Abstract
Waldenström macroglobulinemia (WM), a distinctive subtype of non-Hodgkin lymphoma that features overproduction of immunoglobulin M (IgM), clearly has a familial component; however, no susceptibility genes have yet been identified. We performed a genomewide linkage analysis in 11 high-risk families with WM that were informative for linkage, for a total of 122 individuals with DNA samples, including 34 patients with WM and 10 patients with IgM monoclonal gammopathy of undetermined significance (IgM MGUS). We genotyped 1,058 microsatellite markers (average spacing 3.5 cM), performed both nonparametric and parametric linkage analysis, and computed both two-point and multipoint linkage statistics. The strongest evidence of linkage was found on chromosomes 1q and 4q when patients with WM and with IgM MGUS were both considered affected; nonparametric linkage scores were 2.5 (P=.0089) and 3.1 (P=.004), respectively. Other locations suggestive of linkage were found on chromosomes 3 and 6. Results of two-locus linkage analysis were consistent with independent effects. The findings from this first linkage analysis of families at high risk for WM represent important progress toward identifying gene(s) that modulate susceptibility to WM and toward understanding its complex etiology.
Non-Hodgkin lymphoma (NHL [MIM 605027]) is a common cancer in older adults who live in developed countries, with a lifetime risk of ∼2.0%.1 Lymphomas are heterogeneous in terms of pathology, risk factors, and prognosis, and this heterogeneity has hampered efforts to delineate the genetic determinants of disease. Waldenström macroglobulinemia (WM [MIM 153600]) is a rare subtype of NHL that is highly distinctive on the basis of its hypersecretion of monoclonal immunoglobulin M (IgM).2 The public health burden is substantially increased by the fact that WM behaves in an indolent fashion, requiring treatment for many years before patients ultimately succumb to their disease. Whereas WM is rare, an asymptomatic elevation of monoclonal IgM protein, termed “IgM monoclonal gammopathy of undetermined significance” (IgM MGUS), is more common in the population. Patients with IgM MGUS can progress to develop WM, at the rate of 1.5%–2% per year.3
Little is known about the contribution of either genetic or extrinsic risk factors to the etiology of WM. Descriptions of environmental associations with WM are sparse,4 and WM has not been reported in association with any of several known extrinsic risk factors for NHL.5 Cohort and case-control studies have shown significant familial clustering of NHL and Hodgkin lymphoma (HL [MIM 236000]) and also suggest coaggregation of these B-cell tumors,6,7 and a recent population-based registry study reported significant familial risk specifically for lymphoplasmacytic lymphoma (LPL [the histopathologic correlate of WM]).8 The importance of genetic factors is further suggested by observations of familial clustering of WM.4 In addition, we and others have shown that IgM MGUS, which may serve as a phenotypic marker of susceptibility, can be found with careful screening in a substantial proportion of first-degree relatives of patients with WM.4,9
Identification of inherited susceptibility genes is an important step toward determining pathway(s) that contribute to development of WM. There have been no comprehensive searches of the genome to identify WM-predisposition genes, primarily because of the difficulty of assembling cohorts of informative families. Affected families usually have few living members with WM, because of the late age of onset (median age is ∼65 years in large clinical series10–12 and ∼73 years in population-based registry series [M. L. McMaster, unpublished observations]) and the high related mortality.
We have been accruing a cohort of high-risk families with multiple cases of WM, for phenotypic characterization and genetic evaluation. We studied 11 well-characterized, informative high-risk families from this cohort, by applying a whole-genome search and using densely spaced microsatellite markers to localize predisposition genes.
This study was conducted under institutional review board approval, and all patients gave informed consent for sample collection and analysis. We defined WM as follows: (1) presence of a serum monoclonal IgM component, (2) elevated quantitative serum IgM level, and (3) presence of a morphologically compatible lymphoproliferative infiltrate in the bone marrow. Symptoms attributable to elevated IgM or tumor burden were not required for diagnosis. In contrast, we defined IgM MGUS as the asymptomatic presence of a monoclonal IgM component with elevated quantitative serum IgM level in the absence of a morphologically identifiable lymphocytic infiltrate in the bone marrow. We obtained original pathology material and reports for all individuals with a reported diagnosis of WM, whenever possible. We evaluated all available affected individuals and their first-degree relatives at the National Institutes of Health or in the field, obtaining biospecimens from all study participants. Participants underwent routine serum protein characterization by immunofixation electrophoresis and immunoglobulin quantitation by nephelometry. For those relatives found to have IgM MGUS at screening, further evaluation was recommended. Consenting relatives underwent bone marrow aspiration and biopsy, the results of which were reviewed by an experienced hematopathologist, to distinguish IgM MGUS from asymptomatic WM. At the original screening, 18 individuals were found to have IgM MGUS, of whom 12 consented to bone marrow examination. Of these 12 patients, 7 received a diagnosis of WM, and 5 received a diagnosis of IgM MGUS, on the basis of bone marrow examination results. To be conservative, we classified patients who declined bone marrow examination as having IgM MGUS, for the purpose of this analysis. Eleven families were judged to be informative for linkage analysis (see examples in fig. 1). These families represent a subset of a larger cohort of families ascertained because of the occurrence of multiple cases of WM or the occurrence of a single case of WM in combination with other B-cell disorders within individual families. We chose to limit the current study to the most homogeneous subset, on the basis of the following criteria: families for which we had DNA available for genotyping from at least two patients with WM and from additional affected and/or unaffected first-degree relatives who could provide genotype information suitable for linkage analysis. The presence of other B-cell disorders in a given family did not exclude the family from the study, and each of three families had a single member with another B-cell lymphoproliferative disorder (LPD) (table 1). However, DNA was available from only one of these patients with LPD; thus, for purposes of linkage analysis, this individual was considered unaffected. A total of 122 DNA samples were available from affected and unaffected individuals, including 34 patients with WM and 10 patients with IgM MGUS. Among the patients with WM, the level of diagnostic accuracy was high, on the basis of review of pathology specimens (n=14; 41%), original pathology reports (n=14; 41%), referring physician reports (n=4; 12%), a death certificate (n=1; 3%), or a verbal report by a first-degree relative (n=1; 3%). Table 1 describes the distribution of affected individuals within families. Among all patients with WM and IgM MGUS, DNA samples were available for 39, and genotypes could be inferred for an additional 5. These families have good power to detect linkage if there is one major gene or if a majority of the families are segregating for the same gene. Our largest family (fig. 1B) by itself has an expected LOD score >3.0, with the assumption of dominant transmission of WM and IgM MGUS.
Figure 1. .
A–C, Examples of informative pedigrees, illustrating different pedigree structures among three study families. Note the multigenerational structure (A) with male-to-male transmission consistent with autosomal dominant inheritance. Not shown in panel C is a second-degree cousin also affected with WM.
Table 1. .
Distribution of Individuals Affected with WM, IgM MGUS, or Other LPDs, and Pedigree Configurations among Families with WM
| No. of Patients per Family or Pedigree Configuration |
No. of Families | % |
| WM: | ||
| 2 | 3 | 27 |
| 3 | 1 | 9 |
| 4 | 5 | 45 |
| 5 | 1 | 9 |
| Parent-offspring | 7 | 64 |
| Sibling-sibling ± cousin | 4 | 36 |
| IgM MGUS: | ||
| 0 | 5 | 45 |
| 1 | 4 | 36 |
| ⩾2 | 2 | 18 |
| Other LPDa: | ||
| 0 | 8 | 73 |
| 1 | 3 | 27 |
| ⩾2 | 0 | 0 |
Includes NHL (n=2), chronic lymphocytic leukemia (n=1), HL (n=0), and multiple myeloma (n=0) diagnosed in first-degree relatives of patients with WM. Of these patients with LPD, one patient with NHL was coded as “unaffected” and was included in the analysis; the other patient with NHL and the patient with chronic lymphocytic leukemia were deceased and were not included in the analysis, because our ability to infer genotype was limited.
DNA was extracted from cryopreserved lymphocytes with the use of standard methods. Genotyping was conducted under contract with deCODE Genetics as described elsewhere,13 with the use of their screening set of 1,058 microsatellite markers containing markers from the ABI Linkage Marker (v. 2) screening and intercalating sets and 500 custom-made markers with known allele-size distributions.
The genotype data were checked for Mendelian consistency with the use of the program PEDCHECK.14 The RECODE program was used to prepare the data files for analysis and to estimate allele frequencies from all founders in the pedigrees. Additional genotype errors were detected using the mistyping option of SimWalk2, version 2.8915–17 In total, only a very small number of genotypes (<0.5%) were eliminated because of either Mendelian inconsistencies or high mistyping probability.
Our primary approach to screening the genome for linkage was to compute multipoint NPL score statistics with the use of the program Genehunter (GH).18 This is a conservative approach that does not rely on assumptions about the genetic model. However, since the configuration of most of the families in this sample, which included our largest and most informative families, appeared to be compatible with autosomal dominant transmission of WM and MGUS, we also used GH to compute parametric LOD scores assuming homogeneity and LOD scores assuming heterogeneity (HLOD), with the parameter α defined as the proportion of linked families. Thus, for parametric LOD scores, we assumed WM or the combined WM/MGUS trait to be inherited as a rare dominant gene with a maximum penetrance of 50% and an allele frequency of 0.006. We assumed that penetrance increased with age, and we used age incidence rates in the population to construct liability classes. We analyzed the data under two models of disease classification. For the narrow model, we assumed that only those individuals who received a diagnosis of WM were affected and that all others were unaffected or unknown. For the broad model, we assumed that individuals with either WM or IgM MGUS were affected and that all others were unaffected or unknown. Because some of the pedigrees were too large for GH, the program did eliminate some individuals from the analysis (15 individuals from three families). Regions of the genome with GH P values ⩽.01 were followed up with NPL analyses with the use of the program SimWalk2, to include all individuals in the calculation. To assess interactions among selected locations that came up positive in the initial analysis, we used the program GH 2-locus.19 We computed NPL scores, to avoid specifying parameters for a two-locus disease model. To reduce calculation time, we limited our analyses to 15-bit inheritance vectors and to windows of 60–80 Mb around the most promising peaks from the single-locus analysis.
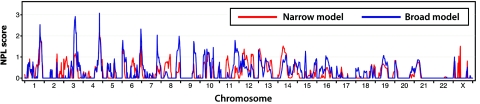
NPL scores from GH for the narrow and broad affection-status models can be seen in figure 2 for chromosomes 1–22. The strongest evidence for linkage was found on chromosomes 1, 3, 4, and 6 (fig. 3). On chromosome 1, several single-point LOD scores in the region from 203 cM to 232 cM were >2.0, with the highest-occurring score at locus D1S205 (LOD score 2.71), under the assumption of the narrow model. The peak signal from multipoint analysis was at map location 207 cM, flanked by loci D1S2615 and D1S2717. There was evidence for linkage under both the narrow and broad models, but the multipoint statistics were higher with the assumption of the broad affection-status model (NPL score 2.5; P=.0089; SimWalk2 P=.004). The HLOD score was 1.70 (α=0.65), indicating evidence of linkage from most of the families. Multipoint analysis of chromosome 4 showed high LOD and NPL scores under the broad model at location 173.6 cM, with flanking markers D4S2910 and D4S1539. Parametric analyses under the narrow model showed a peak in the same location. Under the broad model, the GH NPL score was 3.1 (P=.004), LOD score was 2.5, and HLOD score was 2.9 (α=0.76). However, the P value calculated by SimWalk2 was not significant (P=.07). As can be seen in the figure, the highest statistic is at only one locus, with the surrounding markers showing somewhat weaker evidence of linkage. On chromosome 3, an NPL score of 2.9 (P=.005) was found at location 133 cM, flanked by markers D3S3515 and D3S1267, under the broad model. The highest NPL statistic computed by SimWalk2 was at D3S1558 (location at 129 cM, P=.018). One family had a LOD score of 1.5 in this region, but the HLOD was only 1.1 (α=0.30). On chromosome 6, there was a peak near the q-terminal end under the broad model, with a peak NPL score of 2.3 (P=.01) at location 188 cM, flanked by markers D6S297 and D6S503. The HLOD score was 1.71 (α=0.83). The highest SimWalk2 statistic was at D6S281 (P=.01). There were a few other regions of the genome where the nominal P values were <.05 under either the narrow or the broad model (chromosomes 6, 7, 8, 9, 11, 13, 14, and X), but these may be chance findings. Two-locus NPL scores were computed for pairwise combinations of chromosomes 1, 3, and 4 and resulted in maximal NPL scores of 3.8 (chromosomes 1 and 3), 3.9 (chromosomes 1 and 4) (fig. 4), and 4.1 (chromosomes 3 and 4), in configurations consistent with independent rather than interactive effects. The closest corresponding marker pairs were D1S249 and D4S3030, D1S249 and D4S3030, and D3S1267 and D4S3030, respectively.
Figure 2. .
Multipoint NPL statistics calculated by GH for chromosomes 1–22 under the narrow affection-status model, in which only patients given a diagnosis of WM are designated as “affected,” and under the broad affection-status model, in which patients given a diagnosis of either WM or IgM MGUS are considered affected. GH does not compute multipoint NPL scores for the X chromosome, so statistics shown for the X chromosome are two-point LOD scores.
Figure 3. .
Multipoint LOD and NPL scores under the narrow affection-status model and under the broad affection-status model for chromosomes with suggestive two-point linkage results. Calculations were performed under an assumption of genetic homogeneity for autosomal dominant inheritance with maximum penetrance of 50%. Values <−2 are not shown.
Figure 4. .
Two-locus NPL scores of the regions having strongest evidence for linkage in the initial single-locus parametric and NPL analysis. Results are shown for chromosomes 1 and 4. The arrow labeled “NPL” indicates the point associated with maximum NPL score for the two-locus analysis. The coordinate arrows indicate where the maximum two-locus NPL scores were reached, at markers D1S249 and D4S3030 (207.07 cM and 170.81 cM, respectively).
We have conducted the first, to our knowledge, dense-marker genomewide screen of families at high risk for WM. The two strongest findings are on chromosomes 1q and 4q. These two regions have NPL or LOD scores that are suggestive for linkage, according to the criteria of Lander and Kruglyak.20 However, the 4q finding was less strong when the SimWalk2 statistics were calculated, and the strongest evidence appears to be in a narrow region. Two additional regions on chromosome 3q (NPL score 2.9) and 6q (NPL score 2.3) have slightly lower significance levels and warrant further evaluation. The heterogeneity results indicated that ∼65% of the families show evidence of linkage to the regions on 1, 4, and 6. The two-locus results did not suggest interaction between the linkage peaks, which gives further support to heterogeneity of effects. Since there are no formal tests of significance for the two-locus analysis, however, they should be considered exploratory.19,21
For each of the four most-positive regions for linkage, the evidence was always stronger with assumption of the broad affection-status model—that is, when both WM and IgM MGUS are included as “affected,” which suggests that both conditions share a common susceptibility gene(s). Precise prevalence rates for IgM MGUS in the general population are unknown, but population-based estimates suggest a maximum prevalence of ∼0.5%.22 In contrast, the frequency of IgM MGUS is increased ∼10-fold in first-degree relatives of patients with familial WM,4 and progression of IgM MGUS to WM has been shown to occur in some families.23 These observations are consistent with our results and suggest that the clinical spectrum in familial WM is broad.
It is worth noting that two of the regions identified here (4q and 3q) are bounded by markers that were identified as secondary regions of interest in a similar genomewide linkage study of 44 families at high risk for HL.13 Whereas the clinical and morphological spectra of WM and HL are clearly distinct, they both are malignancies of B-cell origin,24 so it is possible that we have identified regions of common susceptibility. We have diagnosed other LPDs in patients in these and other families with WM that we have studied. Both population and clinicopathological studies suggest some familial overlap among B-cell tumors in general (including NHL, HL, and chronic lymphocytic leukemia [MIM 151400]).6,7,25–27 Furthermore, in a recent case series, 20% of patients with familial WM reported a family history of a B-cell malignancy other than WM.28 Within population-based studies that have reported cross-susceptibility, the relative risk is highest for the same malignancy,6,7,25 suggesting that families enriched for a single disease may provide higher power for detecting linkage to genes with main effects. In particular, a recent registry study calculated the familial risk for B-cell disorders among patients with LPL and reported a standardized incidence ratio of 43.4 for LPL, compared with 4.0 and 4.1 for all NHL and chronic lymphocytic leukemia, respectively. In this analysis, we did not have enough data to test whether counting individuals with other lymphoproliferative cancers as “affected” increases the evidence for linkage; however, this is clearly a question that should be addressed in future studies.
Review of current genome databases—for example, Map Viewer (build 36.1)—reveals that each of the regions identified contains multiple potential candidate genes, including many that are generally involved in cell-cycle control, transcriptional regulation, apoptosis, and/or immune regulation, and others that have roles related specifically to B cells, including cytokines, cell surface markers, and regulators of immunoglobulin gene rearrangement. Because the pathophysiology of WM is uncertain, there could be additional relevant candidate genes. However, none of these genes have been identified as candidates for WM in other studies. One would want to have stronger evidence and narrower regions for these peaks before undertaking a systematic screening for specific candidate genes.
Many hematologic and lymphoid malignancies are characterized by specific cytogenetic abnormalities that have been found to involve genes that are important in oncogenesis. Various cytogenetic aberrations have been reported in sporadic WM, including occasional abnormalities that overlap with our results.29–32 However, few of the reported changes involving chromosomes 1q, 4q, or 3q are recurrent, and none are characteristic of WM. The most common recurrent cytogenetic alteration in WM, del(6)(q21q23),32 is a nonspecific finding also seen in other types of B-cell lymphoma. Furthermore, although several potential candidate genes map to 6q21-q23, these loci are 25–50 cM centromeric of the peak that we identified. Thus, it appears unlikely that the gene(s) involved in del(6q) and seen in sporadic WM are responsible for the predisposition to WM found in our families. We reported elsewhere33 the characterization of a series of patients with familial WM—including 10 of the patients with WM included in the genome scan reported in this article—by standard cytogenetics, spectral karyotyping, and comparative genomic hybridization. None of the cytogenetic changes in familial WM reported by us or by others34–36 occur within any of the regions identified in this study.
There are some limitations to this study. These families were selected to be informative for genetic-mapping studies and are not representative of WM in the general population. If there were substantial genetic heterogeneity, our study would not have sufficient power to detect genes. Larger studies are required to obtain stronger evidence for these locations, to further narrow the regions of interest, and to resolve questions about the spectrum of phenotypes involved, including effects due to age, gender, and other covariates.
In summary, this study is the first systematic genetic analysis of familial WM to apply highly informative, densely spaced markers in a whole-genome search for linkage analysis. The results were most strongly suggestive of linkage to regions on chromosomes 1q and 4q under the assumption that IgM MGUS is a phenotypic marker of susceptibility in these families. Although model-based results were consistent, our conclusions are based on nonparametric analyses and, therefore, do not depend on particular assumptions of a genetic model. Thus, our results can be taken as a conservative estimate of linkage to these loci. Furthermore, findings on chromosomes 3q and 4q overlap with regions that we have previously found to be linked to HL.13 These data, in combination with population and clinicopathologic data, suggest that these B-cell disorders may share susceptibility loci. To confirm and refine these results, we are actively recruiting additional families with WM for study. Our results represent the initial step in the process of discovery of germline WM susceptibility gene(s) and delineation of the pathways that lead to development of WM. Once the genetic determinants of WM susceptibility are defined, the effect of environmental factors in modulating risk can be studied, and more-effective treatment and prevention strategies for WM may be developed. Finally, any genes discovered may play a role in a broader spectrum of B-cell tumors.
Acknowledgments
This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. We thank W. Blattner and G. Shaw, for their contributions to family ascertainment, and E. Jaffe, P. Noel, and M. Rick, for pathology review. We are indebted to V. Byrd, for assistance with the field component of the study, and to B. Hulley, for expert preparation of the figures. The genotyping was conducted under National Cancer Institute contract N02-CP-01108, with a subcontract BRC-1108-35 with deCODE Genetics.
Web Resources
The URLs for data presented herein are as follows:
- Map Viewer database, http://www.ncbi.nlm.nih.gov/mapview/static/humansearch.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NHL, WM, HL, and chronic lymphocytic leukemia)
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ (2006) Cancer statistics, 2006. CA Cancer J Clin 56:106–130 [DOI] [PubMed] [Google Scholar]
- 2.Berger F, Isaacson PG, Piris MA, Harris NL, Müller-Hermelink HK, Nathwani BN, Swerdlow SH (2001) Lymphoplasmacytic lymphoma/Waldenström macroglobulinemia. In: Jaffe ES, Harris NL, Stein H, Vardiman JW (eds) Pathology and genetics of tumours of haematopoietic and lymphoid tissues. IARC Press, Lyon, pp. 132–134 [Google Scholar]
- 3.Kyle RA, Therneau TM, Rajkumar SV, Remstein ED, Offord JR, Larson DR, Plevak MF, Melton LJ (2003) Long-term follow-up of IgM monoclonal gammopathy of undetermined significance. Blood 102:3759–3764 10.1182/blood-2003-03-0801 [DOI] [PubMed] [Google Scholar]
- 4.McMaster ML (2003) Familial Waldenstrom’s macroglobulinemia. Semin Oncol 30:146–152 10.1053/sonc.2003.50063 [DOI] [PubMed] [Google Scholar]
- 5.Fisher SG, Fisher RI (2004) The epidemiology of non-Hodgkin’s lymphoma. Oncogene 23:6524–6534 10.1038/sj.onc.1207843 [DOI] [PubMed] [Google Scholar]
- 6.Goldin LR, Landgren O, McMaster ML, Gridley G, Hemminki K, Li X, Mellemkjaer L, Olsen JH, Linet MS (2005) Familial aggregation and heterogeneity of non-Hodgkin lymphoma in population-based samples. Cancer Epidemiol Biomarkers Prev 14:2402–2406 10.1158/1055-9965.EPI-05-0346 [DOI] [PubMed] [Google Scholar]
- 7.Goldin LR, Pfeiffer RM, Gridley G, Gail MH, Li XJ, Mellemkjaer L, Olsen JH, Hemminki K, Linet MS (2004) Familial aggregation of Hodgkin lymphoma and related tumors. Cancer 100:1902–1908 10.1002/cncr.20189 [DOI] [PubMed] [Google Scholar]
- 8.Altieri A, Bermejo JL, Hemminki K (2005) Familial aggregation of lymphoplasmacytic lymphoma with non-Hodgkin lymphoma and other neoplasms. Leukemia 19:2342–2343 10.1038/sj.leu.2403991 [DOI] [PubMed] [Google Scholar]
- 9.Seligmann M, Danon F, Mihaesco C, Fudenberg HH (1967) Immunoglobulin abnormalities in families of patients with Waldenström’s macroglobulinemia. Am J Med 43:66–83 10.1016/0002-9343(67)90149-0 [DOI] [PubMed] [Google Scholar]
- 10.Gobbi PG, Bettini R, Montecucco C, Cavanna L, Morandi S, Pieresca C, Merlini G, Bertoloni D, Grignani G, Pozzetti U, Caporali R, Ascari E (1994) Study of prognosis in Waldenström’s macroglobulinemia: a proposal for a simple binary classification with clinical and investigational utility. Blood 83:2939–2945 [PubMed] [Google Scholar]
- 11.Dimopoulos MA, Hamilos G, Zervas K, Symeonidis A, Kouvatseas G, Roussou P, Gika D, Karmiris T, Bourantas K, Zomas A, Mitsouli C, Xilouri I, Vervessou E, Matsis K, Anagnostopoulos N, Economopoulos T (2003) Survival and prognostic factors after initiation of treatment in Waldenstrom’s macroglobulinemia. Ann Oncol 14:1299–1305 10.1093/annonc/mdg334 [DOI] [PubMed] [Google Scholar]
- 12.Owen RG, Barrans SL, Richards SJ, O’Connor SJM, Child JA, Parapia LA, Morgan GJ, Jack AS (2001) Waldenstrom macroglobulinemia: development of diagnostic criteria and identification of prognostic factors. Am J Clin Pathol 116:420–428 10.1309/4LCN-JMPG-5U71-UWQB [DOI] [PubMed] [Google Scholar]
- 13.Goldin LR, McMaster ML, Ter Minassian M, Saddlemire S, Harmsen B, Lalonde G, Tucker MA (2005) A genome screen of families at high risk for Hodgkin lymphoma: evidence for a susceptibility gene on chromosome 4. J Med Genet 42:595–601 10.1136/jmg.2004.027433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel E, Sengul H, Weeks DE (2001) Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered 52:121–131 10.1159/000053366 [DOI] [PubMed] [Google Scholar]
- 17.Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- 19.Strauch K, Fimmers R, Baur MP, Wienker TF (2003) How to model a complex trait. 2. Analysis with two disease loci. Hum Hered 56:200–211 10.1159/000076394 [DOI] [PubMed] [Google Scholar]
- 20.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 21.Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP (2000) Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus–trait models: application to mite sensitization. Am J Hum Genet 66:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen HJ, Crawford J, Rao MK, Pieper CF, Currie MS (1998) Racial differences in the prevalence of monoclonal gammopathy in a community-based sample of the elderly. Am J Med 104:439–444 10.1016/S0002-9343(98)00080-1 [DOI] [PubMed] [Google Scholar]
- 23.Fine JM, Lambin P, Massari M, Leroux P (1982) Malignant evolution of asymptomatic monoclonal IgM after seven and fifteen years in two siblings of a patient with Waldenström’s macroglobulinemia. Acta Med Scand 211:237–239 [DOI] [PubMed] [Google Scholar]
- 24.Jaffe ES, Harris NL, Stein H, Vardiman JW (2001) Pathology and genetics of tumours of haematopoietic and lymphoid tissues. IARC Press, Lyon [Google Scholar]
- 25.Goldin LR, Pfeiffer RM, Li XJ, Hemminki K (2004) Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood 104:1850–1854 10.1182/blood-2004-01-0341 [DOI] [PubMed] [Google Scholar]
- 26.Owen RG, Parapia LA, Higginson J, Misbah SA, Child JA, Morgan GJ, Jack AS (2000) Clinicopathological correlates of IgM paraproteinemias. Clin Lymphoma 1:39–43 [DOI] [PubMed] [Google Scholar]
- 27.Pangalis GA, Kyrtsonis MC, Kontopidou FN, Siakantaris MP, Dimopoulou MN, Vassilakopoulos TP, Tzenou T, Kokoris S, Dimitriadou E, Kalpadakis C, Tsalimalma K, Tsaftaridis P, Panayiotidis P, Angelopoulou MK (2005) Differential diagnosis of Waldenstrom’s macroglobulinemia and other B-cell disorders. Clin Lymphoma 5:235–240 [DOI] [PubMed] [Google Scholar]
- 28.Treon SP, Hunter ZR, Aggarwal A, Ewen EP, Masota S, Lee C, Santos DD, Hatjiharissi E, Xu L, Leleu X, Tournilhac O, Patterson CJ, Manning R, Branagan AR, Morton CC (2006) Characterization of familial Waldenstrom’s macroglobulinemia. Ann Oncol 17:488–494 10.1093/annonc/mdj111 [DOI] [PubMed] [Google Scholar]
- 29.Palka G, Spadano A, Geraci L, Fioritoni G, Dragani A, Calabrese G, Franchi PG, Stuppia L (1987) Chromosome changes in 19 patients with Waldenström’s macroglobulinemia. Cancer Genet Cytogenet 29:261–269 10.1016/0165-4608(87)90237-8 [DOI] [PubMed] [Google Scholar]
- 30.Johansson B, Waldenstrom J, Hasselblom S, Mitelman F (1995) Waldenstrom’s macroglobulinemia with the AML/MDS-associated t(1;3)(p36;q21). Leukemia 9:1136–1138 [PubMed] [Google Scholar]
- 31.Mansoor A, Medeiros LJ, Weber DM, Alexanian R, Hayes K, Jones D, Lai R, Glassman A, Bueso-Ramos CE (2001) Cytogenetic findings in lymphoplasmacytic lymphoma/Waldenstrom macroglobulinemia: chromosomal abnormalities are associated with the polymorphous subtype and an aggressive clinical course. Am J Clin Pathol 116:543–549 10.1309/6U88-357U-UKJ5-YPT3 [DOI] [PubMed] [Google Scholar]
- 32.Schop RFJ, Kuehl WM, Van Wier SA, Ahmann GJ, Price-Troska T, Bailey RJ, Jalal SM, Qi Y, Kyle RA, Greipp PR, Fonseca R (2002) Waldenstrom macroglobulinemia neoplastic cells lack immunoglobulin heavy chain locus translocations but have frequent 6q deletions. Blood 100:2996–3001 10.1182/blood.V100.8.2996 [DOI] [PubMed] [Google Scholar]
- 33.McMaster ML, Giambarresi T, Vasquez L, Goldstein AM, Tucker MA (2005) Cytogenetics of familial Waldenstrom’s macroglobulinemia: in pursuit of an understanding of genetic predisposition. Clin Lymphoma 5:230–234 [DOI] [PubMed] [Google Scholar]
- 34.Youinou P, Le Goff P, Saleun JP, Rivat L, Morin JF, Fauchier C, Le Menn G (1978) Familial occurrence of monoclonal gammapathies. Biomedicine 28:226–232 [PubMed] [Google Scholar]
- 35.Taleb N, Tohme A, Abi JD, Kattan J, Salloum E (1991) Familial macroglobulinemia in a Lebanese family with two sisters presenting Waldenstrom’s disease. Acta Oncol 30:703–705 [DOI] [PubMed] [Google Scholar]
- 36.Elves MW, Brown AK (1968) Cytogenetic studies in a family with Waldenström’s macroglobulinaemia. J Med Genet 5:118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]