Abstract
The molecular basis of long-term potentiation (LTP), a long-lasting change in synaptic transmission, is of fundamental interest because of its implication in learning. Usually LTP depends on Ca2+ influx through postsynaptic N-methyl-d-aspartate (NMDA)-type glutamate receptors and subsequent activation of Ca2+/calmodulin-dependent protein kinase II (CaMKII). For a molecular understanding of LTP it is crucial to know how CaMKII is localized to its postsynaptic targets because protein kinases often are targeted to their substrates by adapter proteins. Here we show that CaMKII directly binds to the NMDA receptor subunits NR1 and NR2B. Moreover, activation of CaMKIIα by stimulation of NMDA receptors in forebrain slices increase this association. This interaction places CaMKII not only proximal to a major source of Ca2+ influx but also close to α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA)-type glutamate receptors, which become phosphorylated upon stimulation of NMDA receptors in these forebrain slices. Identification of the postsynaptic adapter for CaMKII fills a critical gap in the understanding of LTP because CaMKII-mediated phosphorylation of AMPA receptors is an important step during LTP.
Ca+/calmodulin-dependent protein kinase II (CaMKII) mediates a variety of different cellular responses to Ca2+ influx (1, 2). An important source of Ca2+ influx into neurons is the N-methyl-d-aspartate (NMDA)-type glutamate receptor, which is activated by the excitatory neurotransmitter glutamate (2). NMDA- and α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMPA)-type glutamate receptors are clustered at postsynaptic sites opposing presynaptic neurotransmitter release sites (3, 4). Brief trains of presynaptic high-frequency stimulation efficiently activate NMDA receptors (5), resulting in postsynaptic Ca2+ influx and long-term potentiation (LTP). LTP is a long-lasting increase in neurotransmission thought to represent the physiological correlate of learning and memory (5, 6). The induction of NMDA receptor-dependent LTP requires activation of CaMKII in the postsynaptic neuron (6, 7). CaMKII is enriched at postsynaptic densities (8, 9), where it is well placed for activation by Ca2+ influx through NMDA receptors and subsequent phosphorylation of neighboring AMPA receptors, an event contributing to LTP (7, 10). A kinase anchoring proteins, usually called AKAPs, place cyclic AMP-dependent protein kinase (PKA) next to selected substrates such as AMPA receptors (11), and receptors for activated C kinase (RACKs) are important for subcellular localization of different protein kinase C (PKC) isozymes (12). Crucial information about the subcellular targeting of CaMKII is lacking. NMDA receptors would be ideal postsynaptic adapter sites for CaMKII, where it would have full access to Ca2+ influx through these receptors. Cortical NMDA receptors consist of one or two NR1 and two or three NR2A and 2B subunits whose C termini are intracellular (13–16). We show that CaMKII is directly associated with NR1 and NR2B.
EXPERIMENTAL PROCEDURES
Materials.
Tetrodotoxin, microcystin-LR, KN62, KN93, and GF109203X were purchased from Calbiochem, MK-801 and CPP were purchased from Research Biochemicals, and enhanced chemiluminescence detection kits were purchased from Amersham. [γ-32P]ATP (111 TBq/mmol) was obtained from New England Nuclear, protein A-Sepharose was from Sigma, recombinant CaMKII was from A. R. Means (Duke University, Durham, NC), and the AC3-I peptide as well as anti-CaMKII antibodies were from H. Schulman (Stanford University, CA).
Immunoprecipitation and Immunoblotting.
Crude rat brain membranes were prepared as described (17) and extracted with deoxycholate (1%) at 0–4°C for solubilization of whole NMDA receptor complexes or with 1% SDS at 50°C, followed by dilution with Triton X-100, to obtain dissociated NMDA receptor subunits, (for details see refs. 17 and 18). Immunoprecipitations and immunoblots were performed with the NR1-, NR2A-, NR2B-, GluR1-, and GluR2/3-specific antibodies αNR1, αNR2A, αNR2B, αGluR1, and αGluR2/3, respectively, and with monoclonal mouse antibodies against CaMKIIα and β (anti-CaMKIIα and anti-CaMKIIβ, respectively) as described (17, 18). SDS treatment of either membrane fractions or immunoprecipitated glutamate receptor complexes at 50°C dissociates the receptor complexes and subsequently allows specific immunoprecipitation of individual subunits after neutralization of SDS by adding an excess of Triton X-100 (17, 18). The specificities of the antibodies have been carefully characterized (17–19). Chromatographically purified nonspecific mouse or rabbit IgG antibodies (Zymed) were used for control immunoprecipitations to test for nonspecific antibody interactions.
CaMKII Assays.
Deoxycholate-extracted NMDA receptor complexes, SDS-dissociated NMDA receptor subunits, or Triton X-100-solubilized AMPA receptor complexes were immunoprecipitated and incubated for 30 min at 32°C in 50 μl phosphorylation buffer (50 mM Hepes-NaOH, pH 7.4/10 mM MgCl2) containing 1 μM microcystin-LR, 50 μM unlabeled ATP, and 0.2 μM [γ-32P]ATP (111 TBq/mmol; DuPont/NEN) (for more details see refs. 17 and 20). To activate CaMKII, 1 mM calmodulin and 0.5 mM CaCl2 were included and, in some experiments, exogenous recombinant CaMKIIα (21) was added to a concentration of 0.5 μM unless stated otherwise. After washing of the immunocomplexes, phosphorylated polypeptides were separated by SDS/PAGE, detected by autoradiography, and quantified by PhosphorImager analysis (17, 20). Tryptic two-dimensional phosphopeptide maps were produced as given in ref. 17.
Stimulation of Cortical Rat Brain Slices.
Acute hippocampal slices (0.4 mm) were prepared from 3-week-old Sprague-Dawley rats using a vibratome and pretreated as described (22). After drug treatment, slices were homogenized in 0.4 ml ice-cold 320 mM sucrose/10 mM Tris⋅Cl, pH 7.4/10 mM EDTA/10 mM EGTA by triturating with an insulin syringe. Membranes were collected by ultracentrifugation (50,000 rpm for 30 min, 70.1 Ti-rotor, 4°C) and solubilized in 1% deoxycholate/50 mM Tris⋅Cl, pH 8.0/10 mM EDTA/10 mM EGTA (17). Unsolubilized material was sedimented by ultracentrifugation as before. Supernatants were used immediately for immunoprecipitation (17) using a mixture of anti-CaMKIIα and anti-CaMKIIβ antibodies. For immunoprecipitation of AMPA receptors, slices were solubilized directly in 1% Triton X-100/0.1% SDS/50 mM Tris⋅Cl, pH 7.4/20 mM EDTA/10 mM EGTA and cleared by ultracentrifugation. The protease inhibitors pepstatin A (1 μM), leupeptin (10 μg/ml), aprotinin (10 μg/ml), and Pefabloc (0.2 mM; Boehringer Mannheim) as well as the phosphatase inhibitors p-nitrophenyl phosphate (1 mM) and microcystin-LR (2 μM) were present in all buffers.
Association of CaMKII with Glutathione S-Transferase (GST) Fusion Proteins.
The recombinant GST fusion protein GST-NR2B839–1482 was expressed from pGHEB, a modified pGEX vector (23), in Escherichia coli in parallel with nonchimeric GST or nonrelevant GST fusion proteins carrying the full-length sequence of cyclin G2 (24) or the SH3 domain of Src or Abl (25) for control, as described (18). GST-NR2B839–1482 contains the C-terminal 644 aa of NR2B (residue 839–1482) fused to the C terminus of GST. Plasmids encoding GST-NR2B839–1346, GST-NR2B839–1120, and GST-NR2B1120–1482 fusion proteins were produced by digesting the GST-NR2B839–1482 plasmid with BamHI/SacI, BamHI/Eco47 III, or Eco47 III/HinD III, respectively. The corresponding fragments were treated with T4 DNA polymerase to obtain blunt ends, gel-purified, re-ligated, and transformed into E. coli (Novablue; Novagen). All constructs were confirmed by DNA sequence analysis. E. coli BL21 (Novagen) was used as host for protein expression.
Twenty-microliter resin samples loaded with about 2 μg GST-NR2B fusion proteins or GST (as estimated by SDS/PAGE and subsequent staining with Coomassie brilliant blue) were incubated with 0.5 μM recombinant CaMKIIα in 50 μl of 150 mM NaCl/10 mM Tris⋅Cl, pH 7.4, for 30 min under tilting at 4°C, washed three times with the same buffer and once with 10 mM Tris⋅Cl, pH 7.4, and then extracted with SDS sample buffer for immunoblotting as described (17, 18).
RESULTS AND DISCUSSION
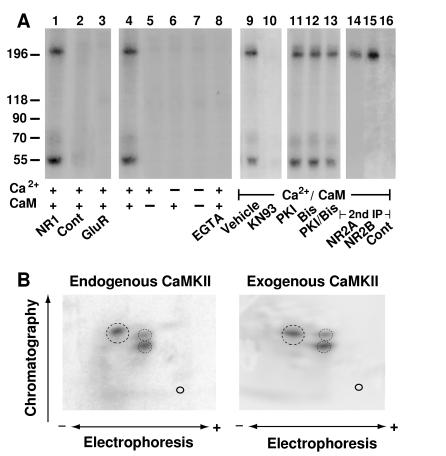
NMDA receptor complexes were solubilized with deoxycholate, immunoprecipitated from rat forebrain, and incubated with [γ-32P]ATP. Two prominent phosphorylated polypeptides were detectable under conditions that activate CaMKII (Fig. 1, lane 1). The endogenous kinase activity required Ca2+ and calmodulin (Fig. 1, lanes 4–8) and was blocked by the CaMKII-selective inhibitor KN93 (Fig. 1, lane 10) but not by a PKA-specific inhibitory PKI peptide (26) or by the bisindolylmaleimide GF109203X, which inhibits multiple PKC isoforms (Fig. 1, lanes 11–13). The efficacy of the two inhibitors for PKA and PKC was confirmed in parallel experiments (M. A. Davare and J.W.H., unpublished results) using the Ca2+ channel α1C subunit as a substrate for purified PKA and PKC, respectively (20). These data indicate that CaMKII is part of the NMDA receptor complex.
Figure 1.
CaMKII activity is associated with NMDA receptors. (A) NMDA receptor complexes were extracted with 1% deoxycholate from crude rat forebrain membrane fractions, and immunoprecipitations were performed with the NR1-specific antibody αNR1 (lanes 1 and 4–16), control mouse IgG (lane 2), or αGluR1 for precipitation of AMPA receptors (lane 3). Immunocomplexes were incubated in phosphorylation buffer containing Ca2+, calmodulin, EGTA (1 mM), KN93 (50 μM), PKI (1 μM), or the bisindolylmaleimide GF109203X (Bis; 1 μM) as indicated. Samples were either subjected directly to SDS/PAGE (lanes 1–13) or dissociated with SDS before a second immunoprecipitation with the antibodies specific for NR2A or 2B or with control rabbit antibodies was performed (lanes 14–16). Phosphorylated polypeptides were detected by autoradiography. Positions of marker proteins are indicated on the left (in kDa). Similar results were obtained in three different experiments. (B) For phosphopeptide mapping of the NR2B subunit phosphorylated by endogenous kinase (Left), NMDA receptors were solubilized, phosphorylated, and dissociated with SDS before reprecipitation of NR2B and SDS/PAGE. To obtain phosphopeptide maps of NR2B phosphorylated by exogenous CaMKII (Right), crude rat forebrain membrane fractions were extracted with SDS. NR2B was immunoprecipitated and phosphorylated with recombinant CaMKII before SDS/PAGE. Of note, no endogenous kinase activity was detectable when NR2B was immunoprecipitated after dissociation with SDS (see Fig. 3A, lane 9). Tryptic two-dimensional phosphopeptide maps were produced in three independent experiments. Main phosphopeptides are circled.
The specificity of the coimmunoprecipitation of the CaMKII activity with NMDA receptors was confirmed with control antibodies (Fig. 1, lane 2). The negative results obtained with the control precipitations exclude the possibility that the phosphorylated polypeptides bound nonspecifically to the immunoprecipitating antibodies or the resin. Because CaMKII is a highly abundant neuronal protein, it is possible that the observed interaction with NMDA receptors may be a result of nonspecific binding of CaMKII to this receptor. Therefore, we carefully evaluated whether CaMKII would also associate with the AMPA receptor complex under our conditions. Like NMDA receptors, AMPA receptors are heteromers formed by GluR1–4 subunits that are homologous to the NMDA receptor subunits (14). However, the phosphorylated polypeptides are not part of or associated with AMPA receptor complexes (Fig. 1, lane 3), further suggesting that the coprecipitation of CaMKII with NMDA receptors is highly specific. Accordingly, CaMKII does not binding to any membrane protein in an indiscriminate fashion.
To evaluate whether the larger 32P-labeled polypeptide was identical to NR2A or 2B, which migrate with an apparent Mr of about 200 kDa in our SDS/PAGE system (17), NMDA receptors were phosphorylated by the endogenous kinase and dissociated with SDS before NR2A and 2B were separately reprecipitated (17). 32P-labeled polypeptides of about 200 kDa were detectable in NR2A and 2B precipitations (Fig. 1, lanes 14–16), demonstrating that both NR2A and NR2B were phosphorylated by the endogenous kinase.
CaMKII is a multimeric complex formed by homologous α and β subunits (Mr = 50 and 60 kDa, respectively) (1). Both subunits undergo rapid autophosphorylation in the presence of Ca2+ and calmodulin, allowing their detection by autoradiography (1). Accordingly, the signal at 55 kDa probably is autophosphorylated CaMKIIα. A phosphorylated polypeptide of 65 kDa also was observed often, suggesting that the β subunit is present in the NMDA receptor complex as well (data not shown). However, the 65-kDa signal was very weak, probably because in the forebrain the β subunit is much less prevalent than the α subunit (1).
To further test whether the endogenous kinase is CaMKII, NR2A and NR2B were phosphorylated either by endogenous kinase or after dissociation with SDS by recombinant exogenous CaMKIIα and analyzed by two-dimensional phosphopeptide mapping. The pattern of the major phosphopeptides derived from NR2A (A.S.L. and J.W.H., data not shown) or NR2B (Fig. 1B) were very similar upon phosphorylation by endogenous or exogenous kinase. These results provide additional evidence for the hypothesis that the endogenous kinase is CaMKII.
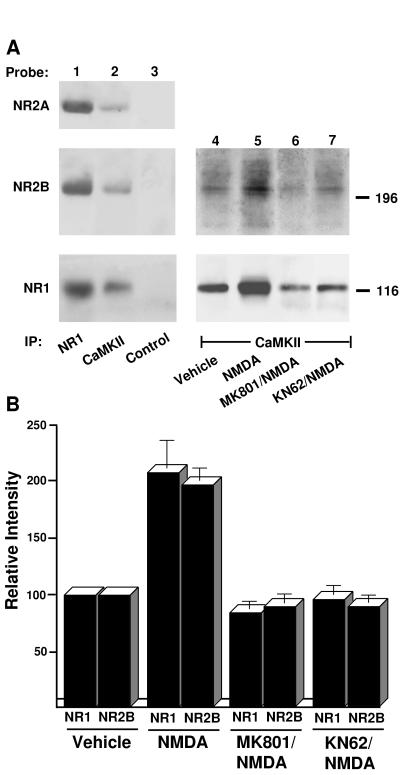
CaMKII complexes were immunoprecipitated from forebrain deoxycholate extracts with anti-CaMKIIα and anti-CaMKIIβ antibodies, which were mixed for higher efficiency. Subsequent immunoblotting with antibodies against NR1, 2A, and 2B subunits demonstrated their presence in CaMKII precipitates but not in control precipitates (Fig. 2A, lanes 1–3), corroborating that CaMKII is associated with NMDA receptor complexes.
Figure 2.
Coimmunoprecipitation of NMDA receptors with CaMKII. (A) To test whether NMDA receptors are associated with CaMKII in vivo, crude membrane fractions were prepared from total rat forebrain and NMDA receptors were solubilized with deoxycholate. After immunoprecipitation with antibodies against NR1, a mixture of the antibodies against CaMKIIα and β (19) or with control mouse IgG, immunoblotting was performed (lanes 1–3) with antibodies against NR2A (Left Top), NR2B (Left Middle), and NR1 (Left Bottom). To measure the effect of NMDA receptor-mediated Ca2+ influx on CaMKII association with NMDA receptors in intact neurons, acute cortical slices were prepared and treated under control conditions (vehicle) or with 200 μM NMDA for 5 min in the absence or presence of 50 μM MK801 or 50 μM KN62. Crude membrane fractions were prepared and solubilized with deoxycholate. Immunoprecipitations were performed with a mixture of the antibodies against CaMKIIα and β (19) before immunoblotting with αNR1 (Right Lower) or a mixture of antibodies against NR2A and NR2B (Right Upper) (lanes 4–7). Similar results were obtained in two other experiments; the results of all three cortical slice experiments were quantified by densitometry of the immunoblotting signals (33) and are summarized in B. Bars = SEM.
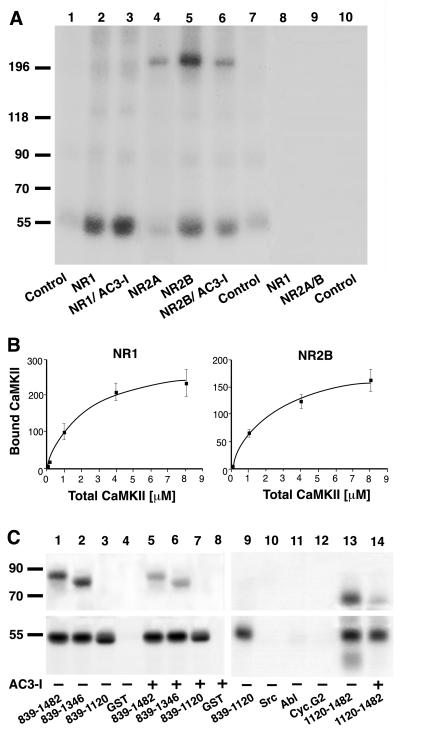
Deoxycholate-solubilized NMDA receptor complexes contain not only various receptor subunits but also structural proteins of the PSD-95 family (18). To investigate whether CaMKII directly interacts with receptor subunits, NMDA receptor complexes were treated with SDS under conditions that completely separate the NMDA receptor subunits from each other and from members of the PSD-95 family (refs. 17 and 18; A.S.L. and J.W.H., unpublished results). Subsequently, NR1, 2A, and 2B subunits were immunoprecipitated individually. Recombinant CaMKIIα was preincubated in [γ-32P]ATP-containing phosphorylation buffer for radioactive labeling by autophosphorylation before the immunocomplexes were added to this mixture. CaMKIIα stably bound to NR1 and NR2B, but not NR2A (Fig. 3A, lanes 2, 4, 5). In similar experiments, CaMKIIα did not coprecipitate with AMPA receptor GluR1 subunits (A.S.L. and J.W.H., data not shown). The finding that CaMKIIα did not form stable complexes with NR2A or GluR1, which possess high degrees of sequence similarities with NR1 and 2B, demonstrates the specificity of the binding of CaMKIIα to NR1 and 2B. Using increasing amounts of CaMKIIα, we observed that binding of CaMKIIα to NR1 as well as to NR2B was saturable (Fig. 3B). NR2A and 2B, but not NR1 (Fig. 3A, lanes 4, 5, and 2, respectively; see also Fig. 1), are phosphorylated under these conditions; however, 32P-labeled NR2A and 2B were absent in NR1 precipitates, further indicating that NR1 and NR2 subunits were efficiently dissociated during the SDS extraction and that they did not reassociate during the immunoprecipitation. When no recombinant CaMKII was added during these experiments, no phosphorylated polypeptide was detectable, arguing that the endogenous CaMKII had been removed completely during dissociation with SDS (Fig. 3A, lanes 8–10).
Figure 3.
CaMKIIα binding to NR1 and NR2B. (A and B) Crude membrane fractions were extracted and NMDA receptor subunits were dissociated with SDS before immunoprecipitation of individual subunits as indicated on the bottom (A) or top (B). Control immunoprecipitations were performed with nonspecific murine (A, lane 1) or rabbit IgG (A, lanes 7 and 10). Recombinant CaMKIIα was preincubated under phosphorylation conditions in the presence of [γ-32P]ATP for labeling by autophosphorylation, and immunocomplexes were incubated with the whole phosphorylation mixture. The signal at 55 kDa (A, lanes 2, 3, 5, and 6) reflects binding of recombinant and autophosphorylated CaMKIIα rather than the presence of the endogenous kinase because no phosphorylation was detectable if recombinant CaMKIIα was omitted (A, lanes 8–10). Phosphorylation of NR2B (A, lane 6) but not association of CaMKIIα with NR1 (A, lane 3) or NR2B (A, lane 6) was inhibited if the competitive catalytic site inhibitor AC3-I (20 μM) was added after autophosphorylation of CaMKIIα (27). Similar results were obtained in two other experiments. Binding of CaMKIIα to NR1 and NR2B was saturable (B; the amounts of CaMKIIα associated with NR1 or NR2B were quantified after SDS/PAGE by using PhosphorImager analysis; values are means ± SEM; n = 3). (C) CaMKIIα was preincubated under phosphorylation conditions with [γ-32P]ATP for labeling by autophosphorylation. Twenty microliters of glutathione Sepharose loaded with approximately 2 μg (as confirmed by staining with Coomassie brilliant blue) of GST or GST fusion proteins (as indicated at the bottom) then was added (together with 20 μM AC3-I when indicated) before samples were washed and analyzed by SDS/PAGE and autoradiog raphy. GST-NR2B839–1482, 839–1346, and 1120–1482, but not 839–1120 or GST alone, were phosphorylated by CaMKII in an AC3-I- sensitive manner (Upper). Autophosphorylated CaMKIIα bound equally well to all four GST-NR2B fusion proteins but not to GST alone or to nonrelevant GST fusion proteins with the Src or Abl SH3 domain or cyclin G2 (Lower).
The synthetic peptide AC3-I is derived from the autoinhibitory domain spanning residues 278–290 of CaMKIIα and inhibits CaMKII by competitively binding to the catalytic domain (27). AC3-I was added to some samples after labeling of CaMKIIα by autophosphorylation and before addition of NMDA receptor subunits to test whether binding of CaMKII to NMDA receptor subunits involved catalytic site interactions. AC3-I did not prevent association of CaMKII with NR1 or NR2B (Fig. 3A, lanes 3, 6). However, it did reduce phosphorylation of NR2B severalfold (Fig. 3A, lane 6), demonstrating that it was an efficient catalytic site inhibitor in our assay. These findings show that CaMKII binding to NR1 as well as to NR2B are not a result of substrate binding to the catalytic site.
In addition to the postsynaptically localized NMDA receptors, a substantial fraction of NMDA receptors is detectable in intracellular organelles such as the endoplasmic reticulum (4). In contrast to mature, postsynaptically localized NMDA receptors, which require ionic detergents such as deoxycholate for solubilization, NR1 subunits can be solubilized efficiently from a microsomal fraction with Triton X-100 (16). We did not observe any CaMKII activity coprecipitating with NR1 subunits after solubilization with Triton X-100 (data not shown). These findings suggest that CaMKII is associated only with mature NMDA receptors present at postsynaptic sites and not with premature receptors found in the endoplasmic reticulum.
Residues 839–1482 form the C-terminal domain of NR2B and constitute the only intracellular portion of substantial size. After labeling by autophosphorylation, recombinant CaMKIIα bound to three different GST fusion proteins carrying residues 839–1482, 839–1346, and 839–1120 equally well but not to GST alone (Fig. 3C Lower, lanes 1–4). Accordingly, residues 839–1120 of NR2B contain a CaMKII-association site. A recent report showed that CaMKII can bind to the NR2B region formed by residues 1260–1309 (28). Serine-1303, which is present in this sequence, is the major phosphorylation site for CaMKII in the C-terminal domain of NR2B in vitro (29). Because of the low KM value of serine-1303 for CaMKII (about 50 nM), CaMKII binding to residues 1260–1309 might have been mediated by serine-1303 binding to the catalytic site. Our shortest fusion protein (839–1120) does not carry this phosphorylation site, nor is it phosphorylated by CaMKII under our conditions (Fig. 3C Upper, lanes 1–4), suggesting that binding of CaMKII to this sequence is not mediated by an interaction between the catalytic site and a substrate site. Furthermore, AC3-I did not affect binding to any of our three GST-NR2B fusion proteins, corroborating that the binding was not due to catalytic site interactions (Fig. 3C Lower, lanes 5–7). Of note, AC3-I did inhibit phosphorylation of GST-NR2B839–1482 and 839-1346 (Fig. 3C Upper, lanes 5 and 6), confirming its efficacy as a catalytic site inhibitor under our conditions. To test whether residues surrounding the serine-1303 phosphorylation site also may contribute to CaMKII binding (28), we made a GST fusion protein containing residues 1120–1482. Interestingly, this fusion protein specifically bound CaMKII as efficiently as 839–1120 (Fig. 3C Lower, lanes 9–13). This interaction was also insensitive to AC3-I (Fig. 3C, lane 14), although AC3-I blocked phosphorylation of GST-NR2B1120–1482. These results suggest that CaMKII binding to 1120–1482 is not mediated by an interaction between the catalytic site of CaMKII and its phosphorylation site at serine-1303. Accordingly, NR2B residues 1260–1309 (28) may contain a second association site for CaMKII close to the serine-1303 phosphorylation site.
Autophosphorylation of CaMKII makes the kinase Ca2+-independent (1) and is important for its binding to residues 1260–1309 (28). To investigate whether binding to 839–1120 can occur independently of autophosphorylation of CaMKII, we incubated GST-NR2B839–1120 with recombinant CaMKIIα in the absence of ATP and Ca2+/calmodulin. After washing of the complex we detected a strong, specific signal for CaMKIIα by immunoblotting with anti-CaMKIIα (A.S.L. and J.W.H., unpublished data). Therefore, association with our binding site does not require autophosphorylation of CaMKII. Accordingly, nonphosphorylated CaMKII can also bind to GST-NR2B839–1482 (A.S.L. and J.W.H., unpublished data). However, because CaMKII has to be autophosphorylated for association with 1260–1309 (28), autophosphorylation increases CaMKII binding to GST-NR2B839–1482 (A.S.L. and J.W.H., unpublished data) and also to a 190-kDa protein in the postsynaptic density, which is most likely NR2B (30).
NMDA receptor-mediated Ca2+ influx and induction of LTP stimulates autophosphorylation of CaMKII (10, 31, 32). Therefore, Ca2+ influx may help to recruit CaMKII to the postsynaptic site by increasing its association with the NMDA receptor. Incubation of rat brain slices with 200 μM of NMDA in the presence of the Na+ channel blocker tetrodotoxin induces NMDA receptor-mediated Ca2+ influx and activation of Ca2+-dependent events at postsynaptic sites without damaging the physiological integrity of the slices (33). Following this protocol, cortical slices were incubated with and without NMDA and crude membrane fractions were prepared and solubilized with deoxycholate. After immunoprecipitation with a mixture of antibodies against CaMKIIα and β, NR1 and 2B were detected by immunoblotting. A strong increase in NR1 and 2B immunoreactivity present in the immunoprecipitated CaMKII complexes is obvious upon stimulation with NMDA (Fig. 2 A, lanes 4 and 5, and B). Pretreatment with the NMDA receptor antagonist MK801 or the CaMKII inhibitor KN62 blocked this NMDA-induced effect (Fig. 2, lanes 7 and 8). Thus, NMDA receptor-mediated Ca2+ influx and subsequent autophosphorylation of CaMKII, which can be inhibited with KN62, strongly increased the association of CaMKII with the NMDA receptor in intact neurons.
Phosphorylation of the AMPA receptor GluR1 subunit by CaMKII plays a central role in LTP that is dependent on Ca2+ influx through NMDA receptors (7, 10). As demonstrated in the previous paragraph, NMDA receptor-mediated Ca2+ influx, which causes autophosphorylation and thereby activation of CaMKII, also results in a strong increase of CaMKII recruitment to postsynaptic sites by its association with NMDA receptors. Because NMDA receptors and AMPA receptors are colocalized, this association may coordinate phosphorylation of AMPA receptors by CaMKII. Therefore, we investigated whether NMDA receptor activation in cortical slices causes CaMKII-dependent phosphorylation of AMPA receptors. All cortical slices were treated in the presence of the bisindolylmaleimide GF109203X to block PKC because serine-831, the main phosphorylation site for CaMKII on the GluR1 subunit, also can be phosphorylated by PKC (34, 35).
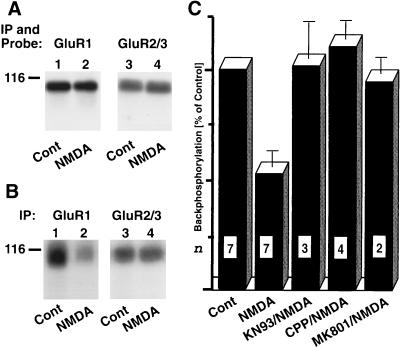
After solubilization, AMPA receptor complexes were immunoprecipitated and analyzed by immunoblotting or back-phosphorylation (22, 36) with recombinant CaMKIIα. Immunoblotting with an antibody against GluR1 and with an antibody that reacts with GluR2 and GluR3 revealed that similar amounts of the respective subunits were present after immunoprecipitation from slices treated either under control conditions or with NMDA (Fig. 4A). After back-phosphorylation, immunoprecipitated AMPA receptor complexes were dissociated with SDS, and GluR1 or a combination of GluR2/3 separately was reprecipitated (18). Incubation with NMDA reduced back-phosphorylation of GluR1 but not GluR2/3 (Fig. 4 B and C). These results show that stimulation with NMDA induces phosphorylation of GluR1 at phosphorylation sites for CaMKII in intact neurons. The effect on back-phosphorylation was inhibited by the NMDA receptor blocker CPP and MK801 and by the CaMKII inhibitor KN93 (Fig. 4C). These findings indicate that NMDA-induced phosphorylation of GluR1 was mediated by NMDA receptor stimulation and activation of CaMKII. CaMKII-mediated phosphorylation of serine-831 in the GluR1 subunit of the AMPA receptor increases its activity (35). Our results indicate that Ca2+ influx through the NMDA receptor can potentiate AMPA receptors by stimulating phosphorylation of GluR1 by CaMKII.
Figure 4.
NMDA receptor activation stimulates phosphorylation of GluR1 by CaMKII. Cortical slices were pretreated with the bisindolylmaleimide GF109203X for 20 min to inhibit PKC-mediated phosphorylation and incubated for 5 min with or without NMDA (200 μM) as indicated (22, 33). After solubilization with Triton X-100, AMPA receptor complexes were immunoprecipitated with a mixture of antibodies against GluR1 and GluR2/3. (A) After dissociated of the immunocomplexes with SDS and reprecipitated with either αGluR1 (lanes 1 and 2) or αGluR2/3 (lanes 3 and 4), immunoblotting was performed with the corresponding antibodies to ensure that equal amounts of GluR1 and GluR2/3 were present in extracts from control and NMDA-treated slices. (B and C) AMPA receptor complexes were back-phosphorylated with recombinant CaMKIIα, dissociated with SDS, and reprecipitated with either GluR1- or GluR2/3-specific antibodies (lanes 1 and 2 and 3 and 4, respectively). After SDS/PAGE, samples were analyzed by autoradiography (B) or quantified by PhosphorImager analysis (C). Some slices were preincubated with 50 μM CPP, MK801, or KN93 before NMDA application. Values are means ± SEM; n is indicated in each column.
In summary, our findings demonstrate that CaMKII directly interacts with NR1 and NR2B. Upon NMDA receptor-mediated Ca2+ influx and subsequent autophosphorylation, binding of CaMKII to NMDA receptors and, thereby, recruitment of CaMKII to the postsynaptic site are increased. This interaction places CaMKII at an ideal location for stimulation by Ca2+ influx through NMDA receptors. It also brings CaMKII in close proximity to AMPA receptors that are phosphorylated and subsequently up-regulated in their activity by CaMKII upon NMDA receptor-mediated Ca2+ influx, especially during LTP (7, 10). Accordingly, NMDA receptors not only are sources for Ca2+ but also serve as the postsynaptic adapter sites for CaMKII. The recruitment of CaMKII by the NMDA receptor is likely to be crucial for the induction of CaMKII-dependent LTP. Our experiments identify the first synapse-specific anchoring protein for CaMKII. It is now possible to identify the exact sequences of the NMDA receptor and CaMKII that mediate the association of CaMKII with NMDA receptor subunits. It then will be feasible to test the physiological relevance of the interaction between CaMKII and NMDA receptor subunits by producing reagents that disrupt that interaction after injection into intact neurons.
Acknowledgments
We thank Dr. R. Jahn, Max Planck Institute for Biophysical Chemistry (Göttingen, Germany), for the antibody 54.2; Dr. R. J. Wenthold, National Institute of Deafness and Other Communication Disorders, National Institutes of Health (Bethesda, MD), for the antibodies against GluR1, GluR2/3, and NR2 subunits; Dr. H. Schulman, Stanford University (Stanford, CA), for the AC3-I peptide and antibodies against CaMKII subunits as well as for many helpful discussions; Dr. A. R. Means, Duke University (Durham, NC), for recombinant CaMKIIα; Dr. C. C. Garner, University of Alabama (Birmingham), for the pGHEB vector encoding GST-NR2B839-1482; Dr. L. M. Graves, University of North Carolina (Chapel Hill), for the PKI peptide; Drs. P. J. Bertics and P. Lipton, both of University of Wisconsin (Madison), for critically reading the manuscript; and Mr. R. J. Creighton and Mr. Brian Haugen for excellent technical assistance. This work was supported by National Institutes of Health Research Grant R01-NS35563 (J.W.H.), American Heart Association Research Grant 97-GS-74 (J.W.H.), a Shaw Scientist Award (J.W.H.), and a grant to the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools.
ABBREVIATIONS
- AMPA
α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- CPP
(±)-3-(2-carboxypiperazine-4-yl)-1-phosphonic acid
- GST
glutathione S-transferase
- LTP
long-term potentiation
- NMDA
N-methyl-d-aspartate
- PKA
cyclic AMP-dependent protein kinase
- PKC
protein kinase C
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Braun A P, Schulman H. Annu Rev Physiol. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh A, Greenberg M E. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 3.Petralia R S, Wenthold R J. J Comp Neurol. 1992;318:329–354. doi: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- 4.Petralia R S, Yokotani N, Wenthold R J. J Neurosci. 1994;14:667–696. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliss T V, Collingridge G L. Nature (London) 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 6.Nicoll R A, Malenka R C. Nature (London) 1995;377:115–118. doi: 10.1038/377115a0. [DOI] [PubMed] [Google Scholar]
- 7.Lisman J, Malenka R C, Nicoll R A, Malinow R. Science. 1997;276:2001–2002. doi: 10.1126/science.276.5321.2001. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy M B, Bennett M K, Erondu N E. Proc Natl Acad Sci USA. 1983;80:7357–7361. doi: 10.1073/pnas.80.23.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly P T, McGuinness T L, Greengard P. Proc Natl Acad Sci USA. 1984;81:945–949. doi: 10.1073/pnas.81.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barria A, Muller D, Derkach V, Griffith L C, Soderling T R. Science. 1997;276:2042–2045. doi: 10.1126/science.276.5321.2042. [DOI] [PubMed] [Google Scholar]
- 11.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 12.Mochly-Rosen D, Gordon A S. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- 13.Seeburg P H. Trends Pharmacol Sci. 1993;14:297–303. doi: 10.1016/0165-6147(93)90047-N. [DOI] [PubMed] [Google Scholar]
- 14.Hollmann M, Heinemann S. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 15.Sheng M, Cummings J, Roldan L A, Jan Y N, Jan L Y. Nature (London) 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 16.Blahos J, II, Wenthold R J. J Biol Chem. 1996;271:15669–15674. doi: 10.1074/jbc.271.26.15669. [DOI] [PubMed] [Google Scholar]
- 17.Leonard A S, Hell J W. J Biol Chem. 1997;272:12107–12115. doi: 10.1074/jbc.272.18.12107. [DOI] [PubMed] [Google Scholar]
- 18.Leonard A S, Davare M A, Horne M C, Garner C C, Hell J W. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- 19.Baitinger C, Alderton J, Poenie M, Schulman H, Steinhardt R A. J Cell Biol. 1990;111:1763–1773. doi: 10.1083/jcb.111.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hell J W, Yokoyama C T, Wong S T, Warner C, Snutch T P, Catterall W A. J Biol Chem. 1993;268:19451–19457. [PubMed] [Google Scholar]
- 21.Cruzalegui F H, Means A R. J Biol Chem. 1993;268:26171–26178. [PubMed] [Google Scholar]
- 22.Hell J W, Yokoyama C T, Breeze L J, Chavkin C, Catterall W A. EMBO J. 1995;14:3036–3044. doi: 10.1002/j.1460-2075.1995.tb07306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller B M, Kistner U, Kindler S, Chung W J, Kuhlendahl S, Fenster S D, Lau L-F, Veh R W, Huganir R L, Gundelfinger E D, Garner C C. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 24.Horne M C, Goolsby G L, Donaldson K L, Tran D, Neubauer M, Wahl A F. J Biol Chem. 1996;271:6050–6061. doi: 10.1074/jbc.271.11.6050. [DOI] [PubMed] [Google Scholar]
- 25.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quilliam L A, Kay B K. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graves L M, Bornfeld K E, Sidhu J S, Argast G M, Raines E W, Ross R, Leslie C C, Krebs E G. J Biol Chem. 1996;271:505–511. doi: 10.1074/jbc.271.1.505. [DOI] [PubMed] [Google Scholar]
- 27.Braun A P, Schulman H. J Physiol. 1995;488:37–55. doi: 10.1113/jphysiol.1995.sp020944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strack S, Colbran R J. J Biol Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- 29.Omkumar R V, Kiely M J, Rosenstein A J, Min K-T, Kennedy M B. J Biol Chem. 1996;271:31670–31678. doi: 10.1074/jbc.271.49.31670. [DOI] [PubMed] [Google Scholar]
- 30.Strack S, Choi S, Lovinger D M, Colbran R J. J Biol Chem. 1997;272:13467–13470. doi: 10.1074/jbc.272.21.13467. [DOI] [PubMed] [Google Scholar]
- 31.Fukunaga K, Muller D, Miyamoto E. J Biol Chem. 1995;270:6119–6124. doi: 10.1074/jbc.270.11.6119. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang Y, Kantor D, Harris K M, Schuman E M, Kennedy M B. J Neurosci. 1997;17:5416–5427. doi: 10.1523/JNEUROSCI.17-14-05416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hell J W, Westenbroek R E, Breeze L J, Wang K K W, Chavkin C, Catterall W A. Proc Natl Acad Sci USA. 1996;93:3362–3367. doi: 10.1073/pnas.93.8.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mammen A L, Kameyama K, Roche K W, Huganir R L. J Biol Chem. 1997;272:32528–32533. doi: 10.1074/jbc.272.51.32528. [DOI] [PubMed] [Google Scholar]
- 35.Barria A, Derkach V, Soderling T. J Biol Chem. 1997;272:32727–32730. doi: 10.1074/jbc.272.52.32727. [DOI] [PubMed] [Google Scholar]
- 36.Forn J, Greengard P. Proc Natl Acad Sci USA. 1978;75:5195–5199. doi: 10.1073/pnas.75.10.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]