Abstract
Familial tumoral calcinosis (FTC) is a rare autosomal recessive disorder characterized by the progressive deposition of calcified masses in cutaneous and subcutaneous tissues, which results in painful ulcerative lesions and severe skin and bone infections. Two major types of FTC have been recognized: hyperphosphatemic FTC (HFTC) and normophosphatemic FTC (NFTC). HFTC was recently shown to result from mutations in two different genes: GALNT3, which codes for a glycosyltransferase, and FGF23, which codes for a potent phosphaturic protein. To determine the molecular cause of NFTC, we performed homozygosity mapping in five affected families of Jewish Yemenite origin and mapped NFTC to 7q21-7q21.3. Mutation analysis revealed a homozygous mutation in the SAMD9 gene (K1495E), which was found to segregate with the disease in all families and to interfere with the protein expression. Our data suggest that SAMD9 is involved in the regulation of extraosseous calcification, a process of considerable importance in a wide range of diseases as common as atherosclerosis and autoimmune disorders.
Extraosseous calcification is considered to play a pivotal role in the pathogenesis of a wide range of common disorders, such as atherosclerosis and renal failure.1,2 Recently, the deciphering of the genetic basis of a number of rare monogenic hereditary syndromes has shed light on the molecular mechanisms regulating this process.3
Familial tumoral calcinosis (FTC) is an uncommon life-threatening disorder characterized by massive periarticular—and seldom visceral—deposition of calcified tumors.4 When associated with hyperphosphatemia, the disease is known as “hyperphosphatemic FTC” (HFTC [MIM #211900]) and can result from mutations in at least two genes: GALNT3, which encodes the glycosyltransferase UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl transferase 3 (ppGalNacT3) that is responsible for initiating O-glycosylation, and FGF23, which encodes a potent phosphaturic protein.5–12 Recent data suggest that FGF23 extracellular secretion may depend on ppGalNacT3-mediated O-glycosylation.13 A bone disorder termed “hyperostosis-hyperphosphatemia syndrome” (MIM #211900) was also shown to result from mutations in GALNT3.14 To complicate the picture even further, we identified a subset of patients with FTC who display normal circulating levels of phosphate. These individuals were found not to carry mutations in GALNT3 or FGF23, which suggests that HFTC and normophosphatemic FTC (NFTC) are nonallelic.5 In the present study, we identified a deleterious mutation in SAMD9 that underlies NFTC in five affected families.
All participants or their legal guardians provided written and informed consent, according to a protocol approved by the local Helsinki Committee and by the Committee for Genetic Studies of the Israeli Ministry of Health. Blood samples were drawn from all family members, and DNA was extracted according to standard procedures. Skin biopsy samples were fixed in formaldehyde. Fibroblast cell cultures were established from punch-skin biopsy samples from patients or healthy control individuals, and were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum.
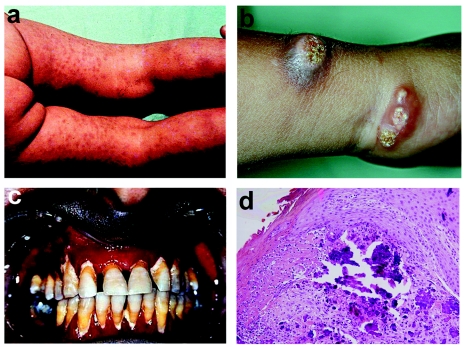
We assessed a total of eight individuals affected with NFTC from five families of Jewish Yemenite origin (fig. 1a). All patients reported reddish-to-hyperpigmented skin lesions during the 1st year of life (fig. 2a), which preceded the later appearance of calcified nodules, distributed mainly over the extremities (fig. 2b). In addition, severe conjunctivitis and gingivitis (fig. 2c) were observed in most of the affected individuals. Results of routine laboratory tests—including calcium, phosphate, vitamin D3 metabolites, and parathyroid-hormone levels—were normal. Histopathological examination of lesional skin biopsies disclosed massive calcium deposition in the mid- and lower dermis (fig. 2d).
Figure 1. .
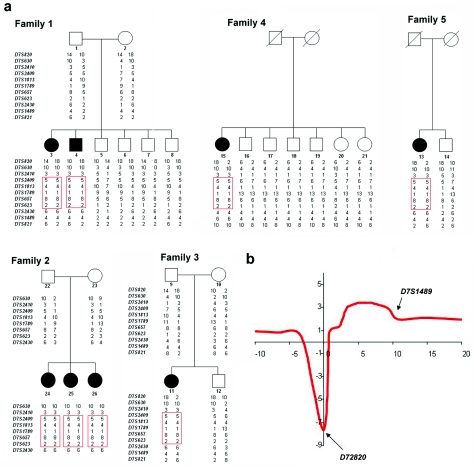
NFTC maps to 7q21-7q21.3. a, Haplotype analysis of five NFTC-affected families, through use of polymorphic markers on chromosome 7q21. Blackened symbols represent affected individuals. The disease-associated haplotype found in the five families is boxed in red. b, Multipoint linkage analysis, which generated a maximum LOD score of 3.4 between markers D7S2409 and D7S623.
Figure 2. .
Clinical features of NFTC—including heralding erythematous papular eruption (a), small calcified tumors ulcerating and discharging chalk-like materials on the surface of the skin (b), and severe gingivitis (c)—and calcified material in the upper dermis, as revealed by histopathological examination (d).
Because all families shared a common ethnic origin and since NFTC is a very rare condition, we postulated the existence of a single homozygous causative mutation and thus used homozygosity mapping to locate the NFTC gene. We performed a genomewide marker screen, using fluorescently labeled primer pairs spanning 731 microsatellite loci (369 markers obtained from PE Biosystems and 362 markers obtained from Research Genetics/Invitrogen) in five affected individuals. We initially identified a 7.6-Mb homozygous interval on 7q21-7q21.3 that was shared by all five patients (not shown). Subsequently, genotypes were obtained for all family members at seven additional microsatellite loci across this region (fig. 1a). Multipoint analysis (with use of Superlink software [Genetic Linkage Analysis Project], as described by Silberstein et al.15) generated a maximum LOD score of 3.4 between markers D7S2409 and D7S623 (fig. 1b). Haplotype analysis revealed a critical recombination event in individual 7 between markers D7S2410 and D7S2409 and a diverging haplotype in patient 15 at marker D7S2430, thereby establishing the centromeric and telomeric boundaries of the disease-associated interval (fig. 1a).
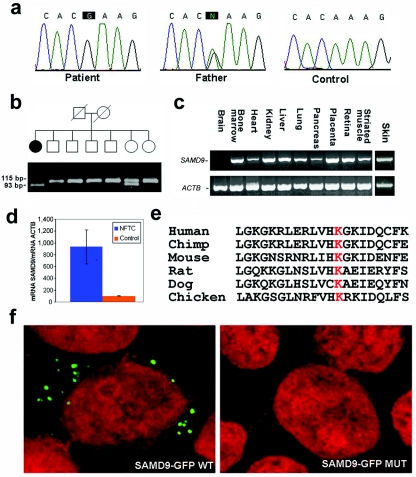
Of 23 genes contained within this interval (Map Viewer [build 36]), we excluded PEX1, since mutations in this gene have been shown to cause peroxismal disorders.16 Twenty-two genes were analyzed for pathogenic mutations by direct sequencing (primer pairs and PCR conditions available on request) of all coding regions, as well as exon-intron boundaries in one or two affected individuals. Only two single-nucleotide substitutions were found to segregate with the disease phenotype in all families. An A→G transition at cDNA position 409 of MGC40405, which codes for a protein of unknown function and results in a valine→methionine substitution, was found in 6% of the general Israeli population (4 of 68 healthy controls). The second mutation is an A→G transition at cDNA position 4483 of SAMD9 (fig. 3a), which is predicted to result in the substitution of a negatively charged glutamic acid residue for a positively charged lysine residue at amino acid position 1495 of the protein sequence. To screen for this mutation, we amplified a 115-bp fragment, using reverse primer 5′-GCAATATAAACATATGCATCGTAC-3′ and allele-specific forward primer 5′-CTTCTTAAAGCACTGGTCAATTTTTTC-3′; the latter creates a recognition site for endonuclease MboII (fig. 3b). Using this assay, we confirmed cosegregation of the mutation with NFTC in all families and excluded the mutation from a panel of 1,042 control chromosomes derived from blood samples obtained from healthy individuals of Jewish origin. We then screened 92 healthy, unrelated Jewish individuals born to couples who immigrated to Israel from Yemen. One individual in this cohort was found to carry both K1495E and the disease haplotype in a heterozygous state, which corresponds to a carrier rate of ∼0.01, fitting the expected prevalence of the disease in the Israeli Jewish Yemenite population.
Figure 3. .
NFTC caused by a mutation in SAMD9. a, Sequence analysis of SAMD9 reveals an A→G transition at cDNA position 4483 in patient 3 (left panel). The father of the patient carries the mutation in a heterozygous state (middle panel). The wild-type sequence is given for comparison (right panel). b, PCR-RFLP analysis, which confirms segregation of the mutation in family 4. PCR amplification was performed as described in the text. Mutation K1495E creates a novel recognition site for MboII. Thus, affected patients display a 93-bp fragment (an additional 22 bp not shown), and healthy individuals show a 115-bp fragment, whereas both fragments are found in heterozygous carriers of the mutation. c, SAMD9 gene expression assessed using Clontech tissue blot cDNA array and using total RNA extracted from skin biopsy samples obtained from healthy individuals. Expression of SAMD9 was compared with that of ACTB. d, RNA was extracted from fibroblast-cell cultures established from two patients with NFTC and two healthy control individuals. SAMD9 mRNA expression, normalized to that of ACTB, was found to be up-regulated in NFTC fibroblasts (blue column) as compared with normal fibroblasts (orange column). e, ClustalW analysis of the SAMD9 protein region encompassing the mutation site, which demonstrates that K1495 is conserved across species. f, HEK293 cells transfected with wild-type (WT) (left panel) and E1495-carrying (right panel) GFP-SAMD9 constructs and examined using confocal microscopy. Note decreased expression in cells transfected with E1495-carrying GFP-SAMD9. MUT = mutant.
SAMD9 encodes a 1,589-aa protein of unknown function and is expressed in a wide range of tissues, including skin (fig. 3c). Quantitative real-time PCR revealed that the gene is strongly expressed in endothelial cells and, to a lesser degree, in fibroblasts (not shown). Moreover, SAMD9 mRNA expression level was found to be sixfold higher in fibroblasts derived from patients with NFTC than in those from healthy controls, which suggests compensatory up-regulation of this gene by a currently unknown mechanism (fig. 3d).
Apart from an N-terminal sterile alpha motif (SAM), a widely distributed putative protein interaction domain,17 SAMD9 bears very little homology to other known proteins, except for a paralogous protein termed “SAMD9L” (SAMD9-like), with which it shares 58% identity at the amino acid–sequence level. K1495 seems to be well conserved across species, suggesting that it is of physiological importance (fig. 3e).
To determine the functional consequences of K1495E mutation in SAMD9, a full-length SAMD9 cDNA was fused to the green fluorescent protein (GFP) sequence in Gateway pDEST53 vector (Invitrogen) and was transfected into HEK293 cells. Confocal microscopy demonstrated clear punctate cytoplasmic expression of the GFP-SAMD9 fusion protein. We then introduced the K1495E mutation into the GFP-SAMD9 cDNA construct, using site-directed mutagenesis (QuikChange JXL Site-Directed Mutagenesis Kit [Stratagene]) according to the manufacturer’s instructions. SAMD9 mRNA levels were shown, by quantitative RT-PCR, to be 4.8-fold higher in HEK293 cells transfected with mutant SAMD9 than in cells transfected with wild-type SAMD9. In seven independent experiments, we observed that K1495E causes loss of punctate expression of the fusion protein after transfection into HEK293 cells, which indicates that this mutation interferes with SAMD9 expression (fig. 3f).
Collectively, the present data indicate that SAMD9 may play a crucial role in the pathogenesis of extraosseous calcification; yet, the detailed mechanisms of action of this protein remain to be determined. The conspicuous inflammatory manifestations reported in NFTC,4 which are absent in HFTC,5 suggest that this protein may be involved in physiological responses to tissue injury.
At this stage, it seems that NFTC represents a clinical entity found specifically in the Jewish Yemenite population. This should not come as a surprise, since it is well known that cultural isolation favors the emergence and clustering of unique genetic alterations, as shown elsewhere for this community.18 In spite of the apparent rarity of NFTC, it encompasses clinical features of relevance to the understanding of important and common disorders. Indeed, two major types of acquired skin calcinosis have been described: (1) metastatic calcinosis, which refers to deposition of calcified materials due to abnormal calcium or phosphate metabolism, as seen in chronic renal failure, and (2) dystrophic calcinosis, in which tissue calcification occurs as a consequence of tissue injury/inflammation, as observed in many unrelated conditions, such as vascular disease, cancer, and autoimmune disorders.19–21 Whereas HFTC models metastatic calcinosis, NFTC bears striking resemblance to acquired dystrophic calcinosis. A growing body of evidence links tissue inflammation to vascular calcification, a major cause of morbidity and mortality in humans.22,23 These observations raise the possibility that alterations in SAMD9 expression may underlie the varying occurrence of dystrophic calcinosis in a spectrum of human diseases.
Acknowledgments
We are grateful to the family members for their enthusiastic, continuous, and generous participation in our study. We thank Vered Friedman, for her help with nucleic acid analysis, and Israel Vlodawski, Dan Kassel, Ami Aronheim, and Mia Horovitz, for providing us with reagents and for stimulating discussions. This study was supported in part by grants from the Israel Science Foundation; the Rappaport Institute for Research in the Medical Sciences, Technion; and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin grant R01 AR052627.
Web Resources
The URLs for data presented herein are as follows:
- Genetic Linkage Analysis Project, http://cbl-fog.cs.technion.ac.il/superlink/ (for Superlink online software)
- Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HFTC and hyperostosis-hyperphosphatemia syndrome)
References
- 1.Atzeni F, Sarzi-Puttini P, Bevilacqua M (2006) Calcium deposition and associated chronic diseases (atherosclerosis, diffuse idiopathic skeletal hyperostosis, and others). Rheum Dis Clin North Am 32:413–426 [DOI] [PubMed] [Google Scholar]
- 2.Jono S, Shioi A, Ikari Y, Nishizawa Y (2006) Vascular calcification in chronic kidney disease. J Bone Miner Metab 24:176–181 10.1007/s00774-005-0668-6 [DOI] [PubMed] [Google Scholar]
- 3.White KE, Larsson TE, Econs MJ (2006) The roles of specific genes implicated as circulating factors involved in normal and disordered phosphate homeostasis: frizzled related protein-4, matrix extracellular phosphoglycoprotein, and fibroblast growth factor 23. Endocr Rev 27:221–241 10.1210/er.2005-0019 [DOI] [PubMed] [Google Scholar]
- 4.Metzker A, Eisenstein B, Oren J, Samuel R (1988) Tumoral calcinosis revisited—common and uncommon features: report of ten cases and review. Eur J Pediatr 147:128–132 10.1007/BF00442209 [DOI] [PubMed] [Google Scholar]
- 5.Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, Zelikovic I, Raimer S, Metzker A, Richard G, Sprecher E (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet 36:579–581 10.1038/ng1358 [DOI] [PubMed] [Google Scholar]
- 6.Benet-Pagès A, Orlik P, Strom TM, Lorenz-Depiereux B (2005) An FGF23 missense mutation causes familial tumoral calcinosis with hyperphosphatemia. Hum Mol Genet 14:385–390 10.1093/hmg/ddi034 [DOI] [PubMed] [Google Scholar]
- 7.Larsson T, Yu X, Davis SI, Draman MS, Mooney SD, Cullen MJ, White KE (2005) A novel recessive mutation in fibroblast growth factor-23 causes familial tumoral calcinosis. J Clin Endocrinol Metab 90:2424–2427 10.1210/jc.2004-2238 [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa S, Lyles KW, Econs MJ (2005) A novel GALNT3 mutation in a pseudoautosomal dominant form of tumoral calcinosis: evidence that the disorder is autosomal recessive. J Clin Endocrinol Metab 90:2420–2423 10.1210/jc.2004-2302 [DOI] [PubMed] [Google Scholar]
- 9.Araya K, Fukumoto S, Backenroth R, Takeuchi Y, Nakayama K, Ito N, Yoshii N, Yamazaki Y, Yamashita T, Silver J, Igarashi T, Fujita T (2005) A novel mutation in fibroblast growth factor 23 gene as a cause of tumoral calcinosis. J Clin Endocrinol Metab 90:5523–5527 10.1210/jc.2005-0301 [DOI] [PubMed] [Google Scholar]
- 10.Chefetz I, Heller R, Galli-Tsinopoulou A, Richard G, Wollnik B, Indelman M, Koerber F, Topaz O, Bergman R, Sprecher E, Schoenau E (2005) A novel homozygous missense mutation in FGF23 causes familial tumoral calcinosis associated with disseminated visceral calcification. Hum Genet 118:261–266 10.1007/s00439-005-0026-8 [DOI] [PubMed] [Google Scholar]
- 11.Campagnoli MF, Pucci A, Garelli E, Carando A, Defilippi C, Lala R, Ingrosso G, Dianzani I, Forni M, Ramenghi U (2006) Familial tumoral calcinosis and testicular microlithiasis associated with a new mutation of GALNT3 in a white family. J Clin Pathol 59:440–442 10.1136/jcp.2005.026369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Specktor P, Cooper JG, Indelman M, Sprecher E (2006) Hyperphosphatemic familial tumoral calcinosis caused by a mutation in GALNT3 in a European kindred. J Hum Genet 51:487–490 10.1007/s10038-006-0377-6 [DOI] [PubMed] [Google Scholar]
- 13.Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H (2006) Polypeptide GaINAc-transferase T3 and familial tumoral calcinosis: secretion of FGF23 requires O-glycosylation. J Biol Chem 281:18370–18377 10.1074/jbc.M602469200 [DOI] [PubMed] [Google Scholar]
- 14.Frishberg Y, Topaz O, Bergman R, Behar D, Fisher D, Gordon D, Richard G, Sprecher E (2005) Identification of a recurrent mutation in GALNT3 demonstrates that hyperostosis-hyperphosphatemia syndrome and familial tumoral calcinosis are allelic disorders. J Mol Med 83:33–38 (erratum 83:240) 10.1007/s00109-004-0610-8 [DOI] [PubMed] [Google Scholar]
- 15.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D (2006) Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet 78:922–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reuber BE, Germain-Lee E, Collins CS, Morrell JC, Ameritunga R, Moser HW, Valle D, Gould SJ (1997) Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat Genet 17:445–448 10.1038/ng1297-445 [DOI] [PubMed] [Google Scholar]
- 17.Schultz J, Ponting CP, Hofmann K, Bork P (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci 6:249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avigad S, Cohen BE, Bauer S, Schwartz G, Frydman M, Woo SL, Niny Y, Shiloh Y (1990) A single origin of phenylketonuria in Yemenite Jews. Nature 344:168–170 10.1038/344168a0 [DOI] [PubMed] [Google Scholar]
- 19.Touart DM, Sau P (1998) Cutaneous deposition diseases. Part II. J Am Acad Dermatol 39:527–544 10.1016/S0190-9622(98)70001-5 [DOI] [PubMed] [Google Scholar]
- 20.Conlin PA, Jimenez-Quintero LP, Rapini RP (2002) Osteomas of the skin revisited: a clinicopathologic review of 74 cases. Am J Dermatopathol 24:479–483 10.1097/00000372-200212000-00004 [DOI] [PubMed] [Google Scholar]
- 21.Boulman N, Slobodin G, Rozenbaum M, Rosner I (2005) Calcinosis in rheumatic diseases. Semin Arthritis Rheum 34:805–812 10.1016/j.semarthrit.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 22.Shao JS, Cai J, Towler DA (2006) Molecular mechanisms of vascular calcification lessons learned from the aorta. Arterioscler Thromb Vasc Biol 26:1423–1430 10.1161/01.ATV.0000220441.42041.20 [DOI] [PubMed] [Google Scholar]
- 23.Stoll G, Bendszus M (2006) Inflammation and atherosclerosis: novel insights into plaque formation and destabilization. Stroke 37:1923–1932 10.1161/01.STR.0000226901.34927.10 [DOI] [PubMed] [Google Scholar]