Abstract
Genetic analysis of a large Indian family with an autosomal dominant cataract phenotype allowed us to identify a novel cataract gene, CRYBA4. After a genomewide screen, linkage analysis identified a maximum LOD score of 3.20 (recombination fraction [θ] 0.001) with marker D22S1167 of the β-crystallin gene cluster on chromosome 22. To date, CRYBA4 was the only gene in this cluster not associated with either human or murine cataracts. A pathogenic mutation was identified in exon 4 that segregated with the disease status. The c.317T→C sequence change is predicted to replace the highly conserved hydrophobic amino acid phenylalanine94 with the hydrophilic amino acid serine. Modeling suggests that this substitution would significantly reduce the intrinsic stability of the crystalline monomer, which would impair its ability to form the association modes critical for lens transparency. Considering that CRYBA4 associates with CRYBB2 and that the latter protein has been implicated in microphthalmia, mutational analysis of CRYBA4 was performed in 32 patients affected with microphthalmia (small eye). We identified a c.242T→C (Leu69Pro) sequence change in exon 4 in one patient, which is predicted here to disrupt the β-sheet structure in CRYBA4. Protein folding would consequently be impaired, most probably leading to a structure with reduced stability in the mutant. This is the first report linking mutations in CRYBA4 to cataractogenesis and microphthalmia.
Cataracts are a leading cause of blindness worldwide, affecting all societies. A significant proportion of cases are genetically determined.1 More than 15 genes for cataracts have been identified, of which the crystallin genes are the most commonly mutated.2 Most of the crystallins have been associated with either murine or human cataracts. Crystallins function primarily as structural proteins of the vertebrate lens, accounting for 80%–90% of the soluble protein fraction. In the human lens, α-crystallin constitutes 40% of the crystallins, β-crystallin 35%, and γ-crystallin 25%.3 The structural stability of crystallins is important to lens transparency, since they last a lifetime with virtually no protein turnover.1,4
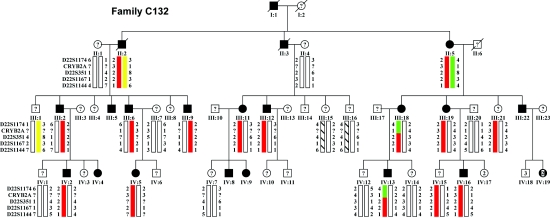
We studied a previously described, four-generation Indian family (family C1325) affected with autosomal dominant congenital lamellar cataract. The pathogenic role of the γ-crystallin genes and that of GJA8 were previously excluded by sequence analysis of the coding sequence.5 Thirteen affected individuals were examined and available for this study. DNA from unaffected individuals and spouses were also available (fig. 1). These unaffected individuals were not all examined and were therefore coded as “unknown” for further genetic analysis. Informed consent was obtained in accordance with the Tenets of the Declaration of Helsinki. Most affected individuals were diagnosed during childhood. However, affected individuals III:2 (late 30s) and III:22 (43 years old) (fig. 1) remain unoperated. This suggests an early insult with variable expressivity.
Figure 1. .
Family C132 pedigree, with haplotypes. Affected individuals (blackened symbols) with a haplotype were clinically examined. “Unaffected” individuals (unblackened symbols) were not examined. Individuals III:18 and IV:13 defined the upper boundary of the disease interval and excluded CRYBB2 (exons 1–3) and CRYBB3 as candidate genes. A lower boundary for the disease interval was not identified. The affected haplotype is depicted in red.
A genomewide linkage scan used 388 microsatellite markers (average resolution 10 cM). Pedigree structure was confirmed using PREST analysis.6,7 Two-point linkage analysis was performed under the assumption of autosomal dominant inheritance with complete penetrance and a disease-allele frequency of 0.001 (MLINK program of the Linkage software package, version 2). A candidate locus was identified on chromosome 22 where a maximum pairwise LOD score of 3.20 at recombination fraction (θ) 0.001 was obtained with D22S1167 (table 1). Location score analysis, performed by use of LINKMAP v5.1, supported the strong evidence for linkage to this region (maximum location score [log10] 3.50, with the interval and flanking markers D22S351 to D22S1167). Fine mapping of the disease interval used STRP markers available through public databases. Direct sequencing of the coding sequence of the four genes of the chromosome 22 crystallin gene cluster (CRYBB2, CRYBB3, CRYBB1, and CRYBA4) identified a heterozygous T→C transition in CRYBA4 (MIM *123631) exon 4. Mutation of the highly conserved Phe 94 (c.317T→C [NCBI accession number NM_001886]) changes the large, hydrophobic, nonpolar amino acid phenylalanine to the small, polar amino acid serine. By use of haplotype analysis and NciI restriction-enzyme digestion, the change was shown to segregate with the disease phenotype. Two individuals who were not available for examination carried the affected haplotype (fig. 1). Of these, case IV:15 was reported not to be eligible for a driving license in Kuwait. The Phe94Ser mutation was not seen in 239 control individuals (160 of mixed ethnicity, 53 Indian, and 26 Pakistani) nor in 50 patients affected with congenital cataracts and 50 patients affected with age-related cataract (data not shown, total of 678 chromosomes).
Table 1. .
Two-Point Linkage Results Between the Cataract Phenotype and Markers on Chromosome 22[Note]
| LOD at θ = |
||||||||||
| Markera | IMDb (cM) |
.0 | .01 | .05 | .1 | .2 | .3 | .4 | Zmax | θmax |
| GATA198E* | … | −2.93 | −1.50 | −.47 | −.08 | .16 | .16 | .09 | .17 | .246 |
| D22S446* | 12.6 | .55 | 1.06 | 1.45 | 1.47 | 1.22 | .82 | .38 | 1.48 | .081 |
| D22S1174 | 4.9 | .27 | .78 | 1.19 | 1.24 | 1.03 | .69 | .31 | 1.24 | .088 |
| CRYB2-CA | 2.1 | −.10 | −.01 | .19 | .29 | .33 | .27 | .16 | .33 | .182 |
| D22S351 | 1.9 | 2.63 | 2.58 | 2.38 | 2.12 | 1.58 | .99 | .41 | 2.63 | .001 |
| D22S1167* | 1.4 | 3.20 | 3.14 | 2.90 | 2.59 | 1.91 | 1.19 | .48 | 3.20 | .001 |
| D22S1144 | 2.7 | 2.90 | 2.85 | 2.62 | 2.33 | 1.71 | 1.05 | .40 | 2.90 | .001 |
| D22S685* | 4.9 | .75 | .74 | .67 | .59 | .43 | .28 | .13 | .75 | .001 |
| D22S445* | 13.4 | 1.83 | 1.79 | 1.62 | 1.41 | .97 | .54 | .19 | 1.83 | .001 |
| D22S444* | 5.7 | .46 | .45 | .41 | .36 | .27 | .17 | .09 | .46 | .001 |
Note.— Individuals presumed “unaffected” were coded as “unknown” for linkage analysis. Boldface type denotes denotes the marker with maximum LOD score (D22S1167).
An asterisk (*) denotes markers genotyped in the initial genome scan.
IMD = intermarker distance, from the Marshfield map.
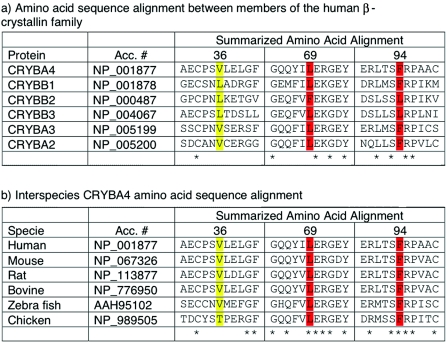
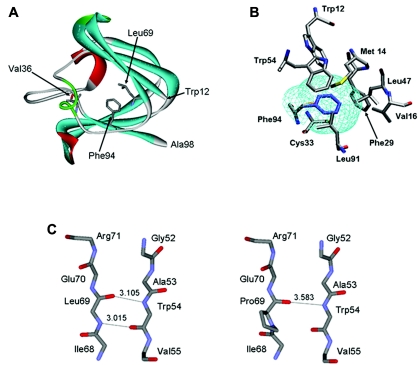
Multiple sequence alignments generated using ClustalW showed that the Phe at position 94 is highly conserved in mouse, rat, bovine, zebrafish, and chicken (fig. 2). In the absence of a known three-dimensional structure for the CRYBA4 protein, homology modeling techniques were used to gain insight into the possible role of Phe94 in maintaining the native protein state. The partial sequence from amino acid 12 to amino acid 98 of CRYBA4 was extracted from the RefSeq resource of the National Center for Biotechnology Information (NCBI) (accession number NP_001877) and was used to search the Protein Databank (PDB)8,9 for a suitable template from among the known crystallin structures. The selected template was crystallin-β B1 (CRYBB1) (PDB entry 1oki_A), which displayed 47% sequence identity with the partial CRYBA4 query sequence. The CRYBA4 sequence was aligned to the template protein by use of Needleman-Wunsch alignment.10 Gaps in the CRYBA4 corresponding to α-helices and β-strands in the template structure were manually displaced toward positions in loop regions. The main-chain atoms in the CRYBA4 were assigned the spatial coordinates of the equivalent residues in the CRYBB1 template. The conformations of the side-chains were built onto the CRYBA4 model backbone by a Metropolis Monte Carlo procedure implemented in the software Modzinger. Insertions and deletions in loop regions were modeled by searching a database of fragments from known protein structures deposited in the PDB for those that are compatible with the target sequence and the template scaffold,11 followed by a structure refinement procedure. The Phe94Ser structure was generated by replacing the Phe side chain with that of Ser at position 94 and optimizing the Ser side chain while keeping all surrounding side chains and backbone atoms fixed at their position in the native CRYBA4 model. (The atomic coordinates of the models built for the wild-type and all mutant proteins analyzed in this study are available at the Web site of the Centre for Computational Biology at the Hospital for Sick Children in Toronto.) Inspection of the CRYBA4 model reveals that Phe94 is positioned in the hydrophobic core formed between the two beta sheets of the second Greek key motif not far from the connecting peptide (amino acids 98–103), as illustrated in figure 3A and 3B. Replacing Phe with Ser in this position introduced a polar group into the hydrophobic core of the protein and created a large packing defect, or cavity, in this region, with a volume, computed by the Prove software, of ∼100 Å3.12 Each of these effects is expected to destabilize the native CRYBA4 structure. On the basis of measures of transfer of free energy of amino acid side chains from organic solvent to water,13 replacement of a completely buried Phe side chain by a Ser would destabilize the native state of the CRYBA4 protein by ∼2.5–2.8 kcal/mol, with the lower value representing the case in which the Ser γ OH would form hydrogen bonds with the backbone of neighboring residues, as is often observed in proteins containing completely buried Ser side chains.14 If we use estimates by Eriksson et al.15 and Xu et al.16—made on the basis of mutagenesis, calorimetric, and structural studies of T4-phage lysozyme mutants—formation of the ∼100 Å3 cavity on the Phe→Ser replacement at position 94 would add as many as 2.2 kcal/mol of destabilization free energy (22 cal/Å3 of cavity volume). This would bring the total destabilization free energy due to the Phe94Ser mutation in CRYBA4 to close to 4.7–5 kcal/mol. This would represent a sizable loss of stability for the protein, considering that the unfolding free energy of wild-type crystallin monomers is probably <10 kcal/mol (a value of 8.7 kcal/mol was reported for βB2 crystallin17), which could well preclude its proper folding, which is required to form the higher-order associations with other crystalline proteins that are critical for the formation and maintenance of lens transparency.
Figure 2. .
Comparative sequence analysis of CRYBA4. Red, pathogenic amino acid change; yellow, change of uncertain pathogenicity.
Figure 3. .
Homology modeling of CRYBA4. A, Modeled structure of CRYBA4. Depicted is the N-terminal domain of the predicted atomic structure of CRYBA4, built using the known structure of 1oki_A as template (see text). The protein backbone is drawn using a solid ribbon model. α-Helix, β-strand, and turn structures are colored in red, cyan, and light green, respectively. Val36, Leu69, and Phe94 residues are shown in full atomic detail. B, Environment of the Phe94 and Ser94 side chains in the modeled CRYBA4 three-dimensional structure. The side chains of Phe94 and Ser94 are colored in blue and orange, respectively. Residues whose atoms are <6.0 Å from the side-chain atoms of Phe94 are shown. The ∼100-Å3 cavity formed by replacing the bulky Phe 94 with Ser is outlined by the mesh colored in cyan. This cavity is lined by the side chains of hydrophobic residues surrounding Ser94. The cavity volume was computed using the ProShape software, with a probe radius of 0.2 Å, which approximates the cavity delimited by the molecular surface of the surrounding residues. C, Portion of the CRYBA4 structure in the vicinity of residue 69, highlighting the differences in the β-sheet backbone structures of the wild-type (Leu69) and mutant (Pro69) proteins. The backbone nitrogen of Leu69 forms a hydrogen bond with the carbonyl group of Trp54, contributing to the stability of the antiparallel β-sheet structure. This hydrogen bond cannot form in the Pro69 mutant. Its backbone nitrogen cannot act as a hydrogen-bond donor, since it is covalently linked to the proline ring.
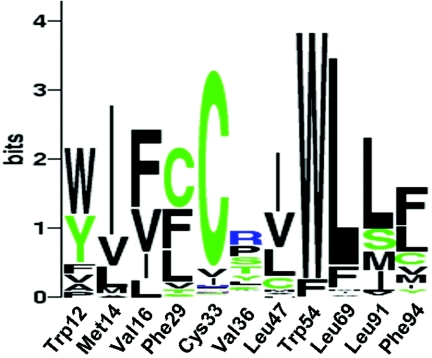
We also noted that, of the eight amino acid residues in contact with Phe94, all are hydrophobic and four are highly conserved in crystalline sequences (Trp12, Cys33, Trp54, and Leu91), with the remaining four partially conserved but always replaced by other hydrophobic residues (fig. 4). This is an additional indication that packing of hydrophobic side chains in this region is important for stability.
Figure 4. .
Sequence logo representation (WebLogo) of the conservation pattern at specific residue positions in CRYBA4. The logo represents the position-specific information content (conservation level), in bits, of the multiple sequence alignments computed using BLAST CRYBA4 sequences. The indicated residue positions follow the numberings of the CRYBA4 sequence. The total height of each column is proportional to the information content (conservation) of the corresponding position, and the height of a letter in a column (representing a given amino acid, using the one-letter code) is proportional to its frequency at that position.33,34
The role of CRYBA4 in eye development was also assessed, because of previous documentation of homozygous changes in CRYBB2 that were associated with severe microphthalmia (small eye) and cataract18 and the interaction of CRYBB2-CRYBA4 monomers.19 In addition, we reported a mutation in CRYBB1 associated with a microcornea cataract phenotype.20 Mutational analysis of 32 patients affected with bilateral microphthalmia, performed using a combination of single-strand conformation polymorphism analysis (SSCP) and direct sequencing, identified several novel sequence changes in the CRYBA4 intron and coding sequence (table 2). The heterozygous c.242T→C transition (Leu69Pro) seen in exon 4 is predicted to be pathogenic. The index case was given a diagnosis of bilateral microphthalmia and cataracts with secondary enophthalmia (sinking of the eyes). No family member was available for clinical assessment. There was no family history of any eye anomaly, suggesting that this change is a de novo mutation. Leu69Pro is highly conserved between species (fig. 2) and is not seen in 368 chromosomes from healthy control individuals. Protein-structure modeling predicts that the change of Leu69 to Pro, in the middle of the β-sheet (fig. 3A and 3C), would likely break the β-sheet. The mutant structure would be very unstable and would not fold appropriately. A 142G→A (Val36Met) sequence change observed in exon 4 was also present in 4 of 320 chromosomes from healthy control individuals (minor-allele frequency 1%). Although this change also involves a highly conserved amino acid, structure modeling suggested that the mutant protein structure maintains a stable conformation with predicted normal folding (fig. 3A). A number of other novel intronic sequence changes were discovered (table 2) and are presumed to be nonpathogenic, because of their presence in healthy controls.
Table 2. .
CRYBA4 Novel Sequence Variants
| No. of Individuals (MAFb) |
||||
| Sequence Change/ Positiona |
Protein Effect | Patients with Microphthalmia |
Healthy Controls |
Screening Methodology |
| c.142 G→A | Val36Met | 1/32 (.02) | 4/160 (.01) | NlaIII |
| IVS3-87, C→T | 2/32 (.03) | 11/56 (.10) | ARMSc assay | |
| IVS3-36, C→T | 2/32 (.03) | 18/118 (.08) | FokI | |
| c.242T→C | Leu69Pro | 1/32 (.02) | 0/184 (.0) | MspI |
| c.317T→Cd | Phe94Ser | 0/32 | 0/239 (.0) | NciI |
| IVS4+3e, A→G | 2/32 (.03) | 20/90 (.1) | ARMS assay | |
| IVS5-18, G→A | 2/32 (.03) | 9/119 (.04) | SSCP/AccI | |
Nucleotide changes are numbered according to NM_001886 (mRNA) or with respect to CRYBA4 intron position on UCSC genome browser May 2004: chr22:25,342,481-5,351,186.
MAF = minor-allele frequency (all heterozygote sequence changes).
ARMS = amplification refractory mutation system.
Family C132 mutation.
This mutation does not significantly change the splice-site score (79.0 wild type, 75.7 mutant), calculated by use of Alex Dong Li's Splice Site Score Calculator.
PCR of cDNA (generated by RT-PCR, with random hexamer primers) from various human tissues (eyes obtained from the local Eye Bank; other tissue RNA from Stratagene) showed that the expression of CRYBA4 is specific to the lens and appears to be higher in the neonatal lens. No expression was detected in retina, cornea, iris, choroid, sclera, or ciliary body (fig. 5). Recent work also suggested that the expression was mostly in lens fiber cells generating the high refractive index and transparency required for lens function.21
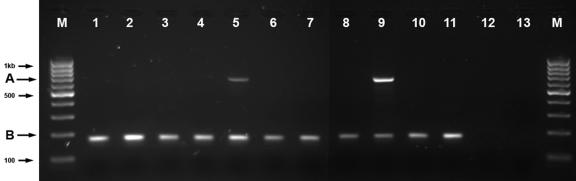
Figure 5. .
Expression profile of CRYBA4 in eye tissues. Lanes 1–7 represent adult eye tissues: 1, sclera; 2, cornea; 3, iris; 4, ciliary body; 5, lens; 6, choroid; 7, retina. Lanes 8–11 are from newborn eye tissues: 8, cornea; 9, lens; 10, choroids; 11, retina. Lane 12 is RT-minus. Lane 13 is no-template control (no cDNA template in the PCR). M, molecular size marker. A duplex PCR was performed on cDNA obtained from different tissues by use of amplification of GAPDH transcript as control for the quality and quantity of RT-PCR templates. Upper band (A) represents the 640-bp PCR product of amplification with CRYBA4-specific primers, exon 2–6 (amplified only in lens tissues), whereas the lower band (B) is the result of amplification with GAPDH-specific primers. Numbers on the left show fragment size in base pairs. Samples 1–7 and 8–11 were amplified at the same time with the same concentration of GAPDH. PCR products were electrophoresed on 2% agarose gels stained with ethidium bromide.
The β-crystallins are major constituents of the mammalian lens, where they associate into dimers, tetramers, and higher oligomers. Appropriate association of crystallins into higher-order complexes is critical to the maintenance of lens transparency.22 The β-crystallin family contains three basic (CRYBB) and four acidic (CRYBA) protein chains, believed to derive from a common ancestor.23 CRYBA4 encodes a predicted 196-aa protein. The conserved human CRYBA4 crystallin domain is 92%–94% identical to rat and bovine βA4. Lampi et al. found that there are 11 major soluble proteins in the young human lens and that βA4 constitutes ∼5% of the total.24
The ability of the normal lens cells to regulate and differentiate proteins in the correct conformation throughout development might rely directly on the ability of accurately translated lens proteins to interact in their properly folded three-dimensional conformation. The temporal expression of crystallin genes vary in development. Different CRYB proteins are found in both prenatal and postnatal developing lens, and their interactions with each other, as well as with other lens proteins, are important in maintenance of lens transparency.25,26 From rat studies, it was shown that the expression of Cryba4 is maximal 2–8 mo after birth, whereas the expression of Crybb2 increases until 6 mo after birth, and the transcript is present in remarkable amounts even 1 year after birth.27 We have shown a higher expression level of CRYBA4 in human neonatal lens versus adult (fig. 5).
Documented varieties of mouse cataract mutants either arose spontaneously or were recovered after parental treatment by chemical mutagen or radiation. However, there is currently no mouse phenotype identified for Cryba4 (Mouse Genome Informatics). To date, the mutations identified in the β-crystallin genes cluster always involved exon 6.5,20,28–32
The present work is, to our knowledge, the first demonstration for the role of CRYBA4 in cataractogenesis and microphthalmia and also the first to show involvement of a highly conserved area of CRYB exon 4. The early expression of CRYBA4 and the predicted protein destabilization by the mutations presented here may explain the early onset of the phenotypes described.
Acknowledgments
The authors are grateful to the families for their enthusiastic participation and to Ms. Yesmino Elia and Erika Bürkle for coordination and technical support. This work was support by grants from the Canadian Institutes for Health Research (CIHR) (MOP-53247, to E.H.), the German Federal Ministry of Research and Technology (BMBF) (IND 03/001, to J.G.), and the Department of Biotechnology, Government of India (project number BT/IN/FRG/JRS/2003–’04, to S.T.S. and P.M.G.). A.D.P. holds a Canada Research Chair in Genetics of Complex Diseases and is funded by Genome Canada. S.W. is CIHR Research Chair in Bioinformatics and Computational Biology and acknowledges support from The Hospital for Sick Children.
Web Resources
The URLs for data presented herein are as follows:
- Alex Dong Li's Splice Site Score Calculator, http://www.genet.sickkids.on.ca/~ali/splicesitescore.html
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/GeneticResearch/compMaps.asp (for Marshfield genetic maps for polymorphic microsatellite genetic locations)
- Centre for Computational Biology, http://www.ccb.sickkids.ca/projIndex.html
- ClustalW, http://www.ebi.ac.uk/clustalw/ (multiple sequence alignment program for DNA or proteins
- Mouse Genome Informatics, http://www.informatics.jax.org/
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
- ProShape, http://nook.cs.ucdavis.edu:8080/~koehl/ProShape/overview.html
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the Human Genome Browser and for physical order and location of markers and SNPs)
- WebLogo, http://weblogo.berkeley.edu/
References
- 1.Hejtmancik JF (1998) The genetics of cataract: our vision becomes clearer. Am J Hum Genet 62:520–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graw J (2004) Congenital hereditary cataracts. Int J Dev Biol 48:1031–1044 10.1387/ijdb.041854jg [DOI] [PubMed] [Google Scholar]
- 3.Wistow GJ, Piatigorsky J (1988) Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem 57:479–504 10.1146/annurev.bi.57.070188.002403 [DOI] [PubMed] [Google Scholar]
- 4.Harding JJ, Crabbe MJC (1984) The lens: development, proteins, metabolism and cataract. In: Davson H (ed) The eye. Vol 1b. Academic Press, London, pp 207–492 [Google Scholar]
- 5.Santhiya ST, Manisastry SM, Rawlley D, Malathi R, Anishetty S, Gopinath PM, Vijayalakshmi P, Namperumalsamy P, Adamski J, Graw J (2004) Mutation analysis of congenital cataracts in Indian families: identification of SNPS and a new causative allele in CRYBB2 gene. Invest Ophthalmol Vis Sci 45:3599–3607 10.1167/iovs.04-0207 [DOI] [PubMed] [Google Scholar]
- 6.McPeek MS, Sun L (2000) Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet 66:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, Wilder K, McPeek MS (2002) Enhanced pedigree error detection. Hum Hered 54:99–110 10.1159/000067666 [DOI] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Montfort RL, Bateman OA, Lubsen NH, Slingsby C (2003) Crystal structure of truncated human βB1-crystallin. Protein Sci 12:2606–2612 10.1110/ps.03265903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48:443–453 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- 11.Claessens M, Van Cutsem E, Lasters I, Wodak S (1989) Modelling the polypeptide backbone with “spare parts” from known protein structures. Protein Eng 2:335–345 [DOI] [PubMed] [Google Scholar]
- 12.EU Biotechnology 3D Validation Project Network (1998) Who checks the checkers? Four validation tools applied to eight atomic resolution structures. J Mol Biol 276:417–436 10.1006/jmbi.1997.1526 [DOI] [PubMed] [Google Scholar]
- 13.Nozaki Y, Tanford C (1971) The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions: establishment of a hydrophobicity scale. J Biol Chem 246:2211–2217 [PubMed] [Google Scholar]
- 14.Miller S, Janin J, Lesk AM, Chothia C (1987) Interior and surface of monomeric proteins. J Mol Biol 196:641–656 10.1016/0022-2836(87)90038-6 [DOI] [PubMed] [Google Scholar]
- 15.Eriksson AE, Baase WA, Wozniak JA, Matthews BW (1992) A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature 355:371–373 10.1038/355371a0 [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Baase WA, Baldwin E, Matthews BW (1998) The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci 7:158–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu L, Liang JJ (2002) Unfolding of human lens recombinant βB2- and γC-crystallins. J Struct Biol 139:191–198 10.1016/S1047-8477(02)00545-2 [DOI] [PubMed] [Google Scholar]
- 18.Kramer P, Yount J, Mitchell T, LaMorticella D, Carrero-Valenzuela R, Lovrien E, Maumenee I, Litt M (1996) A second gene for cerulean cataracts maps to the β crystallin region on chromosome 22. Genomics 35:539–542 10.1006/geno.1996.0395 [DOI] [PubMed] [Google Scholar]
- 19.Cooper PG, Carver JA, Truscott RJ (1993) 1H-NMR spectroscopy of bovine lens β-crystallin: the role of the βB2-crystallin C-terminal extension in aggregation. Eur J Biochem 213:321–328 10.1111/j.1432-1033.1993.tb17765.x [DOI] [PubMed] [Google Scholar]
- 20.Willoughby CE, Shafiq A, Ferrini W, Chan LL, Billingsley G, Priston M, Mok C, Chandna A, Kaye S, Heon E (2005) CRYBB1 mutation associated with congenital cataract and microcornea. Mol Vis 11:587–593 [PubMed] [Google Scholar]
- 21.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A (2004) Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol 86:407–485 10.1016/j.pbiomolbio.2003.11.012 [DOI] [PubMed] [Google Scholar]
- 22.Hejtmancik JF, Wingfield PT, Chambers C, Russell P, Chen HC, Sergeev YV, Hope JN (1997) Association properties of βB2- and βA3-crystallin: ability to form dimers. Protein Eng 10:1347–1352 10.1093/protein/10.11.1347 [DOI] [PubMed] [Google Scholar]
- 23.Piatigorsky J (1987) Gene expression and genetic engineering in the lens. Invest Ophthalmol Vis Sci 28:9–28 [PubMed] [Google Scholar]
- 24.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, David LL (1997) Sequence analysis of βA3, βB3, and βA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem 272:2268–2275 10.1074/jbc.272.4.2268 [DOI] [PubMed] [Google Scholar]
- 25.Bax B, Lapatto R, Nalini V, Driessen H, Lindley PF, Mahadevan D, Blundell TL, Slingsby C (1990) X-ray analysis of βB2-crystallin and evolution of oligomeric lens proteins. Nature 347:776–780 10.1038/347776a0 [DOI] [PubMed] [Google Scholar]
- 26.Norledge BV, Trinkl S, Jaenicke R, Slingsby C (1997) The X-ray structure of a mutant eye lens βB2-crystallin with truncated sequence extensions. Protein Sci 6:1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graw J (1997) The crystallins: genes, proteins and diseases. Biol Chem 378:1331–1348 [PubMed] [Google Scholar]
- 28.Gill D, Klose R, Munier FL, McFadden M, Priston M, Billingsley G, Ducrey N, Schorderet DF, Heon E (2000) Genetic heterogeneity of the Coppock-like cataract: a mutation in CryBB2 on chromosome 22q11.2. Invest Ophthalmol Vis Sci 41:159–165 [PubMed] [Google Scholar]
- 29.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH (1997) Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human β-crystallin gene CRYBB2. Hum Mol Genet 6:665–668 10.1093/hmg/6.5.665 [DOI] [PubMed] [Google Scholar]
- 30.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A (2002) A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet 71:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik JF (2005) Mutations in βB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci 46:2100–2106 10.1167/iovs.04-1481 [DOI] [PubMed] [Google Scholar]
- 32.Yao K, Tang X, Shentu X, Wang K, Rao H, Xia K (2005) Progressive polymorphic congenital cataract caused by a CRYBB2 mutation in a Chinese family. Mol Vis 11:758–763 [PubMed] [Google Scholar]
- 33.Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18:6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]