Abstract
Saccharomyces mating-type switching occurs through a double-strand break-initiated gene conversion event at MAT, using one of two donors located distantly on the same chromosome, HMLα and HMRa. MATa cells preferentially choose HMLα, a decision that depends on the recombination enhancer (RE) that controls recombination along the left arm of chromosome III. We previously showed that an fhk1Δ mutation reduces HMLα usage in MATa cells, but not to the level seen when RE is deleted. We now report that donor preference also depends on binding of the Swi4/Swi6 (SBF) transcription factors to an evolutionarily conserved SCB site within RE. As at other SCB-containing promoters, SBF binds to RE in the G1 phase. Surprisingly, Fkh1 binds to RE only in G2, which contrasts with its cell cycle-independent binding to its other target promoters. SBF and Fkh1 define two independent RE activation pathways, as deletion of both Fkh1 and SCB results in nearly complete loss of HML usage in MATa cells. These transcription factors create an epigenetic modification of RE in a fashion that apparently does not involve transcription. In addition, the putative helicase Chl1, previously involved in donor preference, functions in the SBF pathway.
Mating-type switching in Saccharomyces cerevisiae is initiated by the expression of the HO endonuclease, which creates a double-strand break (DSB) at a specific site within the MAT locus. The DSB is repaired by a gene conversion event using one of two donor loci, HMLα and HMRa, located near the left and right subtelomeric regions, respectively (20, 21). These donors are maintained in a heterochromatic stage; they are not transcribed and are refractory to HO endonuclease cleavage (39, 47, 64).
Although MAT is able to use both HML and HMR to repair the break, there is a strong mating-type-dependent preference in the choice between the two donors. MATa cells preferentially recombine with HML, whereas MATα selects HMR (33, 61, 67, 68). HML usage is mainly regulated, while HMR is used by default (67). Donor preference is independent of whether the donor carries a or α information; moreover, donor selection does not depend on any sequences that uniquely define HML or HMR or any sequences flanking or distal to HML or HMR (61, 67). Preferential selection of HML in MATa cells depends on an approximately 700-bp cis-acting element, the recombination enhancer (RE), which is located 17 kb centromere proximal to HML (66). Deletion of RE causes reversed donor preference in MATa cells: HML usage is reduced to 10% compared to 90% in the wild type (WT) (66). However, RE deletion does not affect donor preference in MATα cells, showing that RE is simply turned off in these cells and is not responsible for the inhibition of HML usage.
In fact, RE regulates the entire left arm of chromosome III for recombination. In MATa, when HML is moved to other locations of the left arm of chromosome III it is still the preferred donor (66). In addition, RE activity is not limited to mating-type switching. The rate of spontaneous recombination between two different leu2 alleles, one replacing HML and the other located near MAT, is ≥10 times higher in MATa versus MATα strains (67). This difference is lost when RE is deleted (66).
Recently we showed that the two arms of chromosome III can be defined as two independent domains and that RE controls the accessibility between these domains (10), probably by increasing the movement and/or conformation of the left arm in the nucleus (4). Thus, when MAT is moved onto the left arm, it becomes the preferred donor in both MATa and MATα cells.
RE is well conserved in the Saccharomyces sensu stricto species (30; Saccharomyces genome database [www.yeastgenome.org]). Comparison of RE sequences from S. cerevisiae, Saccharomyces carlsbergensis, and Saccharomyces bayanus (53, 65), and also from Saccharomyces mikatae and Saccharomyces kudriazvezii, defines five highly conserved regions A, B, C, D, and E. Deletion studies show that each of these regions with the exception of B is necessary for complete RE activity (53, 68). Regions A, D, and E contain Fkh1 binding sites, and this transcription factor has been shown to play an essential role in HML activation in MATa cells (53). Fkh1 was first described as a component of the SFF complex, involved in the regulation of transcription of the CLB2 cluster of genes at the transition between the G2 phase and mitosis.
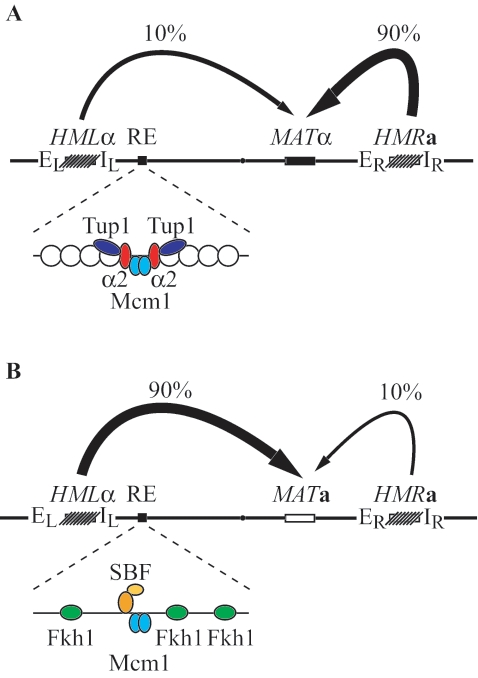
RE is regulated very similarly to a-specific genes (65). Binding of the Matα2-Mcm1 repressor complex to the region C inactivates RE in MATα cells, and binding of Mcm1 activates RE in MATa cells (54, 55, 65, 68). As at a-specific genes, binding of the Matα2-Mcm1 complex to RE in MATα cells is linked to the formation of a highly organized array of nucleosomes, covering the entire 2.5-kb intergenic region containing RE (63) (Fig. 1A). Formation of this chromatin structure is dependent on Tup1 but not Ssn6 (63, 65). This structure is absent in MATa cells when Mcm1 binds RE in the absence of Matα2 (Fig. 1B). Mcm1 function is limited to this opening, which allows trans-acting factors like Fkh1 to bind RE (14, 53). Recently, the DNA repair protein yKu80 has been shown to be involved in the activation of RE (48).
FIG. 1.
RE regulation and donor preference during mating-type switching in Saccharomyces cerevisiae. (A) Donor preference in MATα cells. A schematic representation of the mating-type locus (MAT), the two silent donor loci (HML and HMR), and the RE is shown. In MATα cells, the Matα2-Mcm1 complex recruits the general repressor Tup1 to RE, inactivating RE by formation of highly organized nucleosomes, resulting in the preferred use of HMR. (B) Donor preference in MATa cells. Mcm1 binds to the RE and removes the positioned nucleosomes, allowing binding of trans-acting factors, including Fkh1 and SBF, leading to activation of RE and consequently to the favored usage of HML.
Because fkh1Δ does not reduce MATa donor preference as much as deletion of RE, we looked for additional regulatory elements within RE. We report that an SCB (Swi4/Swi6 cell cycle box), present in region C, is important for RE activity. SCB is usually found in promoters of genes induced at the transition from G1 to S phase. Activation of these genes depends on the SBF complex (Swi4/Swi6 cell cycle box binding factors) (3). SBF is necessary for complete RE activity, but its role in donor preference apparently does not involve its G1/S transcription activity. We found that SBF binds RE in G1/S, while Fkh1 binds in the G2 phase. SBF and Fkh1 activate RE through two independent pathways. Our genetic analysis also shows that the putative ATP-dependent DNA helicase Chl1 (18, 19, 24), previously shown to be involved in donor preference (62), acts through the pathway that involves SBF. We propose that the activation of the left arm of chromosome III for recombination requires the establishment of signals in both G1 and G2 to create a specific organization of chromosome III in the nucleus that leads to the preferential choice of donors present on the left arm.
MATERIALS AND METHODS
Strains.
The swi4 strain was kindly provided by Brenda Andrews and is derived from BY263 (41). Donor preference in this strain was measured in parallel in a wild-type strain of the same background. All other strains used for donor preference experiments were derivatives of DBY745 (ho MATa ade1-100 ura3-53 leu2-3,112). Strains used to monitor MATa donor preference carry HMRα-BamHI, where Ya has been replaced by a Yα allele containing a single-base-pair mutation that creates the BamHI site (67), while strains used to monitor MATα donor preference carry MATα-BamHI. All strains have a galactose-inducible HO endonuclease gene integrated at the ADE3 locus (49) or present on a LEU2 centromeric plasmid (pJH727).
All yeast transformations were done by one-step transformation (7). SWI6, FKH1, MBP1, ACE2, STE12, NDD1, KAR3, CTF4, CTF18, BIM1, NUP170, YKU70, YKU80, and YDR332W deletions have been made by transformation of PCR-amplified fragments obtained from genomic DNA of the Research Genetics strain collection. The chl1Δ::URA3 deletion was made using the split-vectors system (16) with the plasmids pUR-pl008 and pRA-pl008 provided by G.-F. Richard. The cdc7-as3 mutant strain has been described before (26) and is suitable to measure donor preference. The G1::GAL::HO construct (43) was introduced by a cross with JKM95 (42). MATα1 has been disrupted with a PCR fragment containing the KanMX gene flanked by 50-bp sequences identical to those flanking the MATα1 gene. This PCR has been performed using the pFA6 plasmid containing the KanMX4 cassette (60) and mixed primers composed of MATα1 and pFA6 sequences (MATα1KanU, TATGAAATGTATCAACCATATATAATAACTTAATAGACGACATTCACAATCAGCTGAAGCTTCGTACGC; and MATα1KanL, AGTCCCATATTCCGTGCTGCATTTTGTCCGCGTGCCATTCTTCAGCGAGCGCATAGGCCACTAGTGGATCTG). Ya has been deleted in the same way with the primers MatKanP1 (TAGGTAAATTACAGCAAATAGAAAAGAGCTTTTTATTTATGTCTAGTACAGCTGAAGCTTCGTACGC) and MatKanHO+L (AATCATTGAAAACGAATTTATTTAGATCTCATACGTTTATTTATGAACTAGCATAGGCCACTAGTGGATCTG), which conserved the integrity of the HO cut site.
All the modified RE sequences have been reintroduced in strains bearing a deletion of the recombination enhancer as previously described (65).
The strain bearing the Chl1-9xMYC fusion protein was made using the method of Longtine et al. (38) in the DBY745 background. Strains bearing Fkh1-3xHA have been described previously (53). Mcm1-13xMyc was kindly provided by B. Tye (6).
Analysis of donor preference.
Quantification of donor preference on Southern blotting was previously described (65), using ImageQuant V1.2 (Molecular Dynamics). Differences among strains when HML usage is either >90% or <10% are not statistically significant.
Measurement of spontaneous recombination rates.
Spontaneous formation of Leu2+ recombinants was quantified by a fluctuation test based on a minimum of nine independent cultures of each strain, initiated from approximately 200 cells and grown to saturation (36).
ChIP.
Chromatin immunoprecipitation (ChIP) analysis was performed as in reference 53, except for the use of an anti-MYC monoclonal antibody (Sigma) and protein A-agarose (Roche). ChIP of Swi4 and Swi6 used polyclonal antibodies provided by B. Andrews. The following PCR primers were used for amplification: for the RE region, SUN575 and Wu027; the CLB2 promoter region, SUN842 and SUN843; the ARG5,6 coding region, ARG5,6p1 and ARG5,6p2 (53); the PCL1 promoter, prPCL1U (GCATTTGCTTACCAAACTGGC) and pRPCL1L (CAATCCCATTACCATGTAGGC), the SUN4 promoter, SUN4pU (GGTTACCCGACATATATGCTGG) and SUN4pL (CATGCTGAAGGGAACGTGCG) and the PHO3 locus, AW253-PHO35′RT (GGAGAGTTAGCCGATGTTGC) and AW254-PHO33′RT (TAGTCGCCAGGGAAAGAGAA).
Cdc7 inactivation, α-factor, and nocodazole arrest.
Cdc7 was inactivated in cdc7-as3 strains by adding the ATP analogue inhibitor 1-NMPP1 at 10 μM. cdc7-as3 cells were usually arrested 1 doubling time after addition of the drug, and the HO gene was then induced by the addition of galactose to a final concentration of 2%. Cells were arrested in G1 with 10 μg/ml α-factor. Hydroxyurea (HU)-arrested cells were first synchronized with 10 μg/ml α-factor, washed, and then released in the presence of 50 μg/ml pronase and 0.2 M HU. Cells were arrested in G2/M with 20 μg/ml nocodazole.
RESULTS
Search for region C sequences necessary for RE activity.
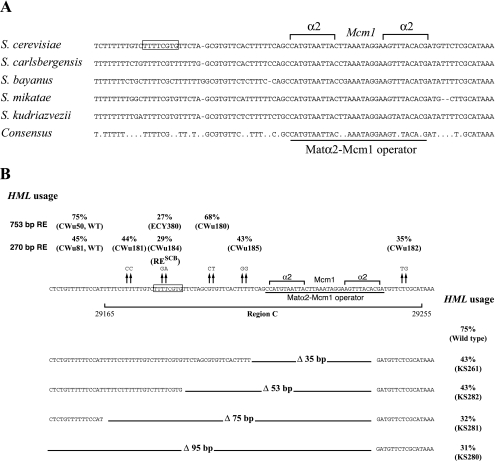
Because the conservation of region C sequences goes well beyond the Matα2-Mcm1 operator among Saccharomyces sensu stricto strains, we examined this region for potential protein binding domains that could be necessary for full RE activity. We took advantage of the newly available genome sequences from several Saccharomyces species (9, 30) to define a more precise consensus of this region. RE is conserved in Saccharomyces sensu stricto species, but not in the more evolutionarily distant Saccharomyces species (sensu lato). Analyzing the five Saccharomyces sensu stricto species available (Fig. 2A), we confirmed that the entire region C is very well conserved. Therefore, sequences other than the Matα2-Mcm1 operator could be important for RE function. We decided to conduct a mutational analysis of the whole region.
FIG. 2.
Deletion and mutational analysis of region C shows that an SCB is necessary for complete RE activity. (A) Sequence comparison of region C among the Saccharomyces sensu stricto species. The MATα2-Mcm1 operator is shown. Sequences enclosed by the box correspond to the SCB necessary for RE activity (reverse complement of the consensus). Only perfectly conserved residues between the five species are indicated in the consensus sequence. (B) (Top) HML usage in strains bearing different mutations in region C. Some of these mutations have been introduced in the 270-bp synthetic RE which lacks region E (lower lane). Others were introduced in the 753-bp minimal RE (upper lane). (Middle) Sequence of region C. The Matα2-Mcm1 operator is underlined, and the SCB is shown in the box. (Bottom) HML usage in strains bearing the 753-bp minimal RE with several deletions of region C sequences.
A 35-bp deletion covering the entire Matα2-Mcm1 operator (Fig. 2B) confers partial RE activity (43% HML usage compared to 5 to 10% when RE is deleted). This partial activity allowed us to carry out a deletion analysis of the region C sequences adjacent to the Matα2-Mcm1 operator. Partial deletions were made in a 753-bp RE in vitro, and the different constructs were inserted in place of a 1.8-kb sequence containing RE and other intergenic sequences between KAR4 and SPB1. A 53-bp deletion confers the same phenotype as the 35-bp deletion; however, 75- and 95-bp deletions show a reduction of HML usage to approximately 30%. Therefore, at least part of the 22-bp sequence between the 53- and 75-bp deletions is necessary for full RE activity. To identify more precisely the key DNA sequences, we made 2-bp mutations at positions perfectly conserved among the Saccharomyces sensu stricto species. Two such mutations were introduced in the 753-bp RE and inserted in place of the 1.8-kb sequence containing RE. A TC→GA mutation at positions 29181 to 29182 (ECY380) in the 22-bp sequences defined above reduced HML usage from 75% in the wild type to 27% in the mutant. We also used a 270-bp minimum RE lacking region E to introduce additional mutations in the chromosome (65). This minimum RE retains significant donor preference activity because strains carrying this insert use HML 45% of the time, whereas strains with the 1.8-kb deletion use HML 5% of the time. A CA→TG mutation in the sequence to the right of the Matα2-Mcm1 operator (Cwu182) does not show a strong reduction in HML usage, suggesting that this part of region C is not fundamental for RE activity. A TT→CC mutation in the 22-bp sequence defined above (Cwu181) does not affect HML usage, but the nearby TC→GA mutation strongly reduced HML usage, confirming our observation with 753-bp RE. These two results allowed us to define a new RE element within a 13-bp fragment. Using MatInspector V2.2 software (46) to look for consensus protein binding sites, we found a perfect match corresponding to the 8-bp reverse complement sequence (TTTTCGTG) of the well-defined Swi4,6-dependent cell cycle box (SCB). This result suggested that Swi4/Swi6 complex (SBF), which binds the SCB (3), could be involved in RE activation.
This SCB is perfectly conserved in S. mikatae and in S. kudriazvezii but somewhat degenerate in S. carlbergensis and in S. bayanus (Fig. 2A). However, genomic studies of the binding sites of SBF in S. cerevisiae show that 49% of the intergenic regions binding SBF do not contained a motif matching the defined SCB consensus (27). In addition, extensive study of the different Swi4 binding sites in the S. cerevisiae genome revealed that the TTTTCGCT sequence found in S. bayanus corresponds to one such site (8). We conclude that this element is probably conserved in these yeasts. The relative conservation of the SCB among the Saccharomyces sensu stricto species reinforces the idea that SBF is important for RE activation.
The Swi4/Swi6 complex is involved in RE activity.
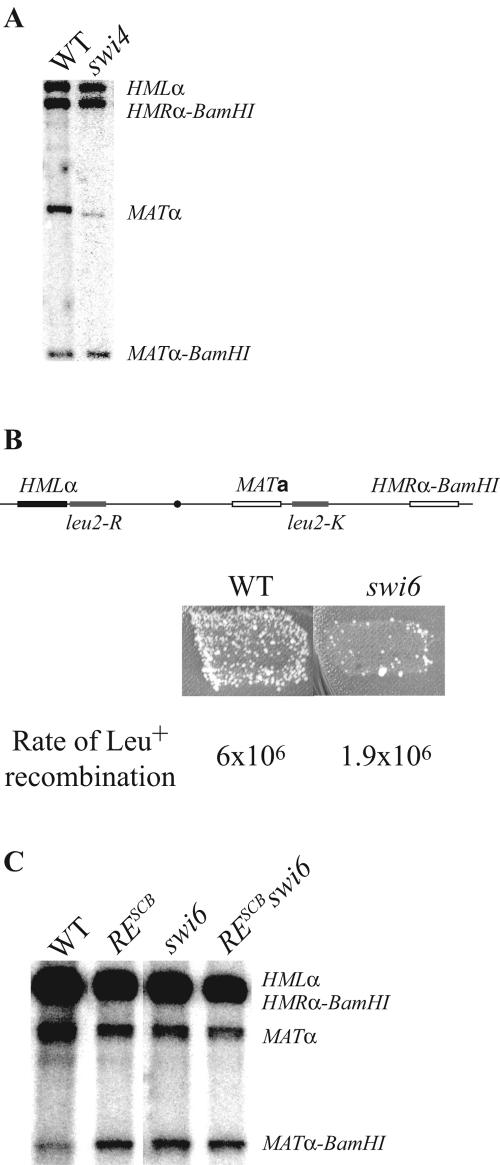
We measured HML usage in strains bearing a replacement of HMRa by HMRα-BamHI (67), allowing the discrimination on Southern blots of MATα and MATα-BamHI products (Fig. 3A). HML usage is significantly reduced in strains with SWI4 deleted (30%) compared to the WT (70%), showing that the SBF complex is involved in RE activation in MATa cells. The effect of the swi4 deletion is similar to the fkh1 deletion we observed previously (53) (Table 1). To confirm the importance of SBF, we tested the effect of swi6Δ on spontaneous intrachromosomal recombination between two leu2 alleles. In this assay, leu2-R was inserted between HML and RE and leu2-K was placed near MAT (Fig. 3B). In wild-type cells, the rates measured in MATa cells are 10-fold greater than in MATα, in an RE-dependent manner. We showed previously that deletion of FKH1 decreases spontaneous leu2 recombination in MATa cells (53). Here, we found that swi6Δ also reduces the formation of Leu+ recombinants. Measurement of the spontaneous recombination rates by a fluctuation test (36) showed that swi6Δ caused a threefold reduction in the rate. Thus, the absence of the SBF complex leads to a reduction of HML usage and of leu2 spontaneous recombination. We conclude SBF takes part in RE activation.
FIG. 3.
Donor preference in swi4 and swi6 mutants. (A) Southern blot of genomic DNA extracted from wild-type (ECY384) and swi4 (ECY411) strains after inducting MAT switching. Genomic DNA was cut with the BamHI and HindIII restriction enzymes, and the Southern blot was probed with Yα sequences. (B) Papillation test measuring recombination between leu2 heteroalleles. In wild-type MATa cells, papillation is strongly reduced in a swi6 mutant (ECY304). Fluctuation tests confirmed these observations. (C) Southern blot of genomic DNA from wild-type (ECY507), RESCB (ECY507), swi6 (ECY508), and RESCB swi6 (ECY509) strains analyzed under the same conditions as in panel A.
TABLE 1.
Epistatic relationships between RESCB, FKH1, CHL1, and yKU80
| Strain | Genotype | % HML usagea |
|---|---|---|
| ECY401 | MATa | 71.5 ± 1.8 |
| XW676 | MATa REΔ | <10 |
| ECY400 | MATa RESCB | 33.1 ± 2.2 |
| ECY399 | MATafkh1 | 34.0 ± 2.8 |
| ECY398 | MATachl1 | 51.8 ± 2.5 |
| ECY494 | MATa yku80 | 64.1 ± 1.1 |
| ECY13 | MATα | <10 |
| ECY111 | MATα chl1 | <10 |
| ECY397 | MATa RESCBfkh1 | 17.7 ± 3.7 |
| ECY396 | MATa RESCBchl1 | 29.7 ± 3.9 |
| ECY495 | MATa RESCByku80 | 34.8 ± 2.1 |
| ECY496 | MATachl1 yku80 | 54.7 ± 4.6 |
| ECY395 | MATafkh1 chl1 | 14.4 ± 2.2 |
| ECY497 | MATafkh1 yku80 | 15.0 ± 1.4 |
| ECY394 | MATa RESCBfkh1 chl1 | <10 |
| ECY499 | MATa RESCB fkh1 yku80 | <10 |
| ECY498 | MATa RESCB chl1 yku80 | 17.8 ± 2.8 |
| ECY500 | MATa fkh1 chl1 yku80 | 14.3 ± 0.7 |
| ECY501 | MATa RESCB fkh1 chl1 yku80 | <10 |
| KS338 | MATa 4A | 65 ± 2 |
| KS379 | MATa 4A fkh1 | <10 |
| ECY177 | MATa 4A chl1 | 54.4 ± 1.3 |
HML usage was determined in MATa strains carrying HMLα and HMRα-BamHI. Genomic DNA extracted from an entire population of cells in which mating-type switching has been induced was cut by BamHI and HindIII and probed with a Yα fragment.
To rule out that any G1/S-specific gene under the control of SBF is involved in donor preference, we measured HML usage (Fig. 3C) in a strain bearing both a deletion of SWI6 and the TC→GA mutations in RESCB (Fig. 2B). The effect of the double mutant (45%) was similar to either single mutant: swi6Δ, 48%; and RESCB, 49%. Therefore, we conclude that SBF acts directly by binding to RE.
Binding of Swi5 to the promoter of the HO gene is necessary to recruit the chromatin remodeling factors Swi2/Snf2 and SAGA, which in turn allow SBF to bind (11). However, deleting SWI5 does not affect HML usage (data not shown), indicating that SBF binding to RE is Swi5 independent. We also observed that ablation of the WHI5 protein, which binds to SBF to repress G1-specific transcription during early G1 (12, 13), does not affect HML usage (data not shown).
Fkh1 and both SBF and Chl1 define two independent pathways involved in RE activation in MATa cells.
Previous studies provided evidence that Fkh1 (53) and Chl1 (62) are involved in MATa donor preference. To determine if these two proteins and SBF act together or in different pathways, we measured HML usage in single, double, and triple mutants derived from diploid strains heterozygous for fkh1Δ and chl1Δ and for the RESCB mutation. We confirmed the involvement of CHL1 in HML activation in MATa cells (Table 1); the reduction in HML usage in the chl1 mutant (to 50%) is less than that in the fkh1 or RESCB mutant (approximately 30%). The difference in HML usage in the RESCB mutant in this experiment (30%) compared with the previous experiment (49%) is unexplained. HML quantification has been performed several times in both strains, and the difference is persistent. Each experiment was conducted with strains derived from two different crosses. The best explanation is that there is an unknown modifier that has arisen in these originally isogenic strains. As shown by reference 62, the involvement of CHL1 in donor preference is specific for MATa since chl1Δ in MATα cells does not affect donor preference (Table 1).
An RESCB chl1Δ double mutant shows the same HML usage as the RESCB single mutants (30%), suggesting that RESCB is epistatic to chl1Δ and that these two factors act in the same pathway. On the other hand, HML usage decreases twofold in the RESCB fkh1Δ (18%) and chl1Δ fkh1Δ (14%) double mutants. This suggests than Fkh1 activates HML through a different pathway from SBF and Chl1. These results define two pathway of HML activation in MATa cells, one involving FKH1 and another depending on SBF and CHL1. However, the RESCB fkh1Δ chl1Δ triple mutant shows a statistically significant reduction in HML usage (<10%), suggesting that SBF and Chl1 are not fully dependent on each other. HML usage in the triple mutant is similar to that observed in MATa cells with RE deleted (66), showing that FKH1, SBF, and CHL1 act in the major HML-activating pathways in MATa cells.
Another way to study the relationship between these three factors is to compare the effects of deleting FKH1 and CHL1 in strains bearing four repeats of region A (4A) instead of RE. The 22-bp region A contains an Fkh1 binding site, and the relatively strong 4A activity (65% HML usage) depends completely on FKH1 (53). This suggests that additional Fkh1 binding sites in tandem can bypass partially the role of SBF. The effect of deleting CHL1 on HML usage in a strain bearing 4A (from 65% to 54%) is similar to the small decrease observed in the RESCB fkh1Δ chl1Δ triple mutant compared to the RESCB fkh1Δ double mutant. This result confirms that CHL1 can act in an SCB-independent manner.
Recently Ruan et al. (48) reported that the DNA end-binding protein yKu80 played an important role in MATa donor preference. We confirmed that a yku80Δ strain reduced MATa donor preference (Table 1), but the roughly 10% reduction to 64% usage of HML was less dramatic than that reported by Ruan et al. (48). yku80Δ did not further reduce the effect of chl1Δ nor of RESCB, though—like chl1Δ—yku80Δ reduced the usage of HML in combination with fkh1Δ. These data suggest that yku80Δ is in the same epistasis group as RESCB (and swi4Δ or swi6Δ) and chl1Δ.
Fkh1 and SBF bind RE at different stages of the cell cycle.
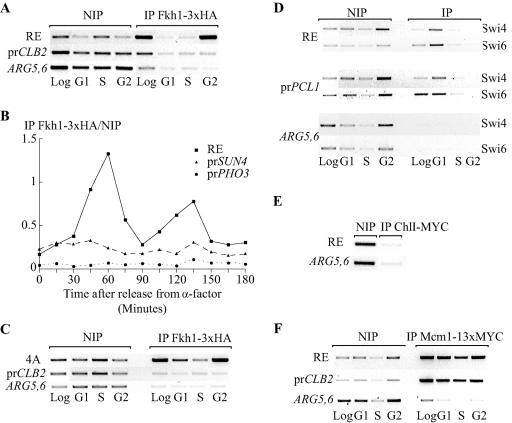
SBF binds to SCB-containing promoters of G1/S-regulated genes during G1 (34). In contrast, Fkh1 binds to its targets all along the cell cycle (35). We examined the binding of these factors to RE during the cell cycle by ChIP. Fkh1 binding to RE was measured in a strain bearing a functional FKH1-3xHA gene integrated in the genome (53). ChIP was performed in extracts from logarithmic phase or from cells arrested in the presence of α-factor (G1/S) or nocodazole (G2/M). To study the binding of Fkh1 to RE in S phase, cells were first arrested with α-factor and then released in the presence of HU. The binding of Fkh1 to the ARG5,6 gene was measured as a negative control. We confirmed the constant, but weak, binding of Fkh1 to the CLB2 promoter all along the cell cycle (Fig. 4A) as previously described (35). The signal observed is two times stronger than the one obtained after amplification of the ARG5,6 open reading frame (ORF). Surprisingly, Fkh1 binds RE mostly, if not exclusively, in the G2/M phase. Fkh1 binding to RE is therefore regulated differently from its binding to the promoters of the CLB2 cluster of genes. The signal generated by the binding of Fkh1 to RE is five times stronger than the signal generated by the binding to the BCL2 promoter.
FIG. 4.
ChIP analysis of Fkh1, SBF, Chl1, and Mcm1 binding to RE during the cell cycle. (A) Fkh1 binds RE in the G2/M phase of the cell cycle. ChIP is shown for primers amplifying RE sequences, the CLB2 promoter, and the unrelated ARG5,6 coding region in a strain bearing the hemagglutinin (HA)-tagged Fkh1 at the natural FKH1 locus (CFY480). ChIP was performed on cells in exponential phase (Log), arrested with α-factor (G1), arrested with α-factor and released in the presence of HU (S), or arrested with nocodazole (G2). Sequence enrichment was determined by PCR prior to (NIP) and after immunoprecipitation (IP). (B) Fkh1-3xHA binding to RE and to the SUN4 and the PHO3 promoters in synchronized cells released from α-factor arrest. The binding efficiency is expressed as the ratio between IP and non-IP values obtained from real-time PCR quantitation. (C) ChIP in a strain bearing four copies of region A instead of RE (KS358). (D) The Swi4/Swi6 complex binds RE in the G1/S phase of the cell cycle. The PCL1 promoter sequence was used as a positive control. (E) Chl1 does not bind RE. ChIP assay with primers amplifying RE sequences and the ARG5,6 ORF on strain ECY266 containing an integrated CHL1-9xMYC gene at the natural CHL1 locus. (F) Mcm1 binds RE all along the cell cycle. ChIP experiment with primers amplifying the RE region and the ARG5,6 coding region in a strain bearing Mcm1 tagged with MYC at the MCM1 locus (MJF190). All these experiments have been reproduced three times, except for that shown in panel B, which is supported by our data obtained with arrested cells.
To exclude an effect of nocodazole arrest on the regulation of Fkh1 binding, we measured Fkh1 binding to RE in synchronized cells released from an α-factor arrest (Fig. 4B). Time points were taken every 15 min, the experiment covering approximately 2 generations. Binding of Fkh1 was also measured at the SUN4 promoter, which is known to bind Fkh1 more efficiently than Fkh2 (22); PHO3 was used as a negative control. A bud count analysis was performed at each time point to follow the cell cycle. Quantifying the formation of the PCR products with a real-time PCR apparatus, we observed constant binding of Fkh1 to the SUN4 promoter through the cell cycle; however, the binding of Fkh1 to the RE is clearly cell cycle regulated, showing two peaks, 60 and 135 min after release (respectively, showing 18- and 15-fold increases over PHO3), where the cells are mostly in the G2 phase. This confirms our observation made with cells arrested with nocodazole.
To test if regulated binding of Fkh1 to RE in the G2 phase is linked to a modification of the chromatin structure of RE dictated by region C, we studied the binding of Fkh1 in a strain bearing four copies of region A instead of RE (Fig. 4C). We also observed a strong enrichment of the RE PCR signal in the population of cells arrested in G2/M. Therefore, the binding of Fkh1 to RE does not fully depend on factors binding to the C domain, such as Mcm1 or SBF.
The same experiment was performed with antibodies directed against Swi4 and Swi6 proteins (Fig. 4D). As a control, we looked at the enrichment of the PCL1 promoter sequence, a target of SBF (44). Both Swi4 and Swi6 bind RE mostly in the G1 phase and to a lesser extent in S phase. This regulation of binding is exactly the same as for the binding at the PCL1 promoter. Thus, we found that Fkh1 and SBF bound RE at two different stages of the cell cycle, which agrees well with the involvement of these two factors in two different pathways of RE activation.
We also studied binding of a Chl1-7xMYC hybrid protein to RE. Addition of the tag to the protein did not affect its function, as no deficiency was observed for donor preference or for chromosome loss, the original phenotype linked to the chl1 mutation (19; data not shown). However, there was no significant binding of Chl1 to RE (Fig. 4E).
Finally, we looked at the binding of Mcm1 to RE and to the BCL2 promoter in a strain bearing a MYC-tagged MCM1 (Fig. 4F). As previously reported (1), Mcm1 occupies the BCL2 promoter at every stage of the cell cycle. Similarly, we found that Mcm1 binds RE throughout the cell cycle. This binding probably could provide an open state of the RE chromatin in MATa cells, allowing SBF to bind in G1/S and Fkh1 to bind in G2/M.
MATa cells arrested in G1/S and in G2/M show reduced donor preference, whereas MATα cells do not.
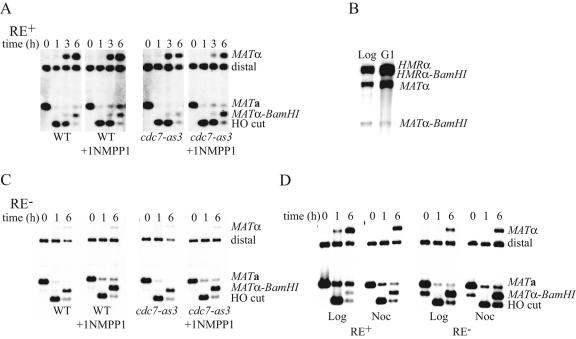
Since SBF binds RE in G1/S and Fkh1 in G2/M, it seems that two independent events are necessary for full activation of RE. To test this idea, we quantified donor preference in cells arrested in G1/S or G2/M. It has been shown recently that HO-induced homologous recombination requires the activation of the cyclin-dependent kinase CDK1 (26); consequently, mating-type switching cannot occur in cells arrested before the “start” point, i.e., under α-factor arrest. However, cells lacking a functional Cdc7 protein kinase move beyond “start” but cannot initiate DNA replication; because CDK1 is activated, these cells perform DSB repair normally (26). Therefore, we quantified donor preference in cells expressing Cdc7-as3, a mutant sensitive to the ATP analogue inhibitor 1-NMPP1 (2, 26). As shown in Fig. 5A, neither the drug nor cdc7-as3 alone affects mating-type switching or donor preference. When HO is induced in mutant cells arrested in the presence of 1-NMPP1, repair efficiency is not affected, but 50% of the cells switched to MATα-BamHI (from HMR), compared to 90% in the same strain without arrest, showing that HML use is reduced by the arrest.
FIG. 5.
Donor preference is affected by an arrest in G1/S and in G2/M in MATa cells but not in cells with RE deleted. (A) Time course experiment of MAT switching in MATa CDC7 (XW652) and MATa cdc7-as3 (GI560) cells arrested or not by the ATP analogue NMPP1. Genomic DNA cut with the restriction enzymes StyI and BamHI and probed with a MAT distal fragment reveals the parental fragment MATa, the fragment resulting from the HO cut, and the MATα and MATα-BamHI products. (B) Expression of HO under a Gal promoter specific for the G1 phase does not affect donor preference in MATa cells. Genomic DNA of the WT strain used in panel A (XW652) and of a G1-Gal-HO strain (ECY273) here digested with HindIII and BamHI and probed with Yα sequences shows the two products, MATα and MATα-BamHI. (C) The same experiment as in panel A but carried out in strains with RE deleted, thus mimicking MATα cells (WT, XW676; cdc7-as3, ECY252). (D) MATa switching in strains with RE deleted or not (XW676 and XW652, respectively). HO is induced in exponential phase (Log) or when cells are arrested in the presence of nocodazole (Noc). Genomic DNA was cut with StyI and BamHI and probed with a MAT distal fragment.
An alternative way to restrict MAT switching to one part of the cell cycle is to use a strain in which HO is under the control of a hybrid promoter which restricts the transcription of the gene to the G1 phase of the cell cycle in the presence of galactose (43). In this strain, HML usage is not affected (89% versus 94% in the wild type; Fig. 5B). Therefore, it is not the restriction of mating-type switching initiation in G1 that causes the defect but rather the arrest itself. It is possible that the arrest modifies chromosome architecture in a way that reduces the accessibility of the left arm of chromosome III. Interestingly, the arrest conferred by the addition of 1-NMPP1 to MATα cdc7-as3 cells does not affect their preferential choice of HMRα-BamHI (Fig. 5C).
HO-induced MATa cells arrested in G2/M in the presence of nocodazole show a similar defect to cells arrested in G1/S (Fig. 5D); HML usage is reduced from 90% to 50%. However, MATα cells arrested in G2/M still use HMR 90% of the time to repair the break at MAT. Thus, the exclusion of HML in MATα cells does not require any cell cycle-regulated mechanism.
We wondered if the defect in HML usage observed in arrested MATa cells was the direct consequence of the lack of Fkh1 binding in G1/S-arrested cells and of SBF binding in G2/M-arrested cells. If so, we would expect to observe a strong deficiency in HML usage in MATa RESCB cells arrested in G1/S but still 50% of HML usage in G2/M-arrested cells, since Fkh1 would activate RE normally. Reciprocally, MATa fkh1Δ cells arrested in G1/S should show 50% of HML usage, since SBF can bind RE, but a complete deficiency when arrested in G2/M. However, we observed approximately 30 to 40% of HML usage in fkh1Δ and RESCB mutants arrested either in G1/S or in G2/M (data not shown). Thus, even if their binding is regulated during the cell cycle, the roles of Fkh1 and SFF in RE activation can be perceived all along the cell cycle. The defect resulting from the arrest in G1/S and in G2/M in wild-type cells does not seem to be linked to the absence of binding of Fkh1 or SBF but rather to a modification of chromosome architecture, which antagonizes RE activation.
KAR3, CTF4, CTF18, and NUP170 genes, genetically related to CHL1, are not involved in donor preference.
To understand the role played by CHL1 in donor preference, we monitored the effects of deletion of four genes synthetic lethal with chl1Δ: KAR3, CTF18, BIM1, and YDR332W (50, 57). We also tested deletions of NUP170 and CTF4, which exhibit chromosome transmission fidelity deficiencies similar to chl1Δ and ctf18Δ (32). There was no modification of HML usage in MATa cells carrying deletions of any of these genes (data not shown).
Mbp1, Ace2, Ste12, and Ndd1 bind to the KAR4-SPB1 intergenic region but are not involved in donor preference.
A study of the association of many transcriptional regulators with intergenic regions across the genome confirmed that Mcm1, Fkh1, Fkh2, Ndd1, and SBF bind to RE (37). In addition, it was found that Mbp1, Ace2, and Ste12 bind the intergenic region containing RE. However, HML usage was not affected in strains carrying deletions of these genes (data not shown). We also measured HML usage in an fkh2Δ ndd1Δ double mutant strain, since fkh2Δ suppresses the lethality associated with the ndd1 mutation (35). We know from previous studies that the fkh2Δ mutation does not affect donor preference in MATa cells (53). The double mutant also does not show any defect (data not shown), arguing that despite the fact that Ndd1 can undergo ChIP with RE, this protein is not involved in its activation.
Does donor preference depend on any a- or α-specific genes?
As discussed above, when four repeats of region A (4A) replace RE, HML is used 65% in MATa but only 50% in MATα (Table 2). Furthermore, in strains harboring mutations of the two Matα2 binding site in the Matα2-Mcm1 operator of region C (65), HML usage was 75% in MATa and only 55% in MATα. Therefore, it appears that an a-specific activator or an α-specific repressor could regulate HML usage. The Matα2-Mcm1 complex represses expression of a-specific genes, whereas Matα1-Mcm1 activates transcription of α-specific genes. Deletion of MATα1 in strains carrying 4A in place of RE or with mutations in the MATα2 binding site of RE shows no increase in HML usage (data not shown). These results exclude involvement of an α-specific repressor in donor preference. Alternatively there could be an α-specific activator of HML as a donor in MATa cells. Deletion of the MAT-Ya sequences in a strain carrying the 4A RE does not affect donor preference (data not shown), ruling out the involvement of Mata1 and Mata2. It is possible that there is an undocumented small open reading frame that plays a role in the process (29).
TABLE 2.
α-specific genes are not required for HML repression as a donor in MATα cells
| Strain | Genotype | % HML usagea |
|---|---|---|
| Cwu150 | MATa | 75 |
| Cwu128 | MATa GT→TG AC→TA (Matα2 sites A and B) | 75 |
| KS338 | MATa 4A | 65 |
| Cwu151 | MATα | 15 |
| Cwu134 | MATα GT→TG AC→TA (Matα2 sites A and B) | 55 |
| KS345 | MATα 4A | 50 |
| ECY119 | MATα1::KanMX4 | 20 |
| ECY120 | MATα1::KanMX4 GT→TG AC→TA (Matα2 sites A and B) | 48 |
| ECY121 | MATα1::KanMX4 4A | 47 |
HML usage has been determined in MATa strains carrying HMLα and HMRα-BamHI, in MATα-BamHI strains carrying HMLα and HMRa, or in MATα1::KanMX4 strains carrying HMLα and HMRa. Genomic DNA extracted from an entire population of cells in which mating-type switching has been induced was cut by BamHI and StyI and probed with a MAT distal fragment.
DISCUSSION
We have found that an evolutionarily conserved SCB within RE plays an important role in donor preference and that the SBF complex, which binds this consensus sequence, is important for RE activity. We also described two independent pathways that govern donor preference: one depends on FKH1, and the other depends on SBF, CHL1, and YKU80.
Although two important transcription factors are involved in MATa donor preference, RE activation does not depend on transcription. Around region E, Szeto et al. (55) did find a weak transcript that does not appear to encode a protein. However, RE activity does not depend on these sequences because they are absent in the 270-bp minimum enhancer (65) or when multimers of region A, D, or E were inserted in place of the 1.8-kb sequence containing RE. Therefore, the role of Fkh1 and SBF in donor preference is very different from their involvement as transcription factors in the G2/M and G1/S transition, respectively.
It is also possible that the binding of SBF and Fkh1 to RE provokes a change in the chromatin structure of the left arm leading in some way to a greater accessibility of the resident donor; however, a recent study of the global chromatin structure of 45 kb of the left arm of chromosome III covering HML and RE did not shown any differences between MATa and MATα cells except for the RE region (15). We propose that the binding of Fkh1 and SBF to RE could reorganize the architecture and the nuclear localization of the left arm of chromosome III. In mammalian cells, the position of a locus can move relative to a heterochromatin domain, in response to a change in its transcriptional state (for review, see references 52 and 56). Also, massive decondensation can occur in the absence of transcription per se (5, 59). We suggest that binding of SBF and Fkh1 to RE could control the nuclear organization of the left arm of chromosome III in a similar way. SBF and Fkh1 could counteract a compact organization of the left arm, making HML more mobile in the nucleus.
Recently, we showed that HML motion is strongly constrained in both MATα and RE-deleted MATa strains, compared with MATa (4). Additionally, the three-dimensional configuration of MAT, HML, and HMR is mating-type dependent, the distance between HML and the other cassettes being greater in MATa cells (4). These data suggest there is constitutive tethering of HML, which is relieved in MATa cells through the binding of SBF and Fkh1 to the RE.
The consequences of cell cycle-dependent regulation of SBF and Fkh1 binding to RE are still unclear. It is possible that the establishment of chromosome III architecture needed for the activation of HML usage relies on two independent events, taking place in G1 through SBF binding to RE and in G2 through Fkh1 binding. Even if HO breaks the DNA at the MAT locus in G1, the absence of binding of Fkh1 in G2 would compromise the accessibility of HML, because the conformation of the left arm has not been properly established. This hypothesis is in agreement with the fact that fkh1Δ and RESCB mutants show the same HML usage defects in both G1/S- and G2/M-arrested cells. It seems that the recruitment of these factors to RE creates an epigenetic state necessary for the organization of the left arm in the nucleus in the subsequent phases of the cell cycle. The defect encountered in the G1 phase in RESCB mutants could lead to the disorganization of chromosome III structure in the subsequent S and G2 phases, explaining the defect also observed in the G2-arrested phase in this mutant. The same idea could apply to the fkh1 mutant. In contrast, arrest in both G1/S and G2/M does not affect donor preference in MATα cells.
Transcriptional activation at G1/S promoters follows a complex ordered series of events first delineated for the developmental and cell cycle-regulated HO promoter which depend on Swi5, SBF, and Whi5 (11-13). At RE, there seems to be no involvement of either Whi5 or Swi5. In addition, the study of the association of Swi5 with intergenic regions across the genome (37) does not show any binding to the RE-containing intergenic region. Finally, we show that the MBF complex, involved in the transcription regulation of another set of genes at the G1/S transition is not involved in RE activation, underlining the specific role of SBF in donor preference.
We have previously suggested that the role of Mcm1 is to open the chromatin structure at RE to allow effector proteins to bind (53, 65). We show here that this protein binds all along the cell cycle. It is therefore possible that the role of Swi5 to recruit SBF at the HO promoter is fulfilled by Mcm1 at RE.
Fkh1 has been shown to bind the promoters of the CLB2 cluster all along the cell cycle (35), an observation that we confirmed. However, binding of Fkh1 to RE is restricted to the G2/M phase of the cell cycle. This property is also observed for the binding to the 4A synthetic RE. This result could mean that the binding of Fkh1 to RE is regulated by a direct modification of the protein. However, as Fkh1 binds the CLB2 cluster promoters all along the cell cycle, it seems that this regulation does not affect directly the DNA binding properties of the protein, but rather its capacity to interact with other proteins binding RE. The cycling properties of Fkh1 binding are not directly linked to Mcm1 or SBF, since these proteins do not have a binding site in the 4A synthetic RE. It is possible that unidentified factors that are involved in Fkh1 regulation bind RE in domains A, D, and E. We are currently working on identifying these factors.
Fkh1 may be more important for RE function than in the regulation of the transcription of the CLB2 cluster. Fkh1 ChIP signals are stronger for RE than for the CLB2 or SUN4 promoters in G2-arrested cells, in logarithmic phase, and in synchronized cells (Fig. 4). The Fkh1 ChIP signal at RE is stronger than the one of Fkh2 or Ndd1 (53). Reciprocally, Fkh2 and Ndd1 ChIP signals are much stronger at the BCL2 cluster promoters than Fkh1 (23, 35, 53). Fkh1 activity can clearly substitute for the one of Fkh2 in the control of the CLB2 cluster expression (35), but Fkh2 cannot replace Fkh1 for RE activation.
We also found that the mutated SCB is epistatic to chl1Δ and to yku80Δ, showing that SBF, CHL1, and YKU80 act in the same pathway of RE activation. Since the deletion of RE reduces the usage of HML in MATa cells to the level observed in MATα cells (66), we know that deleting the SCB element is epistatic to both chl1Δ and yku80Δ. Therefore, as Chl1 does not bind RE, we suggest that SBF bound to RE needs Chl1 to perform its function. Although Chl1 is involved in the establishment of sister chromatid cohesion during S phase (40, 45, 50), none of the other components that genetically interact with Chl1 were found to affect donor preference, including CTF4, CTF18, or KAR3 (31, 40, 51, 58). It is therefore possible that the role of CHL1 in donor preference is not related to its activity in sister chromatid cohesion. It is important to keep in mind that Chl1 acts also in an SBF-independent manner, since the RESCB fkh1 chl1 triple mutant shows a slightly stronger reduction of HML usage than the RESCB fkh1 double mutant.
How Yku80—primarily implicated in DNA end-joining and telomere silencing—acts at RE or in facilitating recombination at HML is unclear, especially given that yku70Δ deletions have little effect (48; our data not shown). Whether it associates directly with RE or binds through Swi4/Swi6 or Fkh1 remains to be established.
Finally, we have ruled out the involvement of other a- and α-specific genes in donor preference. Among the five a-specific genes that have been identified (17), all of them are involved in conjugation and both their cellular localization and their enzymatic properties do not fit with a role in donor preference (25). We cannot fully rule out the involvement of an a-specific sterile RNA or of the product of an unidentified small ORF (29) to explain why HML is slightly less used in MATα cells when the natural RE is replaced by 4A, for example. We cannot also rule out that HMR usage can be slightly increased in MATα cells by an RE-like activity (67, 69).
We still don't know how RE works, but regulation of its activity is very complex. The involvement of SBF and Fkh1 make RE resemble a hybrid-regulated promoter, which recruits SBF in a Swi5-independent manner and Fkh1 in a G2-specific way. Given that RE does not cause modification of chromatin around HML, we speculate that the recruitment of transcription factors can create an epigenetic modification of RE which leads to a change in the regional conformation of the left arm of chromosome III in the nucleus, making it more accessible for recombination, possibly by blocking the tethering of the chromosome arm at several as yet unidentified sites. This hypothesis is supported by the increased mobility of the left arm in MATa cells (4). In Schizosaccharomyces pombe, recent studies have shown that donor preference is dictated by a Swi2-dependent recombination enhancer that controls the choice of the donor by controlling the spreading of a recombination-promoting complex on the donors in a heterochromatin-dependent manner (28). The function of RE in S. cerevisiae appears therefore to be radically different.
Acknowledgments
We are grateful to Brenda Andrews, Kim Nasmyth, Guy-Franck Richard, and Bik Tye for strains, plasmids, and antibodies. We also thank Xuan Wang for technical advice and Miyuki Yamaguchi for technical help.
E.C. was supported in part by grants from l'Association pour la Recherche sur le Cancer and the Philippe Foundation. Research was supported by NIH grant GM20056.
REFERENCES
- 1.Althoefer, H., A. Schleiffer, K. Wassmann, A. Nordheim, and G. Ammerer. 1995. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5917-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. [DOI] [PubMed] [Google Scholar]
- 3.Breeden, L. 1996. Start-specific transcription in yeast. Curr. Top. Microbiol. Immunol. 208:95-127. [DOI] [PubMed] [Google Scholar]
- 4.Bressan, D. A., J. Vazquez, and J. E. Haber. 2004. Mating type-dependent constraints on the mobility of the left arm of yeast chromosome III. J. Cell Biol. 164:361-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter, A. E., S. Memedula, M. J. Plutz, and A. S. Belmont. 2005. Common effects of acidic activators on large-scale chromatin structure and transcription. Mol. Cell. Biol. 25:958-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, V. K., M. J. Fitch, J. J. Donato, T. W. Christensen, A. M. Merchant, and B. K. Tye. 2003. Mcm1 binds replication origins. J. Biol. Chem. 278:6093-6100. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G., N. Hata, and M. Q. Zhang. 2004. Transcription factor binding element detection using functional clustering of mutant expression data. Nucleic Acids Res. 32:2362-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 10.Coïc, E., G.-F. Richard, and J. E. Haber. Saccharomyces cerevisiae donor preference during mating-type switching is dependent on chromosome architecture and organization. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 11.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo, M., J. L. Nishikawa, X. Tang, J. S. Millman, O. Schub, K. Breitkreuz, D. Dewar, I. Rupes, B. Andrews, and M. Tyers. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117:899-913. [DOI] [PubMed] [Google Scholar]
- 13.de Bruin, R. A., W. H. McDonald, T. I. Kalashnikova, J. Yates III, and C. Wittenberg. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117:887-898. [DOI] [PubMed] [Google Scholar]
- 14.Ercan, S., J. C. Reese, J. L. Workman, and R. T. Simpson. 2005. Yeast recombination enhancer is stimulated by transcription activation. Mol. Cell. Biol. 25:7976-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ercan, S., and R. T. Simpson. 2004. Global chromatin structure of 45,000 base pairs of chromosome III in a- and α-cell yeast and during mating-type switching. Mol. Cell. Biol. 24:10026-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairhead, C., B. Llorente, F. Denis, M. Soler, and B. Dujon. 1996. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 12:1439-1457. [DOI] [PubMed] [Google Scholar]
- 17.Galgoczy, D. J., A. Cassidy-Stone, M. Llinas, S. M. O'Rourke, I. Herskowitz, J. L. DeRisi, and A. D. Johnson. 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:18069-18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerring, S. L., F. Spencer, and P. Hieter. 1990. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 9:4347-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber, J. E. 1974. Bisexual mating behavior in a diploid of Saccharomyces cerevisiae: evidence for genetically controlled non-random chromosome loss during vegetative growth. Genetics 78:843-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber, J. E. 2002. Switching of Saccharomyces cerevisiae mating-type genes, p. 927-952. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 21.Herskowitz, I. 1989. A regulatory hierarchy for cell specialization in yeast. Nature 342:749-757. [DOI] [PubMed] [Google Scholar]
- 22.Hollenhorst, P. C., M. E. Bose, M. R. Mielke, U. Muller, and C. A. Fox. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154:1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenhorst, P. C., G. Pietz, and C. A. Fox. 2001. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: implications for regulating the cell cycle and differentiation. Genes Dev. 15:2445-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holloway, S. L. 2000. CHL1 is a nuclear protein with an essential ATP binding site that exhibits a size-dependent effect on chromosome segregation. Nucleic Acids Res. 28:3056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 26.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 28.Jia, S., T. Yamada, and S. I. Grewal. 2004. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119:469-480. [DOI] [PubMed] [Google Scholar]
- 29.Kastenmayer, J. P., L. Ni, A. Chu, L. E. Kitchen, W. C. Au, H. Yang, C. D. Carter, D. Wheeler, R. W. Davis, J. D. Boeke, M. A. Snyder, and M. A. Basrai. 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16:365-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 31.Kenna, M. A., and R. V. Skibbens. 2003. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23:2999-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerscher, O., P. Hieter, M. Winey, and M. A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics 157:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klar, A. J., J. B. Hicks, and J. N. Strathern. 1982. Directionality of yeast mating-type interconversion. Cell 28:551-561. [DOI] [PubMed] [Google Scholar]
- 34.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129-141. [DOI] [PubMed] [Google Scholar]
- 35.Koranda, M., A. Schleiffer, L. Endler, and G. Ammerer. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94-98. [DOI] [PubMed] [Google Scholar]
- 36.Lea, D. E., and C. A. Coulson. 1949. The distribution in the numbers of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 37.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 39.Loo, S., and J. Rine. 1994. Silencers and domains of generalized repression. Science 264:1768-1771. [DOI] [PubMed] [Google Scholar]
- 40.Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas, T. Kwok, R. Newitt, R. Aebersold, C. Boone, G. W. Brown, and P. Hieter. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Measday, V., L. Moore, R. Retnakaran, J. Lee, M. Donoviel, A. M. Neiman, and B. Andrews. 1997. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 17:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasmyth, K. 1987. The determination of mother cell-specific mating type switching in yeast by a specific regulator of HO transcription. EMBO J. 6:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogas, J., B. J. Andrews, and I. Herskowitz. 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66:1015-1026. [DOI] [PubMed] [Google Scholar]
- 45.Petronczki, M., B. Chwalla, M. F. Siomos, S. Yokobayashi, W. Helmhart, A. M. Deutschbauer, R. W. Davis, Y. Watanabe, and K. Nasmyth. 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 117:3547-3559. [DOI] [PubMed] [Google Scholar]
- 46.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravindra, A., K. Weiss, and R. T. Simpson. 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 19:7944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan, C., J. L. Workman, and R. T. Simpson. 2005. The DNA repair protein yKu80 regulates the function of recombination enhancer during yeast mating type switching. Mol. Cell. Biol. 25:8476-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 50.Skibbens, R. V. 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166:33-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skibbens, R. V., L. B. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spector, D. L. 2003. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72:573-608. [DOI] [PubMed] [Google Scholar]
- 53.Sun, K., E. Coic, Z. Zhou, P. Durrens, and J. E. Haber. 2002. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev. 16:2085-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szeto, L., and J. R. Broach. 1997. Role of α2 protein in donor locus selection during mating type interconversion. Mol. Cell. Biol. 17:751-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szeto, L., M. K. Fafalios, H. Zhong, A. K. Vershon, and J. R. Broach. 1998. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev. 11:1899-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taddei, A., F. Hediger, F. R. Neumann, and S. M. Gasser. 2004. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet. 38:305-345. [DOI] [PubMed] [Google Scholar]
- 57.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 58.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Driel, R., and P. Fransz. 2004. Nuclear architecture and genome functioning in plants and animals: what can we learn from both? Exp. Cell Res. 296:86-90. [DOI] [PubMed] [Google Scholar]
- 60.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 61.Weiler, K. S., and J. R. Broach. 1992. Donor locus selection during Saccharomyces cerevisiae mating type interconversion responds to distant regulatory signals. Genetics 132:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiler, K. S., L. Szeto, and J. R. Broach. 1995. Mutations affecting donor preference during mating type interconversion in Saccharomyces cerevisiae. Genetics 139:1495-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, K., and R. T. Simpson. 1997. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 16:4352-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol. 18:5392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, C., K. Weiss, C. Yang, M. A. Harris, B. K. Tye, C. S. Newlon, R. T. Simpson, and J. E. Haber. 1998. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 12:1726-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, X., and J. E. Haber. 1996. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87:277-285. [DOI] [PubMed] [Google Scholar]
- 67.Wu, X., and J. E. Haber. 1995. MATa donor preference in yeast mating-type switching: activation of a large chromosomal region for recombination. Genes Dev. 9:1922-1932. [DOI] [PubMed] [Google Scholar]
- 68.Wu, X., J. K. Moore, and J. E. Haber. 1996. Mechanism of MATα donor preference during mating-type switching of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, X., C. Wu, and J. E. Haber. 1997. Rules of donor preference in Saccharomyces mating-type gene switching revealed by a competition assay involving two types of recombination. Genetics 147:399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]