Abstract
IFN-γ is known as the initial and primary inducer of immunoproteasomes during viral infections. We now report that type I IFN induced the transcription and translation of immunoproteasome subunits, their incorporation into the proteasome complex, and the generation of an immunoproteasome-dependent CD8 T cell epitope in vitro and provide in vivo evidence that this mechanism occurs prior to IFN-γ responses at the site of viral infection. Type I IFN–mediated generation of immunoproteasomes was initiated by either poly(I:C) or HCV RNA in human hepatoma cells and was inhibited by neutralization of type I IFN. In serial liver biopsies of chimpanzees with acute HCV infection, increases in immunoproteasome subunit mRNA preceded intrahepatic IFN-γ responses by several weeks, instead coinciding with intrahepatic type I IFN responses. Thus, viral RNA–induced innate immune responses regulate the antigen-processing machinery, which occurs prior to the detection of IFN-γ at the site of infection. This mechanism may contribute to the high effectiveness (95%) of type I IFN–based therapies if administered early during HCV infection.
Introduction
In the effector phase of a virus-specific adaptive immune response, primed CD8 T cells migrate to the infected organ, where they recognize and lyse infected cells that display viral peptides in the context of MHC class I molecules. Most of these viral peptides are generated from proteins and larger polypeptides by the 26S proteasome, a major cytosolic antigen-processing complex (1).
IFN-γ, a major cytokine in many viral infections, is known to alter the composition and function of the proteasome complex (2, 3). In the absence of IFN-γ, the 26S proteasome complex contains a 20S catalytic core, arranged as 2 heptameric outer rings with 7 α subunits (α1–α7) each and 2 heptameric inner rings with 7 β subunits (β1–β7) each (reviewed in refs. 2, 3). IFN-γ induces the transcription and translation of the 3 immunoproteasome subunits β1i (LMP2), β2i (MECL-1), and β5i (LMP7), which replace their constitutive counterparts, β1, β2, and β5, respectively, during de novo assembly of proteasomes (4). The presence of TNF-α exerts a synergistic effect (5, 6). The resulting immunoproteasomes differ from constitutive proteasomes in qualitative and quantitative aspects of their proteolytic activity (2, 3, 7). Mice that lack the immunoproteasome subunits β1i (7, 8) or β5i (9) have been shown to be unable to efficiently process and present certain CD8 T cell epitopes. One of the best examples for a strictly immunoproteasome-dependent CD8 T cell epitope in humans is the HBV core peptide 141–151 (HBcore141–151), which requires the presence of β5i (10).
Replacement of constitutive proteasomes by immunoproteasomes in the virus-infected organ has been confirmed in a murine model of lymphocytic choriomeningitis virus (LCMV) infection. Within 7 days of LCMV infection, constitutive proteasomes were almost completely replaced by immunoproteasomes at the site of infection, the liver (11). This replacement appeared to depend on IFN-γ, as it was markedly reduced in IFN-γ–/– mice (11). An important role of IFN-γ in the induction of immunoproteasomes was also demonstrated in a murine model of fungal infection (12). In contrast, type I IFN did not appear to play any role. Immunoproteasome induction in LCMV infection was not affected in mice that lacked the IFN-α/β receptor (IFN-α/βR) (11). Likewise, in vitro studies on melanoma cells described that only IFN-γ and not IFN-α induced immunoproteasome subunits and generated functional immunoproteasomes (13).
Since many viruses induce a vigorous type I IFN response much earlier than an IFN-γ response, we here investigated the role of type I IFN in the induction of immunoproteasomes. After demonstrating that type I IFN did indeed trigger induction, assembly, and proteolytic activity of immunoproteasomes in human hepatoma cells and primary hepatocytes in vitro and that this mechanism could be initiated by intracellular double-stranded RNA (dsRNA), poly(I:C), and HCV RNA, we then analyzed the differential contribution of type I IFN and IFN-γ to the induction of immunoproteasomes in HCV-infected chimpanzees. HCV, a single-stranded RNA virus has infected more than 4 million people in the United States and is the main reason for liver transplantation in Western countries (14). Because a vigorous innate immune response occurs within 1–3 weeks and an adaptive immune response becomes detectable much later, i.e., 6–12 weeks after infection (15, 16), we considered HCV infection a suitable model to differentiate between induction of immunoproteasomes by innate and adaptive immune responses.
We now report that the induction of immunoproteasome subunits in this important infection occurred much earlier than the intrahepatic IFN-γ response, thereby challenging the current dogma that IFN-γ is the initial and primary inducer of immunoproteasomes during viral infections. Instead, immunoproteasomes were induced in hepatocytes much earlier, in response to virus-induced type I IFN. Thus, rather than being passive targets, virus-infected hepatocytes actively prepare their antigen-processing machinery for optimal presentation even prior to the arrival of liver-infiltrating T cells.
Results
Type I IFN induces the expression of immunoproteasome subunits.
To investigate a role of type I IFN in the induction of immunoproteasomes, we studied human hepatoma cells and primary hepatocytes, which are targets of many viral infections in humans. Because the group of type I IFNs is composed of 13 different IFN-α subtypes and of IFN-β, we first studied the effect of consensus sequence IFN-α (IFN-α–con1) (17).
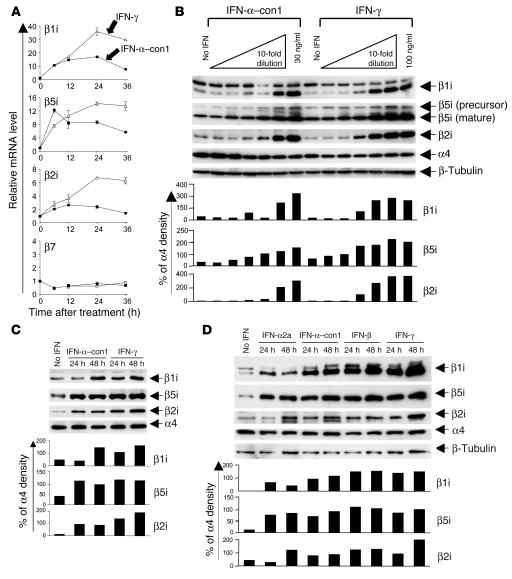
As shown in Figure 1, IFN-α–con1 induced the expression of all 3 immunoproteasome subunits both at the mRNA level (Figure 1A) and at the protein level (Figure 1B) in Huh-7 hepatoma cells. In contrast, the mRNA level of the proteasome β7 subunit (Figure 1A) and the protein level of the proteasome α4 subunit (Figure 1B), which were analyzed as controls and are not inducible by cytokines, remained stable. Type I IFN also induced the expression of immunosubunits in primary human hepatocytes (Figure 1C). In total, hepatocytes from 6 different donors were tested, and in each case, type I IFN exerted the same effect on hepatocytes as IFN-γ (data not shown). Finally, immunosubunits were not only induced by IFN-α–con1, but also by individual, natural members of the type I IFN family such as IFN-α2a and IFN-β, in Huh-7 cells (Figure 1D).
Figure 1. Type I IFN induces the expression of immunoproteasome subunits in vitro.
(A) Huh-7 cells were treated with 3 ng/ml IFN-α–con1 (squares) or 10 ng/ml (200 U/ml) IFN-γ (triangles) for the indicated time periods, after which the mRNA levels of immunoproteasome subunits were quantified by real-time PCR. mRNA levels were normalized to endogenous references (GAPDH and β-actin) and expressed as fold increase over pretreatment levels. The β7 subunit was measured as a control. (B) Huh-7 cells were treated with the indicated doses of IFN-α–con1 or IFN-γ for 48 hours, and Western blot analysis was performed to detect immunoproteasome subunits. The α4 subunit was analyzed as a control. (C) Primary human hepatocytes were treated with 3 ng/ml IFN-α–con1 or 10 ng/ml IFN-γ for the indicated time periods prior to Western blot analysis. (D) Huh-7 cells were treated with 1.8 ng/ml (500 U/ml) IFN-α2a, 3 ng/ml IFN-α–con1, 3 ng/ml IFN-β, or 10 ng/ml IFN-γ for the indicated time periods, and Western blot analysis was performed to detect immunoproteasome subunits. The density of the Western blot bands was quantified and expressed as percent of the density of the α4 control band.
Type I IFN–induced immunoproteasomes exhibit the typical structure and function of IFN-γ–induced immunoproteasomes.
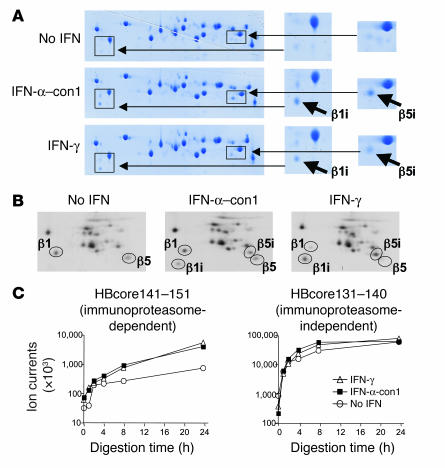
As increased expression of immunoproteasome subunits does not necessarily indicate correct incorporation into the proteasome complex and generation of functional immunoproteasomes, we isolated 20S proteasomes from IFN-α–treated Huh-7 cells and studied their subunit composition by 2-dimensional gel electropheresis. As shown in Figure 2A, 2-dimensional gel electrophoresis revealed the presence of both β1i and β5i in 20S proteasomes from IFN-α–and IFN-γ–treated Huh-7 cells, but these were only marginally observed in proteasomes from untreated Huh-7 cells. These results were confirmed by immunoprecipitation and 2-dimensional gel electrophoresis of metabolically labeled 20S proteasomes (Figure 2B).
Figure 2. Type I IFN–induced immunoproteasomes exhibit the typical structure and function of IFN-γ–induced immunoproteasomes.
(A) Huh-7 cells were treated with 3 ng/ml IFN-α–con1 or 10 ng/ml IFN-γ for 36 hours, and the 20S proteasome complex was biochemically isolated from cell lysates. Isolated 20S proteasomes were analyzed by 2-dimensional gel electropheresis and Coomassie blue staining. The locations of β1i and β5i are indicated in the insets to the right (magnification, ×2). (B) Huh-7 cells were treated with 3 ng/ml IFN-α–con1 or 10 ng/ml IFN-γ for 36 hours and labeled with 35S-methionine. The 20S proteasome complex was immunoprecipitated and analyzed by 2-dimensional gel electropheresis and autoradiography. The locations of β1, β5, β1i, and β5i are indicated. (C) Biochemically isolated 20S proteasome complexes as shown in A were subsequently incubated with the precursor substrate HBcore131–162 for the indicated time periods. In vitro digests were analyzed by HPLC and mass spectrometry at the indicated time points for the presence of the β5i-dependent HBcore141–151 and the β5i-independent HBcore131–140. Type I IFN–induced immunoproteasomes displayed β5i-dependent proteolytic activity.
Next, we studied the function of IFN-α–induced 20S proteasomes and compared it with that of IFN-γ–induced 20S proteasomes. Because specific CD8 T cell epitopes that are generated only by immunoproteasomes and not by constitutive proteasomes are not yet known for HCV, we studied the generation of the well-described HBcore141–151 epitope from HBV, which is strictly immunoproteasome-dependent (i.e., β5i-dependent), as demonstrated in previous studies using mass spectrometry and recognition by peptide-specific cytotoxic T cells as readout (10). The substrate polypeptide HBcore131–162 was in vitro digested with 20S proteasomes isolated from either IFN-γ–treated, IFN-α–treated, or untreated Huh-7 cells, and the digest products were then analyzed for the presence of the immunoproteasome-dependent peptide HBcore141–151 by mass spectrometry (Figure 2C). As a control, the presence of the immunoproteasome-independent HBcore131–140 was also analyzed.
Importantly, IFN-α–induced proteasome complexes generated HBcore141–151 with the same kinetics as IFN-γ–induced immunoproteasomes, while less HBcore141–151 was detectable when digestion was performed with constitutive proteasomes (Figure 2C). The immunoproteasome-independent control HBcore131–140 was generated with the same kinetics by IFN-α– and IFN-γ–induced immunoproteasomes and by constitutive proteasomes (Figure 2C).
Collectively, these data demonstrate that type I IFN not only induced the expression of immunoproteasome subunits at the mRNA level, but also their translation, correct processing, and incorporation into the proteasome complex, resulting in the formation of fully functional immunoproteasomes. Thus, type I IFN–induced immunoproteasomes were indistinguishable from IFN-γ–induced immunoproteasomes not only in their subunit composition, but also in their function.
Intracellular dsRNA induces the expression of immunoproteasome subunits in a type I IFN–dependent manner.
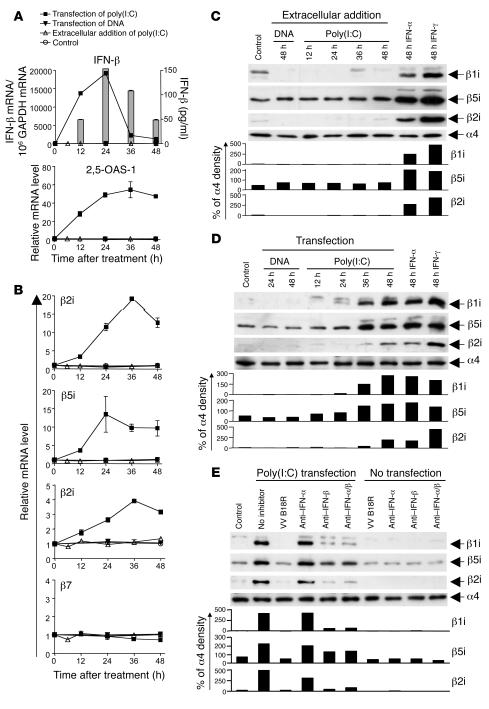
In the above-described in vitro studies, hepatocytes and hepatoma cells were treated with exogenous type I IFN to induce immunoproteasomes. Next, we asked whether endogenous, viral RNA–induced IFN generates immunoproteasomes in hepatoma cells. Sensing of intracellular dsRNA by retinoic acid–inducible gene I (RIG-I) results in the phosphorylation and activation of IRF-3 and subsequent production of IFN-β. Secreted IFN-β binds to the IFN-α/βR in an autocrine and paracrine manner, resulting in activation of the JAK/STAT signaling pathway, transcription of IFN-stimulated genes, and, via production of the IFN-stimulated gene IRF-7, production of various IFN-α subtypes (reviewed in ref. 18). Because Huh-7 cells respond well to intracellular dsRNA via activation of RIG-I, but not to extracellular dsRNA due to the lack of TLR3 (19), Huh-7 cells were transfected with the dsRNA poly(I:C) to simulate the presence of viral dsRNA. Extracellular addition of poly(I:C) or transfection with DNA served as negative controls in this experiment.
As shown in Figure 3, transfection of Huh-7 cells with poly(I:C) induced IFN-β and the downstream response of 2,5-oligoadenylate synthetase (2,5-OAS-1) (Figure 3A) and increased the mRNA levels of all 3 immunoproteasome subunits (Figure 3B) as well as their protein levels (Figure 3D). In contrast, neither extracellular addition of poly(I:C) (Figure 3, B and C) nor transfection of Huh-7 cells with DNA (Figure 3, B and D) had any effect. Collectively, these data demonstrate that immunoproteasome subunits are not only induced by exogenous type I IFN (Figure 1), but also by endogenously produced type I IFN, in response to intracellular dsRNA. To analyze which of the type I IFNs was dominant in this endogenous induction pathway, we added neutralizing antibodies to IFN-α and/or IFN-β to the culture. In separate experiments, we also added the vaccinia virus–encoded B18 receptor protein (VV B18R), which competes with the IFN-α/βR for IFN binding (20). As shown in Figure 3E, anti–IFN-β Ab abrogated the induction of immunoproteasome subunits by intracellular dsRNA. Likewise, the addition of VV B18R abrogated the induction of immunoproteasome subunits. In contrast, neutralization of IFN-α, which is a later step in the endogenous IFN response cascade (18), did not block the induction of immunoproteasome subunits. These results prove that induction of immunoproteasome subunits in response to intracellular dsRNA is indeed mediated by endogenously produced type I IFN. In addition, they demonstrate that early steps in the endogenous type I IFN response cascade, i.e., either neutralization of IFN-β or prevention of binding to the IFN-α/βR, need to be blocked to abrogate the induction of immunoproteasome subunits.
Figure 3. Intracellular dsRNA induces the expression of immunoproteasome subunits by the secretion of type I IFN.
Huh-7 cells were transfected with 10 μg poly(I:C) (squares) or 10 μg pcDNA3.1 plasmid DNA (inverted triangles), treated with 10 μg/ml extracellular poly(I:C) (triangles), or were mock transfected (circles). (A) Quantitation of IFN-β and 2,5-OAS-1 mRNA by real-time PCR. Bars (indicating secreted IFN-β) are shown only for the poly(I:C)-transfected Huh-7 cells because no secreted IFN-β was detectable in the control cultures of DNA-transfected Huh-7 cells or Huh-7 cells treated extracellularly with poly(I:C). (B) Quantitation of mRNA levels of immunoproteasome subunits β1i, β5i, β2i and constitutive subunit β7 by real-time PCR. (C–E) Detection of immunoproteasome subunits β1i, β5i, and β2i by Western blot analysis. (C and D) Huh-7 cells were treated with IFN-α and IFN-γ as well as (C) treated with extracellular DNA and poly(I:C) or (D) transfected with DNA and poly(I:C). (E) Four hours after poly(I:C) transfection of Huh-7 cells, 5,000 U/ml neutralizing anti–IFN-α and 2,000 U/ml neutralizing anti–IFN-β, alone or in combination, as well as 1 μg/ml VV B18R protein were added into culture and incubated for 48 hours, and Western blot was performed to detect immunoproteasome subunits. Anti–IFN-α/β, combination of IFN-α– and IFN-β–neutralizing antibodies. The density of the Western blot bands was quantified and expressed as percent of the density of the α4 control band.
HCV RNA induces the expression of immunoproteasome subunits.
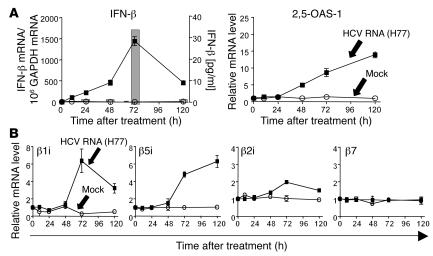
To test whether HCV RNA can induce type I IFN and immunoproteasomes, we transfected Huh-7 cells with HCV RNA. As shown in Figure 4, transfection of Huh-7 cells with HCV RNA induced IFN-β production and the downstream response of 2,5-OAS-1 (Figure 4A) and increased mRNA levels of all 3 immunoproteasome subunits (Figure 4B). These data demonstrate that immunoproteasome subunits can be induced not only by artificial dsRNA [poly(I:C)], but also by HCV RNA itself.
Figure 4. HCV RNA induces the expression of immunoproteasome subunits.
Huh-7 cells were transfected with 2 μg in vitro transcribed H77 HCV RNA (squares) or were mock transfected (circles). (A) mRNA levels of IFN-β and 2,5-OAS-1 were determined by real-time PCR. Secreted IFN-β (bars in left panel) was detectable by ELISA only in the supernatants of HCV RNA-transfected Huh-7 cells, not in supernatants of mock-transfected Huh-7 cells (not shown). (B) mRNA levels of immunoproteasome subunits β1i, β5i, and β2i and the constitutive subunit β7 as determined by real-time PCR.
mRNA levels of immunoproteasome subunits increase early during acute HCV infection and are temporally related to the type I IFN response.
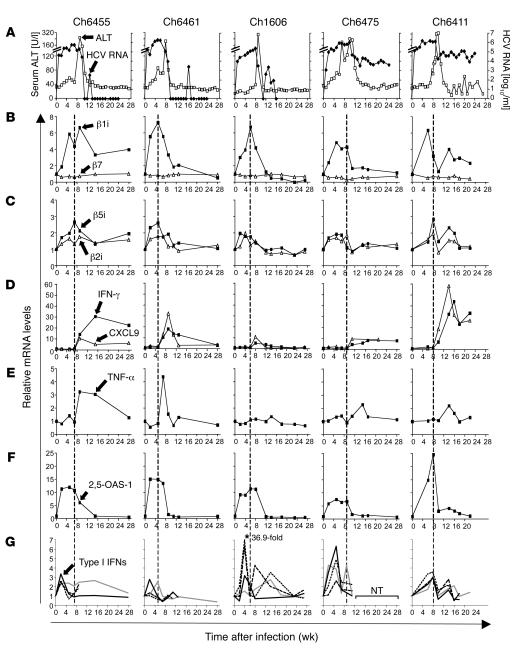
To investigate the differential contribution of type I IFN and IFN-γ to the induction of immunoproteasomes in HCV infection, we prospectively studied 5 chimpanzees with acute HCV infection (Figure 5). Serum alanine aminotransferase (ALT), a marker of liver injury, peaked in all chimpanzees 8–10 weeks after infection. This peak was followed by HCV clearance in chimpanzees with a self-limited course of infection (Ch6455, Ch6461, and Ch1606) or by a partial, 2–3 log10 (i.e., >99%) decrease of serum HCV RNA titers in chimpanzees with a chronically evolving course of infection (Ch6475 and Ch6411) (Figure 5). To study the induction of immunoproteasome subunits at the site of infection, we quantified mRNA levels of the 3 immunoproteasome subunits, β1i, β5i, and β2i, in serial liver biopsies by real-time PCR. As shown in Figure 5, B and C, mRNA levels of all 3 immunoproteasome subunits started to increase as early as 2 weeks after infection and peaked 5–8 weeks after infection. The mRNA level of β7, a nonreplaceable subunit of the proteasome, was studied as a control and remained constant in all chimpanzees at all study time points (Figure 5B).
Figure 5. Immunoproteasome subunit mRNA levels increase early in acute HCV infection and in temporal relation to type I IFN responses.
Five chimpanzees (Ch6455, Ch6461, Ch1606, Ch6475, and Ch6411) were studied prospectively during acute HCV infection. (A) Serum ALT levels (open squares) and serum HCV RNA titers (black diamonds) have previously been reported (44) and are presented for reference purposes. (B–G) Serial liver biopsies were analyzed for mRNA levels of (B) β1i (squares) and β7 (triangles); (C) β5i (squares) and β2i (triangles); (D) IFN-γ (squares) and CXCL9 (triangles); (E) TNF-α; (F) 2,5-OAS-1; and (G) IFN-α2 (black line), IFN-α14 (dashed line), IFN-α21 dotted line), and IFN-β (gray line). mRNA levels were normalized to endogenous references (GAPDH and β-actin) and expressed as fold increase over preinfection levels. In Ch1606, the relative mRNA level of IFN-α21 was 36.9 at week 4. Vertical dashed lines separate the time intervals prior to the first major (greater than 2-fold) increase of IFN-γ or TNF-α mRNA levels. NT, not tested due to the shortage of cDNA.
Next, the kinetics of immunoproteasome subunit induction were compared with the expression levels of IFN-γ and TNF-α, which are well-known inducers of immunoproteasome subunits. Surprisingly, neither IFN-γ nor TNF-α mRNA levels increased substantially (greater than 2-fold) in the liver prior to the increase in immunoproteasome subunit mRNA (Figure 5, D and E). The lack of an early IFN-γ response prior to the induction of immunoproteasome subunits was confirmed by the absent induction of CXC chemokine ligand 9 (CXCL9; also referred to as MIG) mRNA (Figure 5D), which is strictly IFN-γ dependent (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI29832DS1).
Thus, the increase in immunoproteasome subunit mRNA occurred prior to any increase in IFN-γ and TNF-α mRNA levels (dashed vertical lines in Figure 5) in all chimpanzees. Because this temporal relation rendered a role of IFN-γ or TNF-α in the induction of immunoproteasomes highly unlikely, we next examined the intrahepatic type I IFN response. As downstream marker of the type I IFN response, we quantitated 2,5-OAS-1 mRNA. As shown in Figure 5F, an increase of 2,5-OAS-1 mRNA levels was detectable as early as 2 weeks after infection. Its expression kinetics correlated well with those of the 3 immunoproteasome subunits and preceded the increase in IFN-γ mRNA in all chimpanzees. In parallel to the 2,5-OAS-1 response, an increase of mRNA levels of IFN-β mRNA and several IFN-α subtypes (IFN-α2, IFN-α14, and IFN-α21) was detectable (Figure 5G). Collectively, these results indicated a role of type I IFN in the induction of immunoproteasomes during acute HCV infection.
Induction of immunoproteasome subunits in the liver precedes infiltration of CD8 T cells.
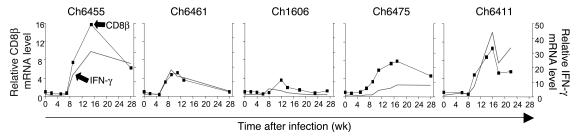
Whereas mRNA levels of all 3 immunoproteasome subunits started to increase as early as 2 weeks after infection, CD8β mRNA levels increased no earlier than 5–8 weeks after infection and peaked 11–16 weeks after infection. As shown in Figure 6, intrahepatic CD8β levels correlated closely with intrahepatic IFN-γ levels, suggesting that liver-infiltrating CD8 T cells produced most of the detectable IFN-γ. The IFN-γ–producing CD8 T cell population included HCV-specific T cells, which targeted multiple epitopes in all structural and nonstructural HCV proteins (data not shown). Induction of immunoproteasome subunits during acute HCV infection therefore occurred much earlier than the intrahepatic IFN-γ response. Indeed, immunoproteasomes were induced in the liver at the same time as type I IFN. Thus, rather than being passive targets, virus-infected hepatocytes actively prepare their antigen-processing machinery for optimal presentation even prior to the arrival of liver-infiltrating T cells.
Figure 6. Induction of immunoproteasome subunits in the liver precedes infiltration of CD8 T cells.
CD8β mRNA levels were quantified in serial liver biopsies of chimpanzees with acute HCV infection. CD8β mRNA amounts (squares) correlated closely with IFN-γ mRNA amounts (gray lines; values reproduced from Figure 5D for reference).
Discussion
The present study demonstrates the role of type I IFN in the induction of functional immunoproteasomes in the target organ of the CD8 T cell response during a viral infection. Not only exogenous IFN-α and IFN-β, but also endogenous production of type I IFN by intracellular HCV RNA or poly(I:C), stimulated induction of functionally active immunoproteasomes in vitro (Figures 1–4). Prospective analysis of liver biopsies from HCV-infected chimpanzees demonstrated that this mechanism was also operative in vivo during the early weeks of HCV infection (Figure 5). Collectively, these results challenge the current dogma of IFN-γ being the sole inducer of immunoproteasomes during a viral infection.
Our findings may represent a more general antiviral response, because dsRNA-induced, RIG-I–mediated type I IFN responses in virus-infected cells have also been described for other RNA viruses such as Newcastle disease virus (21), vesicular stomatitis virus (22), and paramyxovirus (23). Type I IFN–mediated induction of immunoproteasomes was especially evident in HCV infection, where intrahepatic expression of IFN-γ was detectable very late, i.e., not earlier than 5–8 weeks after infection (Figure 5). Thus, the innate type I IFN response not only exerts antiviral functions but also prepares the infected target organ for the adaptive immune response, in particular by immunoproteasome-dependent antigen processing. This mechanism complements a similar role of the innate immune response by type I IFN–induced upregulation of MHC molecules that has been described previously (24, 25). Both mechanisms may increase recognition of infected cells by incoming CD8 T cells and elevate virus-infected cells from passive targets to active facilitators of the adaptive immune response.
The type I IFN–mediated induction of immunoproteasomes described herein differed from the results of a previous publication that described normal induction of immunoproteasomes in LCMV-infected mice that lack the IFN-α/βR (11). In addition to virus and host differences between both models, several additional factors may explain why a type I IFN effect was not observed in the LCMV model. First, the effect of type I IFN may have been masked by an early and vigorous IFN-γ response in the LCMV model. The temporal dissociation between early type I IFN response and late IFN-γ response in the HCV model was essential to recognize the additional and specific effect of type I IFN. Second, human hepatocytes differ from murine hepatocytes in regard to IFN-α/βR expression and responsiveness to type I IFN (26). Whereas human hepatocytes express functional IFN-α/βR 2c, murine hepatocytes predominantly express IFN-α/βR 2a, which exerts inhibitory functions and blunts the effect of type I IFN on murine hepatocytes (26). Third, the new class of IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B) (27, 28), which activates the same JAK/STAT signaling pathway as type I IFN, may have induced immunoproteasomes in IFN-α/βR–/– mice during LCMV infection.
Our findings are supported by microarray studies of gene expression patterns in acute hepatitis C, which also detect early expression of immunoproteasome subunits (29, 30). In addition, they are indirectly supported by data from acute HBV infection (31). In contrast to acute HCV infection, acute HBV infection does not induce any detectable type I IFN response in the liver, as determined by lack of 2,5-OAS-1 mRNA induction in serial liver biopsies of HBV-infected chimpanzees (31). Consistent with the absence of a type I IFN response, no increased expression of immunoproteasome subunits was observed during the early phase of acute HBV infection (31). In fact, immunoproteasome subunits were only induced in the later phase of HBV infection, concomitant with an increase in intrahepatic IFN-γ mRNA levels (31).
What role does the induction of immunoproteasomes by type I IFN play in the course of acute HCV infection? Both the kinetics and the peak levels of intrahepatic immunoproteasome subunit mRNA expression were comparable in all 5 chimpanzees in our study (Figure 5). Likewise, kinetics and peak levels of intrahepatic 2,5-OAS-1 and type I IFN mRNA levels were comparable in all 5 chimpanzees (Figure 5). These findings suggest that differences in the outcome of HCV infection do not solely depend on the presence or absence of type I IFN–mediated induction of immunoproteasomes. However, the timing of type I IFN–mediated induction of immunoproteasomes in relation to the adaptive CD8 T cell response in the liver may be important. An effective CD8 T cell response is an important factor for the outcome of HCV infection, as previously shown by us (32–34) and others (15, 35–43). An increasing gap between transient early type I IFN–mediated induction of immunoproteasomes and late CD8 T cell infiltration in the liver may render the antigen recognition by intrahepatic HCV-specific T cells suboptimal. Interestingly, the timing of the intrahepatic increase of IFN-γ mRNA levels as indicated by the dashed vertical lines in Figure 5 (which correlates closely with intrahepatic CD8β mRNA levels; Figure 6), occurs later in chimpanzees with a chronically evolving course of HCV infection. This is consistent with a previously described delay in ALT peak, another correlate of CD8 T cell activity in the liver, in a larger group of chimpanzees with a chronic outcome of infection (44).
This timely dissociation between early transient immunoproteasome expression in the liver and late infiltration of CD8 T cells may be overcome by repeated exogenous administration of high doses of type I IFN, as impressively shown by the high (95%) HCV clearance rates if recombinant IFN-α is administered during the first 6 months of HCV infection (45–48). The high treatment response rate cannot readily be explained by IFN’s antiviral effects, because clearance rates are much lower in chronic HCV infection (46, 49), when immune escape mechanisms are already established (16, 41) and the HCV-specific T cell response is exhausted (15, 33, 35, 39, 40, 42).
Thus, it is possible that IFN-α–mediated induction of immunoproteasomes and better antigen recognition by liver-infiltrating T cells contribute to the high effectiveness of type I IFN–based therapies (95%) if administered early during HCV infection. Practically, however, it is very difficult to prove this hypothesis in a clinical setting, because it would require multiple liver biopsies per patient during the acute phase of hepatitis C. Chimpanzees are not an appropriate model for this purpose because their HCV titers do not decrease during therapy with recombinant human IFN-α (50, 51).
We conclude that intrahepatic induction of immunoproteasomes precedes the intrahepatic expression of IFN-γ in this clinically relevant model of viral infection in humans. Type I IFN secreted in response to dsRNA in virus-infected hepatocytes induced functional immunoproteasomes with characteristic proteolytic activity.
Methods
Chimpanzees.
Chimpanzees were housed under standard conditions for humane care and in compliance with NIH guidelines at an AALAC-accredited facility and were studied under protocols approved by the Animal Care and Use Committee (Center for Biologics Evaluation and Research) and the Public Health Service Interagency Model Committee (NIH). Four chimpanzees (Ch6455, Ch6461, Ch6475, and Ch6411) were intravenously inoculated with 100 chimpanzee infectious dose 50 (CID50) of monoclonal (H77 p90, genotype 1a) HCV RNA–positive and anti-HCV–negative plasma from Ch1536 (44, 52). Ch1606 was intrahepatically inoculated with in vitro–transcribed clonal HCV RNA (H77 p90, genotype 1a) (44). Serum HCV RNA was quantified by real-time RT-PCR with a sensitivity of 200 RNA copies/ml as previously reported (44). Serial liver biopsies were snap-frozen for RNA extraction.
Extraction of RNA, cDNA synthesis, and TaqMan real-time PCR.
Total RNA was isolated from IFN-treated cells and from snap-frozen and mechanically homogenized liver biopsies using the RNeasy Mini Kit (Qiagen) with an on-column DNase digestion step. RNA (200–400 ng) was reverse transcribed with the First-Strand cDNA Synthesis Kit (Marligen Biosciences) according to the manufacturer’s instructions. TaqMan real-time PCR was performed in duplicates to determine the mRNA levels of β1i, β5i, β2i, β7, CD8β, IFN-γ, CXCL9, TNF-α, 2,5-OAS-1, IFN-α2, IFN-α14, IFN-α21, and IFN-β using primers and probes of TaqMan Gene Expression Assays from Applied Biosystems according to the manufacturer’s instructions. Due to the limited amount of liver biopsy tissue, we could not study more than 4 type I IFN subtypes. The amount of specific mRNA was calculated with Sequence Detector software (version 1.6.3; Applied Biosystems), normalized to endogenous references (GAPDH and β-actin), and expressed as fold increase over the corresponding mRNA levels in preinfection biopsies or untreated cells. The baseline mRNA levels in preinfection liver biopsies represent the mean value obtained from up to 8 chimpanzee preinfection biopsies.
Cell culture.
Huh-7 hepatoma cell lines were grown in DMEM containing 5% FCS, 4.5 g/l glucose, 50 μg/ml streptomycin, 50 IU/ml penicillin, and 2 mM l-glutamine (complete DMEM). Primary human hepatocytes were obtained from BD Biosciences or through the Liver Tissue Procurement and Distribution System, which was funded by NIH contract no. N01-DK-9-2310. Hepatocytes were grown in Hepatocyte Maintenance Medium (Cambrex). All primary human hepatocytes were from HBV-negative and HCV-negative donors. The percentage of contaminating cells in the primary hepatocyte cultures was evaluated by fluorescence-activated cell sorting analysis using anti-CD45 (BD Biosciences) for cells of hematopoietic origin, anti-CD4 (BD Biosciences) for liver sinusoidal endothelial cells (53), and anti-CD14 (BD Biosciences) for Kupffer cells. After exclusion of dead cells by ethidium monoazide staining, the purity of hepatocytes was 86%–98.8%.
IFN treatment and RNA transfection.
Hepatoma cells and primary human hepatocytes were treated with IFN-α–con1 (kindly provided by L. Blatt, InterMune Inc., Brisbane, California, USA), human IFN-α2a (Research Diagnostics), human IFN-β1b (Betaseron; Chiron), or human IFN-γ (PeproTech).
Huh-7 cells were transfected by DMRIE-C reagent (Invitrogen) and Opti-MEM medium (Invitrogen) using either 10 μg poly(I:C) (Sigma-Aldrich) or 10 μg pcDNA3.1 plasmid DNA in a 6-well culture plate or 2 μg proteinase K–treated H77 HCV RNA in vitro transcribed from pHCV-H77 (kindly provided by J. Bukh, National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, USA) in a 48-well culture plate. Four hours after transfection, the culture medium was replaced by complete DMEM. The production of IFN-β was measured in culture supernatant using IFN-β ELISA kit (Biosource International). If blocking experiments were performed, neutralizing IFN-α Ab (R&D Systems) and/or IFN-β Ab (R&D Systems) or VV B18R (eBiosciences) (20) were added when the culture medium was replaced. As controls, 10 μg/ml poly(I:C) were added into Huh-7 cultures without transfection or Huh-7 cells were transfected with 10 μg pcDNA3.1 plasmid DNA.
Western blot analysis.
Cell lysates of 50 μg (for the detection of β5i) or 80 μg (for the detection of β1i, β2i, and α4) were separated on 15% SDS-polyacrylamide gels (54). Immunoblots were probed with human β1i, β5i, β2i, or α4 proteasome subunit–specific polyclonal rabbit antisera (kindly provided by M. Groettrup, University of Konstanz, Konstanz, Germany) or β-tubulin monoclonal Ab (CRP Inc.), and signals were detected with HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc.) or anti-mouse IgG (Seramun Diagnostica GmbH) and chemiluminescence reagent (54). The density of the Western blot bands was quantified using a GS-800 Calibrated Densitometer (Bio-Rad).
Proteasome purification.
20S proteasomes were isolated from Huh-7 hepatoma cells that had been treated either with 3 ng/ml IFN-α–con1 or with 10 ng/ml IFN-γ for 36 hours or, as a control, had been left untreated. Frozen cells were homogenized in lysis buffer containing 10 mM HEPES, pH 7.2; 80 mM KAc; 5 mM MgAc2; and 0,1% Triton X-100. Lysates were applied onto DEAE Sephacel (Amersham Biosciences) and washed with 80 mM KAc until no protein was detected by Ponceau staining. Proteasomes were eluted with 500 mM KAc buffer (10 mM HEPES, pH 7.2; 500 mM KAc; and 5 mM MgAc2) and concentrated by ammonium sulphate precipitation. Protein fractions were separated by ultra centrifugation on sucrose gradients (10%–40%) and ultracentrifuged at 200,000 g for 16 hours using a Beckman Coulter SW 40 rotor. Fractions containing proteasomes were pooled, applied to a Mono Q column (Pharmacia FPLC), and eluted with a gradient of 100–500 mM NaCl in TEAD (20 mM Tris, pH 7.2; 1 mM EDTA; 1 mM NaN3; and 1 mM DTT).
Metabolic labeling and immunoprecipitation of proteasome.
Huh-7 hepatoma cells (1 × 106) were seeded in flasks and cultured in complete DMEM with 3 ng/ml IFN-α–con1 or 10 ng/ml IFN-γ or without any IFN for 36 hours. During the final 12 hours, cells were labeled with translabel 35S-methionine (100 μCi/ml), washed twice, chased for 6 hours in medium with or without IFN, and then lysed in lysis buffer (1% NP40; 20 mM Tris-Cl, pH 7.5; 10 mM EDTA; and 100 mM NaCl with protease inhibitors). After centrifugation (5 min, 20,000 g at 4°C), the lysates were precleared with 25 μl protein A-Sepharose (50% slurry) and 1 mg/ml BSA overnight at 4°C. 20S proteasomes were immunoprecipitated with 50 μl protein A-Sepharose and 5 μl polyclonal 20S proteasome-specific rabbit antiserum for 4 hours at 4°C. Sepharose beads were washed with lysis buffer containing 0.5% NP40, resuspended in sample buffer for nonequilibrium pH gel elctrophoresis (NEPHGE), and subjected to 2-dimensional NEPHGE. Gels were exposed for autoradiography.
Two-dimensional gel electrophoresis.
For separation of 20S proteasomal proteins, isoelectric focusing by carrier ampholytes was combined with SDS-PAGE (54). Protein (25 μg) was applied to a carrier ampholyte isoelectric focussing gel. In the second dimension, proteins were loaded on a 1.5-mm-thick SDS-PAGE (7 × 8 cm) and stained with Coomassie Brilliant Blue G250. Proteasomal subunits were identified on the basis of their migrational behavior in comparison to reference electrophoreses of 20S proteasomes (55).
Peptide digestion and analysis of digests.
Twenty micrograms of the HBV 32-mer polypeptide AYRPPNAPILSTLPETTVVRRRGRSPRRRTPS (HBcore131–162) and 3 μg of purified proteasomes were incubated in 300 μl of assay buffer (20 mM HEPES/KOH, pH 7.8; 2 mM MgAc2; and 1 mM dithiothreitol) at 37°C for the time periods indicated in Figure 2C. The reaction was terminated by the addition of 0.1% trifluoroacetic acid (TFA). Digest products (30 μl) were separated by reverse-phase HPLC (SMART system equipped with a μRPC C2/C18 SC 2.1/10 column; Amersham Biosciences) under the following specific conditions: eluent A, 0.05% TFA; eluent B, 70% acetonitrile containing 0.045% TFA; gradient, 10%–95% eluent B in 15 minutes; flow rate, 70 μl/min. Analysis was performed online using a tandem quadrupol mass spectrometer equipped with an electronspray ion source (LCQ Finnigan MAT; Thermo Electron Corp.). Each scan was acquired over the range m/z = 200–1,400 in 3 seconds. Peptides were identified by their molecular masses and sequence in mass spectrometry/mass spectrometry experiments. Immunoproteasome-dependent generation of HBcore141–151 has also been demonstrated previously using peptide-specific cytotoxic T cells as readout (10).
Supplementary Material
Acknowledgments
The authors thank K. Textoris-Taube for mass spectrometry; U. Zimny-Arndt for 2-dimensional gel electrophoresis; P. Henklein for peptide synthesis; M. Groettrup for antibodies; and T.J. Liang, J.-H. Park, and N. Grandvaux for helpful and stimulating discussion. This study was supported by NIDDK, NIH, and Center for Biologics Evaluation and Research, FDA, intramural research programs; by grant SFB 421 from the Deutsche Forschungsgemeinschaft to U. Seifert and P.-M. Kloetzel; and by U.S. Public Health Service grant CA85883-01 from the NIH to C.M. Rice.
Footnotes
Nonstandard abbreviations used: ALT, alanine aminotransferase; CXCL9, CXC chemokine ligand 9; dsRNA, double-stranded RNA; HBcore, HBV core; IFN-α–con1, consensus sequence IFN-α; IFN-α/βR, IFN-α/β receptor; LCMV, lymphocytic choriomeningitis virus; 2,5-OAS-1, 2,5-oligoadenylate synthetase-1; RIG-I, retinoic acid–inducible gene I; VV B18R, vaccinia virus–encoded B18 receptor protein.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:3006–3014 (2006). doi:10.1172/JCI29832
References
- 1.Rock K.L., et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 2.Van den Eynde B.J., Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr. Opin. Immunol. 2001;13:147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 3.Kloetzel P.M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 4.Fruh K., Yang Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Curr. Opin. Immunol. 1999;11:76–81. doi: 10.1016/s0952-7915(99)80014-4. [DOI] [PubMed] [Google Scholar]
- 5.Foss G.S., Larsen F., Solheim J., Prydz H. Constitutive and interferon-gamma-induced expression of the human proteasome subunit multicatalytic endopeptidase complex-like 1. Biochim. Biophys. Acta. 1998;1402:17–28. doi: 10.1016/s0167-4889(97)00152-3. [DOI] [PubMed] [Google Scholar]
- 6.Loukissa A., Cardozo C., Altschuller-Felberg C., Nelson J.E. Control of LMP7 expression in human endothelial cells by cytokines regulating cellular and humoral immunity. Cytokine. 2000;12:1326–1330. doi: 10.1006/cyto.2000.0717. [DOI] [PubMed] [Google Scholar]
- 7.Van Kaer L., et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen W., Norbury C.C., Cho Y., Yewdell J.W., Bennink J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehling H.J., et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. . Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 10.Sijts A.J.A.M., et al. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J. Exp. Med. 2000;191:503–514. doi: 10.1084/jem.191.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S., et al. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 2001;167:6859–6868. doi: 10.4049/jimmunol.167.12.6859. [DOI] [PubMed] [Google Scholar]
- 12.Barton L.F., Cruz M., Rangwala R., Deepe G.S., Monaco J.J. Regulation of immunoproteasome subunit expression in vivo following pathogenic fungal infection. J. Immunol. 2002;169:3046–3052. doi: 10.4049/jimmunol.169.6.3046. [DOI] [PubMed] [Google Scholar]
- 13.Cangemi G., et al. IFN-alpha mediates the up-regulation of HLA class I on melanoma cells without switching proteasome to immunoproteasome. Int. Immunol. 2003;15:1415–1421. doi: 10.1093/intimm/dxg140. [DOI] [PubMed] [Google Scholar]
- 14.Alter H.J., Seeff L.B. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 15.Thimme R., et al. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehermann B., Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 17.Ozes O.N., Reiter Z., Klein S., Blatt L.M., Taylor M.W. A comparison of interferon-Con1 with natural recombinant interferons-alpha: antiviral, antiproliferative, and natural killer-inducing activities. J. Interferon Res. 1992;12:55–59. doi: 10.1089/jir.1992.12.55. [DOI] [PubMed] [Google Scholar]
- 18.Gale M., Foy E.M. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 19.Li K., Chen Z., Kato N., Gale M., Lemon S.M. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 20.Symons J.A., Alcami A., Smith G.L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M., et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 22.Kato H., et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Melchjorsen J., et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burrone O.R., Milstein C. Control of HLA-A,B,C synthesis and expression in interferon-treated cells. Embo J. 1982;1:345–349. doi: 10.1002/j.1460-2075.1982.tb01172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman R.L., Stark G.R. alpha-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature. 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- 26.Radaeva S., et al. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002;122:1020–1034. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- 27.Sheppard P., et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 28.Kotenko S.V., et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 29.Bigger C.B., Brasky K.M., Lanford R.E. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. . J. Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su A.I., et al. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieland S., Thimme R., Purcell R.H., Chisari F.V. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takaki A., et al. Cellular immune responses persist, humoral responses decrease two decades after recovery from a single source outbreak of hepatitis C. Nat. Med. 2000;6:578–582. doi: 10.1038/75063. [DOI] [PubMed] [Google Scholar]
- 33.Wedemeyer H., et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 2002;169:3447–3458. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 34.Nascimbeni M., et al. Kinetics of CD4+ and CD8+ memory T cell responses during hepatitis C virus rechallenge of previously recovered chimpanzees. J. Virol. 2003;77:4781–4793. doi: 10.1128/JVI.77.8.4781-4793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox A.L., et al. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–112. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thimme R., et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shoukry N., et al. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 2003;197:1645–1655. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grakoui A., et al. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 39.Lauer G.M., et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology. 2004;127:924–936. doi: 10.1053/j.gastro.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 40.Lechner F., et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimoto K., et al. Suppression of HCV-specific T cells without differential hierarchy demonstrated ex vivo in persistent HCV infection. Hepatology. 2003;38:1437–1448. doi: 10.1016/j.hep.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Diepolder H.M., et al. Possible mechanism involving T lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet. 1995;346:1006–1007. doi: 10.1016/s0140-6736(95)91691-1. [DOI] [PubMed] [Google Scholar]
- 43.Chang K.M., et al. Differential CD4 and CD8 T-cell responsiveness in hepatitis C virus infection. Hepatology. 2001;33:267–276. doi: 10.1053/jhep.2001.21162. [DOI] [PubMed] [Google Scholar]
- 44.Major M.E., et al. Hepatitis C virus kinetics and host responses associated with disease and outcome of infection in chimpanzees. Hepatology. 2004;39:1709–1720. doi: 10.1002/hep.20239. [DOI] [PubMed] [Google Scholar]
- 45.Jaeckel E., et al. Treatment of acute hepatitis C with interferon alfa-2b. N. Engl. J. Med. 2001;20:1452–1457. doi: 10.1056/NEJMoa011232. [DOI] [PubMed] [Google Scholar]
- 46.Manns M.P., et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 47.Rahman F., et al. Effects of antiviral therapy on the cellular immune response in acute hepatitis C. Hepatology. 2004;40:87–97. doi: 10.1002/hep.20253. [DOI] [PubMed] [Google Scholar]
- 48.Wiegand J., et al. Long-term follow-up after successful interferon therapy of acute hepatitis C. Hepatology. 2004;40:98–107. doi: 10.1002/hep.20291. [DOI] [PubMed] [Google Scholar]
- 49.Fried M.W., et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. . N. Engl. J. Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 50.Demers G.W., et al. Interferon-alpha2b secretion by adenovirus-mediated gene delivery in rat, rabbit, and chimpanzee results in similar pharmacokinetic profiles. Toxicol. Appl. Pharmacol. 2002;180:36–42. doi: 10.1006/taap.2002.9372. [DOI] [PubMed] [Google Scholar]
- 51.Lanford R., et al. Gene therapy for chronic hepatitis C based on adenoviral-mediated transfer of interferon-2b gene study based on the chimpanzee model. Hepatology. 2000;32(Suppl.):445A. [Google Scholar]
- 52.Kolykhalov A., et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 53.Knolle P.A., et al. Induction of cytokine production in naive CD4(+) T cells by antigen-presenting murine liver sinusoidal endothelial cells but failure to induce differentiation toward Th1 cells. Gastroenterology. 1999;116:1428–1440. doi: 10.1016/s0016-5085(99)70508-1. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y., et al. Expression of the proteasome activator PA28 rescues the presentation of a cytotoxic T lymphocyte epitope on melanoma cells. Cancer Res. 2002;62:2875–2882. [PubMed] [Google Scholar]
- 55.Jungblut P.R., Seifert R. Analysis by high-resolution two-dimensional electrophoresis of differentiation-dependent alterations in cytosolic protein pattern of HL-60 leukemic cells. J. Biochem. Biophys. Methods. 1990;21:47–58. doi: 10.1016/0165-022x(90)90044-d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.