Abstract
CARMA1 is a central regulator of NF-κB activation in lymphocytes. CARMA1 and Bcl10 functionally interact and control NF-κB signaling downstream of the T-cell receptor (TCR). Computational analysis of expression neighborhoods of CARMA1-Bcl10MALT 1 for enrichment in kinases identified calmodulin-dependent protein kinase II (CaMKII) as an important component of this pathway. Here we report that Ca2+/CaMKII is redistributed to the immune synapse following T-cell activation and that CaMKII is critical for NF-κB activation induced by TCR stimulation. Furthermore, CaMKII enhances CARMA1-induced NF-κB activation. Moreover, we have shown that CaMKII phosphorylates CARMA1 on Ser109 and that the phosphorylation facilitates the interaction between CARMA1 and Bcl10. These results provide a novel function for CaMKII in TCR signaling and CARMA1-induced NF-κB activation.
NF-κB activation is important for the maturation, survival, and function of T lymphocytes (20, 50). Much attention has recently focused on molecular scaffolds that organize and facilitate NF-κB activation downstream of antigen receptor engagement. Recent biochemical and genetic studies have identified that the ordered interaction of CARMA1, Bcl10, and mucosa-associated lymphoid tissue 1 (MALT1) is essential for T-cell receptor (TCR)-induced NF-κB activation (9, 10, 12, 17, 27, 32, 34, 48). T cells from mice deficient in any one of these scaffolds/adaptor proteins fail to activate NF-κB following antigen receptor engagement.
CARMA1, a member of the MAGUK family of kinases, encodes a protein that contains a caspase recruitment domain (CARD), a coiled-coil domain, a PDZ domain, a Src homology 3 (SH3) domain, and a C-terminal guanylate kinase domain (42). Upon TCR engagement, CARMA1 is recruited to the immune synapse (6, 11) and CARMA1 recruits Bcl10 via CARD-CARD interaction. The coiled-coil domain is presumed to act as a protein interaction domain, and a Leu-to-Gln substitution (L298Q) in the coiled-coil domain also interferes with antigen receptor-mediated NF-κB activation (17). Under physiological conditions, the PDZ domain is dispensable for NF-κB activation, whereas mutations within the SH3 domain interfere with the membrane recruitment of the CARMA1-dependent trimolecular protein complex (31, 47). Recent studies have suggested that PDK1 plays a central role in linking the proximal TCR activation complex to protein kinase C θ (PKCθ) and the CARMA1/Bcl10/MALT1 (CBM) complex (19). The GUK domain of CARMA1 interacts with PDK1. In addition, mutations within the CARD (the L39R mutation), coiled-coil domain (the L298Q mutation), and SH3 domain (the L808P mutation) also attenuate TCR-mediated NF-κB activation (10, 17, 47). In addition, PKCθ and PKCβ have recently been shown to be essential for CARMA1 activation in T and B lymphocytes, respectively (23, 36). PKCθ in T cells and PKCβ in B cells specifically phosphorylate human serine residue 552/mouse serine residue 564 within the linker region of CARMA1, leading to phosphorylation of IκB and NF-κB activation. However, phosphorylation within the linker region did not alter synaptic recruitment of CARMA1 or its ability to bind Bcl10 in heterologous cell systems (23).
Bcl10, a protein discovered from a recurrent breakpoint in MALT lymphomas, interacts with CARMA1 via its CARD (42, 51). Bcl10 is essential for antigen receptor-induced NF-κB activation, interleukin-2 production, and T-cell proliferation but is not required for TCR-induced tyrosine phosphorylation, calcium flux, or extracellular signal-regulated kinase activation (33). Bcl10 overexpression might be linked to constitutive NF-κB activation and cell proliferation observed with MALT tumors. However, it is unknown to date how the molecular interactions between CARMA1 and Bcl10 are regulated after receptor triggering. The C-terminal Ser/Thr-rich domain of Bcl10 binds to immunoglobulin motifs of MALT1 (46). The oligomerization of MALT1 is sufficient to induce NF-κB activation. Thus, it is suggested that Bcl10 mediates the oligomerization of MALT1 and activates NF-κB (21). Zhou et al. have indicated that Bcl10 promotes NF-κB activation through MALT1- and UBC13-dependent ubiquitination of IκB kinase γ (IKKγ) (52). Furthermore, Sun et al. have demonstrated that TRAF6 and TAK1 mediate IKK activation induced by Bcl10 and MALT1 (40). In addition to the kinase-mediated regulation of NF-κB signaling, increases in calcium levels appear to control NF-κB activity (7).
Large-scale analysis of gene expression data sets provides biologists with a valuable tool for deciphering cellular gene regulatory circuits. It has been repeatedly documented for Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans that broad categories of functionally related genes (e.g., cell cycle genes or components of organelles, such as mitochondria) are often coexpressed (26, 37). Recently, this concept of functional clustering of coexpressed genes has been extended to vertebrates and has been exploited in two examples, to identify a Mendelian disease gene in one case and to identify a mechanism of action of cyclin D1 in cancer (18, 26). To identify physiologically relevant kinases that regulate CARMA1-mediated NF-κB signaling, we examined gene expression signatures of kinases from a large, publicly available microarray data set (38). In this study, we have used pairwise comparisons of the gene expression data to screen for immune-related kinases that may regulate CARMA1-Bcl10-MALT1 assembly in T cells.
Our discovery that calmodulin-dependent protein kinase II gamma (CaMKIIγ) is coregulated with the CBM module led us to explore the role of these kinases in regulating the CARMA1-mediated NF-κB activation. The multifunctional Ca2+/CaMKII family consists of four different genes (α, β, γ, and δ) that mediate cellular responses to Ca2+ in various tissues (13, 44). Previous studies have demonstrated that CaMKII and CaMKIV are also involved in NF-κB activation in T cells (14, 16). Furthermore, a recent study has highlighted the importance of CaMKII in directly modulating NF-κB signaling in neurons (25). In this report, we have demonstrated that CaMKII is recruited to the immune synapse and functions as a key control point in CARMA1-mediated NF-κB activation.
MATERIALS AND METHODS
Reagents.
Anti-CD3 antibody (OKT3) was obtained from Massachusetts General Hospital. Anti-CD28 antibody was purchased from Caltag Laboratories (CA). Tumor necrosis factor alpha (TNF-α) was purchased from R&D Systems (MN). KN93 was purchased from Sigma (St. Louis, MO). Mouse anti-CaMKII antibody (BD Biosciences, CA), which interacts with the C-terminal region, was used to detect endogenous CaMKII. Rabbit anti-CaMKII antibody (Santa Cruz Biotechnology, CA) was used to detect the N-terminal fragment of CaMKII. Anti-Flag M2 monoclonal antibody was purchased from Sigma (St. Louis, MO), and anti-Myc monoclonal antibody (9E10) was purchased from Covance (Berkeley, CA).
Expression constructs.
Human CARMA1 cDNA was a generous gift from J. Tschopp. It was subcloned into the Peak12-Flag vector, which contains an EF1α promoter and a Flag epitope tag, or pCMV-Myc (Clontech, CA). Mouse Bcl10, CARMA2, and CARMA3 cDNAs were obtained from Open Biosystems (CA) and subcloned into the pCMV5-Flag vector, which contains a cytomegalovirus promoter and a Flag epitope tag. The mutations in CARMA1 were introduced with a QuickChange site-directed mutagenesis kit (Stratagene, CA). The construct expressing the N-terminal fragment of rat CaMKIIα from residues 1 to 290 (CaMKII1-290) was a generous gift from R. Maurer. cDNAs encoding human CaMKIIγ (NM_172170) and CaMKIIδ (NM_172127) were obtained from Origene (MD). The NF-κB-dependent firefly luciferase reporter was described previously (43). The construct expressing the IKKβ mutant (with S177A and S181A mutations) was a gift from M. Karin. The serum response element (SRE)-dependent firefly luciferase reporter was a generous gift from M. White. The NFAT-dependent firefly luciferase reporter was provided by A. Ting.
The small interfering RNA (siRNA) expression constructs were made with pSS-H1p vector (a generous gift from D. Billadeau), which expresses a short hairpin type of RNA downstream of the RNA polymerase III-dependent H1 promoter. The siRNA construct D1424 or D1488 targets nucleotides 1424 to 1442 or 1488 to 1506 of CaMKIIδ mRNA (NM_172128), respectively, while the construct G371 or G1660 targets nucleotides 371 to 389 or 1660 to 1678 of CaMKIIγ mRNA (BC_034044), respectively.
Cells.
Jurkat cells, the E6-1 clone, Raji B cells, and 293T cells were obtained from American Type Culture Collection (VA). The CARMA1 Jurkat mutant cell line (JPM50.6) was a generous gift from X. Lin.
Kinome identification.
The complement list of genes encoding kinases in the human genome was obtained from the website http://www.kinase.com/human/kinome, based on recent updates (5, 22). There are 518 human kinases reported by the authors. The public version of the GNF Human Expression Atlas, version 2, was obtained from Novartis (http://wombat.gnf.org/), including the primary cel files, which used the U133a Affymetrix chip and a custom chip (GNF1H) (38). The data set contains the expression values of 33,689 probes, reflecting normalization of each array to a set of 100 housekeeping genes common to both the U133a and the custom GNF1H array. Subsequently, global median scaling across the arrays was performed, resulting in the expression values across samples for each probe set. We were able to identify at least one probe for 490 of the 518 kinases, with a total of 964 probes identified, as a result of some kinases being represented by multiple probes by use of the U133a and custom chips employed with the GNF/Symatlas data set. We used the default filtering parameters in DCHIP to check the quality of expression patterns of each probe, with 824 out of 964 passing these filtering criteria, resulting in at least one probe which passed filtering for 460 of 490 represented kinases (see Table S1 in the supplemental material). We verified that each of these 460 known or predicted kinases encoded a kinase domain as recognized by the PFAM and/or Interpro domain database.
Target set enrichment analysis.
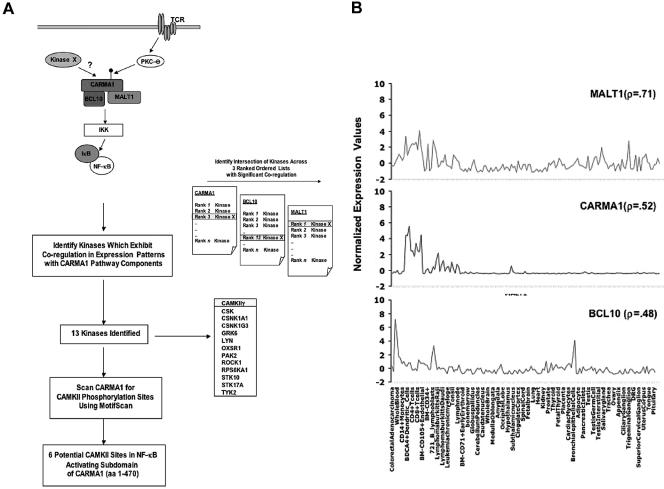
To identify kinases that showed coregulation with the immune-specific components of the TCR signaling cascade, we first ranked all kinase probes (n = 824) in terms of their Pearson's coregulation with respect to CARMA1, MALT1, and Bcl10. As a result, we generated three rank-ordered lists, each having 824 entries. The higher the correlation value, the higher the ranking in the list. We chose a correlation value of r of >0.45 as demonstrating a statistical significant threshold of correlated expression. Genome-wide significance can conservatively be defined using a Bonferroni correction, that is, at a correlation value resulting in 0.05/33,689, or 0.372674584. We then identified kinases which were common to all three lists and which had a correlation cutoff of r of >0.45, with 13 kinases, including CaMKIIγ, meeting these criteria.
Conjugation and immunofluorescent staining.
Conjugate formation was performed as described previously (35). Briefly, Raji cells were loaded with a blue fluorescent cell tracker chloromethylbenzoyl derivative of aminocoumarin (Molecular Probes, OR) according to the manufacturer's instructions and pulsed with 5 μg/ml staphylococcal enterotoxin E (SEE) (Toxin Technology, FL) for 20 min. Jurkat cells were incubated with KN93 (40 μM) or dimethyl sulfoxide (DMSO) for 10 min at 37°C. Thereafter, Raji cells and Jurkat cells were mixed at a ratio of 1:1. The cells were centrifuged at low speed and incubated for 15 min at 37°C. The conjugates were gently resuspended and plated onto poly-l-lysine-coated slides. After fixation with 3.5% paraformaldehyde and 0.1% Tween 20 in phosphate-buffered saline (PBS), the cells were labeled with mouse anti-CaMKII antibody, followed with anti-mouse immunoglobulin G antibody conjugated with Alexa Fluor 488 and phalloidin conjugated with Alexa Fluor 568 (Molecular Probes, OR). The cells were viewed under a fluorescence microscope (Olympus, Japan).
Nuclear extraction and immunoblotting.
Jurkat cells (1 × 107) were resuspended in 400 μl of culture media including KN93 (40 μM) or DMSO, incubated at 37°C for 10 min, and then stimulated with anti-CD3 antibody (5 μg/ml) or TNF-α (5 ng/ml) at 37°C for 10 min. After cells were washed with cold PBS, the nuclear extract was obtained as described previously (15). The nuclear extract (10 μg of total protein) was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane, and blotted with anti-p65 antibody (Upstate Biotechnology, NY) or antiretinoblastoma antibody (NeoMarkers, CA).
Luciferase reporter assay.
Jurkat cells (5 × 106) were resuspended in 250 μl culture medium and electroporated (250 V, 950 μF) with expression constructs, firefly luciferase reporter (3 μg NF-κB, 3 μg SRE, or 10 μg NFAT), and 20 ng pRL-TK construct (Promega, WI) expressing Renilla luciferase. Upon electroporation, the cells were diluted in 2.5 ml culture medium. One day or 3 days later, the cells were lysed and luciferase activities were determined with a dual-luciferase reporter assay system (Promega, WI) according to the manufacturer's instructions. NF-κB, NFAT, or SRE activity was assessed by normalization of firefly luciferase activity to Renilla luciferase activity.
[γ-32P]ATP phosphorylation assay.
293T cells (2 × 106) were seeded in 10-cm plates and grown to approximately 70% confluence. One day later, the cells were transfected with 8 μg of Flag-CARMA1, Flag-CARMA3, and Flag-CARMA1(S109A) with Lipofectamine 2000 (Invitrogen, CA). For recombinant protein generation, cells were allowed to grow for 48 h posttransfection. Recombinant protein was released from 293T cells by washing cells once in PBS and then lysing them in 1 ml radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, and EDTA-free protease inhibitor cocktail [Roche]) on ice for 30 min. Insoluble material was removed by centrifugation (10,000 × g, 10 min, 4°C). For each immunoprecipitation, 30 μl anti-Flag-agarose beads (Sigma) was washed three times in 1 ml RIPA buffer and then added to each lysate. Samples were then rotated for 2 h at 4°C. For elution, anti-Flag-agarose beads were washed first three times in RIPA buffer and then twice in kinase buffer (20 mM MOPS [morpholinepropanesulfonic acid], pH 7.2, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM dithiothreitol, 1 mM CaCl2). The anti-Flag-agarose/immunoprecipitation complex was finally incubated in 30 μl of kinase buffer supplemented with Flag peptide (Sigma) at a final concentration of 200 μg/ml for 10 min. The supernatant was then removed and placed in a centrifuge filter tube (catalog no. 8160; Corning, Inc.) on ice. A second extraction was performed by incubating beads with 200 μg/ml Flag peptide in kinase buffer. Both supernatants were combined and then recovered by centrifugation (6,000 × g, 2 min, 4°C), aliquoted, and flash frozen in liquid nitrogen.
Kinase assays were then performed according to the manufacturer's instructions (Upstate Biotechnology). All substrates were equalized to 1 μg (by Coomassie staining next to known standards) and diluted in kinase buffer to a volume of 10 μl. Then, a combination of CaMKII (50 ng), inhibitor (100 nM final) (Upstate Biotechnology), and kinase buffer was added to a final volume of 40 μl. Finally, 10 μl of 10 μCi [γ-32P]ATP, 75 mM MgCl2, and 500 μM cold ATP was added. Samples were then incubated at 30°C for 10 min. The reaction was stopped by the addition of 20 μl 5× Laemmli buffer. A portion (50 μl) of each sample was then analyzed by SDS-PAGE, stained using Coomassie blue for loading quantification, and dried. γ-32P incorporation was measured by using a phosphorimager system. The remaining 20 μl was analyzed by Western blotting using an antibody raised against CaMKII (Santa Cruz).
Immobilized metal affinity chromatography (IMAC).
293T cells (2 × 105) were seeded in 24-well plates. One day later, the cells were transfected with 0.1 μg of Myc-CARMA1 construct and 1 μg of CaMKII1-290 with 1.5 μl of Lipofectamine 2000. The total amount of DNA was equalized with empty vectors. Eighteen hours later, the cells were lysed in 1 ml of the lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 1% Triton X-100, and 10 μg/ml leupeptin).
Jurkat cells (5 × 106) were electroporated with 10 μg of siRNA construct (vector or D1488) and 1 μg of Myc-CARMA1 construct. Three days later, the cells were incubated with or without anti-CD3 antibody (5 μg/ml) and anti-CD28 antibody (1 μg/ml) for a further 10 min and then lysed in 500 μl of lysis buffer.
Jurkat cells (5 × 106) were electroporated with 1 μg of Myc-CARMA1. One day later, the cells were incubated with or without anti-CD3 antibody (5 μg/ml) and anti-CD28 antibody (1 μg/ml) for a further 10 min and then lysed in 250 μl of the lysis buffer. Phosphoproteins were isolated via IMAC using a PhosphoCruz protein purification system (Santa Cruz Biotechnology, CA) according to the manufacturer's instructions. Briefly, 25 μl of sample lysates was diluted with 1 ml of binding/washing buffer (Santa Cruz Biotechnology) and reacted with 10 μl of PhosphoCruz agarose for 1 h at 4°C. After being washed three times, isolated proteins were eluted in SDS buffer.
RESULTS
Recent genetic and biochemical studies indicate that the trimolecular assembly of CARMA1, Bcl10, and MALT1 is essential for antigen receptor-mediated NF-κB activation (42). Although some of the signaling intermediates that are essential for TCR-mediated NF-κB activation are ubiquitously expressed, the CBM components are restricted in their expression profile. We sought to harness expression data sets to generate a subset of potential CARMA1 candidate kinases based on the premise that a physiologically relevant kinase would exhibit coregulation with the three-component gene set/module (CARMA1, Bcl10, and MALT1). A comparative-genomics approach recently catalogued 510 kinases with 1:1 orthologs in mice, with eight additional human-specific kinases, making it impractical to test each kinase individually for its ability to phosphorylate CARMA1 (5, 22). Thus, we sought to utilize computational analysis of publicly available expression data to identify an enriched list of likely candidates for evaluation. The data set of expression profiles we employed to identify CARMA1 candidate kinases is a compendium of gene profiles drawn across a diverse set of human tissue/cell types (79 normal or transformed tissues/cells; 16,684 genes) known as the GNF tissue compendium or Symatlas (38). To then identify kinases that showed coregulation with the CBM components, we first ranked all kinases in terms of their pairwise Pearson's correlation coefficient (r) with respect to CARMA1, MALT1, and Bcl10. As a result, we generated three rank-ordered lists of kinases. The higher the correlation value, the higher the ranking in the list and the more the kinase mimicked the expression profile of a component, thereby demonstrating that the kinase was in the “expression neighborhood” of the CBM target gene set. Using these three rank-ordered lists, we sought kinases that were significantly coregulated with not just one but all three components. As a result, we generated lists of those kinases whose correlation coefficients exceeded 0.4, 0.45, and 0.5 with respect to all three CBM genes (i.e., the intersection of the three target CBM gene sets at these thresholds). These thresholds were selected as, given the number of genes tested, we estimated that genome-wide significance would correspond to a correlation of between 0.35 and 0.40. As only a single gene was correlated to all three genes (CBM) with an r of >0.5, we elected to pursue the 13 genes which exceeded an r of >0.45 as demonstrating unequivocal coregulation with all three component genes. In addition, many genes with lower correlations likely constitute true coregulation, and, for example, the well-recognized PKCθ and PDK1 would be included only if the threshold were dropped to 0.35 or 0.38, respectively. The list of 13 highly coregulated kinases contains several for which phosphorylation consensus sites are known, such as CaMKII, as well as kinases for which little about phosphorylation target site preference is known, such as oxidative-stress responsive kinase 1. We next combined the expression profiling data with those of Motif Scan, which predicts phosphorylation sites in proteins for a number of kinases. Interestingly, we found predicted CaMKII phopshorylation sites in the amino terminus of CARMA1. Based on the significant coregulation of CaMKIIγ with CARMA1, Bcl10, and MALT1 and the presence of predicted CaMKII phosphorylation sites on CARMA1, we chose to ascertain if CaMKIIγ was a physiologically relevant kinase of CARMA1. Figure 1A illustrates the algorithm used to capture coregulated kinases in the expression neighborhood of CARMA1, Bcl10, and MALT1. The correlation of the CaMKIIγ expression profile with the CBM gene set is shown graphically in Fig. 1B. Previous studies have shown that CaMKIIγ and CaMKIIδ are expressed in the lymphoid tissue compartment (2, 4, 14, 28). Of note, there were several probes for CaMKIIγ in the GNF data set, including one probe representing an immune enriched splice variant, which exhibited the strongest correlation. On further analysis, we noted that probes for the closely related and immune-specific isoform CaMKIIδ were not available in the Symatlas data set. Given that splice variants of CaMKIIδ have been reported to be expressed in T cells, we have included this in follow-up studies. In an attempt to provide experimental evidence for the involvement of CaMKII in T-lymphocyte signaling, we next examined whether CaMKII was recruited to the immune synapse and regulated TCR-specific transcriptional responses.
FIG. 1.
Schema for identification of CaMKII as a potential modulator of CARMA1-mediated NF-κB activation by use of kinase and pathway component expression signatures. (A) (i) Literature analysis of pathway components in TCR-mediated NF-κB activation identified CARMA1, Bcl10, and MALT1 as a functional module whose components exhibit tissue-restricted expression profiles. We hypothesized that an additional CARMA1 regulatory kinase (kinase X) would exhibit a coregulated expression signature with CARMA1, Bcl10, and MALT1. (ii) We derived a list of potential candidate kinases based on expression profiles. Three rank-ordered lists of kinase expression profiles with respect to CARMA1, Bcl10, and MALT1 gene sets were generated. For CARMA1, MALT1, and Bcl10, each of the probes representing the 460 kinases in the data set are ranked from most similar to each to least similar in expression profile across the GNF compendium based on pairwise Pearson's correlation coefficient. Subsequently, 13 kinases were identified among 460, including CaMKIIγ, which exhibited significant correlation with the target gene set of the CARMA1 signaling pathway. (iii) Motif Scan was employed to examine if CARMA1 had potential consensus CaMKII phosphorylation sites. (B) Expression level correlation between CaMKIIγ and the CARMA1-MALT1-Bcl10 gene set. A Z-score normalization (y axis) of microarray intensity values was performed and plotted for CaMKIIγ and the CARMA1-MALT1-Bcl10 target gene set in each of the tissue/cell types (x axis) in the GNF compendium data set (all 79 tissues/cells in tissue compendium are plotted, and a selected subset is labeled). Also indicated are the Pearson's pairwise correlation values (r) between CaMKIIγ and the individual gene set members as a quantification of expression similarity. DRG, dorsal root ganglion.
T-cell receptor engagement redistributes CaMKII.
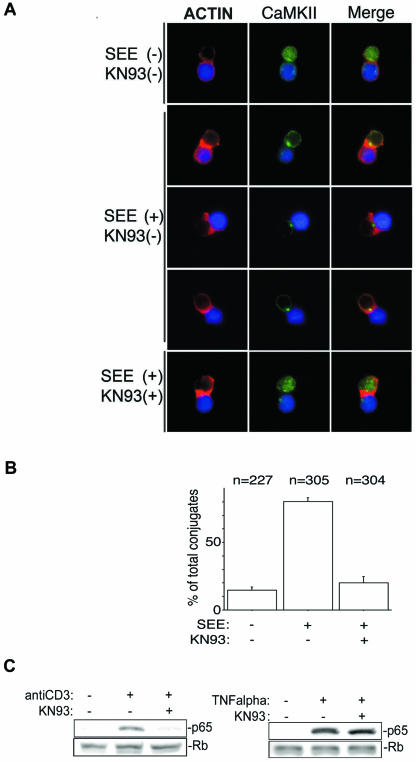
To observe the intracellular distribution of endogenous CaMKII in T cells after antigen stimulation, Jurkat cells were activated with SEE-pulsed Raji cells. Consistent with published observations (8, 35), SEE-pulsed Raji cells induced actin polymerization in Jurkat cells at the contact area (Fig. 2A). Endogenous CaMKII was dynamically redistributed in Jurkat cells to the contact area following superantigen-mediated TCR stimulation (Fig. 2A). KN93, which binds to the calmodulin-binding site of CaMKII and inhibits its activity, interfered with the redistribution (Fig. 2A and B), suggesting that the redistribution of CaMKII is activation dependent (39). A previous study demonstrated that CaMKII is required for IκBα degradation induced by TCR stimulation (14). To confirm that pharmacological inhibition of CaMKII translocation to the immune synapse attenuates NF-κB activation, we assessed nuclear translocation of p65 with/without pharmacological inhibitor KN93 after stimulation with anti-CD3 antibody. Nuclear extracts were isolated from anti-CD3 antibody- or TNF-α-stimulated Jurkat cells in the presence and absence of KN93 to determine whether CaMKII activity was required for p65 nuclear translocation. As shown in Fig. 2C, KN93 inhibited the nuclear translocation of p65 induced by anti-CD3 antibody. Together, these observations suggested that antigen recognition leads to polarized redistribution of CaMKII in T lymphocytes and provided us with an impetus to investigate the signaling outcomes of our computational and cell biological observations.
FIG. 2.
CaMKII redistributes to the immune synapse in an activation-dependent manner. (A) Raji cells were loaded with a fluorescent cell tracker chloromethylbenzoyl derivative of aminocoumarin (blue) and pulsed with SEE for 20 min. See Materials and Methods for details. Alexa Fluor 488 is shown in green, and phalloidin conjugated with Alexa Fluor 568 is shown in red. Representative results from one of three experiments are shown. (B) Quantification of CaMKII redistribution to the contact area in Jurkat cells. Conjugates displaying CaMKII redistribution to the submembranous area were counted under the fluorescence microscope at a magnification of ×100. Results are representative of three independent experiments. n, total number of conjugates examined for each experimental condition. (C) After incubation with KN93 or DMSO, Jurkat cells were stimulated with anti-CD3 antibody or TNF-α and then the nuclear extract was obtained as described in Materials and Methods. The nuclear extract (10 μg of total proteins) was subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and blotted with anti-p65 antibody or antiretinoblastoma (Rb) antibody.
siRNAs of CaMKII interfere with NF-κB activation induced by TCR stimulation but not by TNF-α.
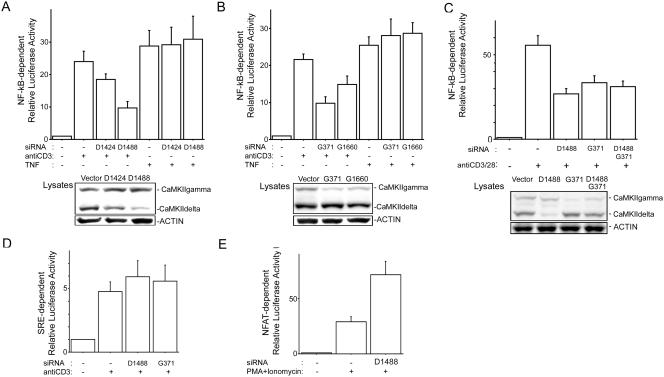
To investigate the biological implications of CaMKII in TCR-mediated NF-κB activation, we generated siRNA constructs targeting CaMKIIδ (D1424 and D1488) and CaMKIIγ (G371 and G1660) isoforms. As shown in Fig. 3A and B, the siRNA constructs D1424 and D1488 suppressed protein expression of endogenous CaMKIIδ and the siRNA constructs G371 and G1660 reduced CaMKIIγ protein expression 3 days after electroporation. We next tested the function of these siRNA constructs by use of NF-κB-dependent reporter assays with Jurkat cells. As shown in Fig. 3A and B, inhibition of CaMKIIδ and CaMKIIγ expression did not alter TNF-α-stimulated NF-κB-directed gene expression but did result in a 50% reduction of anti-CD3 antibody-stimulated NF-κB-directed gene expression. We next explored if knockdown of CaMKIIδ and CaMKIIγ attenuated anti-CD3- and anti-CD28-mediated NF-κB activation. As shown in Fig. 3C, NF-κB activation induced by costimulation was suppressed better by siRNA targeting CaMKIIδ than by siRNA targeting CaMKIIγ. However experiments to silence both CaMKIIδ and CaMKIIγ in the same cell resulted in lower levels of silencing of each gene. Consistent with this result, NF-κB attenuation in the presence of both CaMKIIδ and CaMKIIγ was intermediate compared to attenuation with single transfections of siRNA. This result is consistent with previous reports demonstrating that siRNA-mediated knockdown of multiple target genes in the same cell is titratable (24, 30).
FIG. 3.
siRNAs of CaMKII interfere with NF-κB activation induced by TCR stimulation. (A and B) Jurkat cells were electroporated with 10 μg of siRNA construct (vector, D1424, D1488, G371, or G1660), NF-κB reporter, and pRL-TK construct. Three days later, the cells were incubated with anti-CD3 antibody (1 μg/ml) or TNF-α (1 ng/ml) for 6 h, and then luciferase activities were determined as described in the text. (C) Jurkat cells were electroporated with 10 μg of siRNA construct (vector, D1488, or G371) or 5 μg of D1488 and 5 μg of G371, NF-κB reporter, and pRL-TK. Three days later, the cells were incubated with anti-CD3 antibody (1 μg/ml) or anti-CD28 antibody (0.2 μg/ml) for 6 h, and then luciferase activity was determined. (A to C, lower panels) Equal amounts of protein from lysates used for luciferase assays were blotted for CaMKII and actin. (D) Jurkat cells were electroporated with siRNA construct (vector, D1488, or G371), SRE reporter, and pRL-TK construct. Three days later, the cells were incubated with anti-CD3 antibody (1 μg/ml) for 6 h, and then SRE reporter activity was determined. (E) Jurkat cells were electroporated with siRNA construct (vector or D1488), NFAT reporter, and pRL-TK construct. Three days later, the cells were incubated with phorbol myristate acetate (PMA) (50 ng/ml) plus ionomycin (1 μM) for 6 h, and then NFAT reporter activity was determined. For results in all panels, four independent experiments were performed. Bars represent standard errors.
On the other hand, knockdown of neither CaMKIIδ or CaMKIIγ affected anti-CD3 antibody-induced activation of the SRE, which interacts with the serum response factor and Elk1, suggesting specific involvement of CaMKII in NF-κB activation (Fig. 3D). Consistent with published observations, CaMKII enhanced NFAT activation (Fig. 3E) (28). Taken together, our data demonstrate that interfering with CaMKII expression in T lymphocytes attenuates TCR-mediated NF-κB activation.
CaMKII induces synergistic NF-κB activation with CARMA1.
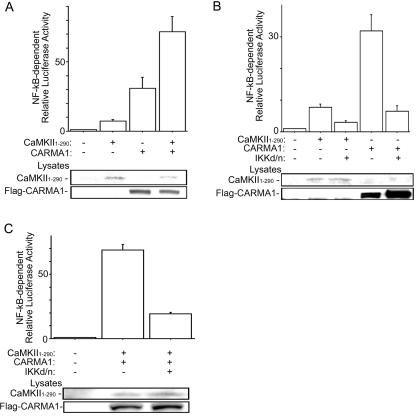
In silico observations suggested that CaMKII may be involved in the regulation of CBM assembly and NF-κB signaling in lymphocytes. To further investigate the involvement of CaMKII in NF-κB activation, we performed NF-κB-dependent reporter assays using the constitutively active fragment of CaMKIIα corresponding to the N-terminal region from residues 1 to 290 (41). The N-terminal residues 1 to 290, which are highly conserved among CaMKII isoforms, activated NF-κB in Jurkat cells and enhanced CARMA1-induced NF-κB activation (Fig. 4A and data not shown) (44). A previous report demonstrated that CaMKII is required for IKK activation induced by phorbol ester (14), suggesting that CaMKII activates NF-κB upstream of IKK. Therefore, we next examined the downstream signals involved in CaMKII-mediated NF-κB activation in T lymphocytes. As shown in Fig. 4B, we found that the dominant-negative mutant of IKKβ (with S177A and S181A mutations) inhibited NF-κB activation induced by CaMKII or CARMA1 and also inhibited activation induced by cotransfection of both CARMA1 and CaMKII (Fig. 4C). Moreover, in IKKγ-deficient Jurkat cells, CaMKII did not activate NF-κB (data not shown). These findings suggest that CaMKII activates NF-κB synergistically with CARMA1 upstream of IKK.
FIG. 4.
Synergistic NF-κB activation of CARMA1 with CaMKII upstream of IKK. (A) Jurkat cells were electroporated with NF-κB reporter, pRL-TK construct, 2 μg of CaMKII1-290, and 2 μg of Flag-CARMA1 construct. One day later, luciferase activities were determined. (B) Jurkat cells were electroporated with NF-κB reporter, pRL-TK construct, 2 μg of the IKKβ dominant-negative (d/n) mutant (with S177A and S181A mutations), and CaMKII1-290 or Flag-CARMA1. One day later, luciferase activities were determined. (C) Jurkat cells were transfected as described for panel B except that CaMKII1-290 and CARMA1 were cotransfected. Western blot analysis using cell lysates was performed with rabbit anti-CaMKII antibody or monoclonal anti-Flag antibody (lower panels). Three independent experiments were performed in duplicate. Bars represent standard errors.
CaMKII phosphorylates CARMA1 on Ser109, and the phosphorylation facilitates the interaction between CARMA1 and Bcl10.
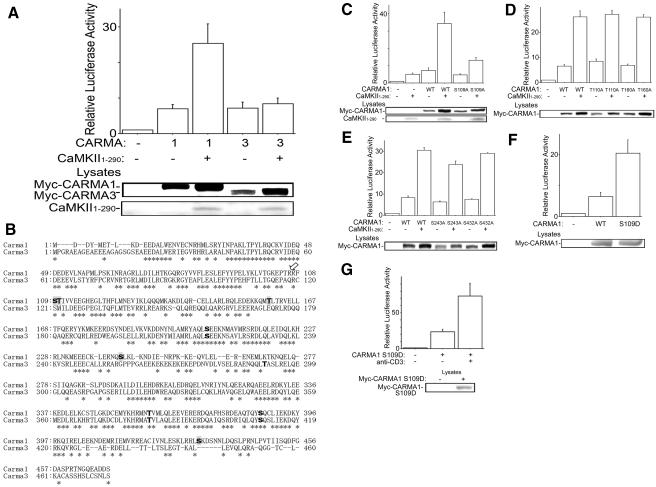
The CARMA family consists of three members, CARMA1 (CARD11), CARMA2 (CARD14), and CARMA3 (CARD10), and all CARMA proteins activate NF-κB (42). Indeed, overexpression of Myc-CARMA1 or Myc-CARMA3 induced NF-κB activation in the reporter assay. Constitutively active CaMKII elevated NF-κB activation induced by CARMA1 but not by CARMA3 (Fig. 5A), indicating that CaMKII is a specific regulator of CARMA1-induced NF-κB activation in T lymphocytes. Although cotransfection with a constitutively active CaMKII construct increased expression levels of both Myc-CARMA1 and Myc-CARMA3, the synergistic NF-κB activation was observed only between Myc-CARMA1 and CaMKII, suggesting that the elevation of CARMA1-induced NF-κB activation is not merely due to increased expression level.
FIG. 5.
Synergistic NF-κB activation of CaMKII with CARMA1 but not with CARMA3. (A) Jurkat cells were electroporated with NF-κB-dependent reporter, pRL-TK construct, 2 μg of CaMKII1-290, and 50 ng of Myc-CARMA1 or Myc-CARMA3. The total amount of DNA was equalized with empty vector. One day later, luciferase activities were determined as described in the text. (B) The Motif Scan program Scansite predicted amino acid residues phosphorylated with CaMKII in the N-terminal regions of CARMA1 and CARMA3 (boldface type). The residues predicted specifically in CARMA1 are shaded. Asterisks indicate residues conserved between CARMA1 and CARMA3. The arrow indicates the P-3 position of CARMA1 Ser109 or CARMA3 Ser121, where CaMKII prefers Arg or Lys for substrates. (C) Jurkat cells were electroporated with NF-κB reporter, pRL-TK construct, 2 μg CaMKII1-290, and 50 ng of Myc-CARMA1 or its mutant with the S109A mutation. The total amount of DNA was equalized with empty vectors. One day later, luciferase activities were determined. (D and E) Results are from an experiment similar to that described for panel C, except with Myc-CARMA1 or its mutant with the (D) T110A or T160A mutation or the (E) S243A or S432A mutation. The total amount of DNA was equalized with empty vectors. One day later, luciferase activities were determined. (F) Jurkat cells were electroporated with NF-κB reporter, pRL-TK construct, and 100 ng of Myc-CARMA1 or its mutant with the S109D mutation. One day later, luciferase activities were determined. (G) Results are from an experiment similar to that described for panel F, except that 18 h later, the cells were incubated with anti-CD3 antibody (1 μg/ml) for a further 6 h, and then luciferase activities were determined. (Lower panels) Cell lysates were blotted with anti-Myc antibody or rabbit anti-CaMKII antibody. Four independent experiments were performed. Bars represent standard errors. WT, wild type.
A previous report has indicated that the CARD and the coiled-coil domain of CARMA1 are sufficient to activate NF-κB (3). As discussed above, we found that the N-terminal fragment (amino acids [aa] 1 to 470) of CARMA1, which contains CARD and the coiled-coil domain, also activated NF-κB synergistically with CaMKII (data not shown). These findings suggested that CaMKII might phosphorylate CARMA1 within the N-terminal region (aa 1 to 470) and modulate NF-κB induced by CARMA1. Therefore, we next analyzed the protein sequence of N-terminal CARMA1 (aa 1 to 470) by using the Motif Scan program Scansite (29), which is provided on the website http://scansite.mit.edu/, to predict the amino acid residues phosphorylated by CaMKII. Scansite predicted Ser109, Thr110, Thr160, Ser205, Ser243, Thr360, Ser387, and Ser432 within the N-terminal region (aa 1 to 470) of CARMA1 and Ser217, Thr291, Thr383, and Ser410 within that (aa 1 to 474) of CARMA3 as phosphorylation residues with CaMKII (Fig. 5B). Ser205, Thr360, and Ser387 of CARMA1 are conserved in CARMA3 (Fig. 5B) and are predicted as phosphorylation residues. Ser109 of CARMA1 is also conserved in CARMA3 (Ser121); however, it was not identified as a CaMKII phosphorylation residue. Previous studies have indicated that CaMKII prefers a basic residue (Arg or Lys) at position P-3 (P-0 is the phosphorylated Ser/Thr residue) for substrates (13). The position P-3 of Ser109 of CARMA1 is Arg, while that of Ser121 of CARMA3 is Gln (Fig. 5B). Thus, we decided to see if the synergistic NF-κB activation with CaMKII is impaired with mutations of the amino acid residues predicted as phosphorylation sites specifically in the N-terminal region of CARMA1: Ser109, Thr110, Thr160, Ser243, and Ser432 (Fig. 5B). Site-directed mutants (with Ser-Ala or Thr-Ala mutations) of the five residues were created, and expression in Jurkat cells was confirmed. The S109A mutation impaired the synergistic NF-κB activation with CaMKII (Fig. 5C), while the T110A, T160A, S243A, and S432A mutations did not alter the NF-κB reporter activity (Fig. 5D and E). Furthermore, NF-κB activation induced by CARMA1 with the S109D mutation was enhanced approximately fourfold compared to that induced by the wild-type protein; this mutation mimics the phosphorylation on Ser109 (Fig. 5F and G). These findings suggest that CaMKII may modulate NF-κB induced by CARMA1 via phosphorylation on Ser109.
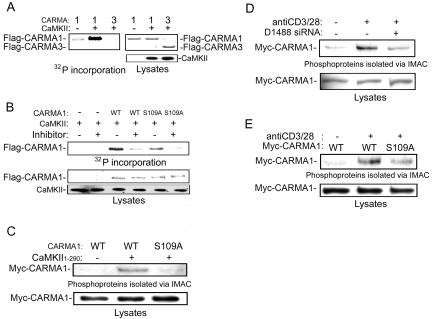
Next, we examined whether CARMA1 is phosphorylated on Ser109 by CaMKII in vitro. To this end, purified CARMA1, CARMA3, and CARMA1 with the S109A mutation were phosphorylated in vitro by CaMKII in the presence or absence of CaMKII inhibitor. We found that 32P was incorporated into CARMA1 with CaMKII in the absence of CaMKII inhibitor and that the incorporation was reduced by the S109A mutation (Fig. 6A and B), suggesting that CaMKII phosphorylates CARMA1 and that Ser109 is one of the phosphorylation sites. In contrast, CARMA3 was not phosphorylated in vitro by CaMKII under similar conditions and serves as a specificity control.
FIG. 6.
Ser109 phosphorylation of CARMA1 with CaMKII. (A) Purified CARMA1 and CARMA3 were phosphorylated in vitro by wild-type CaMKII. (B) Purified CARMA1 and the CARMA1 S109A mutant were phosphorylated in vitro in the presence and absence of a CaMKII inhibitor as indicated. The amount of CARMA1 was determined by Coomassie staining, and then the dried gels were exposed to a phosphorimager. The remainder of each sample was analyzed by Western blotting using an antibody against CaMKII (lower panel). (C) 293T cells were transfected with Myc-CARMA1 or its S109A mutant and CaMKII1-290. The cells were lysed 18 h later. Monoclonal anti-Myc antibody was used for blotting. (D) Jurkat cells were electroporated with siRNA construct (vector or D1488) and Myc-CARMA1 construct. Three days later, the cells were incubated with anti-CD3 antibody (5 μg/ml) and anti-CD28 antibody (1 μg/ml) for 10 min and then lysed. (E) Jurkat cells were electroporated with Myc-CARMA1 or its S109A mutant. One day later, the cells were incubated with anti-CD3 antibody and anti-CD28 antibody for 10 min and then lysed. Phosphoproteins were isolated via IMAC as described in the text. Lower panels show total lysates immunoblotted with anti-Myc antibody. The experiments were performed three times, and representative results are presented. WT, wild type.
As an additional measure to verify that CARMA1 is phosphorylated on Ser109 by CaMKII, we isolated phosphoproteins by using IMAC with 293T cells transfected with constructs of Myc-CARMA1 (1). Wild-type Myc-CARMA1 was isolated in phosphoproteins with cotransfection of CaMKII but the S109A mutant was not (Fig. 6C), indicating that CaMKII phosphorylates CARMA1 on Ser109. Moreover, in Jurkat cells, Myc-CARMA1 was isolated in phosphoproteins with TCR stimulation (Fig. 6D). The amount of phosphorylated Myc-CARMA1 was reduced with cotransfection of the CaMKII siRNA (D1488) construct, while the cotransfection did not affect expression levels of Myc-CARMA1 (Fig. 6D). We next tested whether the S109A mutation altered CARMA1 phosphorylation following anti-CD3 and anti-CD28 stimulation. As shown in Fig. 6E, phosphorylation of CARMA1 with the S109A mutation was reduced compared to that of wild-type CARMA1 following costimulation. These findings suggest that TCR stimulation induces CARMA1 phosphorylation and that CaMKII is involved in the phosphorylation.
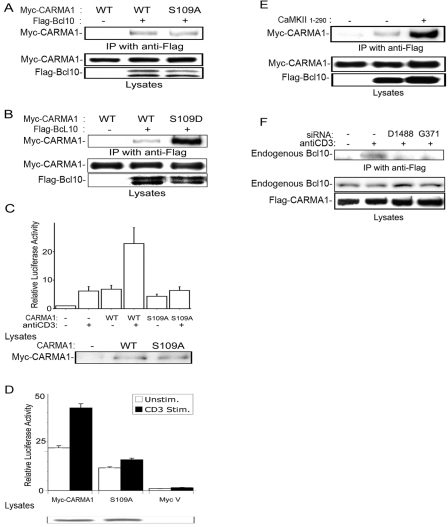
It is well known that CARMA1 activates NF-κB via binding to Bcl10 (42). Thus, we speculated that the phosphorylation on Ser109 of CARMA1 could affect the binding between CARMA1 and Bcl10. The Flag-Bcl10 construct was transfected into cells with Myc-CARMA1 or Ser109 mutant constructs, and then Flag-Bcl10 was immunoprecipitated with anti-Flag antibody. Western blots were probed for Myc-CARMA1. Although the S109A mutation of CARMA1 reduces the binding to Bcl10 only slightly (Fig. 7A), the S109D mutation increased interaction dramatically (Fig. 7B). Consistent with these coimmunoprecipitation findings, the S109A mutation of CARMA1 still activated NF-κB, while the synergistic NF-κB activation with CaMKII or anti-CD3 stimulation was impaired (Fig. 7C and 5C). To further support our observations, we examined the effects of reexpression of CARMA1 and the CARMA1 S109A mutant in CARMA1-deficient Jurkat cells. As shown in Fig. 7D, expression of wild-type CARMA1 restored TCR-mediated NF-κB activation whereas expression of the S109A mutant failed to rescue anti-CD3-mediated NF-κB activation.
FIG. 7.
Regulation of binding between CARMA1 and Bcl10 via Ser109 phosphorylation. (A and B) 293T cells were transfected with 2 μg of Flag-Bcl10 construct and 0.5 μg of Myc-CARMA1 or its S109A mutant (A) or its S109D mutant (B). The total amount of DNA was equalized with empty vectors. The cells were lysed 18 h later. Monoclonal anti-Flag antibody was used for immunoprecipitation (IP). Monoclonal anti-Myc antibody or anti-Flag antibody was used for blotting. Results are representative of three independent experiments. (C and D) Jurkat cells (C) or CARMA1 mutant Jurkat cells (D) were electroporated with NF-κB reporter, pRL-TK construct, and Myc-CARMA1 or its S109A mutant. Eighteen hours later, the cells were incubated with anti-CD3 antibody (1 μg/ml) for a further 6 h, and NF-κB luciferase activities were determined. Four independent experiments were performed. Bars represent standard errors. (E) 293T cells were transfected with 1 μg of Flag-Bcl10, 0.5 μg of Myc-CARMA1, and 2 μg of CaMKII1-290. The total amount of DNA was equalized with empty vectors. The cells were lysed 18 h later. Monoclonal anti-Flag antibody was used for IP. Monoclonal anti-Myc antibody or anti-Flag antibody was used for blotting. (F) Jurkat cells were electroporated with 10 μg of siRNA D1488 or G371 constructs or vector and 5 μg of Flag-CARMA1 construct. Three days later, the cells were incubated with anti-CD3 antibody (5 μg/ml) for 10 min, lysed, and immunoprecipitated with anti-Flag antibody. Total lysates were immunoblotted with anti-Flag or anti-Bcl10 antibody. Results are representative of three independent experiments. WT, wild type; Unstim., unstimulated; Stim., stimulated.
We next examined whether cotransfection of the catalytic domain of CaMKII could enhance wild-type CARMA1 and Bcl10 interaction in 293T cells. Notably, we observed that cotransfection of CaMKII enhanced binding, further validating our results (Fig. 7E). Finally, we determined whether the interaction between endogenous Bcl10 and Flag-CARMA1 in Jurkat cells was disrupted in the presence of RNA interference knockdown of CaMKIIγ or CaMKIIδ. TCR stimulation induced the binding of endogenous Bcl10 to Flag-CARMA1, and the binding was impaired with knockdown of endogenous CaMKII (Fig. 7F). Together, these studies imply that CaMKII is involved in the TCR stimulation-induced interaction between CARMA1 and Bcl10.
DISCUSSION
Our bioinformatics analysis of the microarray expression data allowed us to identify a group of candidate immunologically expressed kinases which could regulate CARMA1 signaling based on their correlated expression with CARMA1, Bcl10, and MALT1. Further analysis suggested that among the 13 kinases with the highest coregulated expression with these signaling effectors, there were consensus CaMKII phosphorylation sites in CARMA1. This approach permitted us to narrow the candidate list of potential CARMA1 kinases that may play a role in TCR-mediated NF-κB activation. Overall, similar approaches may provide a way to harness expression data sets to identify kinases/phosphatases important for immune-cell-specific signaling pathways. In addition, one can also envision the value of interrogating kinase gene expression signatures to identify novel pathway members of Jun N-terminal protein kinase and NFAT activation in lymphocyte signaling and development.
We have elucidated a novel role for CaMKII in TCR-induced NF-κB. Our data show that upon TCR engagement activated CaMKII is redistributed in T cells to the contact area with antigen-presenting cells. Several studies have shown that translocation of CaMKII is an important mechanism for its function at synaptic interfaces for both acute and sustained signaling events (13, 45). It is well known that in neuronal synapses CaMKII is a major constituent of the postsynaptic density and is involved in long-term potentiation leading to learning and memory. Furthermore, a recent report has elegantly demonstrated that CaMKII localization at lipid microdomains enhances substrate phosphorylation and signaling outputs (45). Further studies are required to determine the specific functions of directed movement of CaMKII in T-cell activation. However, it is attractive to speculate that positioning of CaMKII in close proximity to the CBM complex permits access, increases local concentration of the kinase, and limits substrate phosphorylation.
To establish a functional link for CaMKII in T cells, biochemical analysis and reporter assays further revealed that the loss of CaMKII function either by direct pharmacological inhibition of the kinase or by knockdown of the gene attenuated NF-κB activation induced by TCR stimulation. We show here that coexpression of CaMKII synergistically enhances CARMA1-mediated NF-κB activation. In contrast, CaMKII does not enhance CARMA3-mediated NF-κB activation. Only the S109A mutation of CARMA1 impaired the synergistic NF-κB activation with CaMKII, while the S109D mutation mimicking phosphorylation enhanced Bcl10 binding and elevated NF-κB activation. These data provide molecular details of linking CaMKII to NF-κB activation in T cells.
In addition to CaMKII, kinases PKCθ and PDK1 have been implicated in CARMA1 regulation in T cells (19, 23, 36). Together, these observations imply that sequential phosphorylation events regulate CARMA1 activation and interaction with its client proteins in lymphocytes. The recently described model suggests that PKCθ/PKCβ binding to CARMA1 and the ensuing serine phosphorylation within the linker region convert CARMA1 into an active molecule, providing access to the CARD. Our results suggest that CaMKII phosphorylation on CARMA1 serine 109 mediates Bcl10 binding and regulates NF-κB activation. Alternatively, PKCθ may function primarily in the initiation of CARMA1 activation, and subsequent phosphorylation events may be necessary to maintain the active conformation.
Intracellular communication can be fine-tuned by calcium-dependent enzymes at several levels through the modulation of distinct signaling pathways. Another role ascribed to CaMKII is that it serves as a calcium sensor. Previous studies have demonstrated that an increase in Ca2+ positively regulates the magnitude of NF-κB activation. In T cells, TCR engagement elevates Ca2+ concentration via inositol-1,4,5-triphosphate production and activates PKCθ via diacylglycerol (20). PKCθ can phosphorylate CaMKII at Thr286, leading to calmodulin-independent activation (49). Future experiments are needed to determine the exact time scale of this multiple-kinase-mediated regulation of CARMA1 in NF-κB activation. Finally, these findings highlight the power of global gene expression profiling to uncover biological pathways regulating scaffold protein assembly in lymphocyte signaling.
Supplementary Material
Acknowledgments
This work was supported by grants from CCFA, DK43351, and Career Development Funds to R.J.X., and K.I. was supported by the Fellowship of Uehara Memorial Foundation.
We thank Brian Seed, Joe Avruch, Adrian Ting, John Rioux, and Ian Rosenberg for discussions and/or review of the manuscript. We thank Xin Lin, Dan Billadeau, Michael Karin, Richard Maurer, Adrian Ting, and Jürg Tschopp for generous provision of research materials.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersson, L., and J. Porath. 1986. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 154:250-254. [DOI] [PubMed] [Google Scholar]
- 2.Bauch, A., K. S. Campbell, and M. Reth. 1998. Interaction of the CD5 cytoplasmic domain with the Ca2+/calmodulin-dependent kinase IIdelta. Eur. J. Immunol. 28:2167-2177. [DOI] [PubMed] [Google Scholar]
- 3.Bertin, J., L. Wang, Y. Guo, M. D. Jacobson, J. L. Poyet, S. M. Srinivasula, S. Merriam, P. S. DiStefano, and E. S. Alnemri. 2001. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J. Biol. Chem. 276:11877-11882. [DOI] [PubMed] [Google Scholar]
- 4.Bui, J. D., S. Calbo, K. Hayden-Martinez, L. P. Kane, P. Gardner, and S. M. Hedrick. 2000. A role for CaMKII in T cell memory. Cell 100:457-467. [DOI] [PubMed] [Google Scholar]
- 5.Caenepeel, S., G. Charydczak, S. Sudarsanam, T. Hunter, and G. Manning. 2004. The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc. Natl. Acad. Sci. USA 101:11707-11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Che, T., Y. You, D. Wang, M. J. Tanner, V. M. Dixit, and X. Lin. 2004. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-kappaB activation. J. Biol. Chem. 279:15870-15876. [DOI] [PubMed] [Google Scholar]
- 7.Dolmetsch, R. E., R. S. Lewis, C. C. Goodnow, and J. I. Healy. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386:855-858. [DOI] [PubMed] [Google Scholar]
- 8.Dustin, M. L., and J. A. Cooper. 2000. The immunological synapse and the actin cytoskeleton: molecular hardware for T cell signaling. Nat. Immunol. 1:23-29. [DOI] [PubMed] [Google Scholar]
- 9.Egawa, T., B. Albrecht, B. Favier, M. J. Sunshine, K. Mirchandani, W. O'Brien, M. Thome, and D. R. Littman. 2003. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr. Biol. 13:1252-1258. [DOI] [PubMed] [Google Scholar]
- 10.Gaide, O., B. Favier, D. F. Legler, D. Bonnet, B. Brissoni, S. Valitutti, C. Bron, J. Tschopp, and M. Thome. 2002. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat. Immunol. 3:836-843. [DOI] [PubMed] [Google Scholar]
- 11.Hara, H., C. Bakal, T. Wada, D. Bouchard, R. Rottapel, T. Saito, and J. M. Penninger. 2004. The molecular adapter Carma1 controls entry of IkappaB kinase into the central immune synapse. J. Exp. Med. 200:1167-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara, H., T. Wada, C. Bakal, I. Kozieradzki, S. Suzuki, N. Suzuki, M. Nghiem, E. K. Griffiths, C. Krawczyk, B. Bauer, F. D'Acquisto, S. Ghosh, W. C. Yeh, G. Baier, R. Rottapel, and J. M. Penninger. 2003. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18:763-775. [DOI] [PubMed] [Google Scholar]
- 13.Hudmon, A., and H. Schulman. 2002. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 71:473-510. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, K., S. Edin, A. Antonsson, and T. Grundstrom. 2001. Calmodulin-dependent kinase II mediates T cell receptor/CD3- and phorbol ester-induced activation of IkappaB kinase. J. Biol. Chem. 276:36008-36013. [DOI] [PubMed] [Google Scholar]
- 15.Ishiguro, K., and R. Xavier. 2004. Homer-3 regulates activation of serum response element in T cells via its EVH1 domain. Blood 103:2248-2256. [DOI] [PubMed] [Google Scholar]
- 16.Jang, M. K., Y. H. Goo, Y. C. Sohn, Y. S. Kim, S. K. Lee, H. Kang, J. Cheong, and J. W. Lee. 2001. Ca2+/calmodulin-dependent protein kinase IV stimulates nuclear factor-kappa B transactivation via phosphorylation of the p65 subunit. J. Biol. Chem. 276:20005-20010. [DOI] [PubMed] [Google Scholar]
- 17.Jun, J. E., L. E. Wilson, C. G. Vinuesa, S. Lesage, M. Blery, L. A. Miosge, M. C. Cook, E. M. Kucharska, H. Hara, J. M. Penninger, H. Domashenz, N. A. Hong, R. J. Glynne, K. A. Nelms, and C. C. Goodnow. 2003. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18:751-762. [DOI] [PubMed] [Google Scholar]
- 18.Lamb, J., S. Ramaswamy, H. L. Ford, B. Contreras, R. V. Martinez, F. S. Kittrell, C. A. Zahnow, N. Patterson, T. R. Golub, and M. E. Ewen. 2003. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell 114:323-334. [DOI] [PubMed] [Google Scholar]
- 19.Lee, K. Y., F. D'Acquisto, M. S. Hayden, J. H. Shim, and S. Ghosh. 2005. PDK1 nucleates T cell receptor-induced signaling complex for NF-kappaB activation. Science 308:114-118. [DOI] [PubMed] [Google Scholar]
- 20.Li, Z. W., R. C. Rickert, and M. Karin. 2004. Genetic dissection of antigen receptor induced-NF-kappaB activation. Mol. Immunol. 41:701-714. [DOI] [PubMed] [Google Scholar]
- 21.Lucas, P. C., M. Yonezumi, N. Inohara, L. M. McAllister-Lucas, M. E. Abazeed, F. F. Chen, S. Yamaoka, M. Seto, and G. Nunez. 2001. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J. Biol. Chem. 276:19012-19019. [DOI] [PubMed] [Google Scholar]
- 22.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, R., D. Wang, M. Blonska, H. Li, M. Kobayashi, B. Pappu, Y. Chen, and X. Lin. 2005. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-kappaB activation. Immunity 23:575-585. [DOI] [PubMed] [Google Scholar]
- 24.McManus, M. T., B. B. Haines, C. P. Dillon, C. E. Whitehurst, L. van Parijs, J. Chen, and P. A. Sharp. 2002. Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol. 169:5754-5760. [DOI] [PubMed] [Google Scholar]
- 25.Meffert, M. K., J. M. Chang, B. J. Wiltgen, M. S. Fanselow, and D. Baltimore. 2003. NF-kappa B functions in synaptic signaling and behavior. Nat. Neurosci. 6:1072-1078. [DOI] [PubMed] [Google Scholar]
- 26.Mootha, V. K., P. Lepage, K. Miller, J. Bunkenborg, M. Reich, M. Hjerrild, T. Delmonte, A. Villeneuve, R. Sladek, F. Xu, G. A. Mitchell, C. Morin, M. Mann, T. J. Hudson, B. Robinson, J. D. Rioux, and E. S. Lander. 2003. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. USA 100:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton, K., and V. M. Dixit. 2003. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 13:1247-1251. [DOI] [PubMed] [Google Scholar]
- 28.Nghiem, P., T. Ollick, P. Gardner, and H. Schulman. 1994. Interleukin-2 transcriptional block by multifunctional Ca2+/calmodulin kinase. Nature 371:347-350. [DOI] [PubMed] [Google Scholar]
- 29.Obenauer, J. C., L. C. Cantley, and M. B. Yaffe. 2003. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31:3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parrish, S., J. Fleenor, S. Xu, C. Mello, and A. Fire. 2000. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell 6:1077-1087. [DOI] [PubMed] [Google Scholar]
- 31.Pomerantz, J. L., E. M. Denny, and D. Baltimore. 2002. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. EMBO J. 21:5184-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruefli-Brasse, A. A., D. M. French, and V. M. Dixit. 2003. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science 302:1581-1584. [DOI] [PubMed] [Google Scholar]
- 33.Ruland, J., G. S. Duncan, A. Elia, I. del Barco Barrantes, L. Nguyen, S. Plyte, D. G. Millar, D. Bouchard, A. Wakeham, P. S. Ohashi, and T. W. Mak. 2001. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 104:33-42. [DOI] [PubMed] [Google Scholar]
- 34.Ruland, J., G. S. Duncan, A. Wakeham, and T. W. Mak. 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19:749-758. [DOI] [PubMed] [Google Scholar]
- 35.Sancho, D., M. C. Montoya, A. Monjas, M. Gordon-Alonso, T. Katagiri, D. Gil, R. Tejedor, B. Alarcon, and F. Sanchez-Madrid. 2002. TCR engagement induces proline-rich tyrosine kinase-2 (Pyk2) translocation to the T cell-APC interface independently of Pyk2 activity and in an immunoreceptor tyrosine-based activation motif-mediated fashion. J. Immunol. 169:292-300. [DOI] [PubMed] [Google Scholar]
- 36.Sommer, K., B. Guo, J. L. Pomerantz, A. D. Bandaranayake, M. E. Moreno-Garcia, Y. L. Ovechkina, and D. J. Rawlings. 2005. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity 23:561-574. [DOI] [PubMed] [Google Scholar]
- 37.Stuart, J. M., E. Segal, D. Koller, and S. K. Kim. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302:249-255. [DOI] [PubMed] [Google Scholar]
- 38.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumi, M., K. Kiuchi, T. Ishikawa, A. Ishii, M. Hagiwara, T. Nagatsu, and H. Hidaka. 1991. The newly synthesized selective Ca2+/calmodulin dependent protein kinase II inhibitor KN-93 reduces dopamine contents in PC12h cells. Biochem. Biophys. Res. Commun. 181:968-975. [DOI] [PubMed] [Google Scholar]
- 40.Sun, L., L. Deng, C. K. Ea, Z. P. Xia, and Z. J. Chen. 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14:289-301. [DOI] [PubMed] [Google Scholar]
- 41.Sun, P., H. Enslen, P. S. Myung, and R. A. Maurer. 1994. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8:2527-2539. [DOI] [PubMed] [Google Scholar]
- 42.Thome, M. 2004. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat. Rev. Immunol. 4:348-359. [DOI] [PubMed] [Google Scholar]
- 43.Ting, A. T., F. X. Pimentel-Muinos, and B. Seed. 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 15:6189-6196. [PMC free article] [PubMed] [Google Scholar]
- 44.Tombes, R. M., M. O. Faison, and J. M. Turbeville. 2003. Organization and evolution of multifunctional Ca2+/CaM-dependent protein kinase genes. Gene 322:17-31. [DOI] [PubMed] [Google Scholar]
- 45.Tsui, J., M. Inagaki, and H. Schulman. 2005. Calcium/calmodulin-dependent protein kinase II (CaMKII) localization acts in concert with substrate targeting to create spatial restriction for phosphorylation. J. Biol. Chem. 280:9210-9216. [DOI] [PubMed] [Google Scholar]
- 46.Uren, A. G., K. O'Rourke, L. A. Aravind, M. T. Pisabarro, S. Seshagiri, E. V. Koonin, and V. M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6:961-967. [DOI] [PubMed] [Google Scholar]
- 47.Wang, D., R. Matsumoto, Y. You, T. Che, X.-Y. Lin, S. L. Gaffen, and X. Lin. 2004. CD3/CD28 costimulation-induced NF-κB activation is mediated by recruitment of protein kinase C-θ, Bcl10, and IκB kinase β to the immunological synapse through CARMA1. Mol. Cell. Biol. 24:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, D., Y. You, S. M. Case, L. M. McAllister-Lucas, L. Wang, P. S. DiStefano, G. Nunez, J. Bertin, and X. Lin. 2002. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat. Immunol. 3:830-835. [DOI] [PubMed] [Google Scholar]
- 49.Waxham, M. N., and J. Aronowski. 1993. Ca2+/calmodulin-dependent protein kinase II is phosphorylated by protein kinase C in vitro. Biochemistry 32:2923-2930. [DOI] [PubMed] [Google Scholar]
- 50.Weil, R., and A. Israël. 2006. Deciphering the pathway from the TCR to NF-κB. Cell Death Differ. 13:826-833. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, H., M. Q. Du, and V. M. Dixit. 2005. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell 7:425-431. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, H., I. Wertz, K. O'Rourke, M. Ultsch, S. Seshagiri, M. Eby, W. Xiao, and V. M. Dixit. 2004. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427:167-171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.