA (5 μg/ml) for 36 h followed by IL-2 (20 units/ml; Roche) for 36 h before analysis. (F) Alu and hY RNAs cofractionate with HMM A3G complexes in PHA–IL-2 activated primary CD4 T cells. Reactions performed with Pfx polymerase (Pol) but not reverse transcriptase (−RT) served as negative controls in each panel. RNA structures in A and B were adapted in part from refs. 45 and 46.
Recently, hY RNA family members (hY1, hY3, hY4, and hY5) were identified as a novel type of L1-dependent, nonautonomous retroelement (45). All four hY RNAs were detected in the HMM A3G complexes by RT-PCR (Fig. 4B). The detection of both Ro and La proteins in the HMM A3G complexes (Table 1 and Figs. 2 and 3) also supports the presence of hY RNAs in the HMM A3G complexes because Ro forms RNP complexes with La and hY RNAs (34–36). The participation of Alu and hY RNAs in the HMM A3G complexes was further confirmed by co-IP of these retroelements when HA–A3G was expressed in 293T cells and FPLC-derived HMM fractions were subjected to anti-HA immunoprecipitation (IP) (Fig. 4C).
Our studies also identified mRNA sequences associated with the HMM A3G complexes including A3G mRNA (Fig. 4D, lane 2). These complexes also contained both endogenous A3F protein (Table 1) and A3F mRNA (Fig. 4D, lane 2), arguing against artifactual detection of A3G mRNA related to forced NTAP–A3G expression. These findings suggest that A3G and A3F commonly and perhaps coordinately participate in HMM complex assembly.
Based on semiquantitative RT-PCR, Alu and hY RNAs and A3G and A3F mRNAs were estimated to be enriched 20- to 100-fold in the HMM A3G complexes (Fig. 11 A and B). Conversely, mRNAs encoding α-tubulin, β-actin, Hsp70, 60-kDa Ro, and Staufen were not detectable in the HMM A3G complexes. Trace amounts of tRNA–Lys were present (Fig. 11C).
Intriguingly, detection of Alu RNAs and a small subset of hY RNAs in Staufen-containing RNA-transporting granules strictly depended on A3G coexpression (Fig. 4D, compare lane 4 with lane 6). Furthermore, mitogen activation of CD4 T cells induced Alu and hY RNA expression (Fig. 4E) but did not alter the expression of most of the protein cofactors in HMM A3G complexes (Fig. 12). The cofractionation of these nonautonomous retroelement RNAs in mitogen-induced HMM A3G complexes formed in primary CD4 T cells (Fig. 4F) suggests that the Alu and hY RNAs may help nucleate HMM A3G complex assembly.
A3G Restricts L1-Dependent Alu Retrotransposition.
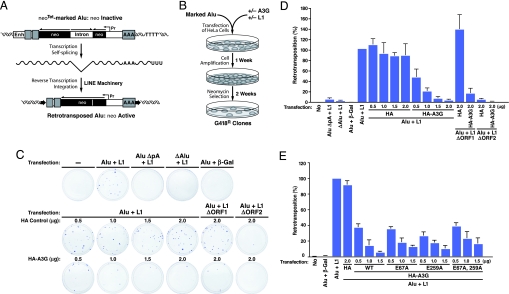
Delineation of the RNA components present in HMM A3G complexes suggests a potential physiological function for these complexes. Specifically, endogenous nonautonomous retroelements might serve as natural cellular targets of A3G. In general, autonomous L1 elements encode their own reverse transcriptase and endonuclease to catalyze their retrotransposition. Nonautonomous elements encode no gene products, depending instead on the L1 machinery for retrotransposition (20, 41, 46). To test whether A3G controls the retrotransposition of Alu RNAs, cell-based L1-dependent Alu retrotransposition assays (46) (Fig. 5A) were performed in the absence or presence of graded amounts of HA vector or HA–A3G DNA (Fig. 5). The number of G418-resistant colonies generated provides a direct measure of successful retrotransposition events (Fig. 5B). To ensure measurement of L1-dependent Alu retrotransposition, additional negative controls were incorporated, including an Alu element lacking its polyA tail or right monomer sequences, vectors encoding truncated, nonfunctional L1 mutants, or the irrelevant β-gal gene. As described (46), the polyA tail and dimeric sequences of Alu and the ORF2, but not ORF1, of L1 are required for Alu retrotransposition (Fig. 5C Top). A3G both assembled into HMM complexes in HeLa cells (Fig. 13A Lower) and greatly restricted L1-mediated retrotransposition of Alu RNAs in a dose-dependent manner (Fig. 5 C and D). Importantly, this inhibitory effect was Alu-sequence-specific because A3G had no effect on retrotransposition of L1 itself (measured by neo-marked L1; data not shown) (21, 24, 27–30). Catalytically inactive (E259A and E67A/E259A) mutants of A3G, which assemble into HMM complexes, effectively inhibited Alu retrotransposition, suggesting a deaminase-independent mechanism of A3G inhibition (Fig. 5E). By using primers bracketing the intron of the neo indicator that were unable to amplify the antisense SV-neo transcript, the recruitment of marked Alu neoTet-derived transcripts into HMM A3G complexes in HeLa cells was detected (Fig. 13A) as was A3G-dependent recruitment of Alu neoTet transcripts into Staufen-containing RNA-transporting granules (Fig. 13 B and C). These studies reveal that A3G inhibits the retrotransposition of nonautonomous Alu retroelements in a nonenzymatic and L1-independent manner involving recruitment of these RNAs into cytoplasmic HMM complexes.
Fig. 5.
A3G restricts L1-dependent Alu retrotransposition. (A) Summary of marked Alu retrotransposition assay. An Alu element (gray boxes) was marked with the neoTet gene (dark box) driven by SV40 promoter (Pr) and placed in reverse orientation (Top). The neoTet gene is rendered inactive by an autocatalytic tetrahymena (Tet) intron inserted in the forward direction. This intron is removed by autosplicing when RNA is produced (Middle), allowing detection of retrotransposition after L1-dependent reverse transcription and integration. The expected structure of the resulting de novo Alu insertion is shown (Bottom). (B) Experimental procedure for detecting the effects of A3G on L1-dependent Alu retrotransposition. A and B were adapted from ref. 46. (C) A3G inhibits Alu retrotransposition. HeLa cells were transfected with Alu neoTet, L1 expression plasmids, and graded amounts of HA–A3G or HA vector DNA. Retrotransposition events were detected by staining G418-resistant (G418R) foci. Additional negative controls included an Alu element deficient in polyA tail (AluΔpA neoTet) or in right monomer sequences (ΔAlu neoTet), vectors encoding truncated, nonfunctional L1 mutants (ΔORF2), and the irrelevant β-gal gene. (D) Number of G418R clones. Data were normalized to the positive control (Alu neoTet + L1). Values represent the means ± SD from three independent experiments. (E) Inhibition of Alu retrotransposition by mutants of A3G (E259A and E67A/E259A) lacking deoxycytidine deaminase activity. Wild-type and E67A A3G retaining enzymatic activity were included for comparison. Values represent the means ± SD from three independent experiments.
Discussion
Our studies demonstrate that Alu, the most prominent nonautonomous retroelement, and likely hY RNAs, serve as natural targets of A3G's restricting activity. The requirement of the L1 machinery for Alu and hY retrotransposition and the identification of these endogenous nonautonomous retroelement RNAs in HMM A3G complexes in the absence of detectable L1 proteins suggest a nonenzymatic mechanism for A3G inhibition of retrotransposition. Specifically through its RNA binding properties (13, 15, 47), A3G sequesters Alu and Y RNAs in cytoplasmic HMM complexes away from the nuclear L1 machinery, thereby interdicting the retrotransposition cycle. This mechanism is quite distinct from the actions of A3A and A3B, which also inhibit Alu retrotransposition. These proteins enter the nucleus and directly interfere with L1 activity (24, 27). The A3G-dependent recruitment of both endogenous Alu RNAs and engineered Alu transcripts to the Staufen-containing RNA-transporting granules further supports this inhibitory mechanism. The lack of inhibitory effect of A3G on autonomous L1 may reflect efficient assembly of retrotransposing L1 RNA with its catalytic machinery encoded by the same RNA (cis preference) (48), which minimizes the chance for A3G to act.
Different classes of RNA granules have been described (49). Germ cell granules and neuronal granules (RNA-transporting granules) harbor highly specific mRNA cargoes, whereas stress granules and processing bodies (PB) are less discriminating. In addition to differing in their mRNA selectivity, RNA-transporting granules contain both the 60S and 40S ribosomal subunits, whereas stress granules contain only small ribosomal subunits, and PBs lack both (49). Although Staufen proteins occur in more than one type of granule, the RNA granules that associate with HMM A3G contain both the 60S and 40S ribosomal subunits and thus are reminiscent of RNA-transporting granules rather than PB proposed to interact with A3G (50). Furthermore, characteristic PB components including endogenous Ago2 and Rck/p54 (DDX6) were not detected in the purified NTAP–A3G complexes (Fig. 14A), and the majority of the Ago2 and Rck/p54-containing complexes are in fractions of lower molecular mass than HMM A3G complexes (Fig. 14B). Nevertheless, whether the Alu RNAs are ultimately degraded by the Staufen-mediated degradation (51) or by transfer to PB (50) merits future study.
RNA-transporting granules control the localization, stability, and translation of resident RNA cargoes (32, 33, 49, 52). The presence of A3G mRNA and translation initiation and elongation factors in the HMM A3G complexes (Table 1 and Fig. 4D) provides an attractive mechanism for how destruction of the HMM complex by Vif (13) might impair de novo synthesis of A3G (6). Interestingly, although Staufen and Ro proteins are in HMM A3G complexes, their mRNAs are not (Fig. 11), consistent with the notion that RNA-transporting granules harbor highly specific mRNA cargoes (49). Future studies are needed to more broadly survey the range of RNA species present in the HMM A3G-containing RNA granules by using microarrays.
In summary, our studies reveal dual physiological functions of A3G in human cells (Fig. 15). In resting CD4 T cells, LMM A3G functions as a potent postentry restricting factor blocking the replication of exogenous HIV-1 retroviruses (13). This restriction may well extend to other exogenous retroviruses. Activation of CD4 T cells induces expression of nonautonomous retroelement RNAs. A3G defends cells from this “threat within” by sequestering these nonautonomous retroelements away from the L1 machinery required for retrotransposition. This action interrupts the retrotransposition cycle of these endogenous retroelements. Unfortunately, the assembly of HMM A3G complexes containing these retroelement RNAs removes the postentry restriction block provided by LMM A3G, rendering the activated CD4 T cells highly permissive for HIV infection. Of note, expression of Alu and hY RNAs is increased in the absence of corresponding changes in most of the various protein cofactors during the activation of CD4 T cells. Thus, these retroelement RNAs may function as a driving force nucleating the assembly of HMM A3G complexes. The use of siRNA to knock down expression of these retroelement RNAs or small molecules that partially impair A3G binding to retroelement RNAs could provide a strategy to preserve both the postentry restricting activity provided by LMM A3G and the protective activity of HMM A3G complexes.
Methods
For TAP, 293T cells plated in T175 flasks were transfected with the indicated plasmids (8 μg for NTAP–A3G; 20 μg for NTAP–Stau and NTAP–Ro) with calcium phosphate and cultured for 48 h. Cells were lysed in buffer containing 50 mM Hepes (pH 7.4), 125–400 mM NaCl, 0.2% Nonidet P-40, 1 mM PMSF, and EDTA-free protease inhibitor mixture (Calbiochem). NTAP-tagged proteins and their associated cofactors in cytoplasmic extracts were isolated by using an InterPlay TAP mammalian purification kit (Stratagene).
Additional methods are presented in Supporting Methods in Supporting Text, including detailed procedures for TAP, tandem MS, IP, RNA extraction and identification, RT-PCR detection of Alu and hY RNAs, L1-mediated marked Alu retrotransposition assay, and sources of antibodies.
Supplementary Material
Acknowledgments
Antibodies reacting with Staufen1, RNA helicase A, Upf1, and 50-kDa La were gifts from Drs. J. Ortin (Spanish National Research Council, Madrid, Spain), F. Grosse (Institute of Molecular Biotechnology, Jena, Germany), J. Steitz (Yale University School of Medicine, New Haven, CT), and G. J. M. Pruijn (University of Nijmegen, Nijmegen, The Netherlands), respectively. We thank Drs. J. Bohuslav, J. Kreisberg, W. Yonemoto, S. Wissing, and K. Stopak for discussions; S. Dixon (University of California, San Francisco) for technical assistance. This work was supported by National Institutes of Health Grants R01 AI065329-01 (to W.C.G.) and RR18928-01; the San Francisco Women's HIV Interdisciplinary Network (National Institutes of Health Grant P01 HD40543 to W.C.G.); American Foundation for AIDS Research Grant 106525-35-RFHF (to Y.-L.C.); University of California San Francisco–Gladstone Institute of Virology and Immunology Center for AIDS Research Grant AI0277635P30; Ligue Nationale Contre le Cancer (C.E. and T.H.); and the Sandler Family Foundation for the University of California San Francisco Biomolecular Resources Center Mass Spectrometry Facility.
Abbreviations
- HMM
high-molecular-mass
- LMM
low-molecular-mass
- RNP
ribonucleoprotein
- A3G
APOBEC3G
- L1
long interspersed nucleotide elements 1
- TAP
tandem affinity purification
- IP
immunoprecipitation
- co-IP
coimmunoprecipitation
- scAlu
small cytoplasmic Alu
- PB
processing body.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 3.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 4.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stopak K, de Noronha C, Yonemoto W, Greene WC. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 7.Conticello SG, Harris RS, Neuberger MS. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 8.Marin M, Rose KM, Kozak SL, Kabat D. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 9.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 10.Sheehy AM, Gaddis NC, Malim MH. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 12.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 13.Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 14.Kreisberg JF, Yonamoto W, Greene WC. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 16.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 17.Sawyer SL, Emerman M, Malik HS. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Webb DM. Hum Mol Genet. 2004;13:1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- 19.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazazian HHJ. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 21.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- 22.Esnault C, Millet J, Schwartz O, Heidmann T. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Curr Biol. 2005;15:661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher AJ, Nissley DV, Harris RS. Proc Natl Acad Sci USA. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Proc Natl Acad Sci USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenglein MD, Harris RS. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 29.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 30.Turelli P, Vianin S, Trono D. J Biol Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 31.Brendel C, Rehbein M, Kreienkamp HJ, Buck F, Richter D, Kindler S. Biochem J. 2004;384:239–246. doi: 10.1042/BJ20040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanai Y, Dohmae N, Hirokawa N. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 33.Villace P, Marion RM, Ortin J. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouraux MA, Bouvet P, Verkaart S, van Venrooij WJ, Pruijn GJ. J Mol Biol. 2002;320:475–488. doi: 10.1016/s0022-2836(02)00518-1. [DOI] [PubMed] [Google Scholar]
- 35.Fabini G, Rutjes SA, Zimmermann C, Pruijn GJ, Steiner G. Eur J Biochem. 2000;267:2778–2789. doi: 10.1046/j.1432-1327.2000.01298.x. [DOI] [PubMed] [Google Scholar]
- 36.Fabini G, Raijmakers R, Hayer S, Fouraux MA, Pruijn GJ, Steiner G. J Biol Chem. 2001;276:20711–20718. doi: 10.1074/jbc.M101360200. [DOI] [PubMed] [Google Scholar]
- 37.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Proc Natl Acad Sci USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss T. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 39.Yong J, Wan L, Dreyfuss G. Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Rowold DJ, Herrera RJ. Genetica. 2000;108:57–72. doi: 10.1023/a:1004099605261. [DOI] [PubMed] [Google Scholar]
- 41.Batzer MA, Deininger PL. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 42.Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. J Mol Biol. 1997;271:222–234. doi: 10.1006/jmbi.1997.1161. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Song L, Gonder MK, Azrak S, Ray DA, Batzer MA, Tishkoff SA, Liang P. Gene. 2006;365:11–20. doi: 10.1016/j.gene.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang DY, Hsu K, Maraia RJ. Nucleic Acids Res. 1996;24:4165–4170. doi: 10.1093/nar/24.21.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perreault J, Noel JF, Briere F, Cousineau B, Lucier JF, Perreault JP, Boire G. Nucleic Acids Res. 2005;33:2032–2041. doi: 10.1093/nar/gki504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewannieux M, Esnault C, Heidmann T. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 47.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 48.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson P, Kedersha N. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wichroski MJ, Robb GB, Rana TM. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim YK, Furic L, Desgroseillers L, Maquat LE. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 52.Miki T, Takano K, Yoneda Y. Cell Struct Funct. 2005;30:51–56. doi: 10.1247/csf.30.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.