Abstract
Mutations in genes encoding either components of the phototransduction cascade or proteins presumably involved in signaling from photoreceptors to adjacent second-order neurons have been shown to cause congenital stationary night blindness (CSNB). Sequence alterations in CACNA1F lead to the incomplete type of CSNB (CSNB2), which can be distinguished by standard electroretinography (ERG). CSNB2 is associated with a reduced rod b-wave, a substantially reduced cone a-wave, and a reduced 30-Hz flicker ERG response. CACNA1F encodes the α1-subunit of an L-type Ca2+ channel (Cav1.4α), which is specific to photoreceptors and is present at high density in the synaptic terminals. Ten of our patients with CSNB2 showed no mutation in CACNA1F. To identify the disease-causing mutations, we used a candidate-gene approach. CABP4, a member of the calcium-binding protein (CABP) family, is located in photoreceptor synaptic terminals and is directly associated with the C-terminal domain of the Cav1.4α. Mice lacking either Cabp4 or Cav1.4α display a CSNB2-like phenotype. Here, we report for the first time that mutations in CABP4 lead to autosomal recessive CSNB. Our studies revealed homozygous and compound heterozygous mutations in two families. We also show that these mutations reduce the transcript levels to 30%–40% of those in controls. This suggests that the reduced amount of CABP4 is the reason for the signaling defect in these patients.
Mutations in genes encoding either different components of the phototransduction cascade (RHO [MIM 180380], GNAT1 [MIM 139330], PDE6B [MIM 163500], GRK1 [MIM 180381], and SAG [MIM 181031]) or proteins presumably involved in signaling from photoreceptors to adjacent second-order neurons (CACNA1F [MIM 300110], NYX [MIM 300278], and GRM6 [MIM 604096]) have been shown to cause congenital stationary night blindness (CSNB), a nonprogressive retinal disorder characterized by impaired night vision and other ocular symptoms such as myopia, hyperopia, nystagmus, and reduced visual acuity (VA).1–21 The different forms of CSNB can be classified as shown in figure 1. Only a few mutations have been described for autosomal dominant and recessive forms with fundus abnormalities, whereas most mutations have been identified in X-linked cases. The X-linked inherited CSNB (XLCSNB) can be divided into two types: a complete form (cCSNB), CSNB1 (MIM 310500), and an incomplete form (icCSNB), CSNB2 (MIM 300071). These two subtypes can be diagnosed by electroretinography (ERG) and by molecular means.22,23 CSNB2 is associated with a reduced rod b-wave, a substantially reduced cone a-wave, and a reduced 30-Hz flicker ERG response, whereas the oscillatory potentials are normal. The disease is associated with mutations in the CACNA1F gene.8,9 In contrast, CSNB1 is characterized by a complete loss of rod b-wave and oscillatory potentials and by largely normal cone a-wave amplitudes, and it is caused by mutations in NYX.10,11 Recently, three families were found to carry mutations in GRM6, the gene encoding the metabotopic glutamate receptor 6 (mGluR6). These mutations lead to an autosomal recessive (ar) form of CSNB (arCSNB), whose clinical phenotype is very similar to that of CSNB1, but the scotopic 15-Hz flicker ERG shows a distinct signal in the rod pathway.20
Figure 1. .
The different forms of human CSNB are classified according to their mode of inheritance, phenotype, and mutated genes. ad = Autosomal dominant; ar = autosomal recessive; cCSNB = complete CSNB; icCSNB = incomplete CSNB. Genes are indicated in italics and are underlined. Chromosomal location is given in parentheses.
When routine screening is performed for CSNB for all known genes associated with this disorder, a certain percentage of cases remain unresolved, which indicates the large degree of genetic heterogeneity in CSNB. These unresolved cases prompted us to use a candidate-gene approach, to search for additional factors that may cause the CSNB phenotype. CACNA1F encodes the α1-subunit of an L-type Ca2+ channel (Cav1.4α), which is specific to photoreceptors and is present at high density in the synaptic terminals.24,25 The Ca2+ influx through Cav1.4 channels triggers the continuous release of glutamate from the photoreceptor synapse in the dark.26,27 CABPs are neuronal Ca2+-binding proteins with similarity to calmodulin and have been shown to modulate voltage-dependent Ca2+ channels.28,29 CABP4 (MIM 608965) is one member of this family and, like Cav1.4α, is specifically located in photoreceptor synaptic terminals. It is directly associated with the C-terminal domain of Cav1.4α and shifts the activation of Cav1.4 to hyperpolarized voltages in transfected cells.29 CABP4 consists of 275 aa encoded by six exons of the CABP4 gene (GenBank accession number NM_145200), which is located on chromosome 11q13.1. Mice lacking Cabp4 showed a thinner outer plexiform layer (OPL), which, in wild-type mice, contains the synapses between the photoreceptor and secondary neurons. In addition, Cabp4−/− retinae also revealed ectopic synapses originating from rod bipolar and horizontal cells that extended into the outer nuclear layer (ONL). ERG studies indicated a reduction in cone and rod synaptic function.30 Recently, similar morphological and functional alterations have been observed in the retina of Cacna1f-mutant mice.31 These studies rendered CABP4 an attractive candidate for arCSNB. Here, we report on the identification of mutations in CABP4 in two families that are indicative for CSNB2, according to clinical examinations.
Subjects and Methods
Patients
Three patients from two unrelated families were investigated clinically and genetically in this study. Genetic studies were also performed in other unaffected family members. Patients and their family members were seen either at the Jules Gonin Eye Hospital in Lausanne, Switzerland, or at the Department for Pathophysiology of Vision and Neuroophthalmology, University Eye Hospital, in Tuebingen, Germany. Written informed consent for molecular genetic testing was obtained from all patients and family members. The study was approved by the ethical committees of the respective institutions. Ophthalmic examination included evaluation of best-corrected VA, kinetic Goldmann perimetry, slit-lamp examination, funduscopy, and ocular coherence tomography; full-field ERG was performed in accordance with International Society for Clinical Electrophysiology of Vision (ISCEV) standards, in both scotopic and photopic conditions.32
Mutation Analysis
We isolated genomic DNA from peripheral blood by standard techniques, using the chemagic Magnetic Separation Module I (Chemagen), the Nucleon extraction kit (Amersham Biosciences), or the manual salting-out procedure.33 All 48 exons of the CACNA1F gene were analyzed for mutations, as described elsewhere.18 Four fragments that cover the six coding exons of CABP4 were PCR amplified with intronic or exonic primers and were sequenced with the appropriate primers (table 1). Buffer conditions for amplification and sequencing of CABP4 were similar to those used for the characterization of GRM6.20 The control panel included >210 alleles from unrelated unaffected individuals.34
Table 1. .
PCR Conditions and Primers Used for the Amplification, Sequencing, and Expression Study of CABP4
| Primer Sequence |
||||
| Primer Name | Forward | Reverse | Annealing Temperature (°C) |
Fragment Size (bp) |
| CABP4_1 | 5′-CCTAGGCTCTCAGCTCTAAG-3′ | 5′-CCAAACCGCAGCAACCCTG-3′ | 60 | 622 |
| CABP4_2_3 | 5′-CCAACATGAGCAGGGGATG-3′ | 5′-GAGAGGCAGGAGCTTGAAC-3′ | 58 | 495 |
| CABP4_4 | 5′-GTGTTTCTTCCTAGGTGCAG-3′ | 5′-GATCTGAACCATCTCTGACC-3′ | 58 | 372 |
| CABP4_5_6 | 5′-AAAAGAGTGGAGCTGGCTGA-3′ | 5′-GATAGAAGTAGTCTGGGGGC-3′ | 60 | 483 |
| RT_CABP4_Exon1_For and RT_CABP4_Exon4_Rev | 5′-GTTGTGACTCCCAAGAGTGA-3′ | 5′-CTCAGCTTTGGGCCTATCAG-3′ | 62 | 524 |
| RT_CABP4_Exon2_For and RT_CABP4_Exon4_Rev | 5′-GAACTGGGCCCCGAGGAG-3′ | 5′-CTCAGCTTTGGGCCTATCAG-3′ | 60 | 224 |
| RT_CABP4_Exon2_For and RT_CABP4_Exon6_Rev | 5′-GAACTGGGCCCCGAGGAG-3′ | 5′-GATAGAAGTAGTCTGGGGGC-3′ | 62 | 581 |
| RT_CSPG2_Exon6_For and RT_CSPG2_Exon9_Rev | 5′-GGTGGTCTACTTGGGGTGAG-3′ | 5′-TCACACTGGTCTCCGCTGTA-3′ | 60 | 208 |
In Silico Analysis
To assess the potential impact of an amino acid substitution on the function of the protein, we used two sequence homology–based programs, Sort Intolerant from Tolerant (SIFT) and Polymorphism Phenotype (PolyPhen). To predict a possible disruption of the calcium-binding EF motif and a posttranslational modification due to the amino acid change or protein elongation, we used ExPASy. Potential splice–donor/acceptor sequence motifs present in reference and mutant genomic sequences were searched by the use of NNSPLICE software35 (Berkeley Drosophilia Genome Project). To predict possible exon-splicing–enhancer binding sites, we used the Web-based tool ESEfinder.
Expression Analysis of CABP4
Venous blood samples from patients, selected family members, and controls were collected in PAXgene tubes. Total RNA was isolated using a commercially available kit (PAXgene Blood RNA kit [Qiagen]) and was processed as described elsewhere.36 Specifically, all blood samples but one were processed within 5 d after storage at ambient temperature. One sample (II-1 from family B) was additionally stored at −20°C after 5 d storage at room temperature, and thawing procedures specified by the manufacturer were followed. For qualitative and quantitative RT-PCR experiments, randomly primed cDNA was generated from 1 μg total RNA with the use of Superscript III (Invitrogen), as well as the High Capacity cDNA Archive kit (Applied Biosystems). RT-PCR experiments were performed using HotStarTaq Polymerase (Qiagen) in the presence of 1.5 mM MgCl2 and Q-solution (Qiagen), with primers in exons 2 and 4 or in exons 2 and 6 for CABP4 and in exons 6 and 9 for CSPG2 (table 1).
Quantitative TaqMan analysis was performed using an assay-on-demand probe (Hs00376966_m1 [Applied Biosystems]) on an ABI 7900 HT sequence detection system. 18S rRNA served as endogenous control.37 For each RNA sample, three independent reverse-transcription reactions were performed, and seven serial dilutions were generated to cover a concentration range of 80–0.62 ng/μl, followed by eight more dilutions ranging from 208 to 0.095 pg/μl. From a control sample, 2-μl aliquots of each dilution step were used to establish a standard curve. For patient and control samples, we chose two different concentrations (40 and 20 ng/μl) to assay CABP4 and one concentration (40 pg/μl) to assay the endogenous control, 18S rRNA, in a reaction volume of 15 μl. Amplification conditions suggested in the standard protocol from the company (Applied Biosystems) were used. Quantification was based on the standard-curve method described elsewhere (User Bulletin #2 [Applied Biosystems]). Together, the three different reverse-transcriptase reactions, each tested with two different dilutions in five separate replicates, represent the bases for the statistical analysis.
RT-PCR products of CABP4 cDNA amplified with primers in exons 2 and 6 were cloned in a commercially available vector (pCR2.1-TOPO [Invitrogen]) and were sequenced with standard M13 primers. The presence of allele-specific transcripts was assessed by sequencing the appropriate RT-PCR products on an ABI PRISM 3100 Genetic Analyzer, with the use of the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) and primers RT_CABP4_Exon1_For and RT_CABP4_Exon4_Rev. Before sequencing, RT-PCR products were purified by the ExoSAP-IT clean-up method (USB Corporation).
Results
Clinical Characteristics
Family A
Family A consists of the index patient (II-5), an affected brother (II-6), four unaffected siblings, and their unaffected parents (fig. 2A). To our knowledge, the parents are not consanguineous. The family history was negative for CSNB. The two patients presented nystagmus and decreased VA in early childhood. In both patients at age 15 years, best-corrected VA was 20/100 in both eyes. The disease course in the index patient (II-5) was stationary for 30 years. Yet, he recently experienced a decrease in VA. Neither patient complained about night blindness, and only patient II-5 had recently developed moderate photophobia accompanied by a mild decrease in VA.
Figure 2. .
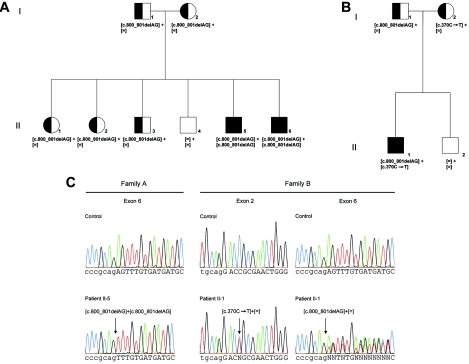
Pedigrees of two families with arCSNB and segregating mutations. A, Pedigree of family A. B, Pedigree of family B. Circles indicate females, squares indicate males, open symbols indicate unaffected individuals, semifilled symbols indicate carriers, and filled symbols indicate affected individuals. Generations and individuals are identified by roman and arabic numerals, respectively. The determined genotype is given below each symbol. C, Electropherograms showing two different CABP4 mutations. Arrows indicate the site of mutation. Patients II-5 and II-6 of family A were homozygous for c.800_801delAG, whereas patient II-1 of family B was compound heterozygous for the missense mutation c.370C→T and for the c.800_801delAG deletion. Lowercase letters indicate intronic sequences, and uppercase letters indicate exonic sequences.
Patient II-5 was 39 years old at the time of this recent examination (in 2005). VA had decreased to 20/200 in both eyes, and the peripheral visual field was moderately constricted (Goldmann kinetic perimetry with size V4e stimulus), with a marked decrease in central sensitivity in both eyes. Results of slit-lamp tests were unremarkable. Results of fundus examination were noticeable for an absent foveal reflex but were otherwise normal. Autofluorescence of the macula was absent, and results of fluorescein angiography were unremarkable. Results of full-field scotopic ERG were markedly abnormal (fig. 3). With the ISCEV standard flash attenuated by 24 dB, the rod b-wave amplitude was moderately reduced, with a marked prolongation of the implicit time (fig. 3A). With the use of the unattenuated ISCEV standard flash, the amplitude and implicit time of the a-wave were normal, but the b-wave remained “negative,” never reaching baseline (b/a–wave ratio 0.5; normal >1.4) (fig. 3B). Under photopic conditions, the b-wave amplitude was moderately to severely reduced, with a slightly delayed implicit time (fig. 3C). Photopic 30-Hz flicker stimulus revealed a decreased amplitude with normal implicit time in both patients (fig. 3D).
Figure 3. .
ERG profile of the patients with CSNB investigated. A–D, Full-field ERG of family A. Results of an unaffected representative subject are shown in the left column. Results from patients II-5 and II-6 are shown in the middle and right columns, respectively. The upper line represents the right eye, the lower line the left eye. A, Scotopic ISCEV standard flash (2.5 candelas [cd]/m2) attenuated by 24 dB. Moderately decreased amplitude and delayed culmination time of the b-wave were present in both patients. B, Scotopic standard flash. Normal a-wave amplitude and implicit time with an absent b-wave (negative ERG) was present in both patients. C, Photopic ISCEV standard flash (30 cd/m2). Decreased amplitude of both a- and b-waves with normal implicit times was present in both patients. D, Photopic 30-Hz flicker stimulus. Decreased b-wave amplitude with normal implicit time was present in both patients. E, Full-field ERG of index patient of family B. OD = right eye; OS = left eye. Top panel, Scotopic ERG. Uppermost trace indicates ERG elicited by ISCEV standard flash (2.5 cd/m2) attenuated by 24 dB, with highly decreased amplitude and delayed culmination time of the b-wave (3 and 9 ms above upper norm for OD and OS, respectively). Middle trace indicates unattenuated scotopic ISCEV standard flash, with normal a-wave amplitude and implicit time with an absent b-wave (negative ERG). Lower trace indicates a repeat of the same conditions as used in the second trace. Bottom panel, Photopic ERG. Upper trace indicates white background light (30 cd/m2) with ISCEV standard flash stimulus and decreased amplitude of both a- and b-waves with delayed implicit time 3 ms above upper norm value; the wavelets toward the end of this trace are blink artifacts in both eyes. Lower trace indicates photopic 30-Hz flicker stimulus with highly decreased amplitude and normal implicit time.
Patient II-6 was 45 years old when recently examined (in 2005). His clinical presentation was similar to that of his younger brother, but without a recent onset of photophobia. VA was 20/200 in the right eye and 20/400 in the left eye. Peripheral visual fields (Goldmann kinetic perimetry with size V4e stimulus) were preserved, with a marked decrease in central sensitivity. Results of slit-lamp and fundus examinations were unremarkable. Results of full-field ERG (fig. 3) were slightly better (i.e., bigger amplitudes and less-delayed implicit times) than those of his younger brother.
Family B
Family B consists of the affected index patient (II-1), his unaffected brother, and the unaffected parents (fig. 2B). The index patient was investigated at age 15 years, to clarify the progressive reduction of his VA. He reported reduced VA and night blindness. The family history was negative for CSNB. Besides microstrabismus divergens, there was no abnormality of eye movements. VA was decreased to 20/30 in both eyes. Automated static perimetry (30°) revealed only very mild relative defects throughout the visual field with normal central thresholds. Results of slit-lamp examination were unremarkable. The fundus showed no pathological findings. Macula reflexes were present. Dark-adaptation threshold after 30 min was elevated by 1 log unit. The panel D15 vision test under saturated conditions resulted in confusions on the tetartan axis in the right eye, whereas no confusions were found in the left eye. Under desaturated conditions, both eyes exhibited numerous confusions in the scotopic axis. Full-field ERG revealed anomalies of both rod and cone systems (fig. 3E). The rod b-wave amplitude, elicited by a white ISCEV standard flash attenuated by 24 dB, was <20% of the normal mean, with slightly prolonged latencies. The maximum response, elicited by the ISCEV standard flash, was dominated by a normal a-wave amplitude and implicit time with markedly reduced b-wave (b/a–wave ratio <0.6; normal values >1.4) (fig. 3E). The oscillatory potentials could not be separated. The single flash cone b-wave response was just outside the lower-norm amplitude, with markedly prolonged implicit time. The 30-Hz flicker ERG amplitude was only ∼10% of the normal mean (fig. 3E). This pattern of ERG is typical for CSNB in general, with a well-developed a-wave but a minimal b-wave. Multifocal ERG studies in this patient revealed that the responses were recordable in all rings up to 40° excentricity, yet amplitudes were markedly reduced to ∼50% of normal values.
Mutation Screening and Expression Analysis
By clinical examination, patients in family A were categorized as suffering from CSNB2 (fig. 2A). The index patient (II-5) and one brother (II-6) showed a phenotype typically observed in patients carrying mutations in CACNA1F (fig. 3A–3D). However, mutation screening was negative for this gene. On the basis of its cellular colocalization and functional interaction with the Cav1.4α channel, we considered CABP4 a good candidate gene.
We screened genomic DNA from all family members by DNA sequence analysis of the ORF of the six exons and flanking intronic sequences of CABP4. In the two affected male patients, a homozygous deletion (c.800_801delAG) of two nucleotides was identified (for transcript analysis, see below) (family A in fig. 2C). Two potential consequences of this c.800_801delAG mutation can be envisioned: (i) a translational frameshift, p.Glu267fs, that gives rise to an elongated protein with 91 additional novel amino acid residues (fig. 4) and (ii) an effect on the splicing efficiency.
Figure 4. .
Sequence alignment of normal CABP4 protein and the two mutant variants. Gray boxes indicate the four predicted Ca2+-binding EF hand motifs; however, EF2 was predicted to be nonfunctional.29 Red letters show the mutated amino acids and the elongated residues. The missense mutation is marked by an arrow. Bold letters highlight known and predicted phosphorylation sites.
Segregation analysis in family A showed that the unaffected parents (I-1 and I-2), two unaffected sisters (II-1 and II-2), and one unaffected brother (II-3) were heterozygous for this mutation, whereas the second unaffected brother (II-4) showed two normal alleles (fig. 2A). None of 216 control alleles showed this deletion.
These results prompted us to analyze the DNA of 32 additional patients with CSNB: 14 patients with uncertain CSNB-type assignment and 18 patients with CSNB2, of whom only 9 had been excluded for mutations in CACNA1F (data not shown). One patient (II-1 from family B) from the group in which CACNA1F had not yet been examined was found to carry two heterozygous alterations in the CABP4 gene (family B in fig. 2B). One allele showed a c.370C→T substitution in exon 2, which may introduce a binding site for SRp55, influencing splicing (Web-based ESEfinder), or may lead to an amino acid exchange (p.Arg124Cys). None of 228 control alleles showed this deletion. The other allele carried the same c.800_801delAG deletion observed in family A (figs. 2B, 2C, and 4). On the basis of conservation of sequence homologies, PolyPhen and SIFT predicted p.Arg124Cys as “functional damaging,” by scores of 2.495 and 0.00, respectively. Results of PolyPhen are reported as “benign” (<0.5), “possibly damaging” or “probably damaging” (0.5–2), “probably damaging” (>2), or “unknown.” Scores depend on the sequences and 3D models available and thus may vary. In SIFT, the results are described as “tolerated” (>0.05) or “affect protein function” (<0.05). None of 210 control alleles showed the c.370C→T substitution. Segregation analysis provided evidence that the father of this patient was heterozygous for the deletion, whereas the mother was heterozygous for the missense mutation. The unaffected brother (II-2) carried no mutation (fig. 2B).
The c.800_801delAG mutation occurred on the same haplotype for two SNPs in intron 3 in both parents of family A. Consequently, a common origin of these alleles is possible: (1) for rs1638564 (c.541+7G→C): I-1, G/G; I-2, G/C; II-1, G/C; II-2, G/C; II-3, G/G; II-4, G/C; II-5, G/G; II-6, G/G; and (2) for rs1638564 (c.541+8A→C): I-1, C/C; I-2, C/A; II-1, C/A; II-2, C/A; II-3, C/C; II-4, C/A; II-5, C/C; II-6, C/C. Family B reported that the father`s family, which carries the c.800_801delAG mutation, immigrated from Switzerland 2 generations previously, which is also where family A originates. Indeed, the c.800_801delAG deletion in family B also occurred on the same haplotype: (1) for rs163864 (c.541+7G→C): I-1, G/C; I-2, G/C; II-1, G/G; II-2, C/C; and (2) for rs1638565 (c.541+8A→C): I-1, C/A; I-2, C/A; II-1, C/C; II-2, A/A. Consequently, a common origin of this mutation in the two families is possible. Thus, we identified mutations in CABP4 in one family with CSNB (family A) previously excluded from the analysis of CACNA1F and in a second family (family B) not yet analyzed.
To exclude the possibility of a CACNA1F mutation in family B, we performed mutation analysis in the index patient (II-1) of this family (fig. 2B). This analysis revealed a hemizygous c.2204A→C (AAC→ACC) transversion in exon 16, which is predicted to lead to a p.Asn735Thr amino acid substitution. PolyPhen and SIFT predicted this sequence variant as possibly damaging, by scores of 1.35 and 0.01, respectively. None of 242 control alleles showed this substitution. However, the unaffected brother (II-2) also showed this sequence variant. As expected for an X-linked mode of inheritance, the mother was heterozygous for this substitution, whereas the father did not carry this sequence variation (data not shown). Thus, we conclude that this CACNA1F sequence variant is not itself disease causing but that it may modify the phenotype. Alternatively, the CACNA1F substitution represents a rare polymorphism not yet described.
To determine the underlying pathogenic effect of the deletion mutation, we investigated two alternatives. The wild-type sequence of CABP4 comprises a duplicate AG dinucleotide at the junction of intron 5 and exon 6: -tctctcccgcagAGTTTGTGAT. The 2-bp–deletion mutation removes one of these AG dinucleotides and, thus, might cause a translational frameshift and/or affect transcript splicing by altering the acceptor site. To distinguish these two possibilities, we performed a computational search for potential splice–donor/acceptor sites within reasonable vicinity of the mutation. The normal splice acceptor -tctctcccgcagAGTTTGTGAT was given a score of 0.98, whereas, for the splice acceptor with a deletion of one AG, -tctctcccgcAGTTTGTGAT, a slightly reduced score of 0.90 was calculated (score 0.0–1.0 reflects increasing splice-site efficiency). We interpreted these findings to mean that the deletion of one of the two tandem AG nucleotides may not dramatically alter splicing efficiency. To test these predictions, we performed RT-PCR experiments. As reported in the literature, CABP4 transcripts had been detected only in retina.29 Here, we were able to show expression of CABP4 also in peripheral blood, using RT-PCR experiments (fig. 5).
Figure 5. .
CABP4 transcript analysis. A, Structure of the CABP4 gene. Boxed symbols represent exons; lines represent introns. Arrows indicate the position of primers used for RT-PCR experiments; the caret indicates the position of the TaqMan probe; dotted lines indicate two mutations in CABP4, c.370C→T and c.800_801delAG. B–D, Ethidium bromide– and GelRed-stained agarose gels with RT-PCR fragments. B, Primers from CABP4 exons 2 and 6. C, Primers from CABP4 exons 2 and 4. D, Primers from CSPG2 exons 6 and 9. E, Relative expression of CABP4 transcripts from quantitative real-time RT-PCR experiments with the use of TaqMan probes. Threshold-cycle values ranged from 19 to 22 for samples probed with 18S rRNA and from 29 to 33 for samples probed with CABP4. Bars represent CIs; P=.05.
Sequence analyses of cloned RT-PCR products revealed the deletion of the AG dinucleotide in the transcripts of patient II-5 in family A, whereas the AG sequence was present in the control. Analysis of four clones from the RT-PCR product of the mother also showed the presence of the AG dinucleotide. Sequencing of RT-PCR products from patient II-1 of family B confirmed the presence of mutant (T) and wild-type (C) alleles at position c.370.
Primers in exons 2 and 6, where the reverse primer is positioned downstream of the deletion mutation in exon 6, yielded a specific fragment of the expected size (581 bp) from control cDNA templates (fig. 5B), and sequence analysis confirmed its identity (data not shown). A fragment of the same size was also obtained from the cDNA of the unaffected mother (I-2) of family A (fig. 2A), her affected son (II-5) (fig. 2A), and the compound heterozygous patient (II-1) of family B (fig. 2B). Using primers in exons 2 and 4 revealed similar results (fig. 5C). A CSNB-unrelated gene, CSPG2, was used as a control (fig. 5D).
Amplification of only these RT-PCR products indicates that no aberrant splice variants accumulate, although the abundance of patients’ transcripts was extremely low. In the case of the amplicon covering exons 2–6, the expected product was very weak and difficult to visualize (fig. 5B). Taken together, the RT-PCR experiments do not indicate that the homozygous deletion (c.800_801delAG) or compound heterozygous mutation (c.800_801delAG and c.370C→T) causes aberrant splicing of CABP4. To compare the transcript levels in cDNA from an unaffected control, from the mother I-2 and the patient II-5 of family A, and from the patient II-1 of family B, we performed real-time quantitative RT-PCR (TaqMan) experiments with a TaqMan probe spanning exons 2 and 3. In comparison to the control (set to 100%), the heterozygous mother of family A (I-2) (fig. 2A) did not show a statistically significant reduction, although a tendency may be indicative (fig. 5E). In contrast, both affected patients (II-5 from family A and II-1 from family B) showed a statistically significant reduction of 60%–70%, compared with the control (fig. 5E).
Discussion
Ten of our patients with the CSNB2 phenotype do not carry mutations in CACNA1F. To search for molecular defects in these patients, we began to investigate the potential role of other factors that are involved in the signal transduction pathway from photoreceptors to adjacent bipolar cells. CABP4 is one such candidate, since mice lacking functional Cabp4 show a phenotype similar to that observed in patients with CSNB2 and with CACNA1F mutations.
Here, we were able to identify two different mutations (c.800_801delAG and c.370C→T) in the CABP4 gene in two families with a phenotype of CSNB2. Interestingly, the deletion c.800_801delAG segregates in both families. On the basis of haplotype reconstruction and the Swiss ancestry of both families, a common origin of this mutation in all three apparently unrelated individuals is possible.
With RT-PCR experiments, we were able to show that CABP4 is expressed in peripheral blood, although previous studies revealed a retina-specific expression in human and mouse, based on northern blot and RT-PCR analyses, respectively.29 The different results observed may be explained by the low abundance of CABP4 transcript in blood. Detection required large amounts of cDNA, 35× amplification cycles, and loading of the entire volume to an agarose gel. In the previous study, expression of the human gene was analyzed only by northern blot hybridization, a method less sensitive than RT-PCR. The expression of the mouse gene was analyzed with RT-PCR experiments. However, cDNA synthesized from blood RNA was not used as a template, and the expression in human and mouse may also vary.29
RT-PCR experiments revealed that the homozygous deletion still yields spliced transcripts, albeit at reduced abundance in comparison with control samples and the heterozygous unaffected mother. Interestingly, lower transcript levels have also been found in the affected patient of family B, who carries the deletion allele and a missense mutation on the second allele. Recently, it has been shown that missense mutations in Cav1.4α can not only influence the gating properties of the L-type Ca2+ channel but can also lead to a reduced protein level.38 Similarly to this example of Cav1.4α, our data suggest that the substitution c.370C→T in our patient may also lead to reduced levels of CABP4 transcripts. The residual amount, 30%–40%, of CABP4 transcripts may explain CSNB2. Even though the scotopic and photopic b-waves and the 30-Hz flicker ERG are reduced, the remaining CABP4 transcript may also result in a low amount of functional proteins, and, thus, signaling is not completely blocked. A possible explanation for the lower expression may be a reduced splicing efficiency, since the deletion altered the context of the splice-acceptor sequence. Additionally, the amino acid substitution (p.Arg124Cys) may also impact the splice machinery, for example, because of the insertion of a novel exon-splicing–enhancer binding site by this substitution. Discovering whether our observations also apply to the retina will require the investigation of affected human retinae.
The c.800_801delAG mutation is predicted to cause a frameshift, p.Glu267fs, which elongates the protein by 91 novel amino acids (fig. 4). Whereas the first of these novel amino acids affects only the last residue of one of the Ca2+-binding motifs (EF4) and, thus, probably does not interfere with this function, the tertiary structure of the extended protein may impair the Ca2+-binding affinity and capacity. The other mutation, p.Arg124Cys, is located upstream of the first EF motif, and, thus, Ca2+ binding seems to not be affected.
Changes in protein structure can alter the interaction with other proteins, influence their intracellular localization, and modify their enzymatic activity and/or protein stability by diverse mechanisms. Recently, it has been shown that CABP4 interacts with a cytoplasmic fragment of Cav1.4α (aa residues 1445–1983).29 Whether the extended CABP4 protein or the amino acid substitution alters the interaction with Cav1.4α needs to be tested.
Another possible mechanism that may result in unstable CABP4 protein level is posttranslational modifications, like phosphorylation of the extended protein. Five novel potential phosphorylation sites are contained within the protein extension. However, it is very likely that the elongated CABP4 protein gets misfolded and, thus, becomes subject to degradation. In summary, in our attempt to understand the complex regulation of nocturnal vision, we have identified yet another protein, CABP4, that is important in this signal transduction pathway.
In humans and mice, mutations in CABP4 lead to a signaling defect from rods and cones to bipolar cells. The photoreceptor synapses in Cabp4-deficient mice were severely disrupted, as shown by the thinning of the OPL, a reduction in the number of synaptic ribbons and photoreceptor terminals, and the deflation of rod spherules and cone pedicles. The absence of Cabp4 also resulted in the formation of ectopic synapses between rods and rod bipolar or horizontal cells in the ONL. To our knowledge, such changes have not yet been described for the human CSNB type and would be better suited to a progressive retinal disease like a cone-rod dystrophy. The severity of the mouse phenotype may be caused by the different type of the mutations. So far, only three patients have been described as showing mutations in CABP4, and data from additional patients carrying mutations in this gene are needed to get a more detailed picture about the phenotype-genotype correlation. However, similar phenotypic differences in human and mice have been described for patients carrying CACNA1F mutations and for mice lacking Cacna1f.31 Interestingly, patients II-5 and II-6 of family A did not complain about night blindness. The disease course was investigated in the index patient, II-5. After CSNB was diagnosed, the disease was stationary over the following 30 years, after which he experienced decreased VA and photophobia. Hence, at least for this patient, a progressive course of the disease seems possible. This aspect still needs to be followed up for patient II-1 from family B and for other patients with novel CABP4 mutations. Compared with CACNA1F, CABP4 is a small gene that can be screened rapidly for mutations in the DNA of patients with CSNB2 and, hence, represents a valuable candidate in diagnostic approaches. This is also of importance since, to date, patients carrying CACNA1F or CABP4 mutations cannot be clinically distinguished, in contrast to patients carrying NYX and GRM6 mutations.20 Mutations in CABP4 and in additional genes implicated in nocturnal vision will help unravel the complexity of signaling from the photoreceptors to the adjacent second-order neurons.
Acknowledgments
The authors thank the patients and families, for their participation in this study, and Mariana Wittmer, who performed the initial CACNA1F screening. This project was financially supported by VELUX Foundation (Switzerland) grant number 162 (to W.B. and C.Z.), Hartmann Müller Foundation (Switzerland) grant number 1002 (to C.Z.), Swiss National Science Foundation grant numbers 3100-067786 (to W.B.) and 32-111948/1 (to F.M. and D.F.S.), and European Union Sixth Framework Programme Integrated Project EVI-GENORET grant number LSHG-CT-2005–512036 (to E.Z.).
Web Resources
The accession number and URLs for data presented herein are as follows:
- Berkeley Drosophilia Genome Project, http://www.fruitfly.org/seq_tools/splice.html
- ESEfinder, http://rulai.cshl.edu/tools/ESE/
- ExPASy, http://www.expasy.org/tools/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CABP4 [accession number NM_145200])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RHO, GNAT1, PDE6B, GRK1, SAG, CACNA1F, NYX, GRM6, CSNB1, CSNB2, and CABP4)
- PolyPhen, http://www.bork.embl-heidelberg.de/PolyPhen/
- SIFT, http://blocks.fhcrc.org/sift/SIFT.html
References
- 1.Dryja TP, Berson EL, Rao VR, Oprian DD (1993) Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat Genet 4:280–283 10.1038/ng0793-280 [DOI] [PubMed] [Google Scholar]
- 2.Rao VR, Cohen GB, Oprian DD (1994) Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature 367:639–642 10.1038/367639a0 [DOI] [PubMed] [Google Scholar]
- 3.al Jandal N, Farrar GJ, Kiang AS, Humphries MM, Bannon N, Findlay JB, Humphries P, Kenna PF (1999) A novel mutation within the rhodopsin gene (Thr-94-Ile) causing autosomal dominant congenital stationary night blindness. Hum Mutat 13:75–81 [DOI] [PubMed] [Google Scholar]
- 4.Dryja TP, Hahn LB, Reboul T, Arnaud B (1996) Missense mutation in the gene encoding the alpha subunit of rod transducin in the Nougaret form of congenital stationary night blindness. Nat Genet 13:358–360 10.1038/ng0796-358 [DOI] [PubMed] [Google Scholar]
- 5.Gal A, Orth U, Baehr W, Schwinger E, Rosenberg T (1994) Heterozygous missense mutation in the rod cGMP phosphodiesterase beta-subunit gene in autosomal dominant stationary night blindness. Nat Genet 7:64–68 10.1038/ng0594-64 [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto S, Sippel KC, Berson EL, Dryja TP (1997) Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet 15:175–178 10.1038/ng0297-175 [DOI] [PubMed] [Google Scholar]
- 7.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A (1995) A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet 10:360–362 10.1038/ng0795-360 [DOI] [PubMed] [Google Scholar]
- 8.Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, Fishman GA, Mets M, Musarella MA, Boycott KM (1998) Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet 19:264–267 10.1038/947 [DOI] [PubMed] [Google Scholar]
- 9.Strom TM, Nyakatura G, Apfelstedt-Sylla E, Hellebrand H, Lorenz B, Weber BH, Wutz K, Gutwillinger N, Ruther K, Drescher B, Sauer C, Zrenner E, Meitinger T, Rosenthal A, Meindl A (1998) An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet 19:260–263 10.1038/940 [DOI] [PubMed] [Google Scholar]
- 10.Bech-Hansen NT, Naylor MJ, Maybaum TA, Sparkes RL, Koop B, Birch DG, Bergen AA, Prinsen CF, Polomeno RC, Gal A, Drack AV, Musarella MA, Jacobson SG, Young RS, Weleber RG (2000) Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet 26:319–323 10.1038/81619 [DOI] [PubMed] [Google Scholar]
- 11.Pusch CM, Zeitz C, Brandau O, Pesch K, Achatz H, Feil S, Scharfe C, Maurer J, Jacobi FK, Pinckers A, Andreasson S, Hardcastle A, Wissinger B, Berger W, Meindl A (2000) The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat Genet 26:324–327 10.1038/81627 [DOI] [PubMed] [Google Scholar]
- 12.Allen LE, Zito I, Bradshaw K, Patel RJ, Bird AC, Fitzke F, Yates JR, Trump D, Hardcastle AJ, Moore AT (2003) Genotype-phenotype correlation in British families with X linked congenital stationary night blindness. Br J Ophthalmol 87:1413–1420 10.1136/bjo.87.11.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zito I, Allen LE, Patel RJ, Meindl A, Bradshaw K, Yates JR, Bird AC, Erskine L, Cheetham ME, Webster AR, Poopalasundaram S, Moore AT, Trump D, Hardcastle AJ (2003) Mutations in the CACNA1F and NYX genes in British CSNBX families. Hum Mutat 21:169 10.1002/humu.9106 [DOI] [PubMed] [Google Scholar]
- 14.Nakamura M, Ito S, Terasaki H, Miyake Y (2001) Novel CACNA1F mutations in Japanese patients with incomplete congenital stationary night blindness. Invest Ophthalmol Vis Sci 42:1610–1616 [PubMed] [Google Scholar]
- 15.Wutz K, Sauer C, Zrenner E, Lorenz B, Alitalo T, Broghammer M, Hergersberg M, de la CA, Weber BH, Wissinger B, Meindl A, Pusch CM (2002) Thirty distinct CACNA1F mutations in 33 families with incomplete type of XLCSNB and Cacna1f expression profiling in mouse retina. Eur J Hum Genet 10:449–456 10.1038/sj.ejhg.5200828 [DOI] [PubMed] [Google Scholar]
- 16.Boycott KM, Maybaum TA, Naylor MJ, Weleber RG, Robitaille J, Miyake Y, Bergen AA, Pierpont ME, Pearce WG, Bech-Hansen NT (2001) A summary of 20 CACNA1F mutations identified in 36 families with incomplete X-linked congenital stationary night blindness, and characterization of splice variants. Hum Genet 108:91–97 10.1007/s004390100461 [DOI] [PubMed] [Google Scholar]
- 17.Jacobi FK, Hamel CP, Arnaud B, Blin N, Broghammer M, Jacobi PC, Apfelstedt-Sylla E, Pusch CM (2003) A novel CACNA1F mutation in a French family with the incomplete type of X-linked congenital stationary night blindness. Am J Ophthalmol 135:733–736 10.1016/S0002-9394(02)02109-8 [DOI] [PubMed] [Google Scholar]
- 18.Zeitz C, Minotti R, Feil S, Matyas G, Cremers F, Hoyng C, Berger W (2005) Novel mutations in CACNA1F and NYX in Dutch families with X-linked congenital stationary night blindness. Mol Vis 11:179–183 [PubMed] [Google Scholar]
- 19.Dryja TP, McGee TL, Berson EL, Fishman GA, Sandberg MA, Alexander KR, Derlacki DJ, Rajagopalan AS (2005) Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci USA 102:4884–4889 10.1073/pnas.0501233102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeitz C, van Genderen M, Neidhardt J, Luhmann UFO, Hoeben F, Forster U, Wycisk K, Mátyás G, Hoyng CB, Riemslag F, Meire F, Cremers FPM, Berger W (2005) Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15 Hz flicker electroretinogram (ERG). Invest Ophthalmol Vis Sci 46:4328–4335 10.1167/iovs.05-0526 [DOI] [PubMed] [Google Scholar]
- 21.Héon E, Musarella MA (1994) Congenital stationary night blindness: a critical review for molecular approaches. In: Wright AF, Jay M (eds) Molecular genetics of inherited eye disorders. Harwood, Chur, pp 277–301 [Google Scholar]
- 22.Miyake Y, Yagasaki K, Horiguchi M, Kawase Y, Kanda T (1986) Congenital stationary night blindness with negative electroretinogram: a new classification. Arch Ophthalmol 104:1013–1020 [DOI] [PubMed] [Google Scholar]
- 23.Boycott KM, Pearce WG, Musarella MA, Weleber RG, Maybaum TA, Birch DG, Miyake Y, Young RS, Bech-Hansen NT (1998) Evidence for genetic heterogeneity in X-linked congenital stationary night blindness. Am J Hum Genet 62:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes S, Kelly ME (2002) Calcium channels at the photoreceptor synapse. Adv Exp Med Biol 514:465–476 [DOI] [PubMed] [Google Scholar]
- 25.Morgans CW (2001) Localization of the alpha(1F) calcium channel subunit in the rat retina. Invest Ophthalmol Vis Sci 42:2414–2418 [PubMed] [Google Scholar]
- 26.Schmitz Y, Witkovsky P (1997) Dependence of photoreceptor glutamate release on a dihydropyridine-sensitive calcium channel. Neuroscience 78:1209–1216 10.1016/S0306-4522(96)00678-1 [DOI] [PubMed] [Google Scholar]
- 27.Rieke F, Schwartz EA (1994) A cGMP-gated current can control exocytosis at cone synapses. Neuron 13:863–873 10.1016/0896-6273(94)90252-6 [DOI] [PubMed] [Google Scholar]
- 28.Lee A, Westenbroek RE, Haeseleer F, Palczewski K, Scheuer T, Catterall WA (2002) Differential modulation of Ca(v)2.1 channels by calmodulin and Ca2+-binding protein 1. Nat Neurosci 5:210–217 10.1038/nn805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, Lee A, Rieke F, Palczewski K (2004) Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci 7:1079–1087 10.1038/nn1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda T, Lem J, Palczewski K, Haeseleer F (2005) A critical role of CaBP4 in the cone synapse. Invest Ophthalmol Vis Sci 46:4320–4327 10.1167/iovs.05-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansergh F, Orton NC, Vessey JP, Lalonde MR, Stell WK, Tremblay F, Barnes S, Rancourt DE, Bech-Hansen NT (2005) Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet 14:3035–3046 10.1093/hmg/ddi336 [DOI] [PubMed] [Google Scholar]
- 32.Marmor MF, Zrenner E (1998) Standard for clinical electroretinography (1999 update): International Society for Clinical Electrophysiology of Vision. Doc Ophthalmol 97:143–156 10.1023/A:1002016531591 [DOI] [PubMed] [Google Scholar]
- 33.Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins JS, Schwartz CE (2002) Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet 71:1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reese MG, Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in Genie. J Comput Biol 4:311–323 [DOI] [PubMed] [Google Scholar]
- 36.Kloeckener-Gruissem B, Bartholdi D, Abdou M-T, Zimmermann DR, Berger W (2006) Identification of the genetic defect in the original Wagner syndrome family. Mol Vis 12:350-355 [PubMed] [Google Scholar]
- 37.Luhmann UF, Lin J, Acar N, Lammel S, Feil S, Grimm C, Seeliger MW, Hammes HP, Berger W (2005) Role of the Norrie disease pseudoglioma gene in sprouting angiogenesis during development of the retinal vasculature. Invest Ophthalmol Vis Sci 46:3372–3382 10.1167/iovs.05-0174 [DOI] [PubMed] [Google Scholar]
- 38.Hoda JC, Zaghetto F, Singh A, Koschak A, Striessnig J (2006) Effects of congenital stationary night blindness type 2 mutations R508Q and L1364H on Ca1.4 L-type Ca channel function and expression. J Neurochem 96:1648–1658 [DOI] [PubMed] [Google Scholar]