Abstract
Protein–protein complexes remain enticing, but extremely challenging, targets for small-molecule drug discovery. In a rare example described earlier, a high-affinity small molecule, SP4206 (Kd ≈ 70 nM), was found to block binding of the IL-2α receptor (IL-2Rα) to IL-2 (Kd ≈ 10 nM). Recently, the structure of the IL-2/IL-2Rα complex was solved [Rickert, M., Wang, X., Boulanger, M. J., Goriatcheva, N., Garcia, K. C. (2005) Science 308:1477–1480]. Using structural and functional analysis, we compare how SP4206 mimics the 83-fold larger IL-2Rα in binding IL-2. The binding free energy per contact atom (ligand efficiency) for SP4206 is about twice that of the receptor because of a smaller, but overlapping, contact epitope that insinuates into grooves and cavities not accessed by the receptor. Despite its independent design, the small molecule has a similar, but more localized, charge distribution compared with IL-2Rα. Mutational studies show that SP4206 targets virtually the same critical “hot-spot” residues on IL-2 that drive binding of IL-2Rα. Moreover, a mutation that enhances binding to the IL-2Rα near these hot spots also enhances binding to SP4206. Although the protein and small molecule do bind the same hot spot, they trap very different conformations of IL-2 because of its flexible nature. Our studies suggest that precise structural mimics of receptors are not required for high-affinity binding of small molecules, and they show that there are multiple solutions to tight binding at shared and adaptive hot spots.
Keywords: drug discovery, protein flexibility, protein–protein interactions
Protein–protein interactions are major regulators of cell biology and important targets in drug discovery. Although antibody therapeutics have been developed that block protein–protein interactions, no approved small-molecule drugs have yet been produced for this important target class (1).
Protein–protein interfaces, which are large, flat, and featureless (2, 3), appear to lack sufficient functionality for small-molecule binding. However, mutational studies suggest that protein–protein interactions are driven by a small set of the contact residues, termed “hot spots,” whose footprints are not significantly larger than those covered by small molecules (4–6). Moreover, there have been several reports of small molecules that disrupt discontinuous protein–protein interactions with reasonable potencies (Ki values <1 μM), and their binding sites have been confirmed by high-resolution structural analysis (7–12).
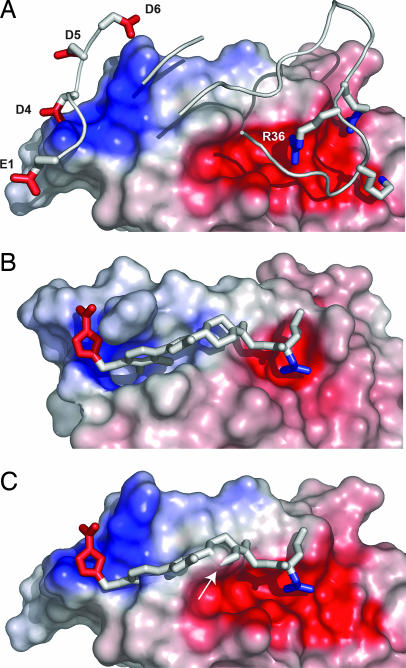
It is important to functionally dissect and analyze successful cases in which a small molecule has mimicked a larger protein partner to comprehend the design principles that transfer from a protein to a small molecule. Recently, SP4206, a small molecule (Structure 1) was discovered that binds with high affinity (Kd ≈ 70 nM) to IL-2 (Fig. 1A) and blocks binding to its natural receptor, IL-2Rα (8, 9). SP4206 was assembled from smaller fragments by using a structure-guided approach with a binding and functional assay (8, 9). Subsequently, the structure of IL-2 bound to the IL-2Rα (Kd ≈ 10 nM) was described (Fig. 1B) (13). This structure showed that the IL-2Rα completely envelops the footprint covered by these small-molecule competitive inhibitors (Fig. 1C).
Structure 1.
Fig. 1.
Contact epitopes for binding of SP4206 or the IL-2Rα. (A) Binding of SP4206 (stick model) with its contact surface (orange, all surface points within 4.5 Å of the ligand) mapped onto IL-2 (gray). (B) Structure of IL-2Rα (yellow ribbons) bound to its contact surface (green, all surface points within 4.5 Å of IL-2Rα) on IL-2 (gray). (C) Binding of SP4206 (stick model) to the contact surface on IL-2 made by IL-2Rα (green). The PDB ID codes are 1PY2 for IL-2 bound to SP4206 (10) and 1Z92 for IL-2 bound to IL-2Rα (13).
In this article, we provide structural and functional analysis to compare these binding epitopes on IL-2. The small molecule binds with twice the ligand efficiency (ΔG per contact atom; refs. 14 and 15) of the receptor because of its smaller size and ability to access cavities not accessed by the receptor. SP4206 shares a similar but more localized electrostatic field, and remarkably it targets the same hot-spot residues that the receptor uses to bind IL-2. However, the contacts to IL-2 from the receptor and small molecule are very different, as is the conformation of IL-2 to bind them. Thus, there are multiple solutions to tight binding at this common and adaptive hot spot on IL-2. Such adaptive hot spots offer more opportunities for drug discovery than is revealed from the static structures of either the individual proteins or their complexes.
Results and Discussion
Structural and Electrostatic Epitopes on IL-2 for Binding Its Receptor Versus Small Molecule.
By inspecting the surface area on IL-2 buried by the IL-2Rα versus SP4206 (Table 1), it is apparent that the receptor contact epitope completely envelops that covered by the small molecule (Fig. 1C). The small molecule is fully covered by the contact epitope from the receptor. This situation is remarkable, given that the small molecule was constructed by using a binding and functional assay without prior structural information for how the receptor bound to IL-2 (7–9).
Table 1.
Calculation of ligand efficiency
| Measurement | IL-2Rα | SP4206 |
|---|---|---|
| Area buried, Å2 | 2,093 | 1,083 |
| Ki, nM | 10.5 | 68.8 |
| ΔG°binding, kcal/mol | −10.9 | −9.8 |
| ΔG°/Å2 buried | 5.2 × 10−3 | 9.0 × 10−3 |
| No. of contact atoms | 134 | 45 |
| Ligand efficiency, kcal per contact atom | 0.08 | 0.22 |
The receptor covers an area about twice the size of that for the small molecule (≈2,100 versus 1,100 Å2) and utilizes a much larger number of contact heavy atoms (134 versus 45) (Table 1). Despite the larger surface area covered, the two molecules bind with similar binding affinities (−10.9 versus −9.8 kcal/mol for the receptor versus SP4206, respectively). Thus, the small molecule binds with 2 to 4-fold greater ligand efficiency when calculated either by free energy per surface area buried or per contact atom used. A traditional measure for ligand efficiency (ΔG binding per contact heavy atom; refs. 14 and 15) shows that the value for SP4206 (0.22 kcal per heavy atom) falls within the range for binding of small molecules to traditional enzyme targets (0.17–1.3 kcal per heavy atom). Moreover, the ligand efficiencies we calculate for other inhibitors to protein–protein interfaces, such as Bcl-xL (11) and Mdm2 (12), are fairly close to those of IL-2 at 0.26 and 0.18 kcal per heavy atom, respectively. Both of these small molecules are more efficient at binding than the helical structures they mimic, which have calculated efficiencies of 0.04 and 0.14 for the BAD helix and p53 helix, respectively. These three examples show that small molecules can be discovered with greater ligand efficiencies than a natural protein partner. As a class these protein–protein interface inhibitors tend to be on the low end of the enzyme inhibitor efficiency range, but not beyond the range. Moreover, these ligands may yet be optimized for ligand efficiency.
One of the notable differences between the binding of the small molecule versus the receptor is a more focused electrostatic epitope. The electrostatic field produced by the atoms on IL-2Rα that are in contact with IL-2 shows a diffuse, but distinctive, zwitterionic character (Fig. 2A). There are two similarly sized and oppositely charged lobes separated by a hydrophobic patch. Remarkably, the small molecule contains a similar and overlapping zwitterionic distribution; however, the electrostatic field is much smaller (Fig. 2B). This difference can be further appreciated by comparing the complementary zwitterionic epitopes on IL-2 for binding these two molecules (Fig. 3). The receptor uses a cluster of three positively charged side chains (R35, R36, and K38) (Fig. 3A) to interact with five negatively charged side chains on IL-2 (E61, E62, E68, E106, and D109). At the other end, the receptor uses five negatively charged side chains (E1, D4, D5 and D6, and E29) to interact with four positively charged side chains on IL-2 (K35, R38, K43, and the more distal K32).
Fig. 2.
Electrostatic fields created by IL-2Rα (A) or SP4206 (B). Interfacial atoms (within 5.3 Å of any IL-2 atom) for both binding partners are shown on the left in stick view. The electrostatic fields created by the ligands in the complexed structures are shown on the right. Blue is positive and red is negative, shown at isopotential levels of ±6 kBT/ec at T = 310 K (ec, electron charge). Note the more localized electrostatic field presented by the small molecule.
Fig. 3.
Binding surfaces and electrostatic fields created by IL-2. (A) IL-2Rα in ribbons and sticks with specific residues labeled binding to the surface of IL-2, which is colored in proportion to the electrostatic field that would be felt by a test atom in contact with the surface. Blue is positive, white is neutral, and red is negative, with a dynamic range of −15 to +15 kBT/ec at T = 310 K, and all potentials are calculated with respect to the dielectric envelope of the fully solvated complex. (B) SP4206 in sticks binding to IL-2 represented as in A. Note the more localized electrostatic field presented by IL-2 in the presence of SP4206. (C) Bound conformation of SP4206 superimposed on the surface of IL-2 that binds the IL-2Rα. Note that in this conformation part of the surface of IL-2 (shown by the arrow) is incompatible with binding.
In sharp contrast, the small molecule uses just one positive charge from its guanido group to form a direct salt bridge to E62 on IL-2 (Fig. 3B). At the other end, the small molecule uses the furonic acid group to interact with a small cluster of positively charged groups from IL-2 (K35 and R38, and the more distal K32 and K43). It is notable that there is only one clearly conserved functionality between the small molecule and receptor; the guanido group from the small molecule and R36 from the receptor both interact with E62 on IL-2. Other than this the contact atoms and corresponding functionalities appear quite different.
Another significant difference between the receptor and the small molecule is that they bind to considerably different conformations of IL-2. The surface of IL-2 that binds the receptor is relatively flat (Fig. 3A), as is typical of most protein–protein complexes (2, 3). In contrast, the small molecule binds to an S-shaped groove that is not evident in the receptor-bound structure. One of the most significant differences is the position of F42 in the center of the IL-2 epitope. It is in an up position to bind the receptor and a down position to bind the small molecule; the down position helps create the binding groove. In addition, there is a substantial reorganization of the loop between residues 31–35 in IL-2 that produces a binding pocket for the furonic acid moiety of the small molecule. This loop is flattened out when binding to the receptor. Thus, the binding conformation of IL-2 for the receptor is sterically incompatible with binding the small molecule (Fig. 3C). The highly adaptive binding surface on IL-2 has been previously noted (7, 10, 16) and suggests how IL-2 is capable of binding two radically different ligands, such as the receptor and the small molecule.
Functional Epitopes on IL-2 for Binding Its Small Molecule Versus Receptor.
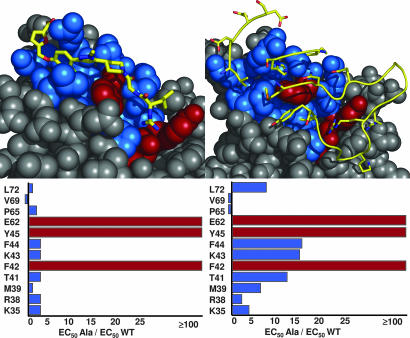
To begin to understand the functional features on IL-2 necessary for binding the small molecule, we mutated to alanine (17) the residues on IL-2 that are known from the x-ray structure to contact SP4206. The mutated positions were K35, R38, M39, T41, F42, K43, F44, Y45, E62, P65, V69, and L72. Each variant was individually cloned, expressed, and purified as described in Materials and Methods. A competitive ELISA was used to test the affinity of the alanine variants versus SP4206 or IL-2Rα (Fig. 4). F42, Y45, and E62 were each found to disrupt binding affinity by at least 100-fold (beyond the detection limit of the assay) for both SP4206 and IL-2Rα. These data are in agreement with a random mutagenesis approach used for murine IL-2, in which mutations at F42, Y45, and E62 were found to reduce cellular activity (18). None of the other positions mutated on IL-2 had a significant impact on the binding affinity of SP4206 (Fig. 4 Left), and of the remaining eight positions, only F44, K43, and T41 exhibited modest (>10-fold) disruptions of binding to IL-2Rα (Fig. 4 Right). Thus, the side chains on IL-2 most important for binding are shared for both SP4206 and IL-2Rα. To confirm that the alanine mutations did not grossly perturb the structure of IL-2, we tested the ability of mutant IL-2 molecules to bind to the IL-2Rβ subunit, which binds at a distinct site (19). None of the alanine substitutions caused a significant change in the ability of IL-2 to bind to IL-2Rβ, suggesting they did not disrupt the correct fold of the protein (data not shown).
Fig. 4.
Hot spots from alanine-scanning of IL-2 for binding SP4206 (Left) or IL-2Rα (Right). (Upper) The two ligands (sticks or ribbons) bound to the surface of IL-2, with residues most critical for binding (F42, Y45, and E62) shown in red and other contacts shown in blue. (Lower) The degree of disruption in affinity (expressed as IC50 Ala/IC50 WT) due to each of the dozen alanine substitutions. Note the similarity of the most critical residues (>100-fold reduction in affinity) indicated by the red bars. Lesser effects are shown in blue bars. See Materials and Methods for further details.
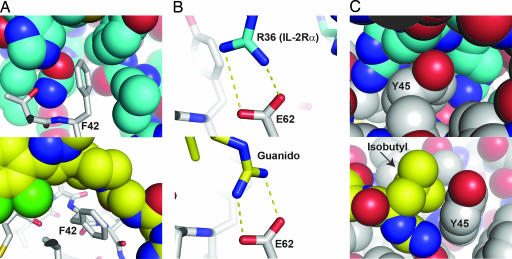
The importance of the three critical residues on IL-2 for binding the IL-2Rα and SP4206 can be further appreciated by the close packing interactions seen in Fig. 5. As noted above, F42 is pointing in an up position and well packed in a cavity in the receptor formed by IL-2Rα side chains M25, N27, L42, Y43, and H120 (Fig. 5A Upper). In the small-molecule structure, F42 is pushed down but making clear van der Waals interactions with the piperidine moiety of SP4206 (Fig. 5A Lower). IL-2 E62 takes part in a salt bridge interaction with R36 of the receptor and the guanido of the SP4206 (Fig. 5B Lower). IL-2 Y45 makes van der Waals interactions with the aliphatic portions of IL-2Rα R35 and R36 (Fig. 5C Upper). As with IL-2 F42, the conformation of IL-2 Y45 shifts to bind to SP4206 and both contacts the isobutyl group and forms a wall with the salt bridge between the guanido moiety on SP4206 and IL-2 E62 (Fig. 5C Lower). Thus, both aromatic side chains on IL-2 are tightly packed when in complex with the receptor or small molecule yet are flexible and exist in different conformations depending on the ligand.
Fig. 5.
Details of contacts between IL-2 (shown in gray) and either IL-2Rα (Upper, shown in blue) or SP4206 (Lower, shown in yellow) at the three hot-spot residues found to be critical for binding: F42 (A), E62 (B), or Y45 (C). In A, note the fully buried nature of F42 in both complexes despite the difference in conformation of this residue between the two complexes. In B, IL-2Rα R36 forms a buried salt bridge with IL-2 E62. This interaction is structurally mimicked by the guanido group on SP4206. In C, Y45 is fully buried upon binding to IL-2Rα and forms a binding pocket for both the guanido group and the isobutyl moiety in SP4206.
Mutations in IL-2 That Improve Affinity Discovered by Phage Display.
It has been possible to systematically improve affinity for a number of other natural interfaces by alanine-scanning (17) or protein display technology (20–23). We wished to determine whether optimizing the hot spot on IL-2 through mutagenesis for enhanced binding to IL-2Rα would show improved affinity for binding the small molecule. Given the uncertainties in which mutations would improve binding, we used a random library-based approach and monovalent phage display (20). IL-2 was displayed on filamentous phage particles (see Materials and Methods). The residues around the binding interface for the compound were randomly mutated and IL-2 variants were selected by successive rounds of binding to plates containing immobilized IL-2Rα. After six rounds of enrichment, individual clones were sequenced and tested for binding to both IL-2Rα and SP4206. Not surprisingly, the critical residues F42, Y45, and E62 were fully conserved (data not shown). Several IL-2 variants were found that improved binding affinity for IL-2Rα by 2- to 10-fold (Table 2). By deconvoluting the most improved mutant (V69A/P65A/Q74P) into single mutants, it was clear that most of the improvement in affinity was derived from the P65A and V69A substitutions, which were 2- and 5-fold improved, respectively. Recently, Rao et al. (21) reported using gene-wide PCR mutagenesis of IL-2 displayed on yeast to select for enhanced affinity toward IL-2Rα. Interestingly, many of their “hits” corresponded to the V69A, Q74P, or a combination of both, as observed here. The V69A improvement was also seen in the alanine-scanning result (Fig. 4A) and a structural interpretation of the result is offered below.
Table 2.
Effect of enhanced affinity mutations on binding of IL-2 to IL-2Rα and SP4206
| Mutant | EC50, nM |
|
|---|---|---|
| IL-2Rα | SP4206 | |
| WT | 10.5 (1) | 68.8 (1) |
| V69A/P65A/Q74P | 1.1 (10) | >100,000 |
| K35L/M39V | 1.9 (5) | 80.1 (1) |
| P65A | 5.2 (2) | 117.0 (2) |
| V69A | 2.0 (5) | 10.4 (7) |
The approximate fold improvement in affinity of each variant versus WT IL-2 for either IL-2Rα or SP4206 is shown in parentheses. Each result is the average of three independent experiments. Experimental details are outlined in Materials and Methods.
Next, we wished to determine how these mutations affected binding to the small molecule. The receptor improved IL-2 triple mutant (P65A/V69A/Q74P) no longer bound SP4206 (EC50 > 100 μM, Table 2), showing that it is possible to generate high selectivity between these binding partners. This capability is not surprising, given the differences in the details of their binding modes. Two of the other receptor-improved variants had little impact on SP4206 binding. For example, P65A slightly enhanced affinity for IL-2 and decreased affinity for SP4206. This result may reflect a biased conformation for binding IL-2Rα versus the small molecule. Interestingly, the V69A variant that was 5-fold enhanced for binding IL-2Rα was 7-fold enhanced for binding SP4206 (EC50 ≈ 10 nM). V69 sits next to the critical F42 and supports its position in the apo form of IL-2.
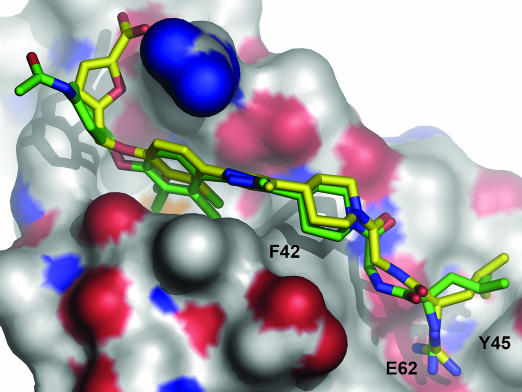
To understand how the V69A mutation affected binding of SP4206, we attempted to crystallize this complex. Unfortunately, diffraction-quality crystals of SP4206 in complex with V69A could not be obtained. It was possible, however, to solve the structure of the complex between SP4160 (Structure 1) and V69A (EC50 = 250 nM) to 2.7-Å resolution (Table 3, which is published as supporting information on the PNAS web site). SP4160 is identical to SP4206 except it contains a terminal benzyl acylamide group in place of the carboxyfuran group. The structure of the SP4160 complex with the V69A mutant reveals several interesting features. First, SP4160 binds in the identical groove as SP4206 except that the loop between helix 3 and 4 is “flattened out” and resembles the receptor bound conformation (Fig. 6). Second, the position of F42 is virtually identical between the two small-molecule-bound conformations. The V69A mutation is smaller and makes it easier for F42 to move to the down position, where it can then create the groove necessary to bind SP4206 or SP4160. Thus, the IL-2 binding site can exist in hybrid conformations in which part of the interface is like SP4206 and the other is like the receptor, suggesting independence between the loop region and that around F42. We observed similar independence between these regions when IL-2 was crystallized with smaller fragments or precursors to SP4206 (7, 14).
Fig. 6.
Structure of SP4160 (green sticks) bound to IL-2. Red and blue represent oxygen and nitrogen atoms, respectively. The structure of SP4206 from its x-ray coordinates (yellow sticks) is overlaid onto SP4160. Note the close superposition except in the region of the benzyl acylamide in SP4160 versus the carboxyfuran in SP4206 on the left. Also, note the absence of the binding pocket in IL-2 for the carboxyfuran moiety of SP4206 seen in Fig. 3B. The positions of hot-spot residues (F42, Y45, and E65) are labeled.
It is notable that the neutral-for-charged substitution in the SP4160 causes a 30-fold reduction in affinity for IL-2 (IC50 = 2 μM) relative to SP4206 (IC50 = 70 nM) (9). When we alanine-scanned the individual basic residues in IL-2 that interacted with the carboxyfuran group, we saw much more modest effects (Fig. 4). This difference for altering the basic groups on IL-2 versus the single negative charge on the compound likely reflects that each member in this cluster of basic groups incrementally contributes to affinity and that neither singly is responsible for the interaction.
A possible explanation for the improved affinity of IL-2 V69A for both SP4206 and the receptor could be that V69 impedes the down or up movement of F42 that is critical to bind SP4206 or the receptor, respectively. V69 packs under F42, so one can imagine that it could hinder the downward movement of F42 needed to form the small-molecule groove (Fig. 7, which is published as supporting information on the PNAS web site). Moreover, attractive van der Waals interactions between F42 and V69 could impede the ability of F42 to spring up and interact with the IL-2Rα. Another possible explanation is that the V69A mutation destabilizes the free IL-2 relative to the bound complexes. Recently, Kossiakoff and coworkers (24) have shown data that affinity-enhanced variants of human growth hormone are significantly less stable than the WT hormone, but equivalently stable when complexed to the receptor. Thus, destabilizing the unbound form while retaining stability of the bound form could account for the overall increase in affinity. It is striking that the same mutation in IL-2 can enhance binding for two different partners but for apparently different structural reasons.
Conclusions
The fact that the small molecule was designed independent of knowledge of the structure of the IL-2R complex provides unbiased comparisons of the basis for binding a big and small molecule. These data show that small molecules and proteins with very different structures can bind the same protein interface with virtually identical functional epitopes. Although the small molecule and receptor bind the same hot spot on IL-2, these residues provide different interactions with each of the partners. The small molecule and receptor bind with very different structural and conformational epitopes. This difference is afforded by the fact that the binding interface on IL-2 can exist in multiple conformations. The epitopes for Bcl-xl and Mdm-2 naturally recognize helical protein partners, yet the small-molecule ligands for these (11, 12) bear no structural resemblance to α-helices, and many of the contacts are different. It has been repeatedly observed when selecting random peptides for protein hormones and receptors that they tend to be selected for functional hot spots at these interfaces; moreover, the structures of the selected peptides bear little resemblance to the structures of the protein they mimic (refs. 25 and 26, for review see ref. 27). Such promiscuous binding sites partially reflect the intrinsic flexibility of the target protein that can accommodate many solutions for tight binding.
The small molecule SP4206 is 1/83 the size of the receptor and uses only about one-third the number of contacts to achieve nearly the same binding affinity as the receptor. The basis for the increased ligand efficiency for the small molecule may derive from at least three factors: more highly focused electrostatic interactions, trapping a conformation of IL-2 that provides deeper cavities and thus increasing surface-to-volume ratio for ligand binding, and better packing with fewer buried waters at the interface.
These studies begin to address several daunting concerns about designing small molecules for protein–protein interfaces. One does not need to precisely graft the structural features of a large receptor into the small molecule to achieve high affinity. It is striking that in this case there was only a single group clearly in common between the small molecule and the receptor (the guanido functionality). The general but diffuse zwitterionic character of the receptor was encapsulated in a small molecule. Adaptive proteins such as IL-2 can create binding grooves that are not evident in either the apo or receptor-bound conformation. Thus, one should not assume the interface observed in a protein–protein complex is all that the small molecule has to bind. Although the ligand efficiencies for the inhibitors of IL-2 and other protein–protein interactions are on the lower end of efficiency relative to inhibitors of enzymes, they provide validation that modest-affinity compounds are possible.
Materials and Methods
Cloning and Expression of IL-2 and Mutants and IL-2Rα.
WT IL-2 and alanine mutants were expressed in Escherichia coli (BL21 DE3 pLysS; Invitrogen, Carlsbad, CA) as insoluble inclusion bodies as described in ref. 7. Constructs corresponding to individual alanine mutations were designed at positions K35, R38, M39, T41, F42, K43, F44, Y45, E62, P65, V69, and L72 and made by using the mutagenesis approach of Kunkel (28). The protein was expressed in BL21 DE3 pLysS cells by inducing a 1-liter culture (in 2× YT medium (Fisher Biotech, Fair Lawn, NJ) plus 100 μg/ml carbenecillin) at OD600 ≈ 1.0 with isopropyl β-d-thiogalactopyranoside (200 μg/ml) for 3 h at 37°C. For a 1-liter culture, inclusion bodies were resuspended in 50 ml of 8 M guanidine hydrochloride, and the soluble material was then slowly dripped over the course of 120 min into a buffer containing 1.1 M guanidine, 110 mM Tris, 6.5 mM cysteamine, and 0.65 mM cystamine, pH 8. This solution was allowed to equilibrate at room temperature all day and was then dialyzed overnight into a buffer of 10 mM ammonium acetate, pH 6/25 mM sodium chloride. Insoluble/aggregated material was removed by centrifugation and discarded. The soluble portion of the refolded protein was filtered through a 0.22-μm filter (Millipore, Billerica, MA) and then purified by chromatography on an S-Sepharose (Amersham Pharmacia, Piscataway, NJ) column using a 25 mM to 1 M NaCl gradient in a buffer of 25 mM ammonium acetate, pH 7. IL-2Rα was prepared as described in ref. 7.
X-Ray Crystallography.
Crystals were grown by the vapor diffusion method using hanging drops on silane-treated glass coverslips and standard trays from Hampton Research (Aliso Viejo, CA). An approximate 1.1 molar excess of SP4160 to IL-2 V69A was used, and the crystals were grown at 10–20 mg/ml IL-2 in 28–31% (vol/vol) PEG 8K/0.1–0.3 M (NH4)2SO4/sodium cacodylate, pH 5.9. Before data collection, crystals were transferred to a reservoir solution supplemented with 20% (vol/vol) glycerol. Diffraction data were collected at −180°C at beamline 7-1 (Stanford Synchrotron Radiation Laboratory) on a MAR345 (Mar Research, Evanston, IL) detector and processed with MOSFLM (λ = 1.08 Å) (29). The structures were determined by molecular replacement using AMORE and refined with REFMAC5 (29). The protein models were adjusted by using O and ligand models were constructed in INSIGHT-II (Accelrys, Waltham, MA).
IL-2/IL-2Rα Inhibition Assay.
Activity of SP4206 against WT IL-2 and each IL-2 variant was measured by the inhibition of the IL-2/IL-2Rα interaction as a function of compound concentration in an ELISA format as described in ref. 9. Approximately 10–20 nM biotinylated IL-2Rα was immobilized in the wells of a streptavidin (Pierce, Rockford, IL)-coated 96-well plate (Maxisorp; Nunc, Rochester, NY). Serial dilutions of SP4206 were prepared in DMSO, added to a solution of IL-2 or IL-2 alanine mutant (2% DMSO final) in Superblock (Pierce) with 0.01% Tween 20, and incubated with the immobilized IL-2Rα. Unbound IL-2 was washed from the plate. Bound IL-2 was measured with 0.65 nM anti-IL-2 antibody labeled with horseradish peroxidase (Pierce) followed by addition of a colorimetric substrate for horseradish peroxidase (3,3′,5,5′-tetramethylbenzidine; Pierce). Inhibition was plotted as a function of compound concentration, and the EC50 was determined by nonlinear regression with Kaleidagraph (Synergy Software, Reading, PA).
IL-2 Phage Display.
A modified form of the low plasmid copy pMal vector from New England Biolabs (Ipswich, MA) formed the basis for construction of the IL-2 phagemid. First, the gene sequence encoding the M13 phage p8 coat protein was cloned into the SacI and HindIII restriction sites by PCR, and its correct sequence was confirmed by DNA sequencing as described in ref. 30. Second, a 5′ PCR primer was designed that encodes an NdeI restriction site containing an ATG start codon site, in frame with DNA encoding the leader sequence from the heat-stable enterotoxin II (STII) of E. coli (protein sequence KKNIAFLLASMFVFSIATNAYA) and IL-2 residues 21–25, was synthesized by Operon Technologies (Huntsville, AL). A 3′ primer was also designed that encoded the reverse complement of the C terminus of IL-2 fused in frame with a SacI restriction site. Using these primers, we generated a PCR product with IL-2 used as a DNA template for the amplification reaction. The resulting PCR product was cloned into the NdeI and SacI sites of the vector, resulting in a construct that encoded the full-length STII-IL-2-M13p8 fusion protein. The libraries encoded NNS codons (where N is any base, and S is G or C) corresponding to the codons at positions K35, R38, M39, T41, F42, K43, F44, Y45, E62, P65, V69, L72, and Q74. Oligo 1 encoded NNS codons for K35, R38, M39, T41, and F42. Oligo 2 encoded mutations for F42, K43, F44, and Y45. Oligo 3 encoded mutations at E62, P65, V69, L72, and Q74. Mutagenesis was performed by using Kunkel mutagenesis (28) as described in detail in ref. 27. In the generation of each library, a diversity of approximately >1 × 109 was achieved by using electrocompetent XL-1 Blue cells (Stratagene, La Jolla, CA). For the selection, each library of IL-2 displayed phage was allowed to incubate for 1 hr on IL-2Rα-coated plates (as described above) in PBS supplemented with 0.2% BSA and 0.05% Tween 20. Unbound phage were washed off with 10 washes of 200 μl PBS/Tris (pH 8) in each well. Bound IL-2 phage were eluted with 100 μl of 100 mM HCl and transferred to an Eppendorf (Westbury, NY) tube containing 30 μl of 1 M Tris, pH 8.0. Approximately 50 μl of the eluted phage solution was then added to 500 μl of actively growing XL-1 Blue cells (OD600 ≈ 0.8). After six rounds of selection, individual clones were sequenced and tested for affinity.
Electrostatic and Surface Area Analysis.
Electrostatic calculations were performed by using the Adaptive Poisson Boltzmann Solver (31) with an interior dielectric constant of 2 and a solvent dielectric constant of 80. Protein charges were assigned by using the Amber force field (32), and the small-molecule charges were assigned by using the Merck Molecular Mechanics Force Field (33). To elucidate the electrostatic contribution of the respective binding partners in the complexed state, all Poisson Boltzmann calculations were performed by using the full dielectric envelope of the complex. Electrostatic potentials on molecular surfaces were visualized by using PyMOL version 0.98 (http://pymol.sourceforge.net/) in a manner such that the coloring of the surface corresponds to the effective potential felt by a probe atom in tangential contact with that surface. Solvent-accessible surface areas were calculated by using the PyMOL program using all-atom models of the binding partners both alone and in complex, using the cocrystal structure conformations of each partner.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Dr. Andrew Braisted. We thank Johan Oslob and Brian Riamundo for making available SP4206 and SP4160 for biochemical studies. We are grateful to our colleagues at Sunesis Pharmaceuticals, who provided inspiration and encouragement for this science. C.D.T. was supported by National Institutes of Health National Cancer Institute Postdoctoral Fellowship 3 F32CA093177-04S1.
Abbreviation
- IL-2Rα
IL-2α receptor.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors corresponding to the crystal structure of SP4160 bound to IL-2 V69A have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1QVN).
References
- 1.Arkin MR, Wells JA. Nat Rev Drug Discovery. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 2.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 3.Stites WE. Chem Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 4.Clackson T, Wells JA. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 5.DeLano WL. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 6.Wells JA. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkin MR, Randal M, DeLano WL, Hyde J, Luong TN, Oslob JD, Raphael DR, Taylor L, Wang J, McDowell RS, et al. Proc Natl Acad Sci USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braisted AC, Oslob JD, DeLano WL, Hyde J, McDowell RS, Waal N, Yu C, Arkin MR, Raimundo BC. J Am Chem Soc. 2003;125:3714–3715. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 9.Raimundo BC, Oslob JD, Braisted AC, Hyde J, McDowell RS, Randal M, Waal ND, Wilkinson J, Yu CH, Arkin MR. J Med Chem. 2004;47:3111–3130. doi: 10.1021/jm049967u. [DOI] [PubMed] [Google Scholar]
- 10.Thanos CD, Randal M, Wells JA. J Am Chem Soc. 2003;125:15280–15281. doi: 10.1021/ja0382617. [DOI] [PubMed] [Google Scholar]
- 11.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 12.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 13.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. Science. 2005;308:1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 14.Kuntz ID, Chen K, Sharp KA, Kollman PA. Proc Natl Acad Sci USA. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins AL, Groom CR, Alex A. Drug Discovery Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 16.Hyde J, Braisted AC, Randal M, Arkin MR. Biochemistry. 2003;42:6475–6483. doi: 10.1021/bi034138g. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham BC, Wells JA. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 18.Zurawski SM, Vega F, Jr, Doyle EL, Huyghe B, Flaherty K, McKay DB, Zurawski G. EMBO J. 1993;12:5113–5119. doi: 10.1002/j.1460-2075.1993.tb06206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Rickert M, Garcia KC. Science. 2005;310:1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 20.Lowman HB, Bass SH, Simpson N, Wells JA. Biochemistry. 1991;30:10832–10838. doi: 10.1021/bi00109a004. [DOI] [PubMed] [Google Scholar]
- 21.Rao BM, Girvin AT, Ciardelli T, Lauffenburger DA, Wittrup KD. Protein Eng. 2003;16:1081–1087. doi: 10.1093/protein/gzg111. [DOI] [PubMed] [Google Scholar]
- 22.Ballinger MD, Jones JT, Lofgren JA, Fairbrother WJ, Akita RW, Sliwkowski MX, Wells JA. J Biol Chem. 1998;273:11675–11684. doi: 10.1074/jbc.273.19.11675. [DOI] [PubMed] [Google Scholar]
- 23.Dwyer JJ, Dwyer MA, Kossiakoff AA. Biochemistry. 2001;40:13491–13500. doi: 10.1021/bi011703b. [DOI] [PubMed] [Google Scholar]
- 24.Horn JR, Kraybill B, Petro E, Coales SJ, Morrow JA, Hamuro Y, Kossiakoff AA. Biochemistry. 2006;45:8488–8498. doi: 10.1021/bi0604328. [DOI] [PubMed] [Google Scholar]
- 25.Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 26.Middleton SA, Barbone FP, Johnson DL, Thurmond RL, You Y, McMahon FJ, Jin R, Livnah O, Tullai J, Farrell FX, et al. J Biol Chem. 1999;274:14163–14169. doi: 10.1074/jbc.274.20.14163. [DOI] [PubMed] [Google Scholar]
- 27.Sidhu SS, Lowman HB, Cunningham BC, Wells JA. Methods Enzymol. 2000;328:333–363. doi: 10.1016/s0076-6879(00)28406-1. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel TA. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 30.Sidhu SS, Weiss GA, Wells JA. J Mol Biol. 2000;296:487–495. doi: 10.1006/jmbi.1999.3465. [DOI] [PubMed] [Google Scholar]
- 31.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J, Cieplak P, Kollman PA. J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 33.Halgren TA. J Comput Chem. 1996;17:490–519. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.