Abstract
The effect of life stress on depression is moderated by a repeat length variation in the transcriptional control region of the serotonin transporter gene, which renders carriers of the short variant vulnerable for depression. We investigated the underlying neural mechanisms of these epigenetic processes in individuals with no history of psychopathology by using multimodal magnetic resonance-based imaging (functional, perfusion, and structural), genotyping, and self-reported life stress and rumination. Based on functional MRI and perfusion data, we found support for a model by which life stress interacts with the effect of serotonin transporter genotype on amygdala and hippocampal resting activation, two regions involved in depression and stress. Life stress also differentially affected, as a function of serotonin transporter genotype, functional connectivity of the amygdala and hippocampus with a wide network of other regions, as well as gray matter structural features, and affected individuals' level of rumination. These interactions may constitute a neural mechanism for epigenetic vulnerability toward, or protection against, depression.
Keywords: amygdala, emotion, environment, gene, hippocampus
Adverse life events can reveal profound interindividual differences, rousing resilience in some and exposing susceptibility to mood disorders, including depression, in others. Diathesis-stress models have sought to explain these individual differences in terms of genetic predispositions interacting with environmental factors (1). Behavioral genetic studies have supported these models (2), with current work focusing on molecular and neural mechanism that may underlie these associations.
Dysfunction of the serotonin (5-hydroxytryptophan, 5-HT) system is implicated in mood disorders, and variation within serotonergic genes has been associated with negative emotional traits such as neuroticism and harm avoidance (3). For example, higher scores in these traits are associated with a common short variant of a repetitive sequence in the transcriptional control region of the 5-HT transporter gene (5-HTT, SERT, SLC6A4), which results in low 5-HT uptake function (4). Two metaanalyses have concluded that presence of the short variant of this repeat (5-HTT-linked polymorphic region, 5-HTTLPR) is associated with higher levels of neuroticism or harm avoidance (5, 6). Although neuroticism itself is a risk factor for depression (7), the link between 5-HTTLPR genotype and depression has been more tenuous, suggesting that 5-HTTLPR genotype does not have a consistent main effect on depression but instead may be moderated through other variables (8).
Caspi et al. (9) conducted a 23-year longitudinal study in a large sample of individuals who were genotyped for the 5-HTTLPR. They found that carriers of the 5-HTTLPR short variant showed more depressive symptoms, diagnosed depression, and suicidality as a function of stressful life events than individuals homozygous for the 5-HTTLPR long variant, thus demonstrating a significant gene-by-environment (GxE) interaction. Several replication studies also have reported a moderating effect of 5-HTTLPR genotype on the effect of life stress on depression (10, 11), although some reported an effect only in females (12, 13), and others failed to find any significant GxE interaction (14, 15). It is possible that methodological differences in subject selection account for some inconsistencies across studies or that clinical assessments or measures of self-report are not sensitive enough to reveal a GxE interaction reliably.
We followed an endophenotype approach (16) to investigate a GxE interaction for 5-HTTLPR genotype. The endophenotype approach seeks to capture gene effects by using dependent variables that are more sensitive than clinical assessment or self-report, such as measures of brain activation. Indeed, the effect size of a given genotype can be one order of magnitude greater when it is measured in terms of brain activation than in terms of traditional behavioral measures (17).
Prior work has shown that 5-HTTLPR genotype modulates brain activation in regions associated with affective processing. The first such study by Hariri et al. (18) reported that amygdala activation in healthy participants during a face-matching task was greater in carriers of the 5-HTTLPR short variant than in individuals homozygous for the 5-HTTLPR long variant. This observation later was replicated by the same group (19, 20) and by others in social phobic patients (21). Other studies extended these reports by showing that 5-HTTLPR genotype modulates functional connectivity between the amygdala and prefrontal cortex (22) and that it plays a role in cognitive processes that go beyond reactivity to affective stimuli (23). Based on these studies, we designated the amygdala as an a priori region of interest.
Prior work also highlighted the effect of stress on neural morphology, particularly in the hippocampus (24). Furthermore, abnormal hippocampal volume has been reported in depressed patients (25–29), suggesting that life stress, possibly in interaction with 5-HTTLPR genotype, may affect hippocampal structure (and perhaps function) in healthy individuals. We therefore designated the hippocampus as a second a priori region of interest.
Our primary goal was to evaluate whether these regions of interest showed evidence of a GxE interaction for the 5-HTTLPR. Our secondary goal was to compare two models of 5-HTTLPR function. The “standard” (or phasic activation) model represents the view that presence of the short variant is associated with increased amygdala reactivity to negative emotional stimuli. It is based on several reports that carriers of the short variant show greater activation to negative than neutral stimuli in the amygdala (18–22). Integrating this model with the GxE literature reviewed above, one would expect that short-variant carriers with more life stress experience should exhibit greater amygdala reactivity to negative stimuli than those with less life stress experience. This association would lead to the prediction that short-variant carriers should exhibit a positive correlation between life stress and amygdala activation to negative (and only negative) stimuli compared with a fixation rest condition. We have proposed an “alternative” (or tonic activation) model (23), which posits that presence of the short variant does not enhance amygdala reactivity to emotional stimuli but rather enhances amygdala activation at rest. This model is based on our demonstration that increased amygdala activation in short-variant carriers to negative-neutral stimuli is not driven by increased activation to negative stimuli but instead is driven by decreased activation to neutral stimuli when compared with a fixation rest condition. Integrating our alternative model with the GxE literature reviewed above, one would expect that short-variant carriers with more life stress experience should exhibit greater increases in amygdala activation at rest than those with less life stress experience. This association would lead to the prediction that short-variant carriers should exhibit a negative correlation between life stress and amygdala activation to neutral test stimuli compared with a fixation rest condition.
Results
Interaction of 5-HTTLPR and Life Stress: Effect on Neural Activation.
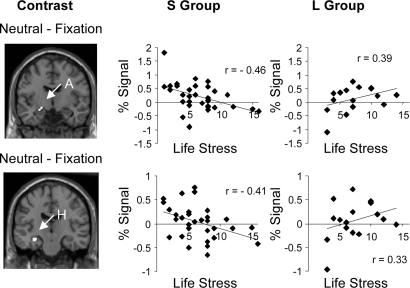
Both a priori regions of interest showed a significant (P < 0.05, familywise-error-corrected) interaction of 5-HTTLPR genotype and life stress (Table 1 and Fig. 1). The pattern of correlations was consistent with our tonic activation model but not with the standard phasic activation model: Compared with a fixation rest condition, activation to face stimuli correlated negatively with life stress in the S group and correlated positively with life stress in the L group. Fig. 1 shows an example from both the amygdala and hippocampus in response to neutral faces, relative to a fixation rest condition (depictions for all significant conditions are available as Figs. 4 and 5, which are published as supporting information on the PNAS web site).
Table 1.
GxE effect on activation
| Condition and loci | Volume, mm3 | Z score | X | Y | Z |
|---|---|---|---|---|---|
| Fear-Fixation | |||||
| Hippocampus | 312 | 2.94 | −32 | −18 | −14 |
| Sad-Fixation | |||||
| Hippocampus | 312 | 2.79 | −32 | −18 | −14 |
| Hippocampus | 200 | 2.81 | 32 | −32 | −6 |
| Happy-Fixation | |||||
| Amygdala | 280 | 2.96 | −22 | −10 | −12 |
| Hippocampus | 408 | 3.11 | −32 | −18 | −14 |
| Neutral-Fixation | |||||
| Amygdala | 296 | 2.91 | −22 | −8 | −12 |
| Hippocampus | 360 | 2.97 | −32 | −18 | −14 |
Negative correlations for S; positive correlations for L.
Fig. 1.
Amygdala and hippocampal activation as a function of 5-HTTLPR genotype and life stress. (Left) Clusters represent voxels in which amygdala (A) and hippocampus (H) activation to neutral faces-fixation varied significantly as an interaction of life stress and 5-HTTLPR genotype. (Center and Right) Columns show scatterplots of percent mean activation across the cluster shown in Left as a function of life stress for the “S Group” (n = 32) and “L Group” (n = 16), respectively.
Recalculation of the data with respect to a potentially functional A/G single-nucleotide polymorphism (SNP rs25531; refs. 30 and 31) within 5-HTTLPR found that the observed interaction may have been driven by the presence of longg carriers in the L group. However, the number of longg carriers (n = 4) was too small to draw any firm conclusions regarding the role of this SNP and, therefore, was not further considered.
Whole-brain analysis identified additional regions in which activation to face stimuli was moderated by a significant (P < 0.005, uncorrected, 20 voxels extent) interaction of 5-HTTLPR genotype and life stress. Among these regions, the left inferior occipital gyrus, a brain region engaged in visual processing, stood out because it showed a significant GxE interaction across all four contrasts. Other regions modulated by the GxE interaction included the superior parietal lobule and superior temporal gyrus, which play a role in imitation and, therefore, may serve as a basis for more complex, socially oriented behaviors (32). A complete listing of these regions is available in Table 2, which is published as supporting information on the PNAS web site.
Interaction of 5-HTTLPR and Life Stress: Effect on Amygdala and Hippocampal Functional Connectivity.
We next evaluated whether the interaction of 5-HTTLPR genotype and life stress modulated functional connectivity between amygdala and hippocampus, based on the clusters listed in Table 1, and other brain regions. These analyses revealed a significant (P < 0.005, uncorrected, 20 voxels extent) GxE effect in a large set of brain regions. For example, a GxE effect altered functional connectivity between hippocampus and putamen, which is of interest because these regions are engaged in separate but interacting memory systems (33). A number of regions believed to represent circuitry required for imitative behavior or its relay to emotion-related systems (32, 34), such as the superior parietal lobule, superior temporal gyrus, precentral gyrus, anterior cingulate, caudate nucleus, and insula, also were moderated by this GxE interaction. A complete listing of these regions is available in Table 3, which is published as supporting information on the PNAS web site.
Interaction of 5-HTTLPR and Life Stress: Effect on Amygdala and Hippocampal Absolute Cerebral Blood Flow (aCBF) at Rest.
The functional MRI (fMRI) data presented above are consistent with our tonic activation model of 5-HTTLPR function. However, a more direct test of the model requires a measure of absolute level of activation at rest. We therefore conducted perfusion scans on 21 subjects to test two predictions made by our model.
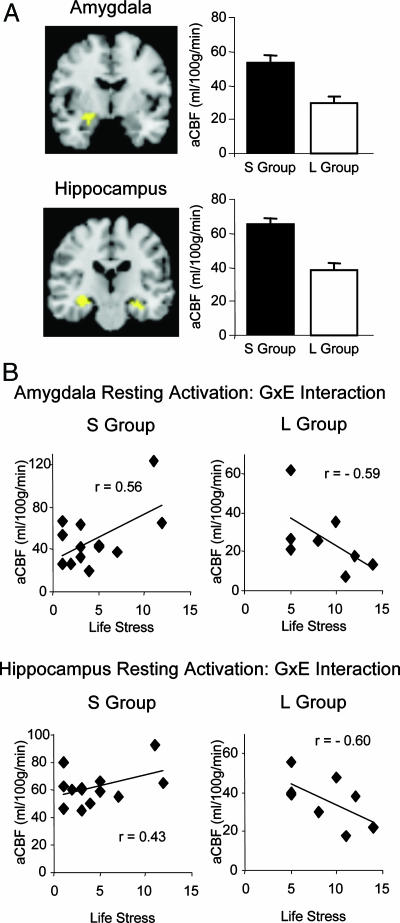
The first prediction was that carriers of the 5-HTTLPR short variant should have higher amygdala activation at rest than noncarriers, based on our previous work (23). This prediction was confirmed in a two-sample t test comparing aCBF between the S and L groups, which found a significant difference in the predicted direction for both the amygdala [cluster 1: 351 mm3, Montreal Neurological Institute (MNI) coordinates: −24, −7, −7; cluster 2: 189 mm3, MNI coordinates: 20, −11, −9; mean activation difference across both clusters: t(19) = −3.24, P < 0.005] and the hippocampus [cluster 1: 1,512 mm3, MNI coordinates: −33, −20, −7; cluster 2: 648 mm3, MNI coordinates: 29, −20, −7; mean activation difference across both clusters: t(19) = −5.28, P < 0.001; Fig. 2A].
Fig. 2.
aCBF in amygdala and hippocampus. (A Left Upper and Lower) Clusters represent voxels where aCBF (measured in ml·100 g−1·min−1) differed significantly between the S Group (n = 13) and L Group (n = 8). (A Right Upper and Lower) Bar graphs show level of activation in amygdala and hippocampus, with error bars depicting SEM. (B) Scatterplots from clusters within amygdala (Upper) and hippocampus (Lower) that showed a significant interaction of life stress and aCBF.
The second prediction was that the interaction of 5-HTTLPR genotype and life stress also should affect the resting activation in the amygdala and hippocampus. Indeed, as shown in Fig. 2B, we observed a significant interaction effect of 5-HTTLPR genotype and life stress on resting level activation in the amygdala (cluster 1: 567 mm3, MNI coordinates: −24, −8 −7; cluster 2: 351 mm3, MNI coordinates: 23, −9, −5; univariate analysis of variance, based on mean of both clusters of GxE interaction: F(1, 17) = 6.49, P = 0.02), and in the hippocampus [cluster 1: 1,782 mm3, MNI coordinates: −33, −20, −7; cluster 2: 540 mm3, MNI coordinates: 29, −20, −9; univariate analysis of variance, based on mean of both clusters of GxE interaction: F(1, 17) = 5.55, P = 0.03].
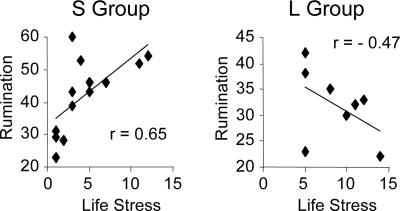
We next investigated the GxE effect on rumination, a form of dysfunctional cognitive reappraisal of negative life events that is a risk factor for depression and anxiety disorders (35). Based on our finding of a significant GxE effect on amygdala resting activation and a prior report associating amygdala activation with trait rumination (36), we predicted a similar GxE effect on self-reported rumination. Indeed, there was a strong GxE interaction [F(1, 17) = 7.38, P < 0.02]): Life stress correlated negatively with rumination for the L group but positively with rumination for the S group (Fig. 3). This pattern is particularly remarkable, given that the S group had experienced significantly fewer life stressors than the L group had [t(19) = −2.67, P < 0.02; mean life stress categories reported: 4.5 vs. 8.8].
Fig. 3.
GxE effect on self-reported rumination. Scatterplots of self-reported rumination as a function of life stress for the S Group (n = 13) and L Group (n = 8), respectively.
Interaction of 5-HTTLPR and Life Stress: Effect on Gray Matter Volume and Concentration.
We (23) and others (20) have described a main effect of 5-HTTLPR genotype on brain structure. Others have documented the effects of stress on brain structural features (24). We therefore conducted a voxel-based morphometry (VBM) analysis on the 48 participants of the fMRI study to evaluate the interaction of 5-HTTLPR genotype and life stress.
There was a significant (P < 0.05, familywise error-corrected) GxE effect on gray matter volume in the left amygdala and hippocampus (amygdala: 353 mm3, maximum P = 0.002, Z = 2.97, MNI coordinates: −18, −5, −26; hippocampus: 261 mm3, maximum P = 0.001, Z = 3.19, MNI coordinates: −25, −10, −27). For both regions, the effect was driven primarily by the L group, which exhibited a positive correlation in gray matter volume with life stress, whereas the S group showed null correlations with life stress.
There was a significant (P < 0.05, familywise error-corrected) GxE effect on gray matter density in the right hippocampus (422 mm3, maximum P = 0.002, Z = 2.93, MNI coordinates: 34, −28, −15). Life stress correlated negatively with gray matter density in the L group and positively in the S group. Thus, measures of gray matter volume and density identified two clusters in the hippocampus that differed in laterality, location within the hippocampus, and direction of the GxE interaction. This variation suggests that the GxE interaction affects gray matter volume and structure through different mechanisms.
There was very little evidence that these GxE effects on amygdala and hippocampal structure can account for their fMRI activation patterns. Activation was not correlated with either gray matter volume or density for the sample overall. The only significant correlation was between hippocampal activation in the L group and gray matter volume (r = .53, P < 0.05), which was limited to the neutral-fixation contrast. These findings suggest that the GxE effect on gray matter structure is not sufficient to account for all observed GxE effects on amygdala and hippocampal activation, although it may account for hippocampal activation in one condition.
Whole-brain analyses identified additional large regions that showed a significant (P < 0.05, corrected at the cluster level) interaction of 5-HTTLPR genotype and life stress. These regions include premotor, motor, and somatosensory regions, as well as the anterior cingulate cortex and caudate nucleus. A complete listing of these regions is available in Table 4, which is published as supporting information on the PNAS web site.
Discussion
Individual differences in 5-HTTLPR genotype are associated with anxiety and depression-related personality traits (3–5) but act as a moderator between life stress and depression (9). We now have begun to identify the neural mechanisms underlying this moderator effect. Prior GxE studies with clinical evaluations and self-report have highlighted the vulnerability of 5-HTTLPR short-variant carriers, creating the impression that carriers homozygous for the long variant are not (or less) affected by life stress. However, our data show that at the neural level of analysis, life stress affects both groups of individuals but in an opposite manner. In our a priori regions of interest, the correlation between life stress and activation to face stimuli (relative to a resting fixation condition) was negative in the S group but positive in the L group. This GxE interaction was significant at corrected levels for the amygdala in response to happy and neutral facial expressions (and near significance at P = 0.59, corrected, in response to fearful and sad facial expressions) and for the hippocampus in response to all facial expressions (all relative to fixation).
This activation pattern is consistent with our tonic activation model but not the standard phasic activation model of 5-HTTLPR function. Specifically, we confirmed our prediction that short-variant carriers should exhibit a negative correlation between life stress and amygdala activation to neutral stimuli compared with a fixation rest condition. We also extended this prediction to the hippocampus, which is our second a priori region of interest. In contrast, the phasic activation model had predicted that short-variant carriers should exhibit a positive correlation between life stress and amygdala activation to negative stimuli compared with a fixation rest condition. We found no evidence for such a positive correlation. Instead, we found a negative correlation that was near corrected significance levels both for the fearful-fixation and sad-fixation contrasts.
The fact that we observed a similar GxE pattern across stimulus conditions is problematic for the phasic, but not for the tonic, activation model. The phasic model is stimulus-dependent, because it explains the association between the 5-HTTLPR short-variant and negative-affective traits (4) in terms of greater amygdala reactivity to negative stimuli. In contrast, the tonic model is stimulus-independent, because it explains the association between the 5-HTTLPR short-variant and negative-affective traits (4) in terms of higher amygdala resting activation, which may engender a chronic state of vigilance, threat, or rumination.
Although the foregoing analysis of our data finds much support for the tonic model, the relative nature of fMRI-based contrast analysis does not allow a definitive conclusion on the validity of these two models. We therefore measured absolute levels of cerebral blood flow by using perfusion imaging. Our perfusion data confirmed that 5-HTTLPR short-variant carriers indeed show elevated levels of amygdala activation at rest compared with carriers who were homozygous for the long variant. Our analysis also revealed a significant GxE effect for amygdala and hippocampus, such that life stress correlated positively with resting activation in the S group and negatively in the L group. Given that amygdala activation is associated with trait rumination (36), we hypothesized and confirmed that life stress correlates positively with rumination in the S group and negatively in the L group. This pattern suggests a mechanism by which genotype differentially affects amygdala-resting activation and rumination levels as a function of life stress. Such a mechanism may imbue L subjects with a protection against, and render S subjects vulnerable toward, mood disorders such as depression (35), although the small perfusion sample size warrants replication.
Both a priori regions of interest also showed evidence for a GxE effect on gray matter structural features. Volume of both the left amygdala and hippocampus increased with greater life stress in the L group but was unaffected by life stress (null correlation) for the S group. Gray matter density was affected by a GxE interaction only in the right hippocampus, where it decreased with life stress in the L group (and increased in the S group). Thus, the interaction of life stress and 5-HTTLPR genotype appears to affect gray matter volume and density differently through mechanisms that are unknown. These complex interactions may explain why prior studies have reported conflicting results relating hippocampal morphology to mood disorders (25–28, 37).
Whole-brain analyses of activation, functional connectivity, and gray matter density and volume revealed additional regions that were moderated by the interaction of 5-HTTLPR genotype and life stress. What is striking regarding these regions is that they belong to a system that mediates imitative behavior, from which social behavior may have evolved (32). Regions involved in imitation, imitative learning or social mirroring (32, 34), and affected by a GxE interaction in this study, include the superior parietal lobule, superior temporal gyrus, inferior frontal gyrus, precentral gyrus, anterior cingulate, striatum (caudate nucleus), insula, and amygdala. Some of these regions contain mirror neurons (38–40), which are activated during goal-directed behavior or the observation of such behavior in others, and Von Economo neurons, which are believed to play a role in social bonding (41). Our study suggests that social behavior may be subject to an interaction of 5-HTTLPR genotype and life stress and that mirror neurons or Von Economo neurons may be the neural substrate of such GxE interactions. Future imaging studies in humans could address whether an interaction of 5-HTTLPR and life stress moderates neural activation during imitation or social processing tasks. Future electrophysiological studies in macaque monkeys could test more directly whether such GxE interactions moderate the behavior of mirror and Von Economo neurons.
In contrast to 5-HTTLPR genotype, which is an objective measure of individual differences, retrospective self-report of life stress history is subjective and less precise. Although we based our measure of life stress events on items included in the life stress calendar method used by Caspi et al. (9), our self-report version did not benefit from the analytical and fact-checking skills of an experienced interviewer nor were data acquired in a longitudinal format. Therefore, we have to be mindful of the possibility that individuals may have differed from one another in their ability to remember events accurately. This concern would be particularly problematic if the measure were based on free recall (which it was not) or when dating or enumerating the number of instances that a particular event occurred (which is information that was not used in the current set of analyses). Instead, we enumerated the number of life stress items (out of a maximum of 28 questionnaire items) that participants endorsed as personal experiences. The sample used in the present study was too small to conduct additional analyses on the role of particular life stressors, because any individual item was endorsed by only a subset of subjects. Increasing the sample size to follow-up with more specialized analyses could address whether chronic stressors have a similar effect as short-term stressors or whether the impact of early childhood stressors differs from that of more recent stressors in adulthood. Because our current sample is too small to take into consideration multiple occurrences of the same type of experience, age at the time of the experience, its recency or duration, or levels of subjectively experienced stress, we regard this enumeration method as a relatively blunt measure of life stress. It is likely that with a significantly larger sample, one could extract additional information regarding neural correlates of epigenesis if these additional variables were taken into account, corroborated by third parties, or if life stress data were collected during the course of a prospective study. As studies begin to refine the methodologies for capturing these epigenetic effects on the brain, we may better understand the mechanisms that render some individuals susceptible and others resilient to depression and other mood disorders.
Materials and Methods
Participants.
Forty-eight healthy adults (mean age = 24.7, SD = 5.6; 26 males) participated in the fMRI and voxel-based morphometry studies, and 21 healthy adults (mean age = 30.4, SD = 8.5; 14 males) participated in the perfusion and rumination studies. Exclusion criteria included history of diagnosed psychopathology, current use of mood-altering medication, substance abuse during the 6 months before the scan, history of severe head trauma, neurosurgery, or neurological condition, pregnancy, or any standard MRI counter indications. All participants gave written informed consent, and the institutional review boards of Stony Brook and Yale universities approved all procedures.
Mood and Personality Measures.
Personality traits of extraversion and neuroticism, and positive and negative mood states were assessed by using standard self-report questionnaires (42, 43). Life stress history was based on a self-report questionnaire developed from items in the life history calendar (44) and contained 28 items related to work, financial and legal problems, death and serious illness, family and relationships, and other stressful life events. Participants checked a box indicating whether (and if so, when or how often) they had ever experienced a particular event. For this current set of analyses, life stress was quantified as the number of categories that were endorsed by each participant (possible range of scores: 0–28). Participants in the perfusion study also completed a rumination questionnaire, the Ruminative Response Scale (45).
Genotyping.
Participants were genotyped according to a protocol published in ref. 4. For all subsequent analyses, the sample was dichotomized according to the presence (s/s and s/l, S group) or absence (l/l, L group) of the 5-HTTLPR short variant. This dichotomy was made a priori, based on the fact that serotonin uptake in human lymphoblastoid cells is comparable for cells that carried either one or two copies of the 5-HTTLPR short variant, whereas cells that were homozygous for the 5-HTTLPR long variant had ≈2-fold higher uptake (4). Within the L group, participants also were genotyped for the presence of a rare A/G single-nucleotide polymorphism (SNP rs25531) that was recently identified in the 5-HTTLPR (31). Demographic information on the S and L groups is available as Supporting Text, which is published as supporting information on the PNAS web site.
fMRI Task Procedures.
Participants viewed four 18-second blocks each of neutral, happy, sad, and fearful face stimuli (6 face stimuli per block), which were presented in semirandom order (each of the four block types was presented before the order was reset and begun again). For each trial, the participant pressed a button indicating whether the face was male or female. Stimuli did not repeat and, thus, there were 24 images per type of facial expression (equal numbers of males and females). Each session began and ended with 30 seconds of fixation cross presentations.
Imaging Procedures.
All participants were scanned in a 3T Siemens Trio scanner located in the Magnetic Resonance Research Center at Yale University.
VBM and fMRI.
High-resolution images for VBM analysis (46) were acquired with a sagittal 3D MPRAGE sequence [inversion time = 1,100, repetition time (TR) = 2,530, echo time = 3.66, flip angle = 7°, matrix = 256 × 256]. For fMRI, 24 functional whole-brain images (5 mm thickness), taken through the plane parallel to the anterior commissurae and posterior commissurae line, were acquired by using a gradient echo T2*-weighted echo-planar imaging scan (TR = 1.5 s; echo time = 30 ms; flip angle = 80°; field of view = 220 × 220 mm).
Perfusion scanning.
Participants were scanned by using the MRI arterial spin labeling perfusion technique to measure the aCBF at rest. We have implemented QUIPSSII (47) with gradient echo-planar imaging for data acquisition. The eight-channel phased-array coil was used for imaging to improve the signal-to-noise ratio. Imaging parameters were as follows: TR, 3,000 ms; echo time, 21 ms; the inversion time for the first slice is 1,400 ms; inversion time 1,700 ms; and delay time (TD) is adjusted to the maximum within each TR; 256 × 192 mm2 field of view, 6.0-mm slice thickness with 2-mm gap, 64 × 48 image matrix, readout bandwidth 2,442 Hz per pixel. Ten anterior commissurae and posterior commissurae-aligned slices were acquired for the top and bottom parts of the brain, respectively. The proton density (PD) weighted image was acquired by using the same sequence as perfusion except that the TR was set to 10 seconds, TD equals 0, and the postinversion recovery time was adjusted to the maximum. The longitudinal relaxation time (T1) mapping was performed with an ultrafast Look-Locker echo-planar imaging T1 mapping sequence (48). Both PD and T1 mapping was performed in the same slice locations as the perfusion weighted imaging, and the PD and T1 maps were used for the aCBF estimation.
Data Analysis.
VBM was conducted according to an optimized VBM protocol (49): age, sex, 5-HTTLPR genotype, life stress, gene-by-life stress, and total gray matter volume were included in the model as covariates to control for confounds. The level of statistical significance was set at P < 0.05, corrected at the cluster level. Perfusion data were analyzed as follows: Functional images were motion corrected by using SPM99. Time series of the perfusion-weighted images were obtained by pairwise “surround” subtraction between interleaved label and control pairs (50–52). The aCBF was calculated from the average perfusion-weighted images for each condition. Other parameters used in cerebral blood flow quantification: T1a, 1,490 ms; απ, 0.90; inversion time, 1,400 ms for the first slice. Difference maps for each individual then were transformed to a reference brain space and for group statistical analysis (53). Analyses of aCBF levels were conducted with customized software and restricted to the hippocampus and amygdala. fMRI data were preprocessed, normalized to the gray matter template, and spatially smoothed with an 8-mm full-width half-maximum isotropic Gaussian filter. Statistical analysis of functional data were conducted in SPM2 and based on fixed-effects models (54) at the individual subject level of analysis and random effects models (55) for group-level analyses. At the individual level of analysis, contrast images were constructed comparing each of the three emotion conditions (happy, sad, and fearful) and the neutral face condition to the fixation-resting condition. At the group level of analysis, we conducted the following analyses to assess epigenetic effects. To evaluate the interaction of genotype and life stress on neural activation, we conducted a multiple regression analysis in SPM2, into which we entered four variables: genotype, life stress, the interaction of genotype and life stress, and age. Because the sample was dichotomized based on 5-HTTLPR genotype, information the presence or absence of the s variants was coded as “+1” and −1“, respectively. The variable coding life stress represented the number of life stress categories endorsed by each individual. The interaction term was the product of the genotype code with the number of endorsed life stress events. By assigning weights of zero to the variables for genotype, life stress, and age and assigning a weight of one to the interaction variable, we were able to conduct a multiple regression analysis within SPM in which we correlated brain activation for each of the contrasts of interest that was uniquely associated with the interaction term, while partialling out any main effects due to genotype, life stress, or age. To evaluate epigenetic effects on functional connectivity, we conducted psychophysiological interaction analyses (56) for each individual, the results of which then were entered into multiple regression group-level analyses as described above. Epigenetic effects on brain structure were assessed by using multiple regression analyses on data derived from a VBM analysis.
The amygdala and hippocampus constituted a priori regions of interest, based on prior publications on the role of 5-HTTLPR genotype on the amygdala (18–23) and studies that showed an effect of stress on hippocampal morphology (24) and hippocampal involvement in depression or anxiety (25–28, 37). For these a priori regions of interest, the level of statistical significance was set at P < 0.05, familywise-error corrected at the voxel level. For whole-brain analyses, search space was confined to the gray matter template mask, and significant clusters were reported if they met or exceeded a significance threshold of P < 0.005 and a cluster size threshold of 20 voxels or greater. All coordinates reported here represent MNI space.
Supplementary Material
Acknowledgments
We thank N. Steigerwald for assistance with genotyping, H. Lin for assistance with the psychophysiological interaction analyses, and A. Aron for advice on statistical analyses. This work was supported by General Clinical Research Center Grant 5-MO1-RR-10710, National Science Foundation Grant BCS-0224221, European Commission Grant NEWMOOD LSHM-CT-2003-503474; and Deutsche Forschungsgemeinschaft Grant SFB 581, KFO 125/1-1.
Abbreviations
- aCBF
absolute cerebral blood flow
- fMRI
functional MRI
- GxE
gene-by-environment
- TR
repetition time
- VBM
voxel-based morphometry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Monroe SM, Simons AD. Psychol Bull. 1991;110:406–425. doi: 10.1037/0033-2909.110.3.406. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Kessler RC, Walters EE, MacLean C, Neale MC, Heath AC, Eaves LJ. Am J Psychiatry. 1995;152:833–842. doi: 10.1176/ajp.152.6.833. [DOI] [PubMed] [Google Scholar]
- 3.Lesch KP. J Psychiatry Neurosci. 2004;29:174–184. [PMC free article] [PubMed] [Google Scholar]
- 4.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 5.Sen S, Burmeister M, Ghosh D. Am J Med Genet. 2004;127:85–89. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- 6.Munafo MR, Clark T, Flint J. Mol Psychiatry. 2005;10:415–419. doi: 10.1038/sj.mp.4001627. [DOI] [PubMed] [Google Scholar]
- 7.Boyce P, Parker G, Barnett B, Cooney M, Smith F. Br J Psychiatry. 1991;159:106–114. doi: 10.1192/bjp.159.1.106. [DOI] [PubMed] [Google Scholar]
- 8.Schinka JA, Busch RM, Robichaux-Keene N. Mol Psychiatry. 2004;9:197–202. doi: 10.1038/sj.mp.4001405. [DOI] [PubMed] [Google Scholar]
- 9.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 10.Kendler KS, Kuhn JW, Vittum J, Prescott CA, Riley B. Arch Gen Psychiatry. 2005;62:529–535. doi: 10.1001/archpsyc.62.5.529. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman J, Yang BZ, Douglas-Palumberi H, Houshyar S, Lipschitz D, Krystal JH, Gelernter J. Proc Natl Acad Sci USA. 2004;101:17316–17321. doi: 10.1073/pnas.0404376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eley TC, Sugden K, Corsico A, Gregory AM, Sham P, McGuffin P, Plomin R, Craig IW. Mol Psychiatry. 2004;9:908–915. doi: 10.1038/sj.mp.4001546. [DOI] [PubMed] [Google Scholar]
- 13.Grabe HJ, Lange M, Wolff B, Volzke H, Lucht M, Freyberger HJ, John U, Cascorbi I. Mol Psychiatry. 2005;10:220–224. doi: 10.1038/sj.mp.4001555. [DOI] [PubMed] [Google Scholar]
- 14.Surtees PG, Wainwright NW, Willis-Owen SA, Luben R, Day NE, Flint J. Biol Psychiatry. 2006;59:224–229. doi: 10.1016/j.biopsych.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Gillespie NA, Whitfield JB, Williams B, Heath AC, Martin NG. Psychol Med. 2005;35:101–111. doi: 10.1017/s0033291704002727. [DOI] [PubMed] [Google Scholar]
- 16.de Geus EJ, Wright MJ, Martin NG, Boomsma DI. Behav Genet. 2001;31:489–495. doi: 10.1023/a:1013360909048. [DOI] [PubMed] [Google Scholar]
- 17.Hamer D. Science. 2002;298:71–72. doi: 10.1126/science.1077582. [DOI] [PubMed] [Google Scholar]
- 18.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 19.Hariri AR, Drabant EM, Munoz KE, Kolachana BS, Mattay VS, Egan MF, Weinberger DR. Arch Gen Psychiatry. 2005;62:146–152. doi: 10.1001/archpsyc.62.2.146. [DOI] [PubMed] [Google Scholar]
- 20.Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, Egan MF, Mattay VS, Hariri AR, Weinberger DR. Nat Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- 21.Furmark T, Tillfors M, Garpenstrand H, Marteinsdottir I, Langstrom B, Oreland L, Fredrikson M. Neurosci Lett. 2004;362:189–192. doi: 10.1016/j.neulet.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 22.Heinz A, Braus DF, Smolka MN, Wrase J, Puls I, Hermann D, Klein S, Grusser SM, Flor H, Schumann G, Mann K, Buchel C. Nat Neurosci. 2005;8:20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- 23.Canli T, Omura K, Haas BW, Fallgatter A, Constable RT, Lesch KP. Proc Natl Acad Sci USA. 2005;102:12224–12229. doi: 10.1073/pnas.0503880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEwen BS, Sapolsky RM. Curr Opin Neurobiol. 1995;5:205–216. doi: 10.1016/0959-4388(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 25.Campbell S, Marriott M, Nahmias C, MacQueen GM. Am J Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- 26.Frodl T, Meisenzahl EM, Zill P, Baghai T, Rujescu D, Leinsinger G, Bottlender R, Schule C, Zwanzger P, Engel RR, et al. Arch Gen Psychiatry. 2004;61:177–183. doi: 10.1001/archpsyc.61.2.177. [DOI] [PubMed] [Google Scholar]
- 27.Hastings RS, Parsey RV, Oquendo MA, Arango V, Mann JJ. Neuropsychopharmacology. 2004;29:952–959. doi: 10.1038/sj.npp.1300371. [DOI] [PubMed] [Google Scholar]
- 28.Rusch BD, Abercrombie HC, Oakes TR, Schaefer SM, Davidson RJ. Biol Psychiatry. 2001;50:960–964. doi: 10.1016/s0006-3223(01)01248-3. [DOI] [PubMed] [Google Scholar]
- 29.Sheline YI, Gado MH, Kraemer HC. Am J Psychiatry. 2003;160:1516–1518. doi: 10.1176/appi.ajp.160.8.1516. [DOI] [PubMed] [Google Scholar]
- 30.Wendland JR, Martin BJ, Kruse MR, Lesch KP, Murphy DL. Mol Psychiatry. 2006;11:224–226. doi: 10.1038/sj.mp.4001789. [DOI] [PubMed] [Google Scholar]
- 31.Sugiura M, Kawashima R, Nakamura K, Okada K, Kato T, Nakamura A, Hatano K, Itoh K, Kojima S, Fukuda H. NeuroImage. 2000;11:36–48. doi: 10.1006/nimg.1999.0519. [DOI] [PubMed] [Google Scholar]
- 32.Iacoboni M. Curr Opin Neurobiol. 2005;15:632–637. doi: 10.1016/j.conb.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Poldrack RA, Packard MG. Neuropsychologia. 2003;41:245–251. doi: 10.1016/s0028-3932(02)00157-4. [DOI] [PubMed] [Google Scholar]
- 34.Carr L, Iacoboni M, Dubeau MC, Mazziotta JC, Lenzi GL. Proc Natl Acad Sci USA. 2003;100:5497–5502. doi: 10.1073/pnas.0935845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolen-Hoeksema S. J Abnorm Psychol. 2000;109:504–511. [PubMed] [Google Scholar]
- 36.Ray RD, Ochsner KN, Cooper JC, Robertson ER, Gabrieli JD, Gross JJ. Cogn Affect Behav Neurosci. 2005;5:156–168. doi: 10.3758/cabn.5.2.156. [DOI] [PubMed] [Google Scholar]
- 37.Sheline YI. Biol Psychiatry. 2003;54:338–352. doi: 10.1016/s0006-3223(03)00347-0. [DOI] [PubMed] [Google Scholar]
- 38.Gallese V, Fadiga L, Fogassi L, Rizzolatti G. Brain. 1996;119:593–609. doi: 10.1093/brain/119.2.593. [DOI] [PubMed] [Google Scholar]
- 39.Rizzolatti G, Fadiga L, Gallese V, Fogassi L. Brain Res Cogn Brain Res. 1996;3:131–141. doi: 10.1016/0926-6410(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 40.Rizzolatti G, Craighero L. Annu Rev Neurosci. 2004;27:169–192. doi: 10.1146/annurev.neuro.27.070203.144230. [DOI] [PubMed] [Google Scholar]
- 41.Allman JM, Watson KK, Tetreault NA, Hakeem AY. Trends Cogn Sci. 2005;9:367–373. doi: 10.1016/j.tics.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Costa PT, McCrae RR. Professional Manual of the Revised NEO Personality Inventory and NEO Five-Factor Inventory. Odessa, FL: PAR; 1992. [Google Scholar]
- 43.McNair ML, Lorr M, Droppleman LF. Profile of Mood States. San Diego, CA: EdITS; 1992. [Google Scholar]
- 44.Caspi A. Int J Methods Psychiatr Res. 1996;6:101. [Google Scholar]
- 45.Treynor W, Gonzales R, Nolen-Hoeksema S. Cognit Ther Res. 2003;27:247–259. [Google Scholar]
- 46.Ashburner J, Friston KJ. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 47.Luh WM, Wong EC, Bandettini PA, Hyde JS. Magn Reson Med. 1999;41:1246–1254. doi: 10.1002/(sici)1522-2594(199906)41:6<1246::aid-mrm22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 48.Freeman AJ, Gowland PA, Mansfield P. Magn Reson Imaging. 1998;16:765–772. doi: 10.1016/s0730-725x(98)00011-3. [DOI] [PubMed] [Google Scholar]
- 49.Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. NeuroImage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer SM, Jackson DC, Davidson RJ, Aguirre GK, Kimberg DY, Thompson-Schill SL. J Cogn Neurosci. 2002;14:913–921. doi: 10.1162/089892902760191135. [DOI] [PubMed] [Google Scholar]
- 51.Wong EC, Buxton RB, Frank LR. NMR Biomed. 1997;10:237–249. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<237::aid-nbm475>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Aguirre GK, Kimberg DY, Roc AC, Li L, Detre JA. Magn Reson Med. 2003;49:796–802. doi: 10.1002/mrm.10437. [DOI] [PubMed] [Google Scholar]
- 53.Backstrom T, Andreen L, Birzniece V, Bjorn I, Johansson IM, Nordenstam-Haghjo M, Nyberg S, Sundstrom-Poromaa I, Wahlstrom G, Wang M, Zhu D. CNS Drugs. 2003;17:325–342. doi: 10.2165/00023210-200317050-00003. [DOI] [PubMed] [Google Scholar]
- 54.Friston KJ. In: Functional Neuroimaging: Technical Foundations. Thatcher R, editor. San Diego: Academic; 1994. pp. 79–93. [Google Scholar]
- 55.Holmes A, Friston K. NeuroImage. 1998;7:754. doi: 10.1006/nimg.1997.0306. [DOI] [PubMed] [Google Scholar]
- 56.Friston KJ, Buechel C, Fink GR, Morris J, Rolls E, Dolan RJ. NeuroImage. 1997;6:218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.