Abstract
Molecular epidemiologic studies of tuberculosis (TB) have focused largely on utilizing molecular techniques to address short- and long-term epidemiologic questions, such as in outbreak investigations and in assessing the global dissemination of strains, respectively. This is done primarily by examining the extent of genetic diversity of clinical strains of Mycobacterium tuberculosis. When molecular methods are used in conjunction with classical epidemiology, their utility for TB control has been realized. For instance, molecular epidemiologic studies have added much-needed accuracy and precision in describing transmission dynamics, and they have facilitated investigation of previously unresolved issues, such as estimates of recent-versus-reactive disease and the extent of exogenous reinfection. In addition, there is mounting evidence to suggest that specific strains of M. tuberculosis belonging to discrete phylogenetic clusters (lineages) may differ in virulence, pathogenesis, and epidemiologic characteristics, all of which may significantly impact TB control and vaccine development strategies. Here, we review the current methods, concepts, and applications of molecular approaches used to better understand the epidemiology of TB.
INTRODUCTION
Consumption, King’s Evil, lupus vulgaris, and phthisis are some of the more colorful names for tuberculosis (TB) that have been used in the last several centuries. Archeological findings from a number of Neolithic sites in Europe and sites from ancient Egypt to the Greek and Roman empires show evidence of a disease consistent with modern TB. TB was described by Hippocrates (400 B.C.) in Of the Epidemics and was clearly documented by Claudius Galen during the Roman Empire. Likewise, TB has been more recently immortalized by artists such as John Keats, D. H. Lawrence, Anton Chekhov, Emily Bronte, Charlotte Bronte, Franz Kafka, Amedeo Modigliani, and Frederick Chopin, all of whom were afflicted by the disease.
In 1882, Robert Koch made the landmark discovery that TB is caused by an infectious agent, Mycobacterium tuberculosis. Although demystifying, Koch's findings introduced the possibility that antimicrobial agents could be developed to combat this age-old scourge (144). Today, despite the availability of effective antituberculosis chemotherapy for over 50 years, TB remains a major global health problem. As the rates of TB infection have fallen dramatically in industrialized countries in the past century, resource-poor countries now bear over 90% of all cases globally. In fact, there are more cases of TB today than ever recorded. As such, there is a need for new therapeutics, diagnostics, and vaccines in conjunction with improved operational guidelines to enhance current TB control strategies. While much is known about the epidemiology of TB, key questions have eluded classical epidemiologists for decades. These include the current rates of active transmission by differentiating disease due to recent or previous infection; the determination of whether recurrent tuberculosis is attributable to exogenous reinfection; whether all M. tuberculosis strains exert similar epidemiologic characteristics in populations; and an understanding of transmission dynamics on a population- or group-specific level, as well as in identifying extensive transmission or outbreaks from what appear to be sporadic, epidemiologically unrelated cases. Molecular epidemiologic methods have facilitated studies that address some of these very questions. In this review, we present the current approaches and issues surrounding the molecular epidemiology of M. tuberculosis and the insights that this relatively new field has contributed to our general understanding of TB epidemiology, pathogenesis, and evolution.
Epidemiology of Tuberculosis
Global incidence and prevalence.
The World Health Organization (WHO) estimates that approximately one-third of the global community is infected with M. tuberculosis (86). In 2000, an estimated 8 to 9 million incident cases and approximately 3 million deaths due to TB occurred worldwide (63). After human immunodeficiency virus (HIV)/AIDS, TB is the second most common cause of death due to an infectious disease, and current trends suggest that TB will still be among the 10 leading causes of global disease burden in the year 2020 (184).
The global distribution of TB cases is skewed heavily toward low-income and emerging economies. The highest prevalence of cases is in Asia, where China, India, Bangladesh, Indonesia, and Pakistan collectively make up over 50% of the global burden. Africa, and more specifically sub-Saharan Africa, have the highest incidence rate of TB, with approximately 83 and 290 per 100,000, respectively. TB cases occur predominantly (approximately 6 million of the 8 million) in the economically most productive 15- to 49-year-old age group (86). Our understanding of TB epidemiology and the efficacy of control activities have been complicated by the emergence of drug-resistant bacilli and by the synergism of TB with HIV coinfection.
Drug resistance.
No sooner were the first antituberculosis agents introduced in humans than the emergence of drug-resistant isolates of M. tuberculosis was observed (172, 190, 293). In vitro studies showed that spontaneous mutations in M. tuberculosis can be associated with drug resistance, while selective (antibiotic) pressure can lead to enhanced accumulation of these drug-resistant mutants (72, 73). The efficient selection of drug resistance in the presence of a single antibiotic led investigators to recommend combination therapy using more than one antibiotic to reduce the emergence of drug resistance during treatment (40, 47, 88). Indeed, when adequate drug supplies are available and combination treatment is properly managed, TB control has been effective (145, 178).
Selection for drug-resistant mutants in patients mainly occurs when patients are treated inappropriately or are exposed to, even transiently, subtherapeutic drug levels, conditions that may provide adequate positive selection pressure for the emergence and maintenance of drug-resistant organisms de novo. One of the contributing factors is the exceptional length of chemotherapy required to treat and cure infection with M. tuberculosis (142). The need to maintain high drug levels over many months of treatment, combined with the inherent toxicity of the agents, results in reduced patient compliance and subsequently higher likelihood of acquisition of drug resistance (74). Therefore, in addition to identifying new antituberculosis agents, the need for shortening the length of chemotherapy is paramount, as it would greatly impact clinical management and the emergence of drug resistance. Since the early 1990s, an alarming trend and a growing source of public health concern has been the emergence of resistance to multiple drugs (MDR-TB), defined as an isolate that is resistant to at least isoniazid (INH) and rifampin (RIF), the two most potent antituberculosis drugs (133, 269). Recent estimates suggest that in 2003 there were 458,000 incident cases (including new and retreatment cases) of MDR-TB globally (95% confidence interval, 321,000 to 689,000) (85, 297). These figures suggest that prevalent cases may be two or three times more numerous than incident cases and that a far greater number of individuals are latently infected (33, 284). While treatment for MDR-TB has greatly improved (mainly in resource-rich settings), it is generally more difficult to treat and has been associated with very high morbidity and mortality, prolonged treatment to cure, and an increased risk of spreading drug-resistant isolates in the community (26, 67, 132, 178).
HIV/AIDS.
HIV infection exerts immense influence on the natural course of TB disease. Individuals with latent M. tuberculosis infection who contract HIV are at risk of developing active TB at a rate of 7 to 10% per year, compared to approximately 8% per lifetime for HIV-negative individuals (219, 220). HIV-infected persons recently infected with M. tuberculosis may progress to active disease at a rate over 35% within the first 6 months, compared to 2 to 5% in the first 2 years among HIV-negative individuals (70). With the introduction of highly active antiretroviral therapy for HIV, the risk of progression to TB among those coinfected with M. tuberculosis, while higher than among HIV-negative cases, is considerably lower (8, 111). The role for CD4+ T cells in protecting against disease progression is underscored by the marked susceptibility to TB in patients with advanced HIV-induced CD4+ T-cell depletion (70, 77, 219). The natural course of HIV disease may also be influenced by M. tuberculosis infection. M. tuberculosis infection results in macrophage activation, which can house resident HIV virions, resulting in active expression of HIV antigens rather than the prolonged latency without antigenic expression of HIV proteins (252). In support of this, Pape et al. observed more rapid progression to AIDS among tuberculin skin test (TST)-positive individuals not given treatment for latent TB infection (INH) than among those who were treated with INH (195). Thus, HIV infection tends to accelerate the progression of TB, while in turn, the host immune response to M. tuberculosis can enhance HIV replication and may accelerate the natural course of HIV/AIDS (252).
Natural Course of Tuberculosis
Historically, much of our understanding of TB has stemmed from descriptive epidemiological studies, limited animal studies, and clinical observations that were made in the early half of the 20th century. These studies have been central to formulating a generalized hypothesis regarding all phases of TB pathogenesis, from exposure to successful infection and subsequent disease (59, 61, 91, 170, 171, 237, 240). Infection is established in approximately one-third of individuals exposed to the tubercle bacillus, and among those infected only 10% ever become symptomatic (61, 134, 203). In most populations, TB involves a long latency period, with symptomatic presentation occurring from 3 months (mainly in the immunocompromised) to decades after the establishment of infection (61, 142). Latency is one of the main hallmarks of M. tuberculosis infection and pathogenesis and has been reviewed specifically elsewhere (4, 118).
TB is spread by aerosolization of droplet nuclei bearing M. tuberculosis particles released from the lungs of patients with cavitary pulmonary or laryngeal disease. Once the particles, of 1 to 5 μm in diameter, are inhaled and phagocytosed by resident alveolar macrophages, a vigorous host cellular immune response involving cytokines and a large number of chemokines ensues (126, 164, 212). This response presumably arrests and limits infection to the primary site of invasion, the lung parenchyma and the local draining lymph nodes (“Ghon complex”), in the majority (90%) of immunocompetent individuals (31, 110). Protective immunity is characterized by granuloma formation that consists primarily of activated M. tuberculosis-infected macrophages and T cells. In 10% of presumed immunocompetent individuals, the infection is not contained and continual bacillary replication (doubling time, 25 to 32 h) results in disease symptoms and associated pathology, including tissue necrosis and cavitation (175). In most instances, patients respond to antibiotic treatment by clearance of the bacilli from tissues and subsequently from sputum, partial reversal of the granulomatous process, and clinical cure (227). When disease ensues, the presentation is variable in regard to severity, duration, therapeutic response, and tissue tropism. Although commonly pulmonary, M. tuberculosis can infect a variety of tissues, such as the meninges, lymph nodes, and tissues of the spine (134, 221). A number of external factors may influence the progression and nature of disease. These include comorbid conditions that dampen the host immune system, such as poorly controlled diabetes mellitus, renal failure, chemotherapy, malnutrition, or intrinsic host susceptibility (19, 281).
Due to the variability in time from infection to disease between individuals, incident cases are comprised of reactivation of a historic infection or the result of a recent transmission event (274, 275). While treatment of reactive and recent transmission cases is similar, the latter may be part of an ongoing outbreak or series of transmission events that warrants control measures. Therefore, a central limitation in understanding the transmission dynamics of M. tuberculosis is that patient links often become obscured as the concentric circles of traditional epidemiological relatedness (contact tracing) are more removed from the index case. In general, cases in low-incidence areas tend to comprise mostly reactive disease, while those in high-incidence regions include both reactive disease and recent transmission.
A crucial aspect in understanding the dynamics of a TB epidemic is the ability to track the spread of specific strains in the population. As discussed below (see “Molecular Epidemiology and Public Health”), over the past two decades, previously unresolved issues, such as population estimates of recent transmission and the ability to distinguish endogenous reactivation from exogenous reinfection, have been made possible by the use of a variety of molecular techniques (15, 35, 46, 109, 224, 263, 275).
MOLECULAR EPIDEMIOLOGY
Molecular epidemiology is a field that has emerged largely from the integration of molecular biology, clinical medicine, statistics, and epidemiology. In essence, molecular epidemiology focuses on the role of genetic and environmental risk factors, at the molecular/cellular or biochemical level, in disease etiology and distribution among populations. More specifically to infectious diseases, molecular epidemiology attempts to utilize a multidisciplinary approach to identify factors that determine disease causation, propagation/dissemination, and distribution (in time and space). This is primarily achieved by associating epidemiologic characteristics with the biologic properties of clinical isolates recovered from symptomatic individuals.
The mid-1980s saw the first integration of molecular methods to discriminate between clinical isolates of M. tuberculosis. While previous methods, such as colony morphology, comparative growth rates, susceptibility to select antibiotics, and phage typing, were useful, they did not provide sufficient discrimination, thus limiting their utility in TB epidemiology. That is, prior to molecular methods, understanding the spread of TB was imprecise and relied on observational data or anecdotal correlations. However, given the plethora of molecular tools available, it is critical to choose an appropriate method(s) to address a particular study question, e.g., transmission dynamics, outbreaks, or phylogenetics. In general, the key aspects in choosing an adequate molecular approach for studying TB epidemiology are the observed rate of polymorphism (stability of biomarker) and the genetic diversity of strains in the population. That is, the rate of change of a biomarker must be adequate to distinguish nonepidemiologically related strains and yet sufficiently “slow” to reliably link related cases. This issue, coupled with general background TB prevalence, should be taken into consideration when choosing molecular epidemiologic methods or in evaluating data.
Genotyping of M. tuberculosis: Current Methods
The TB research community entered the genomic era in 1998 with the publication of the complete annotated genome of M. tuberculosis laboratory strain H37Rv (60). Since then, M. tuberculosis clinical strain CDC1551 and six related mycobacteria, M. leprae, M. ulcerans, M. avium, M. avium paratuberculosis, M. smegmatis, and M. bovis, have been fully sequenced; others, including M. microti, M. marinum, M. tuberculosis strain 210, and M. bovis BCG (bacillus Calmette-Guérin), are nearing completion.
Studies show that the M. tuberculosis complex (i.e., M. tuberculosis, M. bovis, M. microti, M. africanum, M. canettii, and, more recently, M. pinnipedii and M. caprae [7, 64, 103]) genomes are highly conserved: comparative sequence analysis of the 275-bp internal transcribed spacer (ITS) region, an otherwise highly polymorphic region which separates the 16S rRNA and the 23S rRNA, revealed complete conservation between members of the M. tuberculosis complex. Furthermore, sequence analysis of 56 structural genes in several hundred phylogenetically and geographically diverse M. tuberculosis complex isolates suggested that allelic polymorphisms are extremely rare (139, 188, 235). While the members of the M. tuberculosis complex display diverse phenotypic characteristics and host ranges, they represent an extreme example of interspecies genetic homogeneity, with an estimated rate of synonymous nucleotide polymorphisms of 0.01% to 0.03% (60, 98, 123, 235) and no significant evidence for horizontal genetic transfer between genomes, unlike most bacterial pathogens (3, 41, 123, 248).
While the M. tuberculosis complex genome is highly restricted (conserved) in relation to other bacterial pathogens, this monomorphic species does have polymorphic genomic regions. Much like eukaryotic genomes, those of prokaryotes (such as M. tuberculosis) are characteristically punctuated by monomeric sequences repeated periodically (repeated units). There are two types of repetitive units, interspersed repeats (IR) (direct repeats and insertion sequence-like repeats) and tandem repeats (TR) (head-to-tail direct uninterrupted repeats). Prokaryotic microsatellites (1- to 10-bp repeats) and minisatellites (10- to 100-bp repeats, commonly referred to as variable-number tandem repeats [VNTR]) are located in intergenic regions, in regulatory regions, or within open reading frames and are abundant throughout most bacterial genomes. Below, we describe some of the most common genotyping methods currently used. Table 1 summarizes the advantages, limitations, and applications of the various molecular techniques.
TABLE 1.
Evaluation of methods currently used to study the molecular epidemiology of TB
| Typing technique | Advantages | Limitations | Comments |
|---|---|---|---|
| IS6110 RFLP | Gold standard for the molecular | Requires subculturing and DNA isolation | First standardized method |
| analysis | epidemiology of MTCa strains | Slow turnaround time (30-40 days) | Southern blot hybridization-based |
| Patterns can be computerized with | Process is laborious | technique | |
| specialized software | Cannot be used to reliably type isolates with | IS6110 fingerprinting remains the single | |
| Widely utilized; hence, much data | ≤6 IS6110 insertions | most discriminatory technique for the | |
| available for comparison | Poor portability: interlaboratory | analysis of isolates having >6 IS6110 | |
| Biological clock (biomarker stability) | comparative analysis of RFLP patterns | bands | |
| has proven to be very adequate | can be tedious | Provides best resolution for the analysis | |
| for the study of transmission | Strains with no IS6110 insertion (rare) | of W-Beijing isolates | |
| Extensive diversity in patterns for | IS6110 transposition and/or deletion | ||
| isolates with >6 IS6110 insertions | events are always unidirectional and | ||
| Membranes can be rehybridized with | can be considered a form of | ||
| other probes, e.g., for IS mapping | divergent evolution | ||
| or deletion analysis | Some “hot spots,” or preferred | ||
| Mixed infection readily detected by | insertion sites, exist | ||
| varying intensity of the | Can be combined with other molecular | ||
| hybridization bands | techniques for phylogenetic studies | ||
| Applications include molecular epidemiology, evolutionary and phylogeny studies, and detection of laboratory error/cross-contamination | Can be combined with spoligotyping for isolates with ≤6 bands for increased resolution | ||
| Spoligotyping | Simplest technique for MTC strain genotypingData are presented in binary format, allowing inter- and intralaboratory comparisonsCommercial hybridization membranes available for the simultaneous analysis of 45 samplesStandardized analysis for 43 spacersCan be performed directly on cell lysate; no DNA purification requiredCan be performed on nonviable bacteriaTwo large databases available for comparative analysis (see comments)Applications: ideal for a first-step analysis of M. tuberculosis, particularly in regions with diverse populations; molecular epidemiology; and detection of laboratory error/cross-contamination | Less discriminatory than IS6110 RFLP analysis and MIRU-VNTR (12 and 15 loci)Cannot recognize mixed infectionsLess informative in regions with predominant or endemic strains; e.g., W-Beijing in China, Southeast Asia, and Russia | Amplification-hybridization-based analysisTechnique used extensively for phylogeographic studiesFalse-positive strain relatedness may be identified based on convergent evolution of strains; the same patterns may be found on distinct evolutionary branches (due to the fact that the same spacer may be lost independently in different lineages, e.g., strains from clusters III and VII [Fig. 2B])SPOTCLUST website for analysis of spoligofamilies (http://cgi2.cs.rpi.edu/∼vitoli/SPOTCLUST.html)SpolDB4 database (43) and SITVIT database (http://www.pasteur-guadeloupe.fr:8081/SITVITDemo/) |
| MIRU-VNTR (12 loci) | Rapid, high-throughput technique for MTC strain genotypingBetter resolution than spoligotypingDigitized results (number of copies of each repeat) are very portableWell suited for large-scale genotypingCan be performed directly on cell lysate; no DNA purification requiredManual analysis possible by 12 individual PCR amplifications followed by gel electrophoresisAutomated analysis possible with fluorescence-tagged PCR primers and capillary separation (sequencer) or nondenaturing high-performance liquid chromatography | Less discriminatory than IS6110 RFLP genotypingCombined biological clock of 12-locus MIRU-VNTR too slow for the study of endemic strainsSimilar patterns may be found in distinct lineages | Amplification-electrophoresis- based analysisEach locus has different molecular clockStutter bands can occur (DNA replication slippage during PCR amplification of microsatellites)Has also been used for population genetic/evolutionary investigations |
| Labeled primers allow for multiplex PCRs: 4 reactions of 3 multiplex eachCan be used to identify mixed infectionsApplications include molecular epidemiology, the potential for real-time genotyping, and high-throughput typing | |||
| MIRU-VNTR (15 loci) | As for 12-locus MIRU-VNTR typing but with increased resolution (comparable to IS6110 RFLP)Applications include molecular epidemiology, the potential for real-time genotyping, and high-thoughput typing | See 12-locus MIRU-VNTRNo data available yet | See 12-locus MIRU-VNTRThe 15 loci have been selected out of 29 loci testedSelection of loci has yet to be evaluated in different settingsPossible future method of choice for large-scale typing |
| Deletion mapping and deligotyping | Irreversible genetic marker usedHigh throughput with microarray analysisReverse line probe with hybridization membrane possibleResults can be digitalizedMultiplex PCR for 43 loci availableSingle-deletion analysis can identify M. bovis BCGApplications include phylogenetic/evolutionary studies, facilitation of genome structure-function studies and host-pathogen interactions based on specific genomic deletions, and molecular epidemiology | Not yet standardizedRepresentative target deletions need to be determinedTechnique has yet to be evaluated in different settings | Microarray- or amplification-electrophoresis-based analysisAmplification of selected deletions possible by using flanking regionsDiscriminatory power can be greatly increased if direct flanking region is sequencedNeed to discriminate unique deletion events from recurrent deletion events; unique events may be used to determine phylogenetic lineages, while recurrent deletion events are a form of convergent evolution |
| Insertion site mapping and insertion site typing (Insite) | Very precise determination of strain relatednessStrain-specific markers can be used for rapid identification of a particular strain or strain familyAmplification-based investigation for rapid detectionHighly informative when studying strain relatedness and clonalityApplications: best suited for confirmation of clustering of strains; also phylogenetic studies and molecular epidemiology | Need to predetermine IS6110 flanking regionsFor Insite, need to amplify and immobilize target DNA on membrane firstLaborious | Amplification-hybridization-based technique for large-scale analysisVery useful for the confirmation of particular strains or strain familiesInsite also known as Reverse Dot Blot |
| SNP analysis | Most-precise information on strains based on sequencing of polymorphic lociHigh resolutionSome selected SNP can be highly informativeTechnique can be automated for large-scale genotypingApplications include phylogenetic and population genetic investigations, molecular epidemiology, studies of drug resistance, and research on host-pathogen interactions | Requires extensive genomic sequencing of multiple chromosome targets | sSNP do not result in amino acid change and not associated with selective pressure; hence, ideal for population genetic studiesnsSNP create an amino acid change and may be subject to selection pressure; can be used to study drug resistance-determining genetic loci |
MTC, M. tuberculosis complex.
IS6110.
Insertion sequences (IS) are small mobile genetic elements, usually less than 2.5 kb in size, that are widely distributed in most bacterial genomes (52). IS elements are commonly defined as carrying only the genetic information related to their transposition and regulation, unlike transposons, which can also carry genes that encode phenotypic markers (e.g., antibiotic resistance). Transposition of IS elements often causes gene disruptions that can have strong polar effects and in other cases can lead to the activation or alteration of expression of adjacent genes due to the regulatory sequences, including promoters and protein-binding sequences (52, 216a). From an evolutionary perspective, there are at least two distinct hypotheses explaining the role of IS elements in genomes. One regards the elements as genomic parasites that, on balance, harm their hosts (i.e., bacteria) (53). In contrast, others postulate that IS elements are important to their hosts for adaptive evolution, which is maintained by selection of occasional advantageous IS-derived mutations (32).
IS elements in bacterial species are present in varying numbers of copies: IS1 in Escherichia coli strains is present in 2 to 17 copies, whereas the Shigella species contain from 2 to 40 copies (52). Thierry et al. first described IS6110, a 1,355-bp member of the IS3 family that, when intact, is unique to the M. tuberculosis complex (250). IS6110 has an imperfect 28-bp inverted repeat at its ends and generates a 3- to 4-bp target duplication on insertion. Although “hot spots” have been noted (regions in the M. tuberculosis chromosome where IS6110 seems to preferentially insert), IS6110 elements are more or less randomly distributed throughout the genome, with copy numbers ranging from rare clones lacking any IS6110 elements to those with 26 copies (Fig. 1) (152, 174). In 1993, van Embden and colleagues proposed a standardized method for performing IS6110-based Southern blot hybridization analysis (259). The recommendation was based on the use of a common restriction endonuclease (PvuII, which cleaves IS6110 at a single asymmetric site and yields reasonable-size M. tuberculosis chromosomal fragments), a hybridization probe (specific to the right side of IS6110, whereby each hybridizing band corresponds to a PvuII-PvuII chromosomal fragment with a single IS6110 insertion), and standardized molecular weight markers (127). The concurrent development of software applications that assist in the analysis of the resulting IS6110-based restriction fragment length polymorphism (RFLP) patterns has allowed for intra- and interlaboratory comparisons of clinical isolates and the establishment of large national and international strain (and genotype) archives (e.g., Centers for Disease Control and Prevention, Atlanta, GA; Public Health Research Institute, Newark, NJ; National Institute of Public Health and Environment, Bilthoven, The Netherlands) (125, 150, 244, 261).
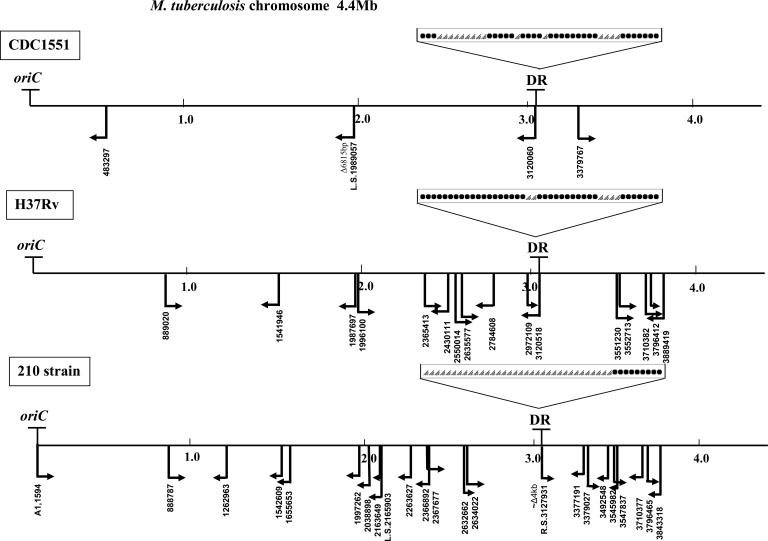
FIG. 1.
Chromosomal maps of three M. tuberculosis strains: CDC1551 (http://tigrblast.tigr.org/cmr-BLAST/), H37Rv (http://genolist.pasteur.fr/TubercuList/index.html), and 210 (http://tigrblast.tigr.org/ufmg/index.cgi?database=m_tuberculosis-strain210%7Cseq). Arrows show the positions and orientations of IS6110 insertions. Left- and right-oriented arrows on the maps indicate IS6110 insertions, confirmed by insertion site mapping at the Public Health Research Institute Tuberculosis Center (unpublished data) and the positions of IS6110 according to Beggs et al. (18). The coordinates of the IS6110 insertions in all three strains correspond to the H37Rv annotated sequence. IS6110 mapping indicates that despite the insertion-preferential loci (“hot spots”), the precise positions (flanking sequences of the IS element) and orientations of the inserted sequences are not repeated in the three strains analyzed; i.e., none of the flanking regions of the four IS6110 copies in CDC1551 correspond to the 17 insertion sites in H37Rv or to the 23 positions (resulting in 21 hybridization bands) of the IS element in strain 210. The structures of the DR loci within these three strains are shown; black dots indicate spacers in the DR locus of corresponding strains, and triangles indicate deleted spacers. The chromosomal loci oriC and DR were described previously (137, 152, 167).
Initially, the dynamics of IS6110 transposition juxtaposed with the stability required for use in epidemiologic investigations was a cause for concern. However, when strains were cultured in vitro (liquid media) for 6 months, in macrophages over a 4-week period, and in a guinea pig model for more than 2 months, their IS6110-based RFLP patterns remained stable (50, 267). These studies attest to the stability of IS6110 over short time periods while transposing over longer time intervals. The IS6110 transposition half-life (t1/2) (the period over which the IS-specific hybridization pattern does not change), taken from sequentially positive culture with sampling intervals ranging from days to months, was estimated to be between 3 and 4 years (75, 291). Warren et al. investigated the stability of IS6110 banding patterns in serial M. tuberculosis isolates collected from patients living in areas of high TB incidence and noted a half-life of 8.74 years when a constant rate of change was assumed (278). The authors note that the rate may be composed of the high rate of change seen during the early disease phase (t1/2 = 0.57 years), when the mycobacterial replication rate is presumably high, and the lower rate in the late disease phase (t1/2 = 10.69 years), when bacterial doubling times are longer during or after treatment. Therefore, they conclude that the observed IS6110 stability is strongly influenced by the time between onset of disease and sample collection. Another investigation of serial patient isolates used deterministic and stochastic simulation models to estimate an IS half-life of 2.4 years for a strain that has 10 IS6110 copies (215). Indeed, IS6110 transposition, which is a replicative process, and half-life may be heavily dependent on strain-specific in vivo replication rates, host-pathogen interactions, or anatomical properties. Nonetheless, IS6110-based RFLP patterns seem to be sufficiently stable (and polymorphic) for studying TB transmission dynamics at the local or population level and over time. For instance, Lillebaek et al. used IS6110 genotyping to demonstrate endogenous reactivation of TB after over 30 years of latency (156).
The utility of any molecular epidemiologic method in population analysis, in addition to adequate stability/polymorphism, is reliant on sufficient biomarker-specific diversity of isolates. Assignment of a genotype is strengthened when there is adequate background strain diversity. In a population-based study in New Jersey, Bifani et al. noted that approximately one-third of the 1,207 clinical isolates subjected to IS6110-based RFLP analysis were unique (or “orphans”) to the sample, while a third of the isolates were categorized into 11 major strain groups that consisted of isolates from 10 or more patients (25). Presumably there is a discrete number of distinct strain types circulating within any given population; classifying a genotype as rare or unique is heavily dependent on the isolate sampling schemes and the size and diversity of the reference database.
As with any genotyping system, there are limitations inherent to IS6110-based RFLP analysis. One such limitation, not partial to IS6110 genotyping, is the interpretation of molecular data in drawing epidemiologic conclusions. That is, genotypic clustering (identical/similar fingerprints of strains isolated from at least two patients) is not synonymous with epidemiologically defined clustering (patient-patient link). This is especially important to keep in mind in areas with low M. tuberculosis genetic diversity or in areas of high endemicity (13, 27, 29, 38). In such situations, strain clustering may involve a number of distinct transmission pathways that finally may not be epidemiologically informative (false-positive links). This shortcoming is similar to that of conventional field epidemiologic investigations where distinct transmission patterns are often elusive, particularly in areas of high TB incidence (29, 112, 224). Therefore, suggested molecular epidemiologic links are greatly strengthened when they are in concordance with conventional methods of TB control (25, 27, 29). A second limitation often cited is the limited resolution in analyzing clinical strains with six or fewer copies of IS6110 (“low-copy-number” strains, clusters I, IIA, IV, and V [Fig. 2A ]) (14, 150, 288, 290). The resolution afforded by the IS6110 RFLP genotyping method is inversely proportional to the number of IS elements, such that identical hybridization patterns may not indicate clonality when six or fewer bands comigrate. Although an IS6110-probed band on a hybridization blot indicates the presence and size of the PvuII-PvuII IS6110-associated DNA fragment, it does not provide the chromosomal location of the IS element. Therefore, identical bands may be from distinct genomic locales. Low-copy-number isolates have been shown to be genetically distinct when secondary independent biomarkers were used (14, 54, 210, 288). In contrast, high-copy-number strains (i.e., bearing more than six IS6110 copies, clusters I to III and VI to VIII [Fig. 2A]) with identical patterns are more likely to be clonal, as the probability of hybridization bands of similar size originating from different IS6110 locations is low. There exist, albeit rarely, strains that lack IS6110, rendering this genotyping method irrelevant (71, 217). Additional limitations of this genotyping system include its inability to distinguish among M. tuberculosis complex members and its labor intensiveness (Table 1) (69).
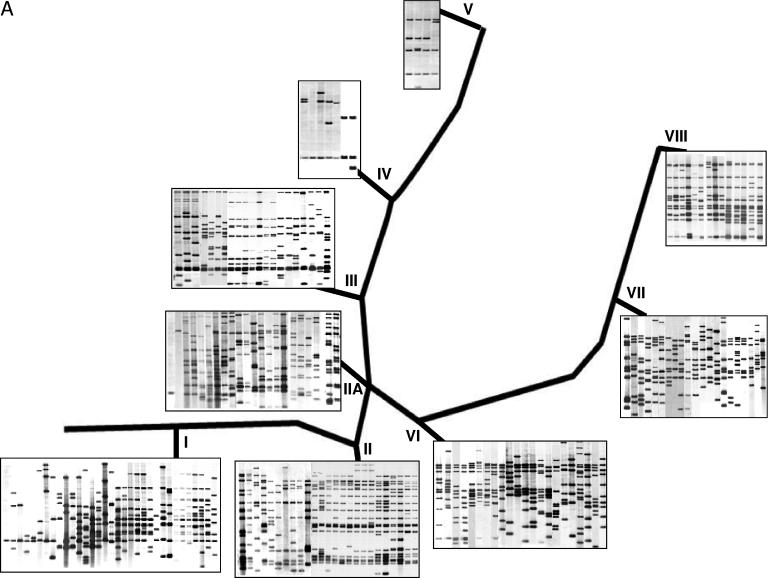
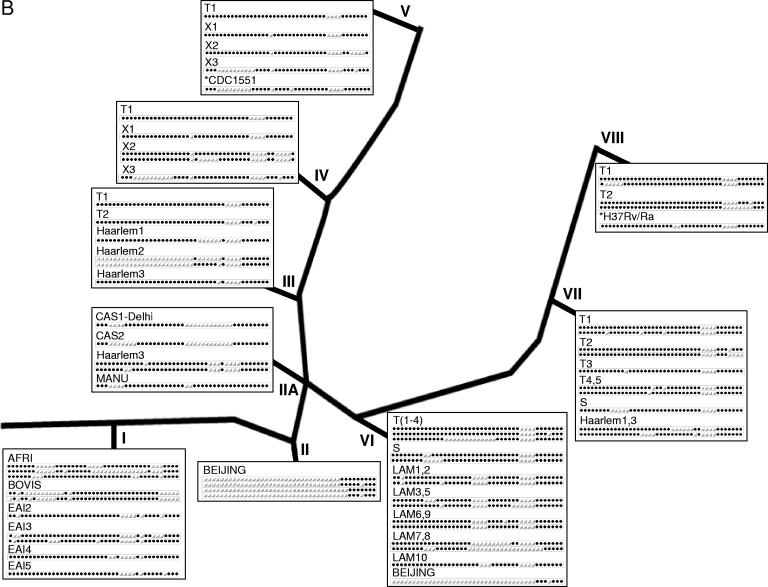
FIG.2.
Representative genotypes superimposed on the SNP-derived phylogenetic framework of M. tuberculosis. Based on SNP analysis of M. tuberculosis clinical isolates (including 1,743 strains from Public Health Research Institute Tuberculosis Center strain collection), a phylogenetic tree with the nine clusters of M. tuberculosis isolates was used to illustrate common genotypic patterns (122). (A) IS6110-based RFLP images. (B) Spoligotype patterns (black dots show spacers present in the chromosomal DR region of strains, and open triangles indicate deleted spacers). The strain spoligofamily definition corresponds to the SpolDB4 (43). Cluster I includes M. tuberculosis complex strains and TbD1+ ancestral isolates. Cluster II is represented by the W-Beijing strain family, including strain 210. Cluster II.A comprises the CAS spoligotype isolates. Clusters I and II belong to PGG1, while II.A comprises both PGG1 and PGG2. The coclustering of isolates from PGG1 and PGG2 in cluster II.A is also shared by some spoligotypes (panel B). PGG2 is further delineated into clusters III, IV, V (including CDC1551), and VI, while PGG3 is represented by clusters VII and VIII (including H37Rv). Isolates with a single IS6110 insertion are found in clusters I, IIA, and IV. Likewise, some spoligotypes appear in more than one cluster. Similar/identical spoligopatterns may be found in unrelated strain clusters (e.g., “Beijing” spoligotypes in cluster VI or “Haarlem” spoligotypes in cluster VII) as a result of independent spacer deletion events; this convergence of spoligotypes could lead to the misinterpretation of genotyping results and illustrates the necessity of using two or more techniques in genotypic analysis. *, annotated laboratory strains (CDC1551 and H37Rv). (Adapted from reference 122 with permission. © 2005 by the Infectious Diseases Society of America. All rights reserved.)
PGRS.
Like IS6110-based RFLP analysis, polymorphic GC-rich repetitive sequence (PGRS) genotyping, first described by Ross et al., is a Southern blot hybridization technique that utilizes the PGRS-specific probe (a 3.4-kb fragment of the PGRS sequence) cloned in plasmid pTBN12 (216). When pTBN12 is used on AluI-digested DNA, it can distinguish strains from unrelated cases of TB and demonstrate identical banding patterns for isolates from epidemiologically related cases (216, 288). In fact, isolates clustered by IS6110-based RFLP analysis were further discriminated by PGRS typing (54). This is particularly the case when IS6110 low-copy-number strains are further analyzed by PGRS genotyping (210, 289). This method, like IS6110 genotyping, is resource intensive, but unlike the IS6110 system, the hybridization patterns generated by PGRS typing are often too complex to computerize for standardization and analysis.
Spacer oligonucleotide typing.
After IS6110-based RFLP analysis, spacer oligonucleotide typing (spoligotyping) is the most commonly used PCR-based technique for subspeciating M. tuberculosis strains (121). M. tuberculosis complex strains contain a distinct chromosomal region consisting of multiple 36-bp direct repeats (DRs) interspersed by unique spacer DNA sequences (35 to 41 bp) (Fig. 1). Two forms of genetic rearrangements have been observed: one type consists of variation in one or a few discrete, contiguous repeats plus spacer sequences (DVRs), which is probably driven by homologous recombination between adjacent or distant chromosomal DRs; the other is driven by transposition of IS6110, which is almost invariably present in the DR locus of M. tuberculosis complex strains (260). As a result of these events, some spacers may be deleted from the genome.
Spoligotyping is based on the detection of 43 interspersed spacer sequences (originally identified in laboratory strain H37Rv and M. bovis BCG vaccine strain P3) in the genomic DR region of M. tuberculosis complex strains. Additional spacers in this region have been reported (260). Membranes spotted with 43 synthetic oligonucleotides are hybridized with labeled PCR-amplified DR locus of the tested strain, resulting in a pattern that can be detected by chemiluminescence (137). The results are highly reproducible, and the binary (present/absent) data generated can be easily interpreted and computerized and are amenable to intralaboratory comparisons. A recent edition of the international spoligotyping database, SpolDB4, contains 1,939 different spoligotypes (ST) identified worldwide that are organized into large ST families (43). ST families are nominated based on the common motif of deleted spacers. Recently, a web-based program has been developed to place spoligotypes into ST families (273). Spoligotyping, unlike IS6110 genotyping, which requires approximately 2 μg of bacterial DNA, can be performed with considerably less DNA and in a fraction of the time; it also allows genotyping of boiling-prepared or impure DNA, nonviable specimens, paraffin-embedded material, and material from slides of Ziehl-Neelsen stainings (82, 205, 258). In some instances, spoligotyping can distinguish among members of the M. tuberculosis complex based on the species-specific presence/absence of spacers (129, 137). It is thought that DR regions irreversibly lose spacers due to homologous recombination or IS6110 transposition events and cannot gain additional DNA fragments. Of note, deletions of DRs and spacers can occur multiple times and independently in unrelated strains, leading to convergent evolution, i.e., the appearance of identical spoligopatterns in phylogenetically unrelated M. tuberculosis strains (Fig. 2B) (277).
Although spoligotyping can be a powerful method to study the molecular epidemiology of M. tuberculosis, its discriminatory power in general is inferior to that afforded by IS6110-based RFLP analysis (150). Strains having identical spoligotype patterns yet distinct IS6110 fingerprint profiles are often encountered (22, 167, 260). For instance, the W-Beijing family of strains, a large phylogenetically related group of M. tuberculosis isolates that comprise hundreds of similar yet distinct IS6110 variations (Fig. 2A, cluster II), all have an almost identical spoligopattern lacking spacers 1 through 34 (Fig. 2B, Beijing) (24, 149, 268). In this case, spoligotyping may be useful in identifying W-Beijing strains in a population; however, this approach will not be able to discern transmission events, especially in regions where these genotypes are noted to be endemic, such as Russia, China, and South Africa (24, 115, 181). In contrast, spoligotyping has been shown to further discriminate IS6110 low-copy-number strains (14, 230). Kremer et al. have shown that spoligotyping together with IS6110 genotyping can provide an accurate and discriminatory genotyping system (150); this approach has been adopted for the universal genotyping program in New York, N.Y. (56). When used alone, the limited discriminatory power of spoligotyping is primarily because it targets a single locus that accounts for less than 0.1% of the M. tuberculosis genome (Fig. 1), unlike IS6110-based RFLP analysis, which examines the distribution of IS6110 throughout the entire genome.
VNTR and MIRU analysis.
Frothingham and Meeker-O'Connell performed a systematic analysis of VNTR loci in M. tuberculosis complex strains and found 11 loci comprising five major polymorphic tandem repeats (MPTR) (A to E) and six exact tandem repeats (ETR) (A to F) ranging in size from 53 to 79 bp (104). Since then, additional VNTR loci have been reported (119, 128, 146, 159, 189, 223, 229). Supply et al. identified 41 VNTR of mycobacterial interspersed repetitive units (MIRU) (tandem repeats of 40 to 100 bp) located in mammalian-like minisatellite regions scattered around the chromosome of H37Rv, CDC1551, and AF2122/97 (169, 247), including loci 4 (VNTR0580) and 31 (VNTR3192), which correspond to ETR D and E, respectively (104). Twelve of the 41 MIRU loci were selected for genotyping of M. tuberculosis clinical isolates and were reported in a 12-digit format corresponding to the number of repeats at each chromosomal locus (169, 247). The digitized data generated by MIRU-VNTR profiling is highly amenable to inter- and intralaboratory comparisons. As additional M. tuberculosis VNTR loci have been included, the various nomenclature from one laboratory to another has created some confusion. As such, Smittipat et al. have proposed a standardization of the VNTR nomenclature based on the four digits of the locus position on the H37Rv genome (for an equivalence table, see reference 228).
The discriminatory power of MIRU-VNTR analysis is typically proportional to the number of loci evaluated; in general, when only the 12 loci are used, it is less discriminating relative to IS6110 RFLP genotyping for isolates with high-copy-number IS6110 insertions but more discriminating than IS6110 RFLP genotyping for isolates with low-copy-number IS6110. When more than 12 loci are used, or MIRU analysis is combined with spoligotyping, the discriminatory power approximates that of IS6110 RFLP analysis. Recently, a comparative study of genotyping methods aimed at evaluating novel PCR-based typing techniques found VNTR analysis to have the greatest discriminatory power among amplification-based approaches (147). MIRU-VNTR genotyping has been used in a number of molecular epidemiologic studies, as well as to elucidate the phylogenetic relationships of clinical isolates (148, 231, 246, 248, 280). VNTR analysis has also been used to evaluate M. bovis transmission (214). A high-resolution MIRU-VNTR genotyping system using an automated sequencer and PCR primers tagged with one of four fluorescent dyes (FAM, NED, VIC, and HEX) has been developed, allowing amplification of four different loci simultaneously by multiplex PCR.
VNTR loci have a variable range of alleles; for example, within the 12 MIRUs, MIRU loci 2 (VNTR0154) and 24 (VNTR2687) have mostly 1 or 2 copies, while VNTR3820 can have from 3 to 32 copies (66, 228, 231, 246). Likewise, the discriminating capacity of a given locus, the molecular clock, or variability in alleles also varies extensively among the loci. For example MIRU10 (VNTR0960) has been found to be the most polymorphic, having mostly 1 to 7 copies or up to 12 alleles in the M. tuberculosis collections analyzed (66, 231, 246; also, unpublished data). Variability at specific MIRU loci often depends on the sample collection (e.g., nationwide, population based, or convenience sampling), geographic origin, and inherent genetic diversity of the strains. For example VNTR2059 has been found to be polymorphic in some studies but not in others (66, 228). An alternative selection of VNTRs should consider the intrinsic differences and variability within different genetic groups and the endemicity or predominance of clones in specific geographic and demographic populations. The use of different sets of VNTR from one collection to another would hamper the ease of interlaboratory analysis, one of the advantages of VNTR analysis. On the other hand, broadly increasing the overall number of loci for genotyping would increase the cost and labor required for analysis and complicate analysis and interpretation, not to mention reducing enthusiasm for routine epidemiological investigations. Presently, there is a concerted effort to select a better combination of VNTR for genotyping (248a). Fifteen of 29 MIRU-VNTR were selected, and >800 clinical isolates of diverse origin were analyzed for discriminatory power relative to IS6110 genotyping. Although promising, this new selection of MIRU-VNTR has yet to be evaluated in different settings.
SNP.
As extensive comparative genomic analysis of M. tuberculosis has revealed remarkable DNA conservation between chromosomes, noted genetic polymorphisms at the nucleotide level have provided researchers with markers to differentiate clinical isolates as well as to study the phylogenetic relatedness of clinical strains. Both nonsynonymous single-nucleotide polymorphisms (nsSNP) and synonymous SNP (sSNP) provide useful genetic information that can be applied to differentiate M. tuberculosis strains; however, they address different biologic questions.
In general, nonsynonymous polymorphisms create an amino acid change that might be subject to internal or external selection pressure. As such, nonsynonymous changes in drug resistance-determining genetic loci can result in phenotypic drug resistance. Accordingly, M. tuberculosis resistance to antituberculosis agents nearly always correlates with genetic alterations (nonsynonymous point mutations, small duplications, or deletions) in resistance-conferring chromosomal regions (Table 2) (168, 206, 208, 295). nsSNP in genes that confer drug resistance can aid in understanding the nature and spread of resistance between and within populations (see “Molecular studies on drug resistance,” below).
TABLE 2.
Genomic regions associated with decreased susceptibility to antituberculosis agentsa
| Antituberculosis agent | Gene | Product | Mutation frequency among drug-resistant clinical isolates (%)b |
|---|---|---|---|
| Streptomycin | rpsL | Ribosomal protein S12 | ∼60 |
| rrs | 16S rRNA | <10 | |
| Rifampin | rpoB | β subunit of RNA polymerase | >95 |
| Isoniazid | katG | Catalase-peroxidase | 60-70 |
| oxyR-ahpC | Alkylhydroreductase | ∼20 | |
| inhA | Enoyl-ACP reductase | <10 | |
| kasA | β-Ketoacyl-ACP synthase | <10 | |
| ndh | NADH dehydrogenase | NA | |
| Ethambutol | embCAB | Arabinosyltransferases | ∼70 |
| Pyrazinamide | pncA | Amidase | 70-100 |
| Ethionamide | inhA | Enoyl-ACP reductase | <10 |
| ethA | Flavoprotein monooxygenase | NA | |
| Kanamycin | rrs | 16S rRNA | ∼65 |
| Fluoroquinolone | gyrA | DNA gyrase α subunit | >90 |
| gyrB | DNA gyrase β subunit | NA | |
| Capreomycinc | tlyA | rRNA methyltransferase | NA |
| rrs | 16S rRNA | NA | |
| Para-aminosalicylic acidd | thyA | Thymidylate synthase | NA |
Mutation frequencies were determined by DNA sequencing and PCR/single-strand conformational polymorphism. Note that the various frequencies of drug resistance genes do not add up to 100% for a compound when compared to phenotypic resistance. This may be due to other, unidentified mechanisms. NA, not available.
See Maus et al. (168).
See Rengarajan et al. (208).
In contrast, synonymous changes, which are considered functionally neutral, do not alter the amino acid profile. These neutral alterations, when in structural or housekeeping genes, can provide the basis to study genetic drift and evolutionary relationships among mycobacterial strains. Sreevatsan et al. exploited two functionally neutral nsSNP in codon 463 (Leu463Arg) of the catalase-peroxidase-encoding gene katG and codon 95 (Thr95Ser) of the A subunit of DNA gyrase gene gyrA to divide the modern M. tuberculosis complex into three principle genetic groups (PGGs), designated PGG1 (katG463 CTC [Leu] gyrA95 ACC [Thr]), PGG2 (katG463 CGG [Arg] gyrA95 ACC [Thr]), and PGG3 (katG463 CGG [Arg] gyrA95 AGC [Ser]) (235). A more robust analysis by Gutacker et al. further divided the three PGGs into nine major clusters (I to VIII and II.A) (122, 123). Other investigators have similarly used sSNP analysis to infer the phylogenetic structure of M. tuberculosis populations and have largely reported consistent findings (3, 9) (see “Phylogeny and Strain Families of M. tuberculosis,” below). While these studies have shed more light on the phylogenetic relatedness of clinical isolates, they also serve as a broad framework to examine whether different lineages display different epidemiologies in populations. Furthermore, SNP analysis is amenable to targeting multiple polymorphisms that are informative in one platform, such as phylogenetic grouping, drug resistance, virulence, and other epidemiologically instructive markers.
Genomic deletion analysis.
Comparative genomic analysis of strains H37Rv and CDC1551 has revealed large-sequence polymorphisms (LSP) in addition to SNP (98). LSP are thought to mainly occur as a result of genomic deletions and rearrangements rather than through recombination following horizontal transfer (42). In the absence of horizontal gene transfer, deletions are irreversible and often unique events and therefore have been proposed for genotyping as well as for constructing phylogenies (41, 117, 255). It was found that up to 4.2% of the entire genome can be deleted in clinical isolates compared to the genome of laboratory strain H37Rv (255). Brosch and colleagues were able to discern the M. tuberculosis complex by deletion analysis by showing that the majority of deletions are not the outcome of independent events but rather are scars of successive deletions (41). Once a deletion occurs in the progenitor strain, the specific deletion can serve as a genetic marker for the genotyping progenies of this strain. For instance, deletion of TbD1 (for “M. tuberculosis specific deletion 1,” a 2,153-bp fragment) was identified in all modern M. tuberculosis strains; in contrast, ancestral strains tested have this locus present (246). Studies of genomic LSP have indicated that deletions are not always randomly distributed in the chromosome but tend to be aggregated (141, 255). Some loci are “hot spots” for DNA deletions and can occur independently in unrelated strains or lineages. Some chromosomal deletions are associated with IS transposition; this is particularly true of loci which are hot spots for IS6110 insertions, such as in the RvD5 and DR regions (41, 218). For other deletions (such as TbD1), the correlation with IS elements has not been determined. Deleted sequences can include putative open reading frames as well as intergenic regions and housekeeping genes (41, 141). Using deleted fragments as genetic markers, this analysis can be performed by a simple PCR-based method or by automated GeneChip techniques (255).
Both ancestral and frequent deletions can correlate with clonal lineages and be used to examine strain relatedness (141, 254, 255). However, careful selection of the deletions should be made when undertaking such studies. For example, deletions within the above-mentioned hot spots for IS insertion can occur independently in different strains, hence a form of convergent evolution. Nonetheless, analysis of chromosomal deletions has proven to be a powerful tool in investigating the global evolution and phylogeny of the M. tuberculosis complex (41, 182, 197). The resolution of deletion analysis can be greatly improved when the exact flanking sequence of lost DNA is determined, especially when analyzing deletions in hot-spot loci. The use of deletion analysis (or deligotyping) for epidemiological investigations is still nascent. This approach has proven very efficient when the presence of a specific deletion associated with a single strain has been predetermined. Under such circumstances, a single PCR may suffice to track down the spread of a single strain (99, 254). However, in studies in which no particular clone or strain is targeted, simultaneous analysis of multiple deletions is required. Recently, a high-throughput method for detecting large polymorphic deletions was developed (117). Here, 43 genomic regions for large-scale deligotyping analysis were selected, and amplicons generated from these 43 deligosites were hybridized to a membrane containing the target sequences of the 43 loci. This approach proved to be highly sensitive and efficient for the rapid screening of clinical isolates. As is the case for other techniques, high-throughput deligotyping needs to be evaluated against different panels of clinical strains and in different epidemiologic and geographic settings.
Identification of strain-specific markers for rapid diagnosis.
Rapid identification of TB transmission is greatly facilitated when strain-specific properties are targeted, as in the case of MDR-TB outbreaks. Genetic markers can comprise any “unique” characteristic that can distinguish target isolates, including unique fragment sequences, duplications, deletions, neutral SNP or polymorphisms associated with IS6110, or a drug resistance phenotype. For example, insertion site mapping (ISM) is a method that can be applied using IS6110 junctions for such purposes. Kurepina et al. used the unique IS6110 insertion site (A1) in the intergenic region in the origin of chromosome replication (oriC) as a marker to identify and classify members of the W-Beijing strain family (Fig. 1, strain 210) (151, 152). Likewise, Plikaytis and coworkers used multiplex PCR to determine two IS6110 insertions within an NTF locus to identify the W-MDR outbreak isolates from New York, N.Y. (201). In addition, an investigation of a strain with a single IS6110 insertion with others possessing two and three insertions was made possible through ISM. This approach (with spoligotyping) allowed the detection of an otherwise unsuspected M. tuberculosis strain cluster (167). In another application, van Rie et al. described a PCR-based method identifying mixed infections from primary samples (262). Other variations on this technique, such as insertion site typing (“Insite”), which uses PCR amplification of IS6110-flanking sequences followed by hybridization against known IS6110-flanking regions, have been reported, allowing for large-scale screening of clinical isolates (241). Therefore, the use of specific markers is highly amenable to studying transmission, aiding in public health activities, and providing valuable evolutionary information.
MOLECULAR EPIDEMIOLOGY AND PUBLIC HEALTH
The field of molecular epidemiology generally aims to investigate whether naturally occurring strains differ in epidemiology. For instance, do specific clinical strains differ in their infectiousness, severity of disease, or susceptibility to antituberculosis agents? In general, the increased resolution afforded by molecular techniques has enabled both short-term (local epidemiological), such as in suspected outbreaks or laboratory error (26, 89, 193), and long-term (global epidemiological) investigations, such as understanding spatiotemporal transmission and evolutionary dynamics (41, 123, 124, 182, 232, 235).
In addition, molecular epidemiology can serve to better inform routine TB control activities. Successful molecular epidemiological investigations have sought to estimate the fraction of cases attributable to recent transmission or reactivation (12, 35, 224), confirm laboratory-based errors (39, 193), distinguish between endogenous reactivation and exogenous reinfection (10, 46, 233, 263), investigate properties and patterns of drug resistance with specific populations or groups of strains (26, 89, 107, 158, 180, 191, 202, 264), and better understand transmission dynamics within specific populations (25, 109, 167, 177). Since molecular techniques do not substitute for classical approaches, the direct utility of molecular epidemiologic investigations for TB control activities are best illustrated when using both molecular and epidemiologic data sources. In addition to use in the study of transmission patterns within populations, molecular markers can be used to evaluate host- and strain-specific risk factors and possible genotypic-specific differences in phenotypes such as virulence, organ tropism, and transmissibility (108, 207, 209, 235, 256). Below we highlight some instances in which the utility of molecular epidemiologic methods has been realized. Table 3 summarizes some of applications of molecular techniques in the study of TB epidemiology.
TABLE 3.
Applications of molecular techniques in studies of TB
| Application |
|---|
| Study of M. tuberculosis transmission dynamics |
| Confirmation of suspected outbreak/transmission |
| Identification of unsuspected transmission |
| Tracing of chains of transmission |
| Evaluation of transmission in specific populations/groups |
| Identification of transmission in a given setting |
| Identification of risk factors and groups at risk of M. tuberculosis infection |
| Discriminating recurrent TB due to exogenous reinfection and reactivation |
| Detection of laboratory error/cross-contamination |
| Determination of geographic spread of strains |
| Monitoring of transmission of drug-resistant strains |
| Determinations of frequency of drug resistance in different settings |
| Investigation of the evolution of drug-resistant TB within and between patients |
| Detection of mixed infections among TB patients |
| Sampling of strain types for further studies |
| Evaluation of TB control programs (level of clustering); e.g., DOTS (direct observed therapy—short course) |
| Identification of strain-specific transmission/infection rates |
| Identification of predominant strain types (clonal strains) in study populations |
| Identification of hypervirulent strains in populations |
| Investigations of the evolution of M. tuberculosis |
Transmission dynamics.
The difficulty in studying the transmission dynamics of M. tuberculosis within a given population stems partly from the natural history of the pathogen itself. Since most successful infections are followed by a variable latency period, the timing of transmission events often remains elusive. Indeed, most immunocompetent individuals (approximately 90%) infected with M. tuberculosis remain disease-free during the course of their lives. Therefore, the long-term persistence of this organism, juxtaposed with the generally high reproductive number (i.e., the average number of new infections that one case causes annually), makes charting transmission pathways within and between communities extremely difficult. Most TB control programs (especially in more developed countries) rely on contact tracing, whereby individuals named by the index case are screened (using purified protein derivative [PPD]-based tuberculin skin testing or chest X rays) and, if indicated, recommended for treatment of latent TB infection (4). While these prevention activities in low-incidence communities have been useful (101), they are often imprecise and tend to underestimate the level of transmission (25, 29, 167, 224).
The imprecision of contact-tracing investigations has been highlighted by several reports that have indicated that limited or casual contact is sufficient for M. tuberculosis transmission. Large population-based molecular epidemiologic studies conducted in San Francisco, Calif., and Baltimore, Md., uncovered, through extensive contact investigations, approximately 10% to 25% epidemiologic links between patients in designated molecular clusters (29, 224). Similarly, Mathema et al., reporting on a molecular cluster of closely related strains identified from a population-based study in New Jersey, was able to uncover only 30% case links within the prescribed cluster with clinical, demographic, and contact-tracing information (167). The Baltimore study reported that within molecular clusters, patients with no known epidemiologic links shared similar risk and demographic profiles. Furthermore, Valway et al., investigating an outbreak of strain CDC1551 in a small, rural community with low TB incidence, documented extensive transmission associated with casual contact (256). Yaganehdoost and colleagues reported on complex transmission patterns among “bar-hopping” patrons in a specific neighborhood in Houston, Tex., as part of a population-based molecular epidemiology study (286). These studies suggest that the imprecision of contact tracing may in part be due to complex transmission patterns in which casual contact may account for a considerable proportion of molecularly clustered yet epidemiologically unlinked cases, highlighting one of the advantages of performing population-based molecular epidemiologic studies: identifying high-risk groups or areas where transmission is ongoing.
Levels of molecular clustering, much like epidemiologic definitions, are subject to the stringency imposed (i.e., identical, related, or unrelated) by the investigators as well as by the specific molecular method used. The criteria of strain relatedness should be assessed with the appropriate study question and population in mind. In regions where the genetic diversity of the bacillary population is limited, for instance, in East Asia, where W-Beijing strains are predominant, clustering based on identical spoligotyping may lead to gross overestimations of true clustering rates since the W-Beijing strains have mostly identical spoligotype patterns. Similarly, clustering based on identical IS6110-based RFLP patterns may yield numerous hybridization profiles with various degrees of similarity and provide an underestimate of true clustering rates, since some strains that have similar IS6110 profiles may in fact be related by a recent common progenitor. For instance, a population-based study conducted in New Jersey identified 68 strains belonging to the W-Beijing family (25). Upon closer inspection, using subtle motifs in the IS6110 banding patterns and secondary molecular methods (VNTR and PGRS), the investigators divided the 68 strains into two groups, A and B. Unlike group B, group A consisted of five closely related IS6110 profiles that shared epidemiologic and demographic characteristics. Here the authors suggested that while groups A and B are phylogenetically related (i.e., both are members of the W-Beijing lineage, PGG-1, and sSNP cluster II [123, 151]), group A represents a clone that has recently spread and evolved in specific U.S.-born communities (i.e., clonal expansion) whereas group B was recovered from mainly non-U.S.-born patients of East Asian origin. Unlike group A, the diverse patterns and contrasting demographic profiles (i.e., mainly HIV seronegative, older age, no known risk factors for TB) noted among group B patients suggest that this group represents mainly reactive cases. Similarly, another study conducted with the same New Jersey population uncovered a large cluster based on a distinct spoligotype that displayed three distinct IS6110 profiles (167). Here, use of ISM and sequencing of “naked” IS6110 flanking regions revealed a stepwise acquisition of IS6110 elements from one to two to three copies. The clonality of the three strains was confirmed by multiple molecular methods. Patients harboring these three IS6110 patterns and distinct spoligotype were similar with respect to geographic and epidemiologic characteristics and differed from patients with unrelated strains, further supporting the molecular method-based clustering. These studies indicate those strains with similar yet nonidentical IS6110 fingerprint patterns may share a recent progenitor and therefore be closely related and part of or an extension of an ongoing series of transmission events.
As highlighted in the New Jersey and Houston studies, population-based studies, when used in conjunction with traditional TB control activities, may facilitate the identification of previously unrecognized transmission events or even outbreaks in populations where there is a high background of reported cases (25, 160, 167, 173, 286). Until recently, it was thought that in low-incidence countries, such as the United States, the majority of TB cases were due to endogenous reactivation. Population-based molecular epidemiologic studies from San Francisco, New York, The Netherlands, and Denmark noted that molecular clustering ranged on average between 35% and 45%, indicating a substantial proportion most probably due to recent transmission (2, 17, 224, 265). This is in contrast to high-incidence populations where clustering is substantially higher, with proportions approaching 70% in some situations (113, 116, 271). It is important to note that molecular clustering is not synonymous with levels of recent transmission. Although molecular clustering may approximate epidemiologic clustering or recent transmission in low-incidence populations, the approximations tend to be more divergent in high-incidence populations due to high rates of infection and multiple transmission pathways. The precise proportion of disease due to recent transmission or endogenous reactivation is variable and heavily dependent on a number of factors, including the annual rate of TB infection, the molecular method employed, effective TB control programs, the size of the infected pool of individuals, age cohort effects (275), immigration history (109, 155), population susceptibility (e.g., genetic susceptibility, HIV prevalence, BCG vaccination), and the sampling strategies employed to derive estimates (114, 185, 186). Thus, when studies account for these independent factors and are used in conjunction with conventional epidemiologic methods, greater resolution and insight into transmission dynamics can be gleaned for the specific communities or populations studied. As such, universal genotyping has been implemented in some cities and countries (15, 56, 65, 136, 265).
Molecular studies on drug resistance.
Over the past decade, much has been learned of the drug targets and mechanisms of resistance to first-line and several second-line antituberculosis agents (Table 2) (168, 206, 208). As mentioned above, M. tuberculosis generally acquires drug resistance via de novo nsSNP, small deletions, or insertions in specific chromosomal loci, unlike most other pathogenic bacteria, which often acquire drug resistance via horizontal transfer. This attribute of M. tuberculosis drug resistance, coupled with fast and efficient DNA sequencing methods, makes studying drug resistance highly amenable for molecular epidemiologic investigations. Molecular epidemiologic studies on drug resistance have generally sought to examine the nature (e.g., genotype-specific mutations, association of specific mutations with phenotypic resistance) and extent (e.g., prevalence of specific mutations in a population) of drug resistance and patient risk factors (e.g., HIV) for acquiring resistance. Some studies have queried the contribution of primary (infection by an already-resistant organism) versus acquired (acquisition of drug resistance within a patient, de novo) drug resistance in specific populations (158, 285), while others have aimed to describe the evolutionary dynamics of drug resistance during clonal expansion or dissemination between and within patients (22, 26, 202). Of note, the terms primary and acquired resistance are not used in the current WHO guidelines, since they cannot be distinguished by most public health programs (unlike molecular epidemiologic studies); rather, they are divided by treatment history. Patients never treated or for treated <1 month and harboring drug-resistant TB are considered primary, and those previously treated or treated for >1 month are labeled acquired (283).
The report by Bifani et al. provides an example of a study of the nature and evolution of drug resistance during a clonal MDR-TB outbreak. Here the investigators describe the genotypic drug resistance profile of strain W and its variants during the outbreak in the early 1990s in New York, N.Y. (26, 101). Of the 357 patient isolates that were invariably resistant to INH, RIF, ethambutol (EMB), streptomycin (STR), and pyrazinamide (PZA) and often resistant to kanamycin (KAN), 253 isolates displayed 18 identical hybridizing bands, strongly indicating clonality and an outbreak. Analysis of five drug resistance chromosomal targets among 50 randomly chosen isolates revealed an identical array of polymorphisms, including a rare dinucleotide substitution in katG315 further supporting clonality and suggesting that the spread of this clone was primary in nature (i.e., acquired resistance prior to dissemination). Since then, at least 11 MDR W variants with subtle variations in IS6110 RFLP profiles have been recovered from New York patients. DNA sequence analyses of drug resistance targets confirm these variants as descendants of the original outbreak strain (i.e., mutations identical to those of strain W); however, in some variants additional resistance to fluoroquinolone and capreomycin was noted (26, 183, 245). Sequence analysis of the fluoroquinolone-resistant strains revealed five different gyrA polymorphisms, indicating that strain W had acquired resistance to first-line agents prior to dissemination and subsequently acquired resistance to fluoroquinolone de novo (285). Here, documentation of primary spread was critical in devising appropriate TB prevention and control measures (1, 100).
Molecular epidemiologic studies of drug resistance have also focused on describing the nature of resistance within patients. For instance, Post et al. sought to better understand the dynamics of drug-resistant subpopulations resident within a patient by characterizing serial isolates recovered from 13 chronic HIV negative MDR-TB patients (202). Serial isolates were characterized by IS6110-based RFLP analysis, spoligotyping, and sequencing of a number of drug resistance-determining regions. It was found that while all cases were infected by a single strain of M. tuberculosis, sputum-derived isolates of 4 of the 13 patients had acquired additional drug resistance mutations during the study. Heterogeneous populations of bacilli with different resistance mutations, as well as mixtures of drug-susceptible and drug-resistant genotypes to specific genetic targets, were noted. This observation was furthered by another study that noted bacilli with additional drug resistance mutations recovered from different human lung lesions (138). Taken together, these studies suggest that a single founder strain of M. tuberculosis may undergo genetic changes during treatment, leading to the accumulation of additional drug resistance independently in discrete physical locales. In addition, it is possible that in patients with mixed infections (more than one infecting strain), the drug resistance profile may be composed of strains with different susceptibilities (e.g., simultaneous infection with mono-INH- and mono-RIF- resistant strains), leading to incorrect MDR resistance profiles (211, 262). Therefore, genetic heterogeneity may require therapeutic targeting of both drug-resistant and drug-susceptible phenotypes, especially with first-line agents.
As shown in the examples above, examinations of target mutations have enabled investigators to determine whether the occurrence of drug resistance among clinical samples is due to primary infection or to de novo acquisition (acquired); the former would implicate active transmission of already-resistant strains in the community studied, and the latter would suggest suboptimal case management or other patient factors, such as poor therapeutic compliance, drug malabsorption, or low drug bioavailability (90, 133, 199, 200). Additionally, drug resistance genetic markers have been used to demonstrate clonality and transmission and to elucidate microevolutionary pathways (22, 26, 183, 272). Other mechanisms or characteristics, not discussed in this review, such as increased expression of specific genes or drug tolerance, which are specific to the infecting strain or restricted to specific phylogenetic lineages may also contribute to reduced susceptibility to antituberculosis agents (36, 79, 198, 276).
Recurrent TB.
Recurrent TB is the reoccurrence of disease after a previous episode has been considered clinically cured or resolved. In general, active TB may develop due to either recent infection or endogenous reactivation of historic infection. However, for the past few decades the role of exogenous reinfection, i.e., caused by a new strain of M. tuberculosis, in TB patients who had previous disease has been heavily debated (48, 236, 243, 274). Stead and others in the 1960s reported that TB always begin with primary infection, and subsequent episodes (of disease) are due to reactivation of these dormant organisms (also known as the unitary concept of pathogenesis) (236). Romeyn suggested that in environments of high TB infectivity, exogenous reinfection does have a role in active TB cases, unlike in communities or countries where TB incidence is low (213). Furthermore, Canetti used the epidemiologic concept of cohort effect to support the notion of exogenous reinfection (48). Between 1962 and 1970 in France, drug resistance to the available antituberculosis agents was 7.8% among patients over the age of 60 years. Since the prevalence of TB infection was as high as 90% 30 years prior (1935), a cohort member aged 60 would have been 30 years old in 1935 and would have invariably been infected with M. tuberculosis. As resistant forms of the bacilli did not appear until the middle to late 1940s, Canetti suggested that these patients harboring drug-resistant organisms acquired it exogenously.
Nevertheless, the long-standing belief was that the majority of recurrent TB is due to endogenous reactivation of the primary M. tuberculosis infection. Although an unsettled issue, clinical cure or successful completion of an appropriate chemotherapy regimen may not result in mycobacterial sterilization (161, 236). Guinea pig studies of M. tuberculosis infection do not support the notion that secondary or tertiary exposure to the bacilli leads to an adverse effect on the host response to the primary infecting strain (296). As such, there have been limited case reports documenting exogenous reinfection occurring among immunocompromised individuals (97, 226); however, host-pathogen factors influencing this seemingly uncommon event have yet to be elucidated (142).
Distinguishing recurrent TB (due to exogenous reinfection or endogenous reactivation) raises several epidemiologic questions regarding the level of active transmission, the infectious burden, and environmental and specific host susceptibility factors in a given population. Clinical, epidemiological, and/or microbiological data cannot conclusively differentiate recurrent TB caused by reactivation or reinfection. Molecular techniques can, when primary (or historical) and secondary samples are available, help distinguish endogenous from exogenous infection. van Rie et al. reported the first comprehensive study of recurrent TB attributable to exogenous reinfection in an ongoing population-based study in Cape Town, South Africa (263). Here, roughly 700 patient isolates were genotyped by IS6110-based RFLP analysis over a 6-year period from a high-TB-incidence region (225 cases/100,000). M. tuberculosis isolates recovered from primary and secondary episodes were available and genotyped for 16 of the 48 recurrent patients with previous TB treatment and documented cure. All but one of the 16 patients was HIV-seronegative. IS6110 analysis indicated that in this group, 75% (12 of 16) of the recurrences were due to an exogenous insult, i.e., a new strain of M. tuberculosis. Concern over the interpretation of data has been noted, as the study included patients with a single positive culture as recurrent cases (76, 95; W. W. Stead and J. H. Bates, Letter, N. Engl. J. Med. 342:1050, 2000; A. van Rie et al., Author's Reply, N. Engl. J. Med 342:1051, 2000). Nevertheless, Vynnycky and Fine suggested, as did Romeyn, that the contribution of exogenous reinfection increases proportionally to the regional incidence of TB, thus supporting the findings of van Rie et al. (213, 274).
Studies conducted in countries with different rates of TB incidence have demonstrated various levels of recurrent disease attributable to exogenous reinfection (46, 135, 233). Sonnenberg et al. reported on the incidence of recurrence among a cohort of HIV-1-infected and uninfected South African mine workers (233). Of the 65 patients with recurrent disease, 39 were available for IS6110 fingerprinting. Of these, 14 patients (36%) were considered exogenously reinfected. The authors found the recurrence rate to be about 2.4 times higher among the HIV-1-infected subgroup than among the uninfected group, suggesting that in regions with a high incidence of TB infection, HIV enhances the rate of recurrence due to high risk of exogenous reinfection. Recently, Verver et al. reported that the age-adjusted incidence rate of TB due to reinfection among patients with successful prior treatment was four times higher than the rate of new TB in Cape Town, South Africa (270). At least in this high-incidence community (313 cases/100,000), individuals with previous TB are strongly associated with an increased risk of developing disease when reinfected. Although potential confounding by HIV status or socioeconomic background may have biased the estimates, this study raises the possibility that there may exists a subgroup of individuals with a predisposition to TB infection and/or that TB disease itself increases the susceptibility to recurrence (19, 49, 292).
Molecular tools have not only facilitated direct evidence for the occurrence of exogenous reinfection among both immunocompetent and immunocompromised individuals but have also provided a platform for studies aimed at assessing the current rates of active transmission in the community, the rate at which this phenomenon occurs in various epidemiologic scenarios (e.g., low TB incidence) among individuals with different risk factors and comorbidities or different race and/or ethnic groups. Although limited, there is evidence for racial variation in the susceptibility or level of innate immunity to M. tuberculosis infection (68, 239); more studies on host susceptibility to reinfection/infection that utilize molecular epidemiologic approaches need to be performed. As such, the finding that M. tuberculosis infection or disease does not afford sufficient acquired immunity against further insults will have profound implications for TB control activities, such as enhanced case finding and preventive therapy among at-risk population groups in areas of high disease prevalence, and for vaccine development. In addition, a study from a high-incidence region reported that multiple infections with different strains are common, suggesting significant reinfection rates and the absence of effective protective immunity conferred by the initial episode, further reinforcing the role of exogenous reinfection in the epidemiology and control of TB (279). Comprehensive reviews of this topic have been published elsewhere (55, 153).
Laboratory error/cross-contamination.
Laboratory error resulting in false-positive cultures of M. tuberculosis can cause erroneous administration of medications, disruption of daily life, and expenditure of resources required for isolation and contact investigations. Mechanisms of laboratory error generally occur when clinical samples are mislabeled or medical devices are contaminated and during the handling or processing of primary patient samples subjected to mycobacterial analyses (laboratory cross-contamination) (39, 45). As this error is often random in nature, the rate of occurrence may be quite variable from one clinical laboratory to another.
Investigating false-positive cultures typically begins by first determining whether the patient has only a single positive acid-fast stain (out of three smears taken on consecutive days) that results in a single positive culture and if the laboratory had processed any other M. tuberculosis isolates during the same time period. When multiple M. tuberculosis patient isolates have been processed in the laboratory during the same time frame, genotyping may be used to determine the possibility of cross-contamination. A study by Small et al. suggested that two identical fingerprints cultured from separate patients within 7 days should be investigated (225). The use of DNA fingerprinting has markedly improved the timeliness and ability to confirm or refute false-positive cultures (16, 56, 96, 192, 225).
Several studies have shown, by molecular techniques, that laboratory error occurs more frequently than previously thought (21, 39, 45). Molecular confirmation of false-positive cultures is often based on sufficient strain diversity in the TB population and distinction between clinical and laboratory strains. That is, when there is sufficient heterogeneity in M. tuberculosis genotypes in a population, the chance of processing two patient isolates with identical fingerprints (within a short time frame) is low and warrants further investigation. This is especially the case in communities with relatively low TB incidence. It is important to note that genotypic heterogeneity and the ability to discriminate genotypes may be dependent on the molecular method. For instance, while most members of the W-Beijing family have identical spoligotypes, they exhibit similar yet distinct IS6110-based RFLP patterns. Therefore, in regions where the W-Beijing family of strains is predominant, such as in East Asia, use of spoligotyping alone to confirm or refute laboratory cross-contamination may lead to unwarranted investigations. This limitation is applicable to most, if not all, molecular methods currently available. To rule out patient isolate-isolate contamination, most clinical laboratories use a laboratory strain (such as H37Ra) for identification and drug susceptibility testing. Here, differentiating between patient isolates and the laboratory control strain is critical to rule out contamination (23). In general, contamination is especially suspected when there are inconsistencies between microbiologic results and clinical findings (179). While this issue does not directly relate to the epidemiology of TB, it serves as a major application of molecular techniques in TB control activities. Nevertheless, laboratory error is an important topic in TB case management and laboratory quality control and has been reviewed specifically elsewhere (45). In one study, false-positive cultures were reported in 13 of 14 molecular epidemiologic studies that examined more than 100 patients. Here, the investigators found a median false-positive rate of 3.1% reported in these studies (45). In another report, a quality assessment study to evaluate the replicability of IS6110-based RFLP analysis and spoligotyping by eight laboratories in the National Tuberculosis Genotyping and Surveillance Network showed variable rates of reproducibility (37). It is important to note that samples may contain heterogeneous populations of M. tuberculosis strains (i.e., mixed infections) and that this does not necessarily point to laboratory cross-contamination. Reports from high-incidence areas suggest that this phenomenon may not be rare (211, 279). Here, RFLP-based methods are generally better suited to identifying patient samples containing more than one strain than are PCR-based methods (Table 1). The impact of mixed infections on the interpretation data from high-incidence settings that primarily use PCR-based methods is unclear and warrants further evaluation.
PHYLOGENY AND STRAIN FAMILIES OF M. TUBERCULOSIS
The remarkable level of DNA conservation of M. tuberculosis housekeeping genes, the paucity of synonymous mutations, and data from genomic deletion analysis has led to the hypothesis that the modern M. tuberculosis strains, which account for the majority of TB worldwide, underwent an evolutionary bottleneck at the time of subspeciation some 15,000 to 20,000 years ago (41, 139, 235). Recently, Gutierrez et al. shed further light into the evolutionary age of M. tuberculosis (124). Here, the investigators classically sequenced portions of six housekeeping genes (katG, gyrB, gyrA, rpoB, hsp65, and sodA) and the complete 16S rRNA gene of nine M. tuberculosis complex and seven smooth (morphologically) tubercle bacilli. The latter group was isolated from patients in Djibouti, East Africa, and based on phylogenetic analysis is believed to have predated the modern M. tuberculosis complex members. The authors estimate that the tubercle bacillus or its ancient predecessor could be 3 million years old and speculate that the smooth bacilli spread/coexisted with early hominids reported to have existed in the same region. The coevolutionary scenario proposed may indeed better explain in part the extent of genetic diversity of clinical strains observed in various regions of the world. Furthermore, such studies may provide insight into the evolution of biomedically significant traits and into the relationships between M. tuberculosis, host populations, and the environment, thus being relevant to the field of molecular epidemiology.
A basic assumption of molecular epidemiology is that lineages of microbial pathogens are for the most part genetically stable over time and geographic distance (154). It follows that genetic polymorphisms that may differentially contribute to a distinct clinical or epidemiologic phenotypes are nonrandomly distributed along clonal lines (187). Molecular epidemiologic analyses of clinical strains of some bacterial pathogens have revealed the presence of specific genotypes associated with distinct phenotypes (30, 131, 154, 187). As such, specific M. tuberculosis clones have been reported to show an unusual degree of outbreak or epidemic potential or marked tissue tropism (26, 102, 234, 256).
A number of studies have sought to examine the phylogenetic structure and relationships within the M. tuberculosis species (3, 9, 93, 123). As mentioned above, Gutacker et al., using over 230 sSNP found in the four M. tuberculosis complex genomes (M. tuberculosis strains H37Rv, CDC1551, and 210 and M. bovis AF2122/97), examined the phylogenetic relationship of M. tuberculosis isolates recovered from geographically distinct populations and found eight distinct M. tuberculosis lineages (123). More recently, analysis of 5,069 M. tuberculosis clinical isolates retrieved from four population-based studies, using the 36 most informative sSNP (from the 230 sSNP previously reported), as well as selected nsSNP and intergenic SNP, found a similar phylogenetic framework of eight clusters (I to VIII), plus an additional cluster (II.A) (122). From these studies, it appears that strains with IS6110 copy numbers ranging from 1 to 6 were restricted to clusters I, II.A, IV, and V (PGG1 and PGG2) (Fig. 2A). Related strains with high numbers of IS6110 insertions (10 to 26) generally belonged to cluster II (PGG1) and represent the W-Beijing strain family, including strain 210. Annotated strain CDC1551 grouped to cluster V (PGG2), while laboratory strain H37Rv was found in cluster VIII (PGG3).
Another study examined 37 neutral mutations in seven drug resistance-determining genes to group M. tuberculosis isolates (9). Those authors identified four major M. tuberculosis lineages in addition to the M. bovis group (related M. bovis strains). Combinations of genetic markers such as PGG, spoligotypes, and the presence of TbD1 (a marker for ancestral M. tuberculosis strains) seem to be restricted to specific lineages. The correlation between sSNP lineages (Baker et al.) and sSNP clusters (Gutacker et al.) is apparent. That is, lineage IV corresponds to cluster I (PGG1, TbD1+); lineage I corresponds to cluster II (PGG1, TbD1−); lineage II combines clusters III, IV, V, and VI (PGG2, TbD1−); and lineage III compiles strains from subcluster II.A (PGG1, TbD1−). It is not clear what lineage corresponds to clusters 7 and 8 (both PGG3). It is important to note that there are inconsistencies between the two reports. For example, Baker et al. suggest that strains CDC1551 and H37Rv are closely related and belong to synonymous sequence type 2 (lineage II), which is contrary to a number of previous reports (3, 98, 123). Figure 1 demonstrates that these two strains do not have any common IS6110 insertions. In another study, Alland et al. performed an analysis using 77 SNP that are polymorphic for H37Rv and CDC1551 and showed that clinical M. tuberculosis and M. bovis isolates could be differentiated into 2 primary branches and 14 secondary branches, with reference strains H37Rv and CDC1551 at the distal ends (3). Recently, Filliol et al. reported on the global phylogeny of M. tuberculosis strains by using 212 SNP markers that showed six deeply branching SNP cluster groups. Overall, the results of this study are consistent with those reported by Gutacker et al. (94, 122). While there are subtle differences between these studies, they nevertheless strongly confirm that M. tuberculosis is highly clonal where distinct lineages can be readily identified and that horizontal gene transfer is nearly absent.
In general, the shape and complexity of phylogenetic trees based on sSNP analysis are heavily dependent on the selected reference strains, the number of polymorphisms analyzed, and the extent of sampling. Indeed, the use of selected and informative sSNP for genotyping affords rapid delineation of relationships among closely related strains of M. tuberculosis and provides a sound framework for examining the differential distribution of biomedically relevant traits, such as virulence and transmissibility (123, 187). Furthermore, phylogenetic studies have provided a basis to examine molecular epidemiologic characteristics and relationships within and between designated lineages.
Grouping of isolates into families based on any commonly used genotyping system is largely reliant on the threshold of similarity imposed by the investigators. Generally, a strain family can be described as a group of isolates that share specific biomarkers or properties indicative of a common recent ancestor. For instance, reports based on spoligotyping have described strains families bearing specific spoligotype patterns (Fig. 2B), such as EAI (deletion of DVRs 29 to 32 and 34), Beijing (deletion of DVRs 1 to 34), CAS (deletion of DVRs 4 to 7 and 23 to 34), Haarlem (deletion of DVRs 8 and 32 to 35), LAM (deletion of DVRs 21 to 24 and 32 to 35), and other smaller groups (92, 93). Here, strains combined into one group or family, based on common spoligotype patterns, often exhibit different yet related IS6110 RFLP profiles (Fig. 2A), with <60% similarity, and probably would not have been associated if RFLP analysis had been used alone. For example, the relatedness of strains in the Beijing family was strongly confirmed by genetic markers other than IS6110 and spoligotyping (151), such as sSNP analysis (122, 123). All W-Beijing strains are found in cluster II (PGG1), and their geographical distribution supports previous reports (Fig. 3) (115, 130). In support, deligotyping analysis noted that members of this group carry large chromosomal deletions, including 207 (7,399-bp deletion in the DR region, leading to the absence DVR 1 to 34), 105 (3,467 bp, Rv0071 to Rv0074 deleted), 149 (9,248 bp, Rv1572 to Rv1586 deleted), and 152 (11,985 bp, 12 genes totally deleted), and most have 181 deleted (711 bp, Rv2262 and Rv2263 deleted) (254).
FIG. 3.
Distribution of M. tuberculosis SNP-derived clusters, based on patient country of origin. CEP, Columbia, Ecuador, and Peru; FIN, Finland; GEH, Guatemala, El Salvador, and Honduras; HDP, Haiti, Dominican Republic, and Puerto Rico; MEX, Mexico; PIB, Pakistan, India, and Bangladesh; SC, South Korea and China; USA, United States; VCP, Vietnam, Cambodia, and the Philippines. (Reprinted from reference 122 with permission. © 2005 by the Infectious Diseases Society of America. All rights reserved.)
Unlike the W-Beijing family of strains, which have been extensively studied, little is known of the molecular characteristics that define the other M. tuberculosis strain families that are globally prevalent. For instance, the Haarlem family, which includes the model virulent Erdman strain, consists of strains isolated mostly from European and South American countries (92). According to sSNP analysis, these strains belong to cluster III (PGG2) and have similar RFLP profiles (Fig. 2A) (123). Other strain families, such as LAM and Delhi (CAS), were grouped to sSNP clusters VI (PGG2) and II.A (PGG1), respectively. LAM family strains exhibit similar IS6110 RFLP patterns, while Delhi strains, often isolated from patients born in the Indian subcontinent (20), represents one of most variable IS6110 groups (122). Another strain family, EAI, represents ancestral strains belonging to cluster I (PGG1), where the TbD1 region is intact (TbD1+) (246). Also, the Manila strain family was identified among isolates from the Philippines that belong to cluster I based on PGG1 and IS6110-based RFLP analysis (81). Much like IS6110-based RFLP analysis and spoligotyping, MIRU-VNTR profiles (based on 12 loci) when superimposed on the SNP-derived M. tuberculosis phylogenetic tree, do indeed display some level of cluster-specific patterns. For instance, strains belonging to the ancestral EAI family (TbD1+, PGG1, cluster I), in addition to their spoligopattern, bear a distinct high copy number at the MIRU23 (≥6), MIRU4 (≥3), and MIRU24 (>1) loci, supporting the hypothesis that these strains are related by descent (246). Furthermore, characteristic 12-loci MIRU-VNTR profiles for the W-Beijing family and the Haarlem family (PGG2, cluster III) are 2 2 1-3 1-3 1-2 5 1 3-7 3 5-6 3 2-4 and 2 2 6-13 2 1-2 5-6 1 1 3 2-3 1-3 2-3, respectively, as well as having a high copy number (6 to 13) at MIRU10 for strains belonging to cluster II.A. Reports have also suggested that strain families and SNP clusters show certain levels of geographic specificity that may in part explain the characteristic genetic markers observed within groups (clonal expansion) and to a lesser degree between groups (115, 122, 255). For example, the cluster I and II strains have been noted to be prevalent among patients originating in Asia (including Russia) while minimally present among patients of South American origin (Fig. 3).
The degree of concordance between SNP-derived clusters or lineages and molecular epidemiologically based (i.e., IS6110-based RFLP, spoligotype, and MIRU) family designations are for the most part consistent (122). Furthermore, it appears that profiles generated by IS6110-based RFLP analysis, spoligotyping, and MIRU-VNTR analysis are not shared between PGG1 and PGG2/PGG3 strains, suggesting early divergence from a common ancestor during M. tuberculosis evolution. However, there are instances in which members of certain strain families are found in multiple PGGs or clusters. For example, the majority of clinical isolates with a single copy of IS6110 are found in cluster I (PGG1); however, they are also noted in cluster II.A (PGG1/2) and cluster IV (PGG2). Also, as shown in Fig. 2B, spoligotype-designated family T1 appears in clusters III to VI (PGG2) as well as in clusters VII and VIII (PGG3). Similarly, strains missing DVRs 1 to 36 and 40 have been noted in clusters II and VI (Fig. 2B); however, these clusters have been deemed unrelated, based on multiple genetic markers (i.e., W-Beijing and LAM families), an observation most likely due to independent deletion events in the DR locus. Such inconsistencies have also been noted elsewhere (122). There may be several explanations for these findings, such as locus-specific evolutionary convergence, especially when only one biomarker is used (e.g., spoligotype, specific deletion), or this may indicate strains undergoing locus-specific transition. However, consistency can be improved when multiple biomarkers, e.g., IS6110 RFLP analysis and spoligotyping or MIRU, are employed. While phylogenetic analysis is used to infer evolutionary processes and molecular epidemiologic approaches are used for examining genetic diversity as it pertains to TB epidemiology, both methods complement one another. That is, molecular approaches can add further resolution to phylogenetically assigned lineages or clusters to indicate microevolutionary processes; in turn, phylogenetic analysis can ground molecular epidemiologic-derived genotypes into broad groups that share informative traits and further our understanding of genotype-phenotype relationships.
STRAIN-SPECIFIC VARIATIONS IN IMMUNITY AND PATHOGENESIS
Recent reports have indicated that different clinical M. tuberculosis strains can induce differential host immune responses, leading to variable differences in pathogenesis and virulence in animal models (80, 157, 162, 164, 165, 256). For instance, Valway et al. reported an outbreak of strain CDC1551 with an unusually high rate of transmission. Based on the large skin test response to PPD in infected individuals, this strain was considered to be extremely virulent (256). However, mice infected with CDC1551 showed an early and vigorous cytokine response compared to infections with other strains, which was associated with longer survival (164). In contrast, mice infected with clinical isolate HN878 showed reduced cytokine induction, higher bacillary load in the lungs, and significantly shorter survival times. Further studies indicated that HN878 preferentially induced the less-protective TH2-associated cytokines (163-165). Genotypic analyses, including IS6110 and spoligotyping, have indicated that HN878 is a member of the globally prevalent W-Beijing phylogenetic lineage (PGG1, cluster II) (24, 163). An earlier study by Zhang and colleagues showed that a W-Beijing strain (strain 210) was able to replicate in human macrophages at a four- to eightfold-higher rate than other unrelated strains (294). Strains 210 and HN878, which are indistinguishable by IS6110-based RFLP analysis, were reported to cause significant disease and were responsible for a series of multistate outbreaks (287).
Recently Reed et al. described a polyketide synthase (pks1-15)-derived phenolic glycolipid (PGL-tb), which is produced by a subset of clinical M. tuberculosis strains that show “hyperlethality” in the murine infection model (HN878 and not H37Rv or CDC1551) (207). Disruption of pks1-15 in HN878 (HN878pks1-15::hyg) resulted in a PGL-tb-deficient strain showing no hypervirulence. Tsenova et al. reported that HN878 and W4 resulted in higher bacillary loads in the cerebrospinal fluid and brain and more-severe pathology, in comparison with the hyperimmunogenic strain CDC1551 in a rabbit model of TB meningitis (253). Initial studies indicate that the presence of an intact pks1-15 gene, and thus the ability to produce PGL-tb, seems to be restricted to members of the W-Beijing strain family (123, 163). It has been hypothesized that poor control of bacillary growth, due to a suboptimal immune response, may amplify the number of stochastic events leading to mutations in target genes, some of which confer antituberculosis resistance. Some studies suggest that W-Beijing family strains may be associated with drug resistance, although these results are not consistent with all reports (83, 84, 115, 196, 242).
A study by Lopez et al. found that genetically distinct M. tuberculosis strains (representing the four major genotype families found globally) resulted in an array of immunopathologies in an murine intratracheal infection model (157). The W-Beijing strain tested, similar to HN878, elicited an inefficient immune response and was the most virulent. Macrophage infections with various M. tuberculosis strain types induced a differential pattern of cytokines in vitro (51). Dormans et al. found in a murine model that 19 different M. tuberculosis complex strains from 11 major genotype families produced responses that varied widely with respect to virulence, pathology, bacterial load, and delayed-type hypersensitivity (80). Thus, considerable evidence indicates, both in vitro and in vivo, that diverse clinical strains of M. tuberculosis can elicit various immune responses and subsequent pathological manifestations. If such differences are reflected in clinical or epidemiologic features (e.g., disease progression, severity, and transmissibility, etc.) then the relevance to molecular epidemiologic studies is apparent. For example, van Crevel et al. reported that TB patients harboring W-Beijing strains, compared to other genotypes, had transient febrile responses shortly after commencing treatment (257). As the disease severity, drug toxicity, and drug resistance were similar between the two groups (W-Beijing versus others), the authors suggested that these differences may be due to the strains' (W-Beijing) ability to induce a different host response, consistent with the studies described above (51, 157, 163, 165, 207). Further studies with human populations are needed.
VACCINES
Globally, BCG remains one of the most widely used vaccines against TB. However, reports on its efficacy to prevent pulmonary TB have been variable and in some instances conflicting (59). New and improved vaccines against TB are critically needed. Over the past decade, with new knowledge about M. tuberculosis- and BCG-specific immune responses and comparative analysis of mycobacterial genomes, novel vaccine design strategies and candidates have been described (120, 143, 194). Strain-specific epidemiologic differences in prevalence, transmissibility, and disease severity (24, 83, 207, 209, 256) and the idea that some M. tuberculosis strains elicit distinct host immune responses have complicated rational vaccine design (157, 162, 163, 165). More-recent studies have examined the level of protection conferred by a candidate vaccine evaluated against infection by clinical strains characterized in molecular epidemiologic studies (120, 143, 282). Furthermore, the extent of immunity against M. tuberculosis, either from early exposure or prior disease, is not well understood, although it is crucial in designing control activities. Thus, examination of whether the protection afforded by candidate vaccines is meaningfully differential by diverse clinical stains is warranted. For instance, will a candidate vaccine efficacious against H37Rv be so against some members of the W-Beijing family of strains that fail to elicit an efficient protective immune response and are reported as globally prevalent (24, 163)? Some murine studies have shown differential efficacy of BCG against different clinical strains (157). Noteworthy in designing candidate vaccines is the growing number of individuals unable to mount an effective T-cell response, such as those infected with HIV or perhaps those with an intrinsic host susceptibility to the disease (19, 49, 62, 222). For a comprehensive review of the current concepts and progress in vaccine development, refer to the recent review by Doherty and Anderson (78).
STRAIN FITNESS
For pathogenic microorganisms, fitness generally refers to heritable variation among members of a given species or phylogenetic lineage (5, 6). Here, fitness may be variably described as the microorganism's capability to survive, replicate, and disseminate. These may include the organism's specific growth properties within its natural host (e.g., bacillary growth within human macrophages), resilience to environmental stress (e.g., nutrient starvation, hypoxia), and transmissibility (249, 251, 256, 294). Some of these characteristics can be assayed in laboratory settings with in vitro and in vivo models, though their direct contributions to molecular epidemiologic processes have yet to be realized. Fitness as it pertains to transmissibility has more direct implications for molecular epidemiology. That is, if strains of M. tuberculosis that bear specific properties enhance their intrinsic transmissibility, the identification of such strains in populations may warrant additional control measures. Some studies have investigated fitness in terms of estimating the average number of secondary infections or secondary cases as the transmissibility of a particular strain (or genotype) in a susceptible population (44, 185). In estimating epidemic potentials, parameter estimates are heavily dependent on empirical epidemiologic data from specific human populations, such as molecular and/or epidemiologic cluster investigations and model-based estimates of average fitness (33, 34, 57). To date, most molecular epidemiologic studies have examined fitness in relation to drug resistance (33, 57, 58).
Despite the impact of drug-resistant M. tuberculosis on transmission dynamics, the consequences for fitness remain poorly understood. Following the outbreaks of MDR-TB in the early 1990s in the United States, it was widely feared that MDR isolates would continually spread and become endemic. Fifteen years later, while outbreaks associated with a given MDR genotype tended to “wax and wane,” in general they have remained restricted to specific populations (87). This observation may be due to appropriate public health activities or to the cyclical nature of TB outbreaks; however, it is also possible that the acquisition of drug resistance in M. tuberculosis has a biological cost. As noted for other pathogens, the loss of fitness resulting from acquired resistance can translate into reduced growth rate or transmissibility or loss of virulence and hence the organism can be outcompeted by drug-susceptible counterparts in the absence of selection (6). Alternatively, studies with M. smegmatis have indicated that resistant isolates may acquire compensatory mutations that restore the organism's reproductive potential or fitness (140). Several studies have noted that the relative in vitro fitness of RIF-resistant stains decreased in some spontaneous rpoB mutants, while other frequently observed rpoB mutations found in clinical isolates were similar to those of wild-type (i.e., RIF-susceptible) parental strains (28, 106). Other studies, using RIF resistance as a model, have noted a decrease in bacillary growth rates in macrophages (166). A study in the mid-1950s similarly noted a loss of fitness associated with INH resistance (176). However, Pym et al. noted that the impact of katG mutations on fitness may be codon specific (204), as a common mutation such as S315T in the katG locus does not appear to alter the organism's virulence or transmissibility (57, 266). Nonetheless, a population-based study conducted in San Francisco, Calif., has shown that INH-resistant isolates are less likely to result in secondary cases (44). A recent study by Gagneux et al. showed that the fitness cost of specific mutations associated with INH resistance may in part be influenced by strain genetic diversity (105). The heterogeneity of mutations, coupled with multiple single-locus mutations conferring different antibiotic resistance profiles in different genetic backgrounds (i.e., genotypes), has made evaluating fitness gain or loss complex and difficult to interpret (58), although some mathematical models suggest that even small populations of fit MDR strains have the potential to outcompete drug-susceptible or less-fit MDR strains (57).
If the biological traits that confer a fitness advantage are nonrandomly distributed along clonal lines, there may be certain M. tuberculosis clonal families that are more fit than others. Some strain families, such as the W-Beijing family, that have been noted to be globally prevalent or have been associated with outbreaks, extensive transmission, and/or dominant phenotypes may indeed bear some fitness advantage (11, 24, 115). Although the success of the W-Beijing family has been extensively documented, there are other genotypes, such as Haarlem, EAI, and LAM, which also display success in terms of prevalence in worldwide or specific populations. However, the reasons for specific strain family predominance or association with certain epidemiologic characteristics, such as outbreaks or severity of disease, have not been well elucidated. As mentioned above, factors that may enhance fitness or “success” in a given population may include a strain's ability to better withstand and replicate within the hostile environment of the host, modulate the host response, and up- or down-regulate expression of specific genes so as to alter the antigenic profile and virulence or even to resist the effects of antimycobacterial agents. However, due to the myriad confounding and effect-modifying factors, such as HIV prevalence, current TB control activities, and genetic susceptibility of the host, the evaluation of fitness based on either epidemiologic parameters or laboratory assays alone is difficult. Clearly, more work is needed to better define and subsequently measure the relative fitness of the pathogen as it relates to infection, transmission, and disease potential within human populations.
CONCLUSION
M. tuberculosis is an obligate pathogen that does not naturally replicate outside of its host environment. As such, M. tuberculosis complex members are believed to have coevolved with hominids for millions of years. Consequently, it is very possible that, unlike other opportunistic pathogens, viable tubercle bacilli encode the minimum ensemble of virulence genes required for successful infection, replication, and dissemination. Thus, the relative success of one clonal M. tuberculosis family over another might rely on the interplay between levels of gene expression and environmental factors (e.g., host). The differential gene expression may also be influenced by variations resulting from chromosomal rearrangements, such as recombinations/deletions, transposition of IS elements, or SNP, aspects which may be studied through the multidisciplinary nature of molecular epidemiological studies. Such genomic events may lead to adaptive advantages and hence determine a successful clonal lineage, as indicated by some studies.
Much of our current knowledge of TB epidemiology has stemmed from numerous observational and anecdotal studies done in the early part of the 1900s; molecular epidemiology as a field has had a similar initiation. Nevertheless, molecular epidemiologic studies over the past 10 years have added significantly to our understanding of TB epidemiology and biology. The use of molecular methods, coupled with classical epidemiologic approaches, has afforded greater resolution and accuracy in describing both the local and global epidemiology of TB, including the detection of unsuspected transmission events and direct evidence for exogenous reinfection in recurrent disease. Molecular epidemiologic methods have added further to population genetic approaches which have suggested biomedically and epidemiologically relevant characteristics specific to phylogenetic lineages or strain families. These include strains that exhibit specific virulence properties, epidemic potential, and various replication rates. Furthermore, immunologic studies have shed insight on the differential pathogenesis elicited by distinct clinical strains. There is a great need for the development of more methodological approaches (e.g., novel study design, mathematical models) to analyze molecular epidemiologic data that account for confounding, effect modification, and statistical power. Still, the continual integration of information generated by the current advances in genomic, transcriptomic, and proteomic analyses in designing molecular epidemiologic studies, as well as an understanding of host-pathogen interactions and identification of host susceptibility genes, will invariably enhance our understanding of M. tuberculosis biology and epidemiology and perhaps even aid in demystifying this elusive pathogen.
Acknowledgments
We thank Dorothy Fallows for critical review of the manuscript and Elena Shashkina and Kozue Kishii for technical assistance.
REFERENCES
- 1.Agerton, T. B., S. E. Valway, R. J. Blinkhorn, K. L. Shilkret, R. Reves, W. W. Schluter, B. Gore, C. J. Pozsik, B. B. Plikaytis, C. Woodley, and I. M. Onorato. 1999. Spread of strain W, a highly drug-resistant strain of Mycobacterium tuberculosis, across the United States. Clin. Infect. Dis. 29:85-95. [DOI] [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Thoracic Society. 2000. Targeted tuberculin testing and treatment of latent tuberculosis infection. Am. J. Respir. Crit. Care Med. 161:S221-S247. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, R. M., and R. M. May. 1991. Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford, United Kingdom.
- 6.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 7.Aranaz, A., E. Liebana, E. Gomez-Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and L. Dominguez. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 8.Badri, M., D. Wilson, and R. Wood. 2002. Effect of highly active antiretroviral therapy on incidence of tuberculosis in South Africa: a cohort study. Lancet 359:2059-2064. [DOI] [PubMed] [Google Scholar]
- 9.Baker, L., T. Brown, M. C. Maiden, and F. Drobniewski. 2004. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg. Infect. Dis. 10:1568-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandera, A., A. Gori, L. Catozzi, A. Degli Esposti, G. Marchetti, C. Molteni, G. Ferrario, L. Codecasa, V. Penati, A. Matteelli, and F. Franzetti. 2001. Molecular epidemiology study of exogenous reinfection in an area with a low incidence of tuberculosis. J. Clin. Microbiol. 39:2213-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barczak, A. K., P. Domenech, H. I. Boshoff, M. B. Reed, C. Manca, G. Kaplan, and C. E. Barry III. 2005. In vivo phenotypic dominance in mouse mixed infections with Mycobacterium tuberculosis clinical isolates. J. Infect. Dis. 192:600-606. [DOI] [PubMed] [Google Scholar]
- 12.Barnes, P. F., H. el-Hajj, S. Preston-Martin, M. D. Cave, B. E. Jones, M. Otaya, J. Pogoda, and K. D. Eisenach. 1996. Transmission of tuberculosis among the urban homeless. JAMA 275:305-307. [PubMed] [Google Scholar]
- 13.Barnes, P. F., Z. Yang, S. Preston-Martin, J. M. Pogoda, B. E. Jones, M. Otaya, K. D. Eisenach, L. Knowles, S. Harvey, and M. D. Cave. 1997. Patterns of tuberculosis transmission in Central Los Angeles. JAMA 278:1159-1163. [PubMed] [Google Scholar]
- 14.Bauer, J., A. B. Andersen, K. Kremer, and H. Miorner. 1999. Usefulness of spoligotyping to discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J. Clin. Microbiol. 37:2602-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer, J., A. Kok-Jensen, P. Faurschou, J. Thuesen, E. Taudorf, and A. B. Andersen. 2000. A prospective evaluation of the clinical value of nation-wide DNA fingerprinting of tuberculosis isolates in Denmark. Int. J. Tuberc. Lung Dis. 4:295-299. [PubMed] [Google Scholar]
- 16.Bauer, J., V. O. Thomsen, S. Poulsen, and A. B. Andersen. 1997. False-positive results from cultures of Mycobacterium tuberculosis due to laboratory cross-contamination confirmed by restriction fragment length polymorphism. J. Clin. Microbiol. 35:988-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer, J., Z. Yang, S. Poulsen, and A. B. Andersen. 1998. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J. Clin. Microbiol. 36:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellamy, R. 2006. Genome-wide approaches to identifying genetic factors in host susceptibility to tuberculosis. Microbes Infect. 8:1119-1123. [DOI] [PubMed] [Google Scholar]
- 20.Bhanu, N. V., D. van Soolingen, J. D. van Embden, L. Dar, R. M. Pandey, and P. Seth. 2002. Predominace of a novel Mycobacterium tuberculosis genotype in the Delhi region of India. Tuberculosis (Edinburgh) 82:105-112. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya, M., S. Dietrich, L. Mosher, F. Siddiqui, B. E. Reisberg, W. S. Paul, and J. R. Warren. 1998. Cross-contamination of specimens with Mycobacterium tuberculosis: clinical significance, causes, and prevention. Am. J. Clin. Pathol. 109:324-330. [DOI] [PubMed] [Google Scholar]
- 22.Bifani, P., B. Mathema, M. Campo, S. Moghazeh, B. Nivin, E. Shashkina, J. Driscoll, S. S. Munsiff, R. Frothingham, and B. N. Kreiswirth. 2001. Molecular identification of streptomycin monoresistant Mycobacterium tuberculosis related to multidrug-resistant W strain. Emerg. Infect. Dis. 7:842-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bifani, P., S. Moghazeh, B. Shopsin, J. Driscoll, A. Ravikovitch, and B. N. Kreiswirth. 2000. Molecular characterization of Mycobacterium tuberculosis H37Rv/Ra variants: distinguishing the mycobacterial laboratory strain. J. Clin. Microbiol. 38:3200-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 25.Bifani, P. J., B. Mathema, Z. Liu, S. L. Moghazeh, B. Shopsin, B. Tempalski, J. Driscol, R. Frothingham, J. M. Musser, P. Alcabes, and B. N. Kreiswirth. 1999. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA 282:2321-2327. [DOI] [PubMed] [Google Scholar]
- 26.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreiswirth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 27.Bifani, P. J., B. Shopsin, P. Alcabes, B. Mathema, B. N. Kreiswirth, Z. Liu, J. Driscoll, R. Frothingham, and J. M. Musser. 2000. Molecular epidemiology and tuberculosis control. JAMA 284:305-307. [PubMed] [Google Scholar]
- 28.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishai, W. R., N. M. Graham, S. Harrington, D. S. Pope, N. Hooper, J. Astemborski, L. Sheely, D. Vlahov, G. E. Glass, and R. E. Chaisson. 1998. Molecular and geographic patterns of tuberculosis transmission after 15 years of directly observed therapy. JAMA 280:1679-1684. [DOI] [PubMed] [Google Scholar]
- 30.Blaser, M. J., and J. M. Musser. 2001. Bacterial polymorphisms and disease in humans. J. Clin. Investig. 107:391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloom, B. R., and C. J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 32.Blot, M. 1994. Transposable elements and adaptation of host bacteria. Genetica 93:5-12. [DOI] [PubMed] [Google Scholar]
- 33.Blower, S. M., and T. Chou. 2004. Modeling the emergence of the “hot zones”: tuberculosis and the amplification dynamics of drug resistance. Nat. Med. 10:1111-1116. [DOI] [PubMed] [Google Scholar]
- 34.Blower, S. M., A. R. McLean, T. C. Porco, P. M. Small, P. C. Hopewell, M. A. Sanchez, and A. R. Moss. 1995. The intrinsic transmission dynamics of tuberculosis epidemics. Nat. Med. 1:815-821. [DOI] [PubMed] [Google Scholar]
- 35.Borgdorff, M. W., N. Nagelkerke, D. van Soolingen, P. E. de Haas, J. Veen, and J. D. van Embden. 1998. Analysis of tuberculosis transmission between nationalities in the Netherlands in the period 1993-1995 using DNA fingerprinting. Am. J. Epidemiol. 147:187-195. [DOI] [PubMed] [Google Scholar]
- 36.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 37.Braden, C. R., J. T. Crawford, and B. A. Schable. 2002. Quality assessment of Mycobacterium tuberculosis genotyping in a large laboratory network. Emerg. Infect. Dis. 8:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 39.Braden, C. R., G. L. Templeton, W. W. Stead, J. H. Bates, M. D. Cave, and S. E. Valway. 1997. Retrospective detection of laboratory cross-contamination of Mycobacterium tuberculosis cultures with use of DNA fingerprint analysis. Clin. Infect. Dis. 24:35-40. [DOI] [PubMed] [Google Scholar]
- 40.British Medical Research Council. 1949. Treatment of pulmonary with para-aminosalicylic acid and streptomycin. Br. Med. J. 2:1521-1525. [PMC free article] [PubMed] [Google Scholar]
- 41.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:452-458. [DOI] [PubMed] [Google Scholar]
- 43.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, C. Martin, I. Mokrousov, O. Narvskaia, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgos, M., K. DeRiemer, P. M. Small, P. C. Hopewell, and C. L. Daley. 2003. Effect of drug resistance on the generation of secondary cases of tuberculosis. J. Infect. Dis. 188:1878-1884. [DOI] [PubMed] [Google Scholar]
- 45.Burman, W. J., and R. R. Reves. 2000. Review of false-positive cultures for Mycobacterium tuberculosis and recommendations for avoiding unnecessary treatment. Clin. Infect. Dis. 31:1390-1395. [DOI] [PubMed] [Google Scholar]
- 46.Caminero, J. A., M. J. Pena, M. I. Campos-Herrero, J. C. Rodriguez, O. Afonso, C. Martin, J. M. Pavon, M. J. Torres, M. Burgos, P. Cabrera, P. M. Small, and D. A. Enarson. 2001. Exogenous reinfection with tuberculosis on a European island with a moderate incidence of disease. Am. J. Respir. Crit. Care Med. 163:717-720. [DOI] [PubMed] [Google Scholar]
- 47.Canetti, G., and J. Grossett. 1961. Teneur des souches sauvages de mycobacterium tuberculosis envariants resistants a l'isoniazide et en variants resistants a la streptomycine sur milieu de Loewenstein-Jensen. Ann. Inst. Pasteur 101:28. [PubMed] [Google Scholar]
- 48.Canetti, G., I. Sutherland, and E. Svandova. 1972. Endogenous reactivation and exogenous reinfection: their relative importance with regard to the development of non-primary tuberculosis. Bull. Int. Union Tuberc. 47:116-134. [PubMed] [Google Scholar]
- 49.Casanova, J. L., E. Schurr, L. Abel, and E. Skamene. 2002. Forward genetics of infectious diseases: immunological impact. Trends Immunol. 23:469-472. [DOI] [PubMed] [Google Scholar]
- 50.Cave, M. D., K. D. Eisenach, G. Templeton, M. Salfinger, G. Mazurek, J. H. Bates, and J. T. Crawford. 1994. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 32:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chacon-Salinas, R., J. Serafin-Lopez, R. Ramos-Payan, P. Mendez-Aragon, R. Hernandez-Pando, D. Van Soolingen, L. Flores-Romo, S. Estrada-Parra, and I. Estrada-Garcia. 2005. Differential pattern of cytokine expression by macrophages infected in vitro with different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 140:443-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 53.Charlesworth, B., P. Sniegowski, and W. Stephan. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215-220. [DOI] [PubMed] [Google Scholar]
- 54.Chaves, F., Z. Yang, H. el Hajj, M. Alonso, W. J. Burman, K. D. Eisenach, F. Dronda, J. H. Bates, and M. D. Cave. 1996. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J. Clin. Microbiol. 34:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiang, C. Y., and L. W. Riley. 2005. Exogenous reinfection in tuberculosis. Lancet Infect. Dis. 5:629-636. [DOI] [PubMed] [Google Scholar]
- 56.Clark, C. M., C. R. Driver, S. S. Munsiff, J. R. Driscoll, B. N. Kreiswirth, B. Zhao, A. Ebrahimzadeh, M. Salfinger, A. S. Piatek, and J. Abdelwahab. 2006. Universal genotyping in tuberculosis control program, New York City, 2001-2003. Emerg. Infect. Dis. 12:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen, T., and M. Murray. 2004. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nat. Med. 10:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen, T., B. Sommers, and M. Murray. 2003. The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect. Dis. 3:13-21. [DOI] [PubMed] [Google Scholar]
- 59.Colditz, G. A., T. F. Brewer, C. S. Berkey, M. E. Wilson, E. Burdick, H. V. Fineberg, and F. Mosteller. 1994. Efficacy of BCG vaccine in the prevention of tuberculosis. Meta-analysis of the published literature. JAMA 271:698-702. [PubMed] [Google Scholar]
- 60.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. (Erratum, 396:190.) [DOI] [PubMed] [Google Scholar]
- 61.Comstock, G. W. 1975. Frost revisited: the modern epidemiology of tuberculosis. Am. J. Epidemiol. 101:363-382. [DOI] [PubMed] [Google Scholar]
- 62.Cooke, G. S., and A. V. Hill. 2001. Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2:967-977. [DOI] [PubMed] [Google Scholar]
- 63.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 64.Cousins, D. V., R. Bastida, A. Cataldi, V. Quse, S. Redrobe, S. Dow, P. Duignan, A. Murray, C. Dupont, N. Ahmed, D. M. Collins, W. R. Butler, D. Dawson, D. Rodriguez, J. Loureiro, M. I. Romano, A. Alito, M. Zumarraga, and A. Bernardelli. 2003. Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int. J. Syst. Evol. Microbiol. 53:1305-1314. [DOI] [PubMed] [Google Scholar]
- 65.Cowan, L. S., L. Diem, T. Monson, P. Wand, D. Temporado, T. V. Oemig, and J. T. Crawford. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crofton, J., P. Chaulet, D. Maher, J. Grosset, W. Harris, H. Norman, M. D. Iseman, and B. Watt. 1997. Guidelines for the management of multidrug-resistant tuberculosis. World Health Organization, Geneva, Switzerland.
- 68.Crowle, A. J., and N. Elkins. 1990. Relative permissiveness of macrophages from black and white people for virulent tubercle bacilli. Infect. Immun. 58:632-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dale, J. W., D. Brittain, A. A. Cataldi, D. Cousins, J. T. Crawford, J. Driscoll, H. Heersma, T. Lillebaek, T. Quitugua, N. Rastogi, R. A. Skuce, C. Sola, D. Van Soolingen, and V. Vincent. 2001. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis. 5:216-219. [PubMed] [Google Scholar]
- 70.Daley, C. L., P. M. Small, G. F. Schecter, G. K. Schoolnik, R. A. McAdam, W. R. Jacobs, Jr., and P. C. Hopewell. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N. Engl. J. Med. 326:231-235. [DOI] [PubMed] [Google Scholar]
- 71.Das, S., C. N. Paramasivan, D. B. Lowrie, R. Prabhakar, and P. R. Narayanan. 1995. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, South India. Tuberc. Lung Dis. 76:550-554. [DOI] [PubMed] [Google Scholar]
- 72.David, H. L. 1971. Resistance to d-cycloserine in the tubercle bacilli: mutation rate and transport of alanine in parental cells and drug-resistant mutants. Appl. Microbiol. 21:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.David, H. L., and C. M. Newman. 1971. Some observations on the genetics of isoniazid resistance in the tubercle bacilli. Am. Rev. Respir. Dis. 104:508-515. [DOI] [PubMed] [Google Scholar]
- 74.Davies, J. 1998. Antibiotic resistance in mycobacteria, p. 195-208. Wiley, Chichester, United Kingdom. [PubMed]
- 75.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 76.de Boer, A. S., and D. van Soolingen. 2000. Recurrent tuberculosis due to exogenous reinfection. N. Engl. J. Med. 342:1050-1051. [PubMed] [Google Scholar]
- 77.Di Perri, G., M. Cruciani, M. C. Danzi, R. Luzzati, G. De Checchi, M. Malena, S. Pizzighella, R. Mazzi, M. Solbiati, E. Concia, et al. 1989. Nosocomial epidemic of active tuberculosis among HIV-infected patients. Lancet ii:1502-1504. [PubMed] [Google Scholar]
- 78.Doherty, T. M., and P. Andersen. 2005. Vaccines for tuberculosis: novel concepts and recent progress. Clin. Microbiol. Rev. 18:687-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Domenech, P., M. B. Reed, and C. E. Barry III. 2005. Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73:3492-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dormans, J., M. Burger, D. Aguilar, R. Hernandez-Pando, K. Kremer, P. Roholl, S. M. Arend, and D. van Soolingen. 2004. Correlation of virulence, lung pathology, bacterial load and delayed type hypersensitivity responses after infection with different Mycobacterium tuberculosis genotypes in a BALB/c mouse model. Clin. Exp. Immunol. 137:460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Douglas, J. T., L. Qian, J. C. Montoya, J. M. Musser, J. D. Van Embden, D. Van Soolingen, and K. Kremer. 2003. Characterization of the Manila family of Mycobacterium tuberculosis. J. Clin. Microbiol. 41:2723-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Driscoll, J. R., M. A. McGarry, and H. W. Taber. 1999. DNA typing of a nonviable culture of Mycobacterium tuberculosis in a homeless shelter outbreak. J. Clin. Microbiol. 37:274-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Drobniewski, F., Y. Balabanova, V. Nikolayevsky, M. Ruddy, S. Kuznetzov, S. Zakharova, A. Melentyev, and I. Fedorin. 2005. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA 293:2726-2731. [DOI] [PubMed] [Google Scholar]
- 84.Drobniewski, F., Y. Balabanova, M. Ruddy, L. Weldon, K. Jeltkova, T. Brown, N. Malomanova, E. Elizarova, A. Melentyey, E. Mutovkin, S. Zhakharova, and I. Fedorin. 2002. Rifampin- and multidrug-resistant tuberculosis in Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg. Infect. Dis. 8:1320-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dye, C., M. A. Espinal, C. J. Watt, C. Mbiaga, and B. G. Williams. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 185:1197-1202. [DOI] [PubMed] [Google Scholar]
- 86.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 282:677-686. [DOI] [PubMed] [Google Scholar]
- 87.Dye, C., B. G. Williams, M. A. Espinal, and M. C. Raviglione. 2002. Erasing the world's slow stain: strategies to beat multidrug-resistant tuberculosis. Science 295:2042-2046. [DOI] [PubMed] [Google Scholar]
- 88.East African/British Medical Research Council. 1972. Controlled clinical trial of short-course (6-month) regimens of chemotherapy for treatment of pulmonary tuberculosis. Lancet i:1079-1085. [PubMed] [Google Scholar]
- 89.Edlin, B. R., J. I. Tokars, M. H. Grieco, J. T. Crawford, J. Williams, E. M. Sordillo, K. R. Ong, J. O. Kilburn, S. W. Dooley, K. G. Castro, et al. 1992. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 326:1514-1521. [DOI] [PubMed] [Google Scholar]
- 90.Espinal, M. A., S. J. Kim, P. G. Suarez, K. M. Kam, A. G. Khomenko, G. B. Migliori, J. Baez, A. Kochi, C. Dye, and M. C. Raviglione. 2000. Standard short-course chemotherapy for drug-resistant tuberculosis: treatment outcomes in 6 countries. JAMA 283:2537-2545. [DOI] [PubMed] [Google Scholar]
- 91.Ferebee, S. 1969. Controlled chemoprophylaxis trials in tuberculosis: A general review. Adv. Tuberc. Res. 17:29-106. [PubMed] [Google Scholar]
- 92.Filliol, I., J. R. Driscoll, D. Van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. D. Anh, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, G. Kallenius, E. Kassa-Kelembho, T. Koivula, H. M. Ly, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. De Waard, C. Sola, and N. Rastogi. 2002. Global distribution of Mycobacterium tuberculosis spoligotypes. Emerg. Infect. Dis. 8:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Filliol, I., J. R. Driscoll, D. van Soolingen, B. N. Kreiswirth, K. Kremer, G. Valetudie, D. A. Dang, R. Barlow, D. Banerjee, P. J. Bifani, K. Brudey, A. Cataldi, R. C. Cooksey, D. V. Cousins, J. W. Dale, O. A. Dellagostin, F. Drobniewski, G. Engelmann, S. Ferdinand, D. Gascoyne-Binzi, M. Gordon, M. C. Gutierrez, W. H. Haas, H. Heersma, E. Kassa-Kelembho, M. L. Ho, A. Makristathis, C. Mammina, G. Martin, P. Mostrom, I. Mokrousov, V. Narbonne, O. Narvskaya, A. Nastasi, S. N. Niobe-Eyangoh, J. W. Pape, V. Rasolofo-Razanamparany, M. Ridell, M. L. Rossetti, F. Stauffer, P. N. Suffys, H. Takiff, J. Texier-Maugein, V. Vincent, J. H. de Waard, C. Sola, and N. Rastogi. 2003. Snapshot of moving and expanding clones of Mycobacterium tuberculosis and their global distribution assessed by spoligotyping in an international study. J. Clin. Microbiol. 41:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. B. del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fine, P. E., and P. M. Small. 1999. Exogenous reinfection in tuberculosis. N. Engl. J. Med. 341:1226-1227. [DOI] [PubMed] [Google Scholar]
- 96.Fitzpatrick, L., C. Braden, W. Cronin, J. English, E. Campbell, S. Valway, and I. Onorato. 2004. Investigation of laboratory cross-contamination of Mycobacterium tuberculosis cultures. Clin. Infect. Dis. 38:e52-e54. [DOI] [PubMed] [Google Scholar]
- 97.Fitzpatrick, L. K., A. Okwera, R. Mugerwa, R. Ridzon, J. Ehiner, and I. Onorato. 2002. An investigation of suspected exogenous reinfection in tuberculosis patients in Kampala, Uganda. Int. J. Tuberc. Lung Dis. 6:550-552. [DOI] [PubMed] [Google Scholar]
- 98.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Freeman, R., M. Kato-Maeda, K. A. Hauge, K. L. Horan, E. Oren, M. Narita, C. K. Wallis, D. Cave, C. M. Nolan, P. M. Small, and G. A. Cangelosi. 2005. Use of rapid genomic deletion typing to monitor a tuberculosis outbreak within an urban homeless population. J. Clin. Microbiol. 43:5550-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Frieden, T. R., P. I. Fujiwara, R. M. Washko, and M. A. Hamburg. 1995. Tuberculosis in New York City—turning the tide. N. Engl. J. Med. 333:229-233. [DOI] [PubMed] [Google Scholar]
- 101.Frieden, T. R., L. F. Sherman, K. L. Maw, P. I. Fujiwara, J. T. Crawford, B. Nivin, V. Sharp, D. Hewlett, Jr., K. Brudney, D. Alland, and B. N. Kreisworth. 1996. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA 276:1229-1235. [PubMed] [Google Scholar]
- 102.Friedman, C. R., G. C. Quinn, B. N. Kreiswirth, D. C. Perlman, N. Salomon, N. Schluger, M. Lutfey, J. Berger, N. Poltoratskaia, and L. W. Riley. 1997. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis. J. Infect. Dis. 176:478-484. [DOI] [PubMed] [Google Scholar]
- 103.Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 105.Gagneux, S., M. V. Burgos, K. DeRiemer, A. Encisco, S. Munoz, P. C. Hopewell, P. M. Small, and A. S. Pym. 2006. Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog. 2:e61. [Online.] http://dx.doi.org/10.1371/journal.ppat.0020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gagneux, S., C. D. Long, P. M. Small, T. Van, G. K. Schoolnik, and B. J. Bohannan. 2006. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312:1944-1946. [DOI] [PubMed] [Google Scholar]
- 107.Garcia-Garcia, M. L., M. E. Jimenez-Corona, A. Ponce-de-Leon, A. Jimenez-Corona, M. Palacios-Martinez, S. Balandrano-Campos, L. Ferreyra-Reyes, L. Juarez-Sandino, J. Sifuentes-Osornio, H. Olivera-Diaz, J. L. Valdespino-Gomez, and P. M. Small. 2000. Mycobacterium tuberculosis drug resistance in a suburban community in southern Mexico. Int. J. Tuberc. Lung Dis. 4:S168-S170. [PubMed] [Google Scholar]
- 108.Garcia de Viedma, D., G. Lorenzo, P. J. Cardona, N. Alonso Rodriguez, S. Gordillo, M. J. Ruiz Serrano, and E. Bouza. 2005. Association between the infectivity of Mycobacterium tuberculosis strains and their efficiency for extrarespiratory infection. J. Infect. Dis. 192:2059-2065. [DOI] [PubMed] [Google Scholar]
- 109.Geng, E., B. Kreiswirth, C. Driver, J. Li, J. Burzynski, P. DellaLatta, A. LaPaz, and N. W. Schluger. 2002. Changes in the transmission of tuberculosis in New York City from 1990 to 1999. N. Engl. J. Med. 346:1453-1458. [DOI] [PubMed] [Google Scholar]
- 110.Ghon, A. 1923. The primary complex in human tuberculosis and its significance. Am. Rev. Tuberc. 7:314-317. [Google Scholar]
- 111.Girardi, E., C. A. Sabin, A. d'Arminio Monforte, B. Hogg, A. N. Phillips, M. J. Gill, F. Dabis, P. Reiss, O. Kirk, E. Bernasconi, S. Grabar, A. Justice, S. Staszewski, G. Fatkenheuer, and J. A. Sterne. 2005. Incidence of tuberculosis among HIV-infected patients receiving highly active antiretroviral therapy in Europe and North America. Clin. Infect. Dis. 41:1772-1782. [DOI] [PubMed] [Google Scholar]
- 112.Glynn, J. R., J. Bauer, A. S. de Boer, M. W. Borgdorff, P. E. Fine, P. Godfrey-Faussett, and E. Vynnycky. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 3:1055-1060. [PubMed] [Google Scholar]
- 113.Glynn, J. R., A. C. Crampin, M. D. Yates, H. Traore, F. D. Mwaungulu, B. M. Ngwira, R. Ndlovu, F. Drobniewski, and P. E. Fine. 2005. The importance of recent infection with Mycobacterium tuberculosis in an area with high HIV prevalence: a long-term molecular epidemiological study in Northern Malawi. J. Infect. Dis. 192:480-487. [DOI] [PubMed] [Google Scholar]
- 114.Glynn, J. R., E. Vynnycky, and P. E. Fine. 1999. Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques. Am. J. Epidemiol. 149:366-371. [DOI] [PubMed] [Google Scholar]
- 115.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Godfrey-Faussett, P., P. Sonnenberg, S. C. Shearer, M. C. Bruce, C. Mee, L. Morris, and J. Murray. 2000. Tuberculosis control and molecular epidemiology in a South African gold-mining community. Lancet 356:1066-1071. [DOI] [PubMed] [Google Scholar]
- 117.Goguet de la Salmoniere, Y. O., C. C. Kim, A. G. Tsolaki, A. S. Pym, M. S. Siegrist, and P. M. Small. 2004. High-throughput method for detecting genomic-deletion polymorphisms. J. Clin. Microbiol. 42:2913-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gomez, J. E., and J. D. McKinney. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinburgh) 84:29-44. [DOI] [PubMed] [Google Scholar]
- 119.Goyal, M., D. Young, Y. Zhang, P. A. Jenkins, and R. J. Shaw. 1994. PCR amplification of variable sequence upstream of katG gene to subdivide strains of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:3070-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grode, L., P. Seiler, S. Baumann, J. Hess, V. Brinkmann, A. N. Eddine, P. Mann, C. Goosmann, S. Bandermann, D. Smith, G. J. Bancroft, J. M. Reyrat, D. van Soolingen, B. Raupach, and S. H. Kaufmann. 2005. Increased vaccine efficacy against tuberculosis of recombinant Mycobacterium bovis bacille Calmette-Guerin mutants that secrete listeriolysin. J. Clin. Investig. 115:2472-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 122.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193:121-128. [DOI] [PubMed] [Google Scholar]
- 123.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gutierrez, M. C., S. Brisse, R. Brosch, M. Fabre, B. Omais, M. Marmiesse, P. Supply, and V. Vincent. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 1:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Heersma, H. F., K. Kremer, and J. D. van Embden. 1998. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol. Biol. 101:395-422. [DOI] [PubMed] [Google Scholar]
- 126.Henderson, R. A., S. C. Watkins, and J. L. Flynn. 1997. Activation of human dendritic cells following infection with Mycobacterium tuberculosis. J. Immunol. 159:635-643. [PubMed] [Google Scholar]
- 127.Hermans, P. W., D. van Soolingen, J. W. Dale, A. R. Schuitema, R. A. McAdam, D. Catty, and J. D. van Embden. 1990. Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hermans, P. W., D. van Soolingen, and J. D. van Embden. 1992. Characterization of a major polymorphic tandem repeat in Mycobacterium tuberculosis and its potential use in the epidemiology of Mycobacterium kansasii and Mycobacterium gordonae. J. Bacteriol. 174:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Heyderman, R. S., M. Goyal, P. Roberts, S. Ushewokunze, S. Zizhou, B. G. Marshall, R. Makombe, J. D. Van Embden, P. R. Mason, and R. J. Shaw. 1998. Pulmonary tuberculosis in Harare, Zimbabwe: analysis by spoligotyping. Thorax 53:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iseman, M. D. 2000. A clinician's guide to tuberculosis, p. 323-324. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 133.Iseman, M. D., and L. A. Madsen. 1989. Drug-resistant tuberculosis. Clin. Chest Med. 10:341-353. [PubMed] [Google Scholar]
- 134.Jagirdar, J., and, and D. Zagzag. 1996. Pathology and insights into pathogenesis of tuberculosis, p. 467-482. In W. N. Rom and S. M. Garay (ed.), Tuberculosis, 1st ed. Little, Brown and Company, New York, N.Y.
- 135.Jasmer, R. M., L. Bozeman, K. Schwartzman, M. D. Cave, J. J. Saukkonen, B. Metchock, A. Khan, and W. J. Burman. 2004. Recurrent tuberculosis in the United States and Canada: relapse or reinfection? Am. J. Respir. Crit. Care Med. 170:1360-1366. [DOI] [PubMed] [Google Scholar]
- 136.Jasmer, R. M., J. A. Hahn, P. M. Small, C. L. Daley, M. A. Behr, A. R. Moss, J. M. Creasman, G. F. Schecter, E. A. Paz, and P. C. Hopewell. 1999. A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991-1997. Ann. Intern. Med. 130:971-978. [DOI] [PubMed] [Google Scholar]
- 137.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaplan, G., F. A. Post, A. L. Moreira, H. Wainwright, B. N. Kreiswirth, M. Tanverdi, B. Mathema, S. V. Ramaswamy, G. Walther, L. M. Steyn, C. E. Barry III, and L. G. Bekker. 2003. Mycobacterium tuberculosis growth at the cavity surface: a microenvironment with failed immunity. Infect. Immun. 71:7099-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kapur, V., T. S. Whittam, and J. M. Musser. 1994. Is Mycobacterium tuberculosis 15,000 years old? J. Infect. Dis. 170:1348-1349. [DOI] [PubMed] [Google Scholar]
- 140.Karunakaran, P., and J. Davies. 2000. Genetic antagonism and hypermutability in Mycobacterium smegmatis. J. Bacteriol. 182:3331-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaufmann, S. H. 2001. How can immunology contribute to the control of tuberculosis? Nat. Rev. Immunol. 1:20-30. [DOI] [PubMed] [Google Scholar]
- 143.Kaufmann, S. H. 2005. Recent findings in immunology give tuberculosis vaccines a new boost. Trends Immunol. 26:660-667. [DOI] [PubMed] [Google Scholar]
- 144.Koch, R. 1882. Die Aetiologie der Tuberculose. Berl. Klinische Wochenschr. 19:221-230. [Google Scholar]
- 145.Kochi, A., B. Vareldzis, and K. Styblo. 1993. Multidrug-resistant tuberculosis and its control. Res. Microbiol. 144:104-110. [DOI] [PubMed] [Google Scholar]
- 146.Kovalev, S. Y., E. Y. Kamaev, M. A. Kravchenko, N. E. Kurepina, and S. N. Skorniakov. 2005. Genetic analysis of Mycobacterium tuberculosis strains isolated in Ural region, Russian Federation, by MIRU-VNTR genotyping. Int. J. Tuberc. Lung Dis. 9:746-752. [PubMed] [Google Scholar]
- 147.Kremer, K., C. Arnold, A. Cataldi, M. C. Gutierrez, W. H. Haas, S. Panaiotov, R. A. Skuce, P. Supply, A. G. van der Zanden, and D. van Soolingen. 2005. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J. Clin. Microbiol. 43:5628-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kremer, K., B. K. Au, P. C. Yip, R. Skuce, P. Supply, K. M. Kam, and D. van Soolingen. 2005. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J. Clin. Microbiol. 43:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kurepina, N., E. Likhoshvay, E. Shashkina, B. Mathema, K. Kremer, D. van Soolingen, P. Bifani, and B. N. Kreiswirth. 2005. Targeted hybridization of IS6110 fingerprints identifies the W-Beijing Mycobacterium tuberculosis strains among clinical isolates. J. Clin. Microbiol. 43:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Sooligen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuberc. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 153.Lambert, M. L., E. Hasker, A. Van Deun, D. Roberfroid, M. Boelaert, and P. Van der Stuyft. 2003. Recurrence in tuberculosis: relapse or reinfection? Lancet Infect. Dis. 3:282-287. [DOI] [PubMed] [Google Scholar]
- 154.Levin, B. R., M. Lipsitch, and S. Bonhoeffer. 1999. Population biology, evolution, and infectious disease: convergence and synthesis. Science 283:806-809. [DOI] [PubMed] [Google Scholar]
- 155.Lillebaek, T., A. B. Andersen, A. Dirksen, E. Smith, L. T. Skovgaard, and A. Kok-Jensen. 2002. Persistent high incidence of tuberculosis in immigrants in a low-incidence country. Emerg. Infect. Dis. 8:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lillebaek, T., A. Dirksen, I. Baess, B. Strunge, V. O. Thomsen, and A. B. Andersen. 2002. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J. Infect. Dis. 185:401-404. [DOI] [PubMed] [Google Scholar]
- 157.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lutfey, M., P. Della-Latta, V. Kapur, L. A. Palumbo, D. Gurner, G. Stotzky, K. Brudney, J. Dobkin, A. Moss, J. M. Musser, and B. N. Kreiswirth. 1996. Independent origin of mono-rifampin-resistant Mycobacterium tuberculosis in patients with AIDS. Am. J. Respir. Crit. Care Med. 153:837-840. [DOI] [PubMed] [Google Scholar]
- 159.Magdalena, J., A. Vachee, P. Supply, and C. Locht. 1998. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 36:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Malakmadze, N., I. M. Gonzalez, T. Oemig, I. Isiadinso, D. Rembert, M. M. McCauley, P. Wand, L. Diem, L. Cowan, G. J. Palumbo, M. Fraser, and K. Ijaz. 2005. Unsuspected recent transmission of tuberculosis among high-risk groups: implications of universal tuberculosis genotyping in its detection. Clin. Infect. Dis. 40:366-373. [DOI] [PubMed] [Google Scholar]
- 161.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nat. Med. 6:1327-1329. [DOI] [PubMed] [Google Scholar]
- 162.Manabe, Y. C., A. M. Dannenberg, Jr., S. K. Tyagi, C. L. Hatem, M. Yoder, S. C. Woolwine, B. C. Zook, M. L. Pitt, and W. R. Bishai. 2003. Different strains of Mycobacterium tuberculosis cause various spectrums of disease in the rabbit model of tuberculosis. Infect. Immun. 71:6004-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Manca, C., M. B. Reed, S. Freeman, B. Mathema, B. Kreiswirth, C. E. Barry III, and G. Kaplan. 2004. Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect. Immun. 72:5511-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Manca, C., L. Tsenova, C. E. Barry III, A. Bergtold, S. Freeman, P. A. Haslett, J. M. Musser, V. H. Freedman, and G. Kaplan. 1999. Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J. Immunol. 162:6740-6746. [PubMed] [Google Scholar]
- 165.Manca, C., L. Tsenova, A. Bergtold, S. Freeman, M. Tovey, J. M. Musser, C. E. Barry III, V. H. Freedman, and G. Kaplan. 2001. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proc. Natl. Acad. Sci. USA 98:5752-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mariam, D. H., Y. Mengistu, S. E. Hoffner, and D. I. Andersson. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mathema, B., P. J. Bifani, J. Driscoll, L. Steinlein, N. Kurepina, S. L. Moghazeh, E. Shashkina, S. A. Marras, S. Campbell, B. Mangura, K. Shilkret, J. T. Crawford, R. Frothingham, and B. N. Kreiswirth. 2002. Identification and evolution of an IS6110 low-copy-number Mycobacterium tuberculosis cluster. J. Infect. Dis. 185:641-649. [DOI] [PubMed] [Google Scholar]
- 168.Maus, C. E., B. B. Plikaytis, and T. M. Shinnick. 2005. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.McCune, R. M., F. M. Feldmann, H. P. Lambert, and W. McDermott. 1966. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J. Exp. Med. 123:445-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McCune, R. M., F. M. Feldmann, and W. McDermott. 1966. Microbial persistence. II. Characteristics of the sterile state of tubercle bacilli. J. Exp. Med. 123:469-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.McDermott, W., S. J. Hadley, H. Hull-Smith, and A. Tracy. 1947. Streptomycin in the treatment of tuberculosis in humans. Ann. Intern. Med. 27:769-822. [DOI] [PubMed] [Google Scholar]
- 173.McElroy, P. D., T. R. Sterling, C. R. Driver, B. Kreiswirth, C. L. Woodley, W. A. Cronin, D. X. Hardge, K. L. Shilkret, and R. Ridzon. 2002. Use of DNA fingerprinting to investigate a multiyear, multistate tuberculosis outbreak. Emerg. Infect. Dis. 8:1252-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McHugh, T. D., and S. H. Gillespie. 1998. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J. Clin. Microbiol. 36:1410-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Medlar, E. M. 1955. Necropsy studies of human pulmonary tuberculosis. Am. Rev. Tuberc. 71(2):29-55. [Google Scholar]
- 176.Middlebrook, G. 1954. Isoniazid-resistance and catalase activity of tubercle bacilli: a preliminary report. Am. Rev. Tuberc. 69:471-472. [DOI] [PubMed] [Google Scholar]
- 177.Milan, S. J., K. A. Hauge, N. E. Kurepina, K. H. Lofy, S. V. Goldberg, M. Narita, C. M. Nolan, P. D. McElroy, B. N. Kreiswirth, and G. A. Cangelosi. 2004. Expanded geographical distribution of the N family of Mycobacterium tuberculosis strains within the United States. J. Clin. Microbiol. 42:1064-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.MMWR. 2003. American Thoracic Society, CDC, and Infectious Disease Society of America: recommendations and reports. Treatment of tuberculosis. Morb. Mortal. Wkly. Rep. 52:1-77. [PubMed] [Google Scholar]
- 179.MMWR. 2000. Misdiagnoses of tuberculosis resulting from laboratory cross-contamination of Mycobacterium tuberculosis cultures—New Jersey, 1998. Morb. Mortal. Wkly. Rep. 49:413-416. [PubMed] [Google Scholar]
- 180.Mokrousov, I., I. Filliol, E. Legrand, C. Sola, T. Otten, E. Vyshnevskaya, E. Limeschenko, B. Vyshnevskiy, O. Narvskaya, and N. Rastogi. 2002. Molecular characterization of multiple-drug-resistant Mycobacterium tuberculosis isolates from northwestern Russia and analysis of rifampin resistance using RNA/RNA mismatch analysis as compared to the line probe assay and sequencing of the rpoB gene. Res. Microbiol. 153:213-219. [DOI] [PubMed] [Google Scholar]
- 181.Mokrousov, I., O. Narvskaya, T. Otten, A. Vyazovaya, E. Limeschenko, L. Steklova, and B. Vyshnevskyi. 2002. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 153:629-637. [DOI] [PubMed] [Google Scholar]
- 182.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 183.Munsiff, S. S., B. Nivin, G. Sacajiu, B. Mathema, P. Bifani, and B. N. Kreiswirth. 2003. Persistence of a highly resistant strain of tuberculosis in New York City during 1990-1999. J. Infect. Dis. 188:356-363. [DOI] [PubMed] [Google Scholar]
- 184.Murray, C. J., and J. A. Salomon. 1998. Modeling the impact of global tuberculosis control strategies. Proc. Natl. Acad. Sci. USA 95:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Murray, M. 2002. Determinants of cluster distribution in the molecular epidemiology of tuberculosis. Proc. Natl. Acad. Sci. USA 99:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Murray, M. 2002. Sampling bias in the molecular epidemiology of tuberculosis. Emerg. Infect. Dis. 8:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Musser, J. M. 1996. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg. Infect. Dis. 2:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Musser, J. M., A. Amin, and S. Ramaswamy. 2000. Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Namwat, W., P. Luangsuk, and P. Palittapongarnpim. 1998. The genetic diversity of Mycobacterium tuberculosis strains in Thailand studied by amplification of DNA segments containing direct repetitive sequences. Int. J. Tuberc. Lung Dis. 2:153-159. [PubMed] [Google Scholar]
- 190.Newton, J. K. 1949. Presented at the Minutes of the Sixth Streptomycin Conference, VA-Tuberculosis Section.
- 191.Niemann, S., E. Richter, S. Rusch-Gerdes, H. Thielen, and H. Heykes-Uden. 1999. Outbreak of rifampin- and streptomycin-resistant tuberculosis among homeless in Germany. Int. J. Tuberc. Lung Dis. 3:1146-1147. [PubMed] [Google Scholar]
- 192.Nivin, B., J. Driscoll, T. Glaser, P. Bifani, and S. Munsiff. 2000. Use of spoligotype analysis to detect laboratory cross-contamination. Infect. Control Hosp. Epidemiol. 21:525-527. [DOI] [PubMed] [Google Scholar]
- 193.Nivin, B., P. I. Fujiwara, J. Hannifin, and B. N. Kreiswirth. 1998. Cross-contamination with Mycobacterium tuberculosis: an epidemiological and laboratory investigation. Infect. Control Hosp. Epidemiol. 19:500-503. [DOI] [PubMed] [Google Scholar]
- 194.Orme, I. M. 2005. Tuberculosis vaccines: current progress. Drugs 65:2437-2444. [DOI] [PubMed] [Google Scholar]
- 195.Pape, J. W., S. S. Jean, J. L. Ho, A. Hafner, and W. D. Johnson, Jr. 1993. Effect of isoniazid prophylaxis on incidence of active tuberculosis and progression of HIV infection. Lancet 342:268-272. [DOI] [PubMed] [Google Scholar]
- 196.Park, Y. K., S. Shin, S. Ryu, S. N. Cho, W. J. Koh, O. J. Kwon, Y. S. Shim, W. J. Lew, and G. H. Bai. 2005. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J. Microbiol. Methods 63:165-172. [DOI] [PubMed] [Google Scholar]
- 197.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pasca, M. R., P. Guglierame, E. De Rossi, F. Zara, and G. Riccardi. 2005. mmpL7 gene of Mycobacterium tuberculosis is responsible for isoniazid efflux in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 49:4775-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Peloquin, C. A. 2001. Tuberculosis drug serum levels. Clin. Infect. Dis. 33:584-585. [DOI] [PubMed] [Google Scholar]
- 200.Peloquin, C. A., A. T. Nitta, W. J. Burman, K. F. Brudney, J. R. Miranda-Massari, M. E. McGuinness, S. E. Berning, and G. T. Gerena. 1996. Low antituberculosis drug concentrations in patients with AIDS. Ann. Pharmacother. 30:919-925. [DOI] [PubMed] [Google Scholar]
- 201.Plikaytis, B. B., J. L. Marden, J. T. Crawford, C. L. Woodley, W. R. Butler, and T. M. Shinnick. 1994. Multiplex PCR assay specific for the multidrug-resistant strain W of Mycobacterium tuberculosis. J. Clin. Microbiol. 32:1542-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Post, F. A., P. A. Willcox, B. Mathema, L. M. Steyn, K. Shean, S. V. Ramaswamy, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and G. Kaplan. 2004. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J. Infect. Dis. 190:99-106. [DOI] [PubMed] [Google Scholar]
- 203.Puffer, R. R., H. C. Steward, and R. S. Gass. 1945. Tuberculosis in household contacts associates: the influence of age and relationship. Am. Rev. Tuberc. 52:89-103. [DOI] [PubMed] [Google Scholar]
- 204.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qian, L., J. D. Van Embden, A. G. Van Der Zanden, E. F. Weltevreden, H. Duanmu, and J. T. Douglas. 1999. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J. Clin. Microbiol. 37:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 207.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84-87. [DOI] [PubMed] [Google Scholar]
- 208.Rengarajan, J., C. M. Sassetti, V. Naroditskaya, A. Sloutsky, B. R. Bloom, and E. J. Rubin. 2004. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol. Microbiol. 53:275-282. [DOI] [PubMed] [Google Scholar]
- 209.Rhee, J. T., A. S. Piatek, P. M. Small, L. M. Harris, S. V. Chaparro, F. R. Kramer, and D. Alland. 1999. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rhee, J. T., M. M. Tanaka, M. A. Behr, C. B. Agasino, E. A. Paz, P. C. Hopewell, and P. M. Small. 2000. Use of multiple markers in population-based molecular epidemiologic studies of tuberculosis. Int. J. Tuberc. Lung Dis. 4:1111-1119. [PubMed] [Google Scholar]
- 211.Richardson, M., N. M. Carroll, E. Engelke, G. D. Van Der Spuy, F. Salker, Z. Munch, R. P. Gie, R. M. Warren, N. Beyers, and P. D. Van Helden. 2002. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J. Clin. Microbiol. 40:2750-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Roach, D. R., A. G. Bean, C. Demangel, M. P. France, H. Briscoe, and W. J. Britton. 2002. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J. Immunol. 168:4620-4627. [DOI] [PubMed] [Google Scholar]
- 213.Romeyn, J. A. 1970. Exogenous reinfection in tuberculosis. Am. Rev. Respir. Dis. 101:923-927. [DOI] [PubMed] [Google Scholar]
- 214.Roring, S., A. N. Scott, R. Glyn Hewinson, S. D. Neill, and R. A. Skuce. 2004. Evaluation of variable number tandem repeat (VNTR) loci in molecular typing of Mycobacterium bovis isolates from Ireland. Vet. Microbiol. 101:65-73. [DOI] [PubMed] [Google Scholar]
- 215.Rosenberg, N. A., A. G. Tsolaki, and M. M. Tanaka. 2003. Estimating change rates of genetic markers using serial samples: applications to the transposon IS6110 in Mycobacterium tuberculosis. Theor. Popul. Biol. 63:347-363. [DOI] [PubMed] [Google Scholar]
- 216.Ross, B. C., K. Raios, K. Jackson, and B. Dwyer. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 30:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216a.Safi, H., et al. 2004. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol. Microbiol. 52:999-1012. [DOI] [PubMed] [Google Scholar]
- 217.Sahadevan, R., S. Narayanan, C. N. Paramasivan, R. Prabhakar, and P. R. Narayanan. 1995. Restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, India, by use of direct-repeat probe. J. Clin. Microbiol. 33:3037-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sampson, S. L., R. M. Warren, M. Richardson, T. C. Victor, A. M. Jordaan, G. D. van der Spuy, and P. D. van Helden. 2003. IS6110-mediated deletion polymorphism in the direct repeat region of clinical isolates of Mycobacterium tuberculosis. J. Bacteriol. 185:2856-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Selwyn, P. A., D. Hartel, V. A. Lewis, E. E. Schoenbaum, S. H. Vermund, R. S. Klein, A. T. Walker, and G. H. Friedland. 1989. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N. Engl. J. Med. 320:545-550. [DOI] [PubMed] [Google Scholar]
- 220.Selwyn, P. A., B. M. Sckell, P. Alcabes, G. H. Friedland, R. S. Klein, and E. E. Schoenbaum. 1992. High risk of active tuberculosis in HIV-infected drug users with cutaneous anergy. JAMA 268:504-509. [PubMed] [Google Scholar]
- 221.Sepkowitz, K. A., J. Raffalli, L. Riley, T. E. Kiehn, and D. Armstrong. 1995. Tuberculosis in the AIDS era. Clin. Microbiol. Rev. 8:180-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Skamene, E., E. Schurr, and P. Gros. 1998. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu. Rev. Med. 49:275-287. [DOI] [PubMed] [Google Scholar]
- 223.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 224.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 225.Small, P. M., N. B. McClenny, S. P. Singh, G. K. Schoolnik, L. S. Tompkins, and P. A. Mickelsen. 1993. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J. Clin. Microbiol. 31:1677-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Small, P. M., R. W. Shafer, P. C. Hopewell, S. P. Singh, M. J. Murphy, E. Desmond, M. F. Sierra, and G. K. Schoolnik. 1993. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N. Engl. J. Med. 328:1137-1144. [DOI] [PubMed] [Google Scholar]
- 227.Smith, I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 16:463-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Smittipat, N., P. Billamas, M. Palittapongarnpim, A. Thong-On, M. M. Temu, P. Thanakijcharoen, O. Karnkawinpong, and P. Palittapongarnpim. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 43:5034-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuberc. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 230.Soini, H., X. Pan, L. Teeter, J. M. Musser, and E. A. Graviss. 2001. Transmission dynamics and molecular characterization of Mycobacterium tuberculosis isolates with low copy numbers of IS6110. J. Clin. Microbiol. 39:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sola, C., I. Filliol, E. Legrand, S. Lesjean, C. Locht, P. Supply, and N. Rastogi. 2003. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect. Genet. Evol. 3:125-133. [DOI] [PubMed] [Google Scholar]
- 232.Sola, C., I. Filliol, E. Legrand, I. Mokrousov, and N. Rastogi. 2001. Mycobacterium tuberculosis phylogeny reconstruction based on combined numerical analysis with IS1081, IS6110, VNTR, and DR-based spoligotyping suggests the existence of two new phylogeographical clades. J. Mol. Evol. 53:680-689. [DOI] [PubMed] [Google Scholar]
- 233.Sonnenberg, P., J. Murray, J. R. Glynn, S. Shearer, B. Kambashi, and P. Godfrey-Faussett. 2001. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 358:1687-1693. [DOI] [PubMed] [Google Scholar]
- 234.Soto, C. Y., M. C. Menendez, E. Perez, S. Samper, A. B. Gomez, M. J. Garcia, and C. Martin. 2004. IS6110 mediates increased transcription of the phoP virulence gene in a multidrug-resistant clinical isolate responsible for tuberculosis outbreaks. J. Clin. Microbiol. 42:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Stead, W. W. 1967. Pathogenesis of a first episode of chronic pulmonary tuberculosis in man: recrudescence of residuals of the primary infection or exogenous reinfection? Am. Rev. Respir. Dis. 95:729-745. [DOI] [PubMed] [Google Scholar]
- 237.Stead, W. W. 1967. Pathogenesis of the sporadic case of tuberculosis. N. Engl. J. Med. 277:1008-1012. [DOI] [PubMed] [Google Scholar]
- 238.Reference deleted.
- 239.Stead, W. W., J. W. Senner, W. T. Reddick, and J. P. Lofgren. 1990. Racial differences in susceptibility to infection by Mycobacterium tuberculosis. N. Engl. J. Med. 322:422-427. [DOI] [PubMed] [Google Scholar]
- 240.Stead, W. W., W. A. Slagel, and J. Noble. 1972. INH prophylaxis. N. Engl. J. Med. 286:159-160. [PubMed] [Google Scholar]
- 241.Steinlein, L. M., and J. T. Crawford. 2001. Reverse dot blot assay (insertion site typing) for precise detection of sites of IS6110 insertion in the Mycobacterium tuberculosis genome. J. Clin. Microbiol. 39:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Streicher, E. M., R. M. Warren, C. Kewley, J. Simpson, N. Rastogi, C. Sola, G. D. Van Der Spuy, P. D. Van Helden, and T. C. Victor. 2004. Genotypic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from rural districts of the Western Cape Province of South Africa. J. Clin. Microbiol. 42:891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Styblo, K. 1980. Recent advances in epidemiological research in tuberculosis. Adv. Tuberc. Res. 20:1-63. [PubMed] [Google Scholar]
- 244.Suffys, P. N., M. E. Ivens de Araujo, M. L. Rossetti, A. Zahab, E. W. Barroso, A. M. Barreto, E. Campos, D. van Soolingen, K. Kremer, H. Heersma, and W. M. Degrave. 2000. Usefulness of IS6110-restriction fragment length polymorphism typing of Brazilian strains of Mycobacterium tuberculosis and comparison with an international fingerprint database. Res. Microbiol. 151:343-351. [DOI] [PubMed] [Google Scholar]
- 245.Sullivan, E. A., B. N. Kreiswirth, L. Palumbo, V. Kapur, J. M. Musser, A. Ebrahimzadeh, and T. R. Frieden. 1995. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet 345:1148-1150. [DOI] [PubMed] [Google Scholar]
- 246.Sun, Y. J., R. Bellamy, A. S. Lee, S. T. Ng, S. Ravindran, S. Y. Wong, C. Locht, P. Supply, and N. I. Paton. 2004. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J. Clin. Microbiol. 42:1986-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 248.Supply, P., R. M. Warren, A. L. Banuls, S. Lesjean, G. D. Van Der Spuy, L. A. Lewis, M. Tibayrenc, P. D. Van Helden, and C. Locht. 2003. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol. Microbiol. 47:529-538. [DOI] [PubMed] [Google Scholar]
- 248a.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 27 September 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 249.Theus, S. A., M. D. Cave, and K. D. Eisenach. 2005. Intracellular macrophage growth rates and cytokine profiles of Mycobacterium tuberculosis strains with different transmission dynamics. J. Infect. Dis. 191:453-460. [DOI] [PubMed] [Google Scholar]
- 250.Thierry, D., M. D. Cave, K. D. Eisenach, J. T. Crawford, J. H. Bates, B. Gicquel, and J. L. Guesdon. 1990. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 18:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Timm, J., F. A. Post, L. G. Bekker, G. B. Walther, H. C. Wainwright, R. Manganelli, W. T. Chan, L. Tsenova, B. Gold, I. Smith, G. Kaplan, and J. D. McKinney. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA 100:14321-14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Toossi, Z., J. G. Sierra-Madero, R. A. Blinkhorn, M. A. Mettler, and E. A. Rich. 1993. Enhanced susceptibility of blood monocytes from patients with pulmonary tuberculosis to productive infection with human immunodeficiency virus type 1. J. Exp. Med. 177:1511-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tsenova, L., E. Ellison, R. Harbacheuski, A. L. Moreira, N. Kurepina, M. B. Reed, B. Mathema, C. E. Barry III, and G. Kaplan. 2005. Virulence of selected Mycobacterium tuberculosis clinical isolates in the rabbit model of meningitis is dependent on phenolic glycolipid produced by the bacilli. J. Infect. Dis. 192:98-106. [DOI] [PubMed] [Google Scholar]
- 254.Tsolaki, A. G., S. Gagneux, A. S. Pym, Y. O. Goguet de la Salmoniere, B. N. Kreiswirth, D. Van Soolingen, and P. M. Small. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 43:3185-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Valway, S. E., M. P. Sanchez, T. F. Shinnick, I. Orme, T. Agerton, D. Hoy, J. S. Jones, H. Westmoreland, and I. M. Onorato. 1998. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N. Engl. J. Med. 338:633-639. [DOI] [PubMed] [Google Scholar]
- 257.van Crevel, R., R. H. Nelwan, W. de Lenne, Y. Veeraragu, A. G. van der Zanden, Z. Amin, J. W. van der Meer, and D. van Soolingen. 2001. Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg. Infect. Dis. 7:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.van der Zanden, A. G., A. H. Hoentjen, F. G. Heilmann, E. F. Weltevreden, L. M. Schouls, and J. D. van Embden. 1998. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol. Pathol. 51:209-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.van Embden, J. D., T. van Gorkom, K. Kremer, R. Jansen, B. A. van Der Zeijstqq, and L. M. Schouls. 2000. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 182:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.van Embden, J. D., D. Van Soolingen, H. F. Heersma, A. J. De Neeling, M. E. Jones, M. Steiert, V. Grek, F. R. Mooi, and J. Verhoef. 1996. Establishment of a European network for the surveillance of Mycobacterium tuberculosis, MRSA and penicillin-resistant pneumococci. J. Antimicrob. Chemother. 38:905-907. [DOI] [PubMed] [Google Scholar]
- 262.van Rie, A., T. C. Victor, M. Richardson, R. Johnson, G. D. van der Spuy, E. J. Murray, N. Beyers, N. C. Gey van Pittius, P. D. van Helden, and R. M. Warren. 2005. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 172:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 264.van Rie, A., R. M. Warren, N. Beyers, R. P. Gie, C. N. Classen, M. Richardson, S. L. Sampson, T. C. Victor, and P. D. van Helden. 1999. Transmission of a multidrug-resistant Mycobacterium tuberculosis strain resembling “strain W” among noninstitutionalized, human immunodeficiency virus-seronegative patients. J. Infect. Dis. 180:1608-1615. [DOI] [PubMed] [Google Scholar]
- 265.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 266.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 267.van Soolingen, D., P. W. Hermans, P. E. de Haas, D. R. Soll, and J. D. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Vareldzis, B. P., J. Grosset, I. de Kantor, J. Crofton, A. Laszlo, M. Felten, M. C. Raviglione, and A. Kochi. 1994. Drug-resistant tuberculosis: laboratory issues. World Health Organization recommendations. Tuberc. Lung Dis. 75:1-7. [DOI] [PubMed] [Google Scholar]
- 270.Verver, S., R. M. Warren, N. Beyers, M. Richardson, G. D. van der Spuy, M. W. Borgdorff, D. A. Enarson, M. A. Behr, and P. D. van Helden. 2005. Rate of reinfection tuberculosis after successful treatment is higher than rate of new tuberculosis. Am. J. Respir. Crit. Care Med. 171:1430-1435. [DOI] [PubMed] [Google Scholar]
- 271.Verver, S., R. M. Warren, Z. Munch, E. Vynnycky, P. D. van Helden, M. Richardson, G. D. van der Spuy, D. A. Enarson, M. W. Borgdorff, M. A. Behr, and N. Beyers. 2004. Transmission of tuberculosis in a high incidence urban community in South Africa. Int. J. Epidemiol. 33:351-357. [DOI] [PubMed] [Google Scholar]
- 272.Victor, T. C., A. M. Jordaan, A. van Rie, G. D. van der Spuy, M. Richardson, P. D. van Helden, and R. Warren. 1999. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuberc. Lung Dis. 79:343-348. [DOI] [PubMed] [Google Scholar]
- 273.Vitol, I., J. Driscoll, B. Kreiswirth, N. Kurepina, and K. P. Bennett. 2. May 2006, posting date. Identifying Mycobacterium tuberculosis complex strain families using spoligotypes. Infect. Genet. Evol. [Online.] doi: 10.1016/j.meegid.2006.03.003. [DOI] [PubMed]
- 274.Vynnycky, E., and P. E. Fine. 1997. The natural history of tuberculosis: the implications of age-dependent risks of disease and the role of reinfection. Epidemiol. Infect. 119:183-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vynnycky, E., N. Nagelkerke, M. W. Borgdorff, D. van Soolingen, J. D. van Embden, and P. E. Fine. 2001. The effect of age and study duration on the relationship between “clustering” of DNA fingerprint patterns and the proportion of tuberculosis disease attributable to recent transmission. Epidemiol. Infect. 126:43-62. [PMC free article] [PubMed] [Google Scholar]
- 276.Wallis, R. S., S. Patil, S. H. Cheon, K. Edmonds, M. Phillips, M. D. Perkins, M. Joloba, A. Namale, J. L. Johnson, L. Teixeira, R. Dietze, S. Siddiqi, R. D. Mugerwa, K. Eisenach, and J. J. Ellner. 1999. Drug tolerance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Warren, R. M., E. M. Streicher, S. L. Sampson, G. D. van der Spuy, M. Richardson, D. Nguyen, M. A. Behr, T. C. Victor, and P. D. van Helden. 2002. Microevolution of the direct repeat region of Mycobacterium tuberculosis: implications for interpretation of spoligotyping data. J. Clin. Microbiol. 40:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Warren, R. M., G. D. van der Spuy, M. Richardson, N. Beyers, M. W. Borgdorff, M. A. Behr, and P. D. van Helden. 2002. Calculation of the stability of the IS6110 banding pattern in patients with persistent Mycobacterium tuberculosis disease. J. Clin. Microbiol. 40:1705-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, N. Beyers, N. C. Gey Van Pittius, and P. D. Van Helden. 2003. Patients with active tuberculosis often have different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169:610-614. [DOI] [PubMed] [Google Scholar]
- 280.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, G. D. van der Spuy, R. Johnson, V. N. Chihota, C. Locht, P. Supply, and P. D. van Helden. 2004. Clonal expansion of a globally disseminated lineage of Mycobacterium tuberculosis with low IS6110 copy numbers. J. Clin. Microbiol. 42:5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wilkinson, R. J., M. Llewelyn, Z. Toossi, P. Patel, G. Pasvol, A. Lalvani, D. Wright, M. Latif, and R. N. Davidson. 2000. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet 355:618-621. [DOI] [PubMed] [Google Scholar]
- 282.Williams, A., G. J. Hatch, S. O. Clark, K. E. Gooch, K. A. Hatch, G. A. Hall, K. Huygen, T. H. Ottenhoff, K. L. Franken, P. Andersen, T. M. Doherty, S. H. Kaufmann, L. Grode, P. Seiler, C. Martin, B. Gicquel, S. T. Cole, P. Brodin, A. S. Pym, W. Dalemans, J. Cohen, Y. Lobet, N. Goonetilleke, H. McShane, A. Hill, T. Parish, D. Smith, N. G. Stoker, D. B. Lowrie, G. Kallenius, S. Svenson, A. Pawlowski, K. Blake, and P. D. Marsh. 2005. Evaluation of vaccines in the EU TB vaccine cluster using a guinea pig aerosol infection model of tuberculosis. Tuberculosis (Edinburgh) 85:29-38. [DOI] [PubMed] [Google Scholar]
- 283.World Health Organization. 2006. Definitions: case registrations, bacteriology and treatment otucomes, p. 18-23. Guidelines for the programmatic management of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland.
- 284.World Health Organization. 1996. TB: groups at risk. WHO report on the tuberculosis epidemic. World Health Organization, Geneva, Switzerland.
- 285.Xu, C., B. N. Kreiswirth, S. Sreevatsan, J. M. Musser, and K. Drlica. 1996. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J. Infect. Dis. 174:1127-1130. (Erratum, 175:1027, 1997.) [DOI] [PubMed] [Google Scholar]
- 286.Yaganehdoost, A., E. A. Graviss, M. W. Ross, G. J. Adams, S. Ramaswamy, A. Wanger, R. Frothingham, H. Soini, and J. M. Musser. 1999. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J. Infect. Dis. 180:1245-1251. [DOI] [PubMed] [Google Scholar]
- 287.Yang, Z., P. F. Barnes, F. Chaves, K. D. Eisenach, S. E. Weis, J. H. Bates, and M. D. Cave. 1998. Diversity of DNA fingerprints of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 36:1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yang, Z., F. Chaves, P. F. Barnes, W. J. Burman, J. Koehler, K. D. Eisenach, J. H. Bates, and M. D. Cave. 1996. Evaluation of method for secondary DNA typing of Mycobacterium tuberculosis with pTBN12 in epidemiologic study of tuberculosis. J. Clin. Microbiol. 34:3044-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yang, Z. H., J. H. Bates, K. D. Eisenach, and M. D. Cave. 2001. Secondary typing of Mycobacterium tuberculosis isolates with matching IS6110 fingerprints from different geographic regions of the United States. J. Clin. Microbiol. 39:1691-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yang, Z. H., K. Ijaz, J. H. Bates, K. D. Eisenach, and M. D. Cave. 2000. Spoligotyping and polymorphic GC-rich repetitive sequence fingerprinting of Mycobacterium tuberculosis strains having few copies of IS6110. J. Clin. Microbiol. 38:3572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yeh, R. W., A. Ponce de Leon, C. B. Agasino, J. A. Hahn, C. L. Daley, P. C. Hopewell, and P. M. Small. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J. Infect. Dis. 177:1107-1111. [DOI] [PubMed] [Google Scholar]
- 292.Yew, W. W., and C. C. Leung. 2005. Are some people not safer after successful treatment of tuberculosis? Am. J. Respir. Crit. Care Med. 171:1324-1325. [DOI] [PubMed] [Google Scholar]
- 293.Youmans, G. P., E. H. Williston, W. H. Feldman, and C. H. Hinshaw. 1946. Increase in resistance of tubercle bacilli to streptomycin. A preliminary report. Proc. Mayo Clin. 21:126-127. [PubMed] [Google Scholar]
- 294.Zhang, M., J. Gong, Z. Yang, B. Samten, M. D. Cave, and P. F. Barnes. 1999. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J. Infect. Dis. 179:1213-1217. [DOI] [PubMed] [Google Scholar]
- 295.Zhang, Y., C. Vilcheze, and W. R. J. Jacobs. 2005. Mechanisms of drug resistance in Mycobacterium tuberculosis, p. 115-140. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. J. Jacobs (ed.), Tuberculosis and the tubercle bacillus. American Society for Microbiology, Washington, D.C.
- 296.Ziegler, J. E., M. L. Edwards, and D. W. Smith. 1985. Exogenous reinfection in experimental airborne tuberculosis. Tubercle 66:121-128. [DOI] [PubMed] [Google Scholar]
- 297.Zignol, M., et al. 2006. Global incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 194:479-485. [DOI] [PubMed] [Google Scholar]