Abstract
BAL1 is a transcription modulator that is overexpressed in chemoresistant, diffuse large B-cell lymphomas (DLBCLs). BAL1 complexes with a recently described DELTEX family member termed BBAP. Herein, we characterized BAL1 and BBAP expression in primary DLBCL subtypes defined by their comprehensive transcriptional profiles. BAL1 and BBAP were most abundant in lymphomas with a brisk host inflammatory response, designated host response (HR) tumors. Although these DLBCLs include significant numbers of tumor-infiltrating lymphocytes and interdigitating dendritic cells, BAL1 and BBAP were expressed primarily by malignant B cells, prompting speculation that the genes might be induced by host-derived inflammatory mediators such as gamma interferon (IFN-γ). In fact, IFN-γ induced BAL1 and BBAP expression in DLBCL cell lines; doxycycline-induced BAL1 also increased the expression of multiple IFN-stimulated genes, directly implicating BAL1 in an IFN signaling pathway. We show that BAL1 and BBAP are located on chromosome 3q21 in a head-to-head orientation and are regulated by a IFN-γ-responsive bidirectional promoter. BBAP regulates the subcellular localization of BAL1 by a dynamic shuttling mechanism, highlighting the functional requirement for coordinated BBAP and BAL1 expression. IFN-γ-induced BAL1/BBAP expression contributes to the molecular signature of HR DLBCLs and highlights the interplay between the inflammatory infiltrate and malignant B cells in these tumors.
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults. Although approximately 50 to 55% of DLBCL patients can be cured with modern therapy, the remaining patients succumb to their disease (1). In a broad-based screen for genes and pathways associated with inferior DLBCL outcome, we previously identified a novel, risk-related gene, termed BAL1 (B-aggressive lymphoma 1). In pilot and confirmatory series of DLBCLs, BAL1 expression was significantly higher in chemoresistant tumors (3, 4).
BAL1 encodes a nuclear protein with C-terminal similarities to the catalytic domain of Tankyrase and polyADP ribose polymerase (PARP) and a duplicated N-terminal sequence homologous to the nonhistone region of histone macro-H2A termed the macro domain. Recent studies indicate that BAL1 is a member of a larger gene family with C-terminal PARP homology and multiple N-terminal macro domains (3). Several lines of evidence implicate macro domain-containing proteins in the regulation of transcription, including structural similarities to DNA binding domains and interference with transcriptional factor binding in a positioned nucleosome (3, 6, 9, 28). Consistent with these observations, we recently demonstrated that the BAL1 full-length protein and N-terminal macro domain modulate transcription when tethered to a promoter (3).
Of interest, BAL1 complexes with a recently described DELTEX family member and E3 ubiquitin ligase termed BBAP (B-aggressive lymphoma and BAL1 binding partner) (34). Both genes are localized to a 46-kb region on chromosome 3q, prompting speculation regarding coordinated regulation. Despite the clear link between BAL1 overexpression and poor outcome in DLBCL, the signals modulating the expression of BAL1 and its binding partner, BBAP, in high-risk tumors remain unknown.
Recent studies indicate that discrete subsets of DLBCLs can be identified by their unique transcriptional profiles and associated clinical and genetic features—oxidative phosphorylation (OXP), B-cell receptor/proliferation (BCR), and host response (HR) tumors (27). HR tumors have a brisk host immune/inflammatory response and increased expression of T/natural killer (NK) cell receptor and activation pathway components, complement cascade members, and macrophage/dendritic cell markers. Consistent with this signature, primary HR DLBCLs contain significantly higher numbers of morphologically distinct tumor-infiltrating lymphocytes and interdigitating dendritic cells (27). The T-cell and dendritic cell infiltrates in HR tumors resemble those of a histologically defined provisional WHO subtype of DLBCL, T-cell/histiocyte-rich B-cell lymphoma (T/HRBCL). Similar to T/HRBCLs, HR tumors have fewer known genetic abnormalities than non-HR tumors and occur in younger patients, who often present with splenomegaly and bone marrow involvement (27). In spite of the brisk inflammatory infiltrates in HR (and T/HRBCL) tumors, patients with these DLBCLs do not have a more favorable outcome.
Primary HR tumors have increased expression of multiple inflammatory mediators and downstream targets, including IFN-γ and IFN-γ-induced transcripts. IFN-γ is typically secreted by activated tumor-infiltrating cytotoxic T lymphocytes and NK and NK/T cells. IFN-γ signaling leads to the activation of JAK1 and -2 kinases and subsequent phosphorylation of STAT-1 (signal transducer and activator of transcription 1). Following homodimerization, STAT-1 translocates to the nucleus and induces the transcription of IFN-stimulated genes (ISG) with STAT-1 binding sites in their promoters. A major STAT-1 target is IRF-1 (IFN regulatory factor 1), a transcription factor that activates additional secondary IFN-γ target genes with IRF-1 binding sites. Recent studies suggest that IFN-γ modulates the host response to tumors in two different ways. Although the cytokine aids in the protection of hosts against tumor development (immunosurveillance), IFN-γ also promotes the outgrowth of tumors with reduced immunogenicity (immunoediting) (11, 13, 15, 16, 26, 31-33, 35).
Herein, we demonstrate that BAL1 and BBAP are highly expressed in HR DLBCLs with a brisk inflammatory infiltrate and IFN-γ signature. In addition, BAL1 and BBAP are coordinately regulated by a single IFN-γ-responsive bidirectional promoter that includes canonical IRF and STAT binding sites. Highlighting the importance of BAL1 and BBAP coregulation, we find that BBAP regulates the subcellular localization of BAL1 by a dynamic shuttling mechanism.
MATERIALS AND METHODS
Generation of BAL1 Tet-On inducible clones.
The BAL1-inducible lymphoma cell line was generated using a Tet-On gene expression system (BD Biosciences Clontech, Palo Alto, CA). Briefly, the FLAG-tagged BAL1 open reading frame was generated by PCR amplification and ligated into the pTRE2 vector. An aggressive B-cell lymphoma cell line (BJAB) was transfected with the pTET-On regulatory plasmid and selected by neomycin resistance. pTET-On-positive clones were screened for low background and high inducibility with pTRE2-Luc. Selected clones were supertransfected with pTRE2-FLAG.BAL1 or an empty pTRE2 vector and selected by puromycin resistance. Thereafter, cells were treated with doxycycline at a final concentration of 1 μg/ml (BD Biosciences Clontech). After 3 to 48 h of doxycycline treatment, candidate clones were lysed, size fractionated, and immunoblotted with anti-FLAG (Sigma-Aldrich, St. Louis, MO) as described below. Two clones with low baseline BAL1 background expression and high BAL1 inducibility (clones 1 and 2 [C1 and C2]) and one empty vector control clone (Vector) were selected for further experiments. All clones were maintained in RPMI 1640 medium supplemented with 10% tetracycline (Tet)-approved fetal calf serum (BD Biosciences Clontech), 10 mM HEPES buffer, 4 mM l-glutamine, 50 U/ml penicillin, 50 U/ml streptomycin, 1.5 mg/ml G418, and 0.75 μg/ml puromycin.
Microarray analysis of BAL1 inducible clones.
To eliminate variability in gene expression caused by doxycycline and/or selection drugs, BAL1-inducible clones and empty vector controls were maintained in culture medium containing the same concentrations of selection drugs (puromycin and neomycin). For the microarray experiments, BAL1-inducible clones and the empty vector control clone were treated with doxycycline for 0 to 48 h. Thereafter, total RNA was extracted, processed, hybridized to U133A and U133B Affymetrix oligonucleotide microarrays, and scanned as previously described (27). All experiments were performed in duplicate. After replicate arrays were pooled, the top 4,112 genes meeting variation index criteria (0.3 < standard deviation/mean < 10) (24) were selected for further analysis. Genes that were induced or repressed following BAL1 induction were selected by two-way analysis of variance (ANOVA) and rank correlation test with a P value cutoff of <0.001. All analyses were done with the dChip v1.3 program (24). Selected transcripts (IRF-7 and STAT-1) were validated by quantitative reverse transcriptase PCR (RT-PCR) using predesigned gene expression assays (catalog no. Hs00185375_m1 and Hs00234829_m1; Applied Biosystems, Foster City, CA), the Applied Biosystems 7500 real-time PCR system, and the manufacturer's recommended relative quantification method (with GAPDH [glyceraldehyde-3-phosphate dehydrogenase] as the reference).
Primary DLBCL microarray data and cluster designation.
RNA microarray data from 176 primary DLBCLs was obtained as previously described (27). Based on their gene expression profiles, tumors were assigned to comprehensive clusters (OXP, BCR/proliferation, or HR) and cell-of-origin categories (Germinal Center, activated B, “other”) (27, 37).
Quantitative PCR-TaqMan analysis of BAL1 and BBAP transcripts.
RNAs from 106 (51 BCR, 28 OXP, and 27 HR tumors) of the above-mentioned primary DLBCLs (27) were used to evaluate BAL1 and BBAP transcript abundance by quantitative TaqMan PCR. In brief, cDNAs were transcribed from total RNA (27) using Superscript II reverse transcriptase (Invitrogen). The transcribed cDNA was diluted in water to a volume of 200 μl; 5 μl of this diluted template was used in each 20-μl reaction mixture. All reactions were carried out in an ABI 7700 thermal cycler using TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA). Each BAL1 or BBAP reaction mixture contained a 100 nM concentration of the TaqMan probe and 200 nM concentrations of upstream and downstream primers (probe and primer sequences are available upon request) in a total reaction volume of 20 μl. The thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of a denaturing step at 95°C for 15 seconds and an extension step at 60°C for 1 min. Reactions were performed in triplicate for each primer and probe set. PCR was also performed with a serial dilution curve of plasmid DNA containing the PCR target sequence. Cycle threshold (CT) values were generated using Sequence Detection Software, version 1.7 (Applied Biosystems), and the values from triplicate reactions were averaged. CT values from the serial dilution were plotted against log (number of plasmid copies) to generate standard curves for each primer and probe set. With these curves, tumor specimen CT values were used to calculate the amount of template present for each transcript. The level of gene expression of each target was calculated relative to that of the housekeeping gene, cyclophilin A (GenBank accession no. NM_021130). Multiple comparisons of median expression values were done by Kruskal-Wallis ANOVA. All statistical analyses were done using Statistica 6.0 software (Statistica, Tulsa, OK).
Analysis of BAL1 and BBAP induction in IFN-γ-treated lymphoma cell lines by immunoblotting.
Lymphoma cell lines (DHL4 and DHL10) were maintained as previously described (4) After 2 to 48 h of IFN-γ treatment (10 to 1,000 U/ml; Roche Applied Science, Indianapolis, IN), cells were lysed in buffer containing 150 mM NaCl, 50 mM Tris (pH 7.4), 1% Nonidet P-40, and protease inhibitor mixture (Complete; Roche Applied Science). Whole-cell lysate protein concentrations were determined by Bradford assay, and equal amounts of protein were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) sample buffer, heated at 95°C, size fractionated by sodium dodecyl sulfate-PAGE, and transferred to polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA). Membranes were immunostained with the appropriate primary antibody (anti-BAL1 [4] or anti-BBAP [34]) and horseradish peroxidase-conjugated mouse anti-rabbit or goat anti-mouse secondary antibody (Amersham Biosciences, Piscataway, NJ) and developed using a chemiluminescence method (ECL; Amersham Biosciences).
Analysis of the BAL1/BBAP promoter and generation of BAL1/BBAP promoter constructs.
Computational analysis of the BAL1/BBAP promoter was performed with publicly available programs: Genomatix (http://www.genomatix.de) (30), FirstEF (http://rulai.cshl.org/tools/FirstEF) (10), GrailEXP (http://compbio.ornl.gov/grailexp) (38), rVISTA (http://rvista.dcode.org/) (25), and CONSITE (http://mordor.cgb.ki.se/cgi-bin/CONSITE/consite) (29). To generate a series of BAL1 promoter reporter constructs, fragments spanning nucleotides −2452 to +458, −2020 to +458, −1559 to +458, −544 to +458, −109 to +458, −2452 to +58, and −198 to +58 from the previously identified BAL1 transcription start site (TSS) (4) were PCR amplified and cloned into the promoterless pGL3 luciferase vector (Promega, Madison, WI). The BBAP promoter region (+1648 to −109 from the BAL1 TSS) was PCR amplified and cloned into the same reporter vector. A fragment containing the BAL1/BBAP predicted shared minimal promoter (−109 to +458 bp from BAL1 TSS) was cloned bidirectionally.
Generation of mutant BAL1/BBAP promoter constructs.
The predicted IRF and STAT binding sites or the entire minimal promoter sequence was deleted from the BAL1/BBAP regulatory region using a PCR-based mutagenesis approach. Briefly, the BAL1−109+458Luc plasmid served as a template to PCR amplify promoter fragments: nucleotides +80 to +239, +254 to +458, +80 to +264, +283 to +458, −109 to +79, and +308 to +458. These PCR products were excised from agarose gel and purified; thereafter, the following pairs were blunt-end ligated: (i) +80 to +239 and +254 to +458, (ii) +80 to +264 and +283 +458, (iii) +80 to +239 and +283 to +458, and (iv) −109 to +79 and +308 to +458. The ligation products (i) BAL1+80+458dIRF, (ii) BAL1+80+458dSTAT, (iii) BAL1+80+458dIRE-IFN response element, and (iv) BAL1−109+458dMP-minimal promoter, respectively, were reamplified, gel purified, and cloned into the pGL3 vector. Fragments +80 to +458, −109 to +79, +80 to +307, and +308 to +458 were amplified and cloned into the same reporter vector and used as controls.
Analysis of BAL1/BBAP promoter constructs and luciferase assays.
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (Cellgro Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (Cellgro Mediatech), 10 mM HEPES buffer, 4 mM l-glutamine, 50 U/ml penicillin, and 50 U/ml streptomycin. HEK293 cells were seeded on 96-well plates, grown to 60 to 80% confluence, and cotransfected with 80 ng/well of the appropriate promoter pGL3 construct (wild-type or mutant BAL1 or BBAP) and 20 ng/well of the control reporter plasmid, pRL-TK (Promega), using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol. Six hours after transfection, cells were treated with 1,000 U/ml of IFN-γ. After 24 h of incubation, cells were lysed and luciferase activities were determined by chemiluminescence assay using the dual luciferase assay kit (Promega) and a Luminoskan Ascent luminometer (Thermo Lab Systems, Franklin, MA). Luciferase activities are presented as means from three experiments ± standard deviations.
Inhibition of IFN-γ-mediated BAL1 induction with AG490.
HEK293 cells were transfected with 0.4 μg of the BAL1/BBAP promoter reporter plasmid BAL1−109+458Luc and 0.1 μg of plasmid pRL-TK and treated with IFN-γ (1,000 U/ml) with or without the JAK2 kinase inhibitor AG490 (Calbiochem, San Diego, CA) (10, 25, or 50 μM) or vehicle (dimethyl sulfoxide) alone. After a 24-h incubation, luciferase activities were measured as described above.
Analyses of IRF and STAT binding sites in the BAL1/BBAP promoter electrophoretic mobility shift assay.
Nuclear extracts from HEK293 or HeLa cells treated with IFN-γ (1,000 U/ml) for 0, 2, or 6 h were prepared with a nuclear extraction kit (Active Motif, Carlsbad, CA) and stored at −80°C. Double-stranded, PAGE-purified wild-type and mutant probes (wild-type IRF [5′-CAAAGTTTCAGTTTCGCTTCCCTGGAGGTC-3′] and STAT [5′-TCCCTCCCTGTTCCCGGCAGAGCCGCTTCC-3′]; mutant IRF _MUT [5′-CAAAGTTTCAtacgCGCTTCCCTGGAGGTC] and STAT_MUT [5′-TCCCTCCCTGcgatCGGCAGAGCCGCTTCC] [mutant bases are in lowercase letters]) were end labeled with [γ-32P]ATP, purified on Sephadex G-25 columns (Roche Applied Science), and used in binding reactions. DNA binding was carried out using 5 μg of nuclear extracts and ∼10,000 cpm of radiolabeled probe in 20 μl of binding buffer [4% glycerol, 1 mM MgCl2, 50 μM EDTA, 50 μM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 50 mg/ml of poly(dI-dC)]. The reaction mixtures were incubated at room temperature for 30 min, loaded on a 5% polyacrylamide gel, and electrophoresed at 30 mA for 2 h. Gels were vacuum dried and exposed to X-ray films overnight at −80°C. For competitor studies, a 100× molar excess of unlabeled wild-type or mutant probe was included in the binding reaction mixtures. For supershift studies, 1 μl of IRF-1, IRF-2, or STAT-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA; Abcam, Cambridge, MA) was added to the reaction mixture 15 min prior to the probe.
IRF-1 and -2 transient transfections.
The cDNAs for the IRF-1 and IRF-2 genes were generated by RT-PCR using RNA isolated from IFN-γ-treated Raji cells with TRIzol (Invitrogen, Carlsbad, CA). One microgram of RNA was reverse transcribed and PCR amplified using a one-step RT-PCR kit (Invitrogen). Amplification products were gel purified and ligated into the linearized vector pFLAG-CMV2 (Sigma-Aldrich). HEK293 cells were transfected with 0.4 μg of the BAL1/BBAP promoter reporter plasmid BAL1−109+458Luc, 0.1 μg of plasmid pRL-TK, and 0.4 μg of pFLAG-IRF1, pFLAG-IRF2, or pFLAG-CMV2 alone as described above. Thereafter, cells were lysed and luciferase activities determined by chemiluminescence assay (as described above).
Immunohistochemistry.
Immunohistochemistry was performed using 5-μm-thick formalin-fixed, paraffin-embedded tissue sections of HR-type DLBCLs. Slides were deparaffinized, pretreated with 10.0 mM citrate buffer (pH 6.0; Zymed, South San Francisco, CA) in a steam pressure cooker (Decloaking Chamber; BioCare Medical, Walnut Creek, CA), and washed in distilled water. All further steps were performed at room temperature in a hydrated chamber. Slides were initially treated with peroxidase block (DAKO USA, Carpinteria, CA) for 5 min to quench endogenous peroxidase activity and subsequently incubated with primary rabbit polyclonal anti-BAL1 (1:2,000) (4) or rabbit polyclonal anti-BBAP (34) (1:100) antibodies in antibody diluent (DAKO) for 1 h. Thereafter, slides were washed in 50 mM Tris-Cl-0.05% Tween 20, pH 7.4, and anti-rabbit horseradish peroxidase-conjugated antibody (Envision Plus; DAKO) was applied for 30 min. After further washing, immunoperoxidase staining was developed using diaminobenzidine chromogen (DAKO) per the manufacturer's instructions and counterstained with Harris hematoxylin.
BAL1 and BBAP immunofluorescence staining and fluorescence microscopy.
COS-7 cells were seeded on glass coverslips and transfected with enhanced green fluorescent protein (eGFP)-BAL1 and/or FLAG-BBAP plasmids described previously (4, 34) and FuGENE6 (Roche Applied Science). Forty-four hours after transfection, cells were treated with 10 ng/ml leptomycin B (LMB; Sigma-Aldrich) or vehicle (methanol) alone for 4 h as indicated in Fig. 6. Thereafter, coverslips were rinsed with ice-cold phosphate-buffered saline (PBS), cells were fixed in 4% paraformaldehyde in PBS for 20 min, and eGFP-BAL1-expressing cells were analyzed. For FLAG-BBAP studies, cells were permeabilized in 0.1% NP-40 in PBS for 15 min and nonspecific binding was blocked with 5% nonimmune mouse serum in PBS for 15 min. Cells were then incubated for 60 min with PBS supplemented with 0.1% NP-40 and 5% mouse serum containing primary antibody (mouse anti-FLAG monoclonal antibody [M2; Sigma-Aldrich]), washed extensively with PBS containing 0.1% NP-40, and then incubated for 60 min with secondary antibody [Rhodamine Red-X-conjugated F(ab′)2 fragment goat anti-mouse antibody; Jackson ImunoResearch Laboratories, West Grove, PA] in PBS containing 0.1% NP-40 and 5% mouse serum. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). After being washed extensively, coverslips were mounted in gel mount medium (Biomeda, Foster City, CA) and analyzed by fluorescence microscopy with a SPOT camera (Diagnostic Instrument, Sterling Heights, MI). Composite images were generated by Adobe Photoshop software.
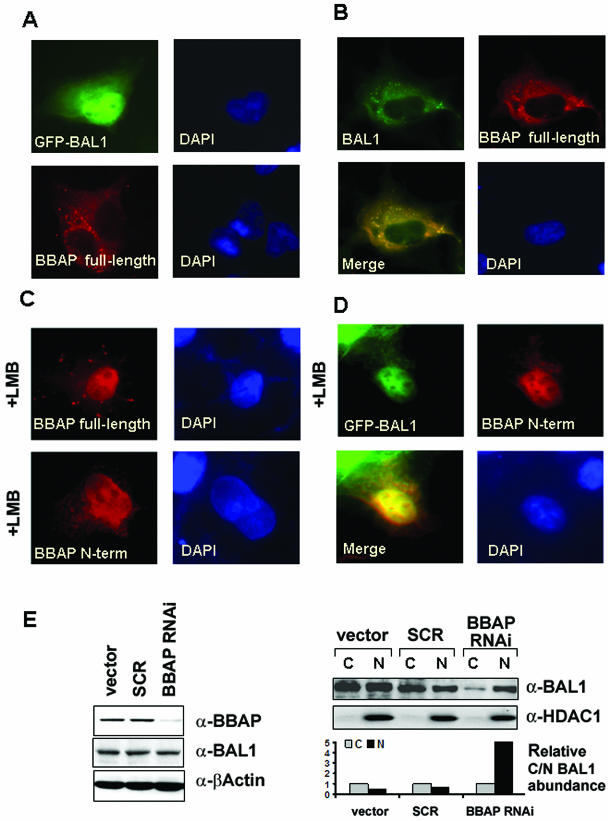
FIG. 6.
Subcellular localization of transfected BAL1 and BBAP in COS-7 cells. COS-7 cell monolayers were grown on coverslips and transfected with the following expression vectors: either eGFP-BAL1 or full-length FLAG-BBAP (A), eGFP-BAL1 and FLAG-BBAP (B), FLAG-tagged full-length or N-terminal BBAP (C), and eGFP-BAL1 and FLAG-tagged N-terminal BBAP (D). Cells in panels C and D were treated with LMB (10 ng/ml) as indicated. After 24 h, cells were fixed and subjected to indirect immunofluorescence. Nuclei were visualized by counterstaining with DAPI. (E) shRNA-mediated knockdown of BBAP. HeLa cells were transduced with a retrovirus expressing BBAP-specific shRNA (BBAP-RNAi) or virus encoding no shRNA (vector) or expressing a negative-control scrambled sequence (SCR). Seventy-two hours after retroviral transduction, whole-cell lysates or nuclear and cytosolic fractions were obtained, size fractionated, and analyzed by immunoblotting (left panel) or immunoblotting and scanning densitometry (right panel). (Left panel) Immunoblots of whole-cell lysates analyzed with anti-BBAP (a-BBAP), anti-BAL1, and anti-β-actin (loading control). (Upper right panel) Immunoblots of nuclear and cytosolic fractions analyzed with anti-BAL1 and anti-HDAC1 (nuclear-protein control). (Lower right panel) Densitometric analysis of relative nuclear and cytosolic BAL1 protein abundances in vector-, SCR-, or BBAP RNAi-treated cells.
RNA interference-mediated BBAP knockdown.
BBAP-specific small interfering RNA (siRNA) was designed using siRNA Selection Program (39) (http://jura.wi.mit.edu/bioc/siRNAext/), synthetized as single-stranded DNA oligonucleotides by Integrated DNA Technologies (Coralville, IA), and annealed. A BBAP-specific, annealed DNA oligonucleotide (BBAP RNAi, GATCCGGCGAAGCAGTATGTTCTATTCAAGAGATAGAACATACTGCTTCGCCTTTTTTG) or scrambled oligonucleotide (SCR, GATCCCCUCCAUAUCUCGCGCGUCTTCAAGAGAGACGCGCGAGATATGGAGGTTTTTTG) with BamHI and EcoRI overhangs was ligated into the linearized retroviral vector pSIREN-RetroQ-ZsGreen (BD Clontech). Thereafter, 15 μg of the BBAP RNAi-pSIREN-RetroQ-ZsGreen or SCR-pSIREN-RetroQ-ZsGreen retroviral vector was cotransfected with 8 μg of pKAT and 15 μg of a vesicular stomatitis virus G protein vector into 293T cells using FuGENE 6 transfection reagent (Roche Applied Science). Following 60 h, retroviral supernatants were collected, mixed with hexadimethrine bromide (Sigma-Aldrich) at an 8-μg/ml final concentration, and used to infect HeLa cells. Following 12 h of retroviral transduction, HeLa cells were incubated for 60 h and used to prepare whole-cell extracts in the lysis buffer described above or used to prepare nuclear and cytoplasmic fractions (nuclear extraction kit; Active Motif). Extracts were size fractionated on polyacrylamide gels and immunoblotted with anti-BAL1, anti-BBAP, anti-β-actin (Sigma-Aldrich), or anti-HDAC1 (Santa Cruz Biotechnology) antibodies as described above. Densitometric analysis of X-ray films was performed with ChemiImager 4400 software (Alpha Innotech, San Leandro, CA).
RESULTS
BAL1 induces IFN-responsive genes.
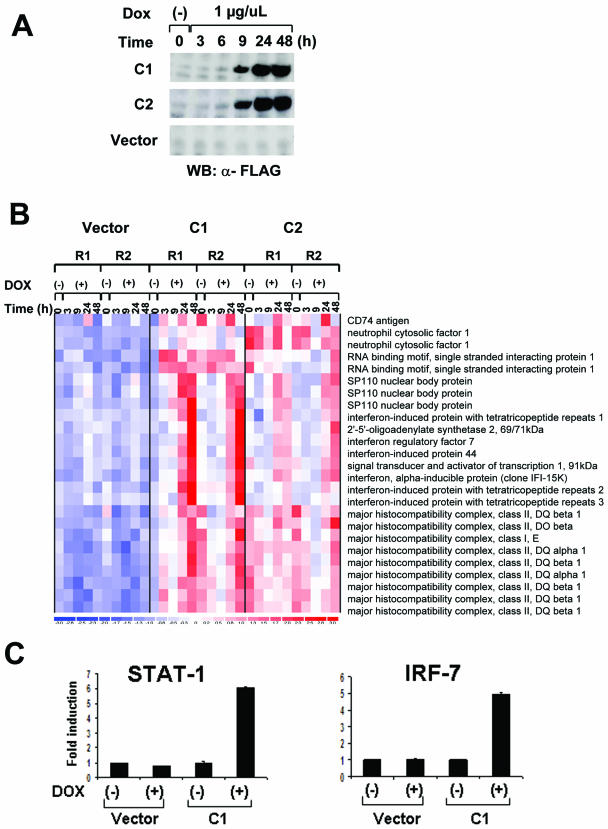
The nuclear localization of BAL1 and its role as a modulator of transcription prompted us to identify BAL1 target genes. Using a Tet-inducible system in a lymphoma cell line with low levels of endogenous BAL1, we selected two BAL1-inducible clones (C1 and C2) and a vector-only clone for further experiments. Following doxycycline treatment, BAL1 expression was detectable at 6 h and maximal at 24 to 48 h (Fig. 1A). At sequential time points following doxycycline treatment, total RNA from BAL1-inducible clones and empty vector controls was obtained for transcriptional profiling. Genes that were induced or repressed by BAL1 were selected by two-way ANOVA and rank correlation tests (P < 0.001). With these criteria, 110 genes were induced and 92 genes were repressed following the expression of BAL1. Of the 110 upregulated genes (represented by 130 probe sets), 17 genes (represented by 25 probe sets [15%]) were bona fide type 1 and 2 ISG (e.g., 2′-5′ OAS, IFIT1, IFIT2, STAT-1, IRF-7, and IFI-15K) or genes indirectly modulated by IFN (HLA class II) (chi-square test, P < 0.001) (Fig. 1B). Using quantitative RT-PCR, we then confirmed BAL1 induction of two selected ISG, IRF-7 and STAT-1 (Fig. 1C).
FIG. 1.
BAL1 induction of ISG. (A) BAL1 expression in a doxycycline-inducible lymphoma cell line. BAL1-inducible and empty vector control lines were treated with doxycycline (Dox) for the indicated periods of time. Thereafter, whole-cell lysates were size fractionated, immunoblotted, and analyzed with anti-FLAG antibody. WB, Western blotting; α-FLAG, anti-FLAG. (B) ISG induced by BAL1. Seventeen genes (represented by 25 probe sets) were bona fide type 1 and 2 ISG or genes indirectly modulated by IFN-γ. (C) Quantitative RT-PCR analysis of STAT-1 and IRF-7 induction by BAL. RNAs from a BAL1-inducible clone (C1) and control vector-only cells that were untreated (−) or treated with doxycycline (+) for 36 h were analyzed for STAT-1 and IRF-7 abundance by quantitative RT-PCR.
IFN-γ induces BAL1 expression in DLBCLs.
Next, we treated B-lymphoma cell lines with 10 to 1,000 units/ml IFN-γ and evaluated subsequent BAL1 expression by immunoblotting. The BAL1 binding partner, BBAP, was similarly assessed. Both BAL1 and BBAP were induced in B-lymphoma cell lines treated with increasing concentrations of IFN-γ (Fig. 2A).
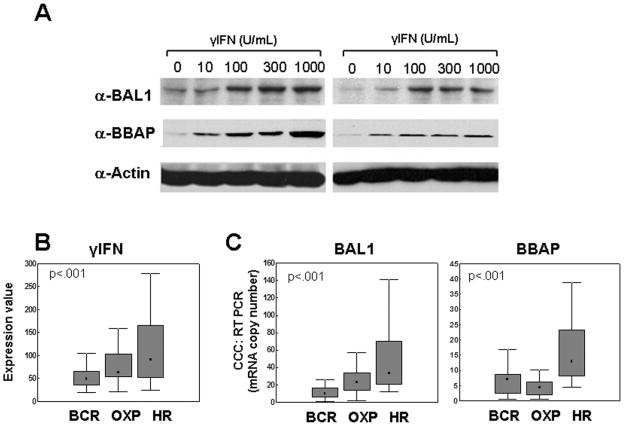
FIG. 2.
BAL1 and BBAP induction by IFN-γ in lymphoma cell lines and overexpression in primary DLBCLs. (A) Induction of BAL1 and BBAP by IFN-γ in two lymphoma cell lines (DHL4 [right panel] and DHL10 [left panel]). Cells were treated with the indicated doses of IFN-γ (0 to 1,000 U/ml) for 24 h. Thereafter, whole-cell lysates were size fractionated, immunoblotted, and analyzed with anti-BAL1 or anti-BBAP antibodies (α-BAL1 or α-BBAP, respectively). (B and C) Overexpression of IFN-γ, BAL1, and BBAP transcript abundance in DLBCL comprehensive clusters. Primary DLBCLs were identified as BCR, OXP, and HR tumors, and levels of IFN-γ, BAL1, and BBAP transcript abundance in the clusters were compared using a Kruskal-Wallis test. (B) IFN-γ transcript abundances were compared using microarray data (U133A probe set 210354_at [arbitrary units]). (C) BAL1 and BBAP transcript abundance was evaluated using real-time quantitative RT-PCR. Data are developed using graphs which denote the medians (▪), 25 to 75% values (shaded bars), and nonoutlier ranges (I bars).
IFN-γ, BAL1, and BBAP are more abundant in HR DLBCLs than in non-HR DLBCLs.
We then assessed BAL1 and BBAP expression in primary DLBCLs with a brisk host immune/inflammatory response and increased IFN-γ signaling (HR tumors) (Fig. 2B and C). In newly diagnosed DLBCLs with available transcriptional profiles and comprehensive cluster designations, γ-IFN, BAL1, and BBAP transcripts were significantly more abundant in HR tumors than in BCR and OXP tumors (P < 0.001, Kruskal-Wallis test) (Fig. 2B and data not shown) and highly correlated across the data set (correlation coefficient R, 0.72; P < 0.001 [data not shown]). Similar results were obtained when BAL1 and BBAP transcripts were directly analyzed in 106 of the same DLBCLs using quantitative PCR (Fig. 2C) (P < 0.001). In addition, BAL1 and IFN-γ transcript levels were closely correlated across the DLBCL series (correlation coefficient R, 0.44; P < 0.0001 [data not shown]). Therefore, BAL1 and BBAP are most abundant in DLBCLs defined by their brisk inflammatory infiltrate and signature of IFN-γ signaling. No significant differences in BAL1/BBAP expression were observed when the same tumor series was segregated into the developmentally defined cell-of-origin categories (data not shown) (27, 37).
In silico prediction of the BAL1/BBAP promoter.
To elucidate potential mechanisms of IFN-γ-mediated BAL1 and BBAP induction, we analyzed the 5′ regulatory regions of BAL1 and BBAP in silico and demonstrated a head-to-head orientation of the BAL1 and BBAP loci on chromosome 3q21 (Fig. 3A). Next, the sequence 4 kb upstream to 1 kb downstream from the BAL1 TSS was evaluated with two independent promoter prediction algorithms (10, 30). A single CpG-related minimal promoter, shared by both genes, was identified at +80 to +307 bp from the BAL1 TSS (Fig. 3A). Comparison of the predicted human, mouse, and rat BAL1/BBAP sequences (25, 29) revealed one highly conserved regulatory module that included IRF (+239 to +254) and STAT (+264 to +283) binding sites (data not shown).
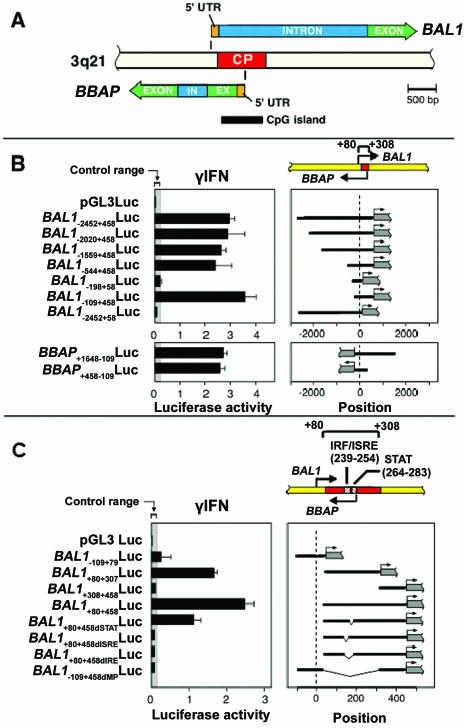
FIG. 3.
Identification and characterization of the IFN-γ-inducible, bidirectional BAL1/BBAP promoter. (A) Schematic representation of the bidirectional genomic organization of BAL1 and BBAP genes and their shared, CpG-related promoter (common promoter [CP], indicated by a red bar). In, intron; Ex, exon; 5′UTR, 5′ untranslated region. (B) Identification of a minimal, bidirectionally active BAL1/BBAP promoter. (Upper panel) BAL1 promoter fragments including or lacking the predicted minimal promoter sequence (+80 to +307, represented by a red bar) were cloned into a luciferase reporter vector, transiently transfected into HEK293 cells, and assayed for responsiveness to IFN-γ. (Upper right panel) Respective BAL1 promoter constructs (black lines) were cloned upstream of the firefly luciferase gene (gray bar with arrow). (Upper left panel) Representative luciferase activities from three independent experiments are normalized to Renilla luciferase activity and are presented as black bars ± standard deviations. (Lower panel) The reversed minimal promoter construct (BBAP+458+109Luc) or constructs with an additional 1.2 kb of 5′ upstream sequence (BBAP+1648−109Luc) were transiently transfected into HEK293 cells and assessed as for the upper panel. (C) The bidirectional BAL1/BBAP promoter requires intact IRF-1 and STAT binding sites for IFN-γ-induced activity. A series of deletion constructs lacking predicted individual IRF or STAT binding sites or both sites (dIRF, dSTAT, and dIRE, respectively) were cloned into the pGL3 vector, transiently transfected into HEK293 cells, and assessed as described for panel B. ISRE, IFN-stimulated response element.
The BAL1/BBAP promoter is bidirectionally activated by IFN-γ.
To elucidate the mechanism of IFN-γ-mediated BAL1/BBAP induction and locate critical cis-acting elements in the shared regulatory regions, a set of BAL1 promoter fragments was cloned into a luciferase reporter vector, transiently transfected into HEK293 cells, and assessed for responsiveness to IFN-γ (Fig. 3B). Promoter constructs including the predicted BAL1/BBAP minimal promoter sequence (+80 to +307) increased IFN-γ-responsive luciferase activity >20-fold, whereas constructs lacking the minimal promoter sequence were unresponsive to IFN-γ treatment (Fig. 3B, top panel). A series of 5′ deletions of the longest BAL1−2452+458Luc promoter construct revealed that sequences −2452 to −109 bp from the BAL1 TSS were dispensable for IFN-γ-mediated BAL1 activation (Fig. 3B, top panel). Similar results were obtained with COS-7 cells (data not shown).
To assess bidirectional BAL1/BBAP promoter activity, a fragment containing the minimal shared promoter sequence (−109 to +458 from the BAL1 TSS) was cloned into the pGL3 luciferase vector in the opposite direction. The reversed minimal promoter also increased luciferase activity >20-fold following IFN-γ treatment (Fig. 3B, bottom panel). BBAP promoter constructs with an additional 1.2 kb of 5′ upstream sequence (BBAP+1648−109Luc) were similarly responsive to IFN-γ, indicating that there were no additional IFN-γ regulatory elements within this region. Similar results were obtained with COS-7 cells (data not shown).
IFN-γ-induced BAL1/BBAP transactivation requires JAK2 kinase and an intact IRF element.
We next analyzed the downstream signaling molecules required for IFN-γ-mediated BAL1/BBAP induction. For this reason, HEK293 cells were transfected with a luciferase reporter vector driven by the BAL1/BBAP minimal promoter (BAL1−109+458Luc) and treated with IFN-γ in the presence or absence of the chemical JAK2 inhibitor AG490 (used at doses that specifically inhibit JAK2 kinase activity without nonspecific toxicity [12, 22]). AG490 inhibited IFN-γ-induced BAL1/BBAP promoter activity in a dose-dependent manner, directly implicating JAK2 and the associated STAT-1 and IRF transcription factors in this process (data not shown).
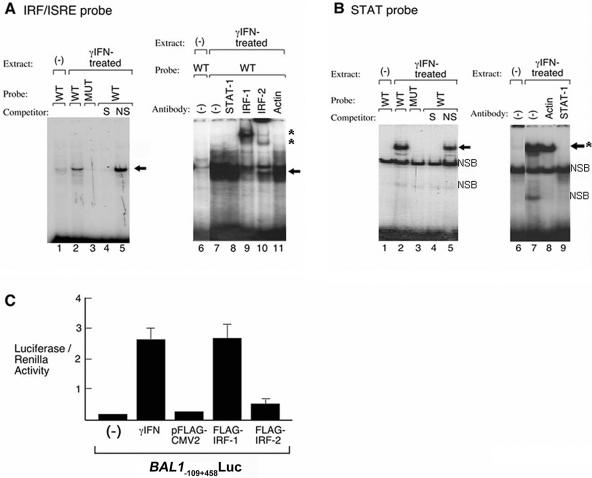
Next, we directly analyzed the requirement for IRF and STAT transcription factors in electrophoretic mobility shift assays (Fig. 4). Nuclear extracts from IFN-γ-treated HEK293 (or HeLa) cells were incubated with radiolabeled wild-type or mutant probes corresponding to IRF and STAT elements in the BAL1/BBAP promoter. BAL1/BBAP IRF and STAT wild-type probes, but not mutant probes, directly bound to IFN-γ-induced nuclear proteins (Fig. 4A and B, left panels). The complexes formed with BAL1/BBAP IRF and STAT probes were displaced by unlabeled wild-type but not by mutant competitors, further confirming binding specificity. In supershift assays, the BAL1/BBAP IRF complex was retarded by IRF-1 and IRF-2 antibodies (Fig. 4A, right panels). In similar assays, the BAL1/BBAP STAT complex was specifically disrupted by a STAT-1 antibody (Fig. 4B). The complex associated with a control STAT-1 probe was also disrupted with a STAT-1 antibody, as previously described (data not shown) (8).
FIG. 4.
BAL1/BBAP IRF and STAT promoter elements directly bind IFN-γ-induced proteins. (A) IRF shift assay. (Left panel) Nuclear extracts from IFN-γ-treated (lanes 2 to 5) or untreated (lane 1) cells were incubated with a wild-type (WT) (lanes 1, 2, 4, and 5) or mutant (MUT) (lane 3) 32P-labeled, double-stranded DNA probe corresponding to the IRF binding site in the BAL1/BBAP promoter. Specific (S) or nonspecific (NS) competitor was included in certain assays (lane 4 or 5, respectively). The gel shift band corresponding to the probe-protein complex is indicated with an arrow. (Right panel) Nuclear extracts from IFN-γ-treated (lanes 7 to 11) or untreated (lane 6) cells were incubated with the 32P-labeled IRF probe. Antibodies against STAT-1, IRF-1, IRF-2, or β-actin were included in lane 8, 9, 10, or 11, respectively. The gel shift band corresponding to probe-protein complex is indicated with an arrow, and supershift bands corresponding to probe-protein-antibody complex are noted with asterisks. (B) STAT-1 shift assay. (Left panel) Nuclear extracts from IFN-γ-treated (lanes 2 to 5) or untreated (lane 1) cells were incubated with a wild-type (lanes 1, 2, 4, and 5) 32P-labeled, double-stranded DNA probe corresponding to the STAT binding site in the BAL1/BBAP promoter. Specific (S) or nonspecific (NS) competitor was included in lane 4 or 5, respectively. The gel shift band corresponding to the formed probe-protein complex is indicated with an arrow; the lower shift band is nonspecific (NSB). (Right panel) Nuclear extracts from IFN-γ-treated (lanes 7 to 9) or untreated (lane 6) cells were incubated with the wild-type, 32P-labeled STAT probe. Antibodies against STAT-1 or β-actin were included in lane 9 or 8, respectively. The gel shift band corresponding to formed probe-protein complex is indicated by an arrow, and disruption of the complex by the STAT-1 antibody is noted by asterisks. (C) Roles of IRF-1 and -2 in BAL1 promoter activation. The BAL1 minimal promoter construct (BAL1−109+458Luc) was transiently cotransfected with the empty pFLAG-CMV2 vector or with the IRF-1 or IRF-2 pFLAG-CMV2 expression vector. Control cells were transfected with BAL1−109+458Luc (alone) and treated with IFN-γ or left untreated as indicated. Thereafter, cells were lysed and assessed for luciferase activity as described in the legend to Fig. 3.
The individual roles of IRF-1 and IRF-2 proteins in inducing BAL1/BBAP expression were then evaluated in cotransfection studies (Fig. 4C). Specifically, the cDNAs for IRF-1 and IRF-2 were cloned, placed under the transcriptional control of the cytomegalovirus promoter, and transiently cotransfected into HEK293 cells with the BAL1/BBAP minimal promoter reporter vector (BAL1−109+458Luc). Cotransfection of FLAG-tagged IRF-1 significantly increased BAL1/BBAP promoter activity, whereas FLAG-tagged IRF-2 had only a minor effect (Fig. 4C).
BAL1 and BBAP expression and subcellular localization in HR-type DLBCLs.
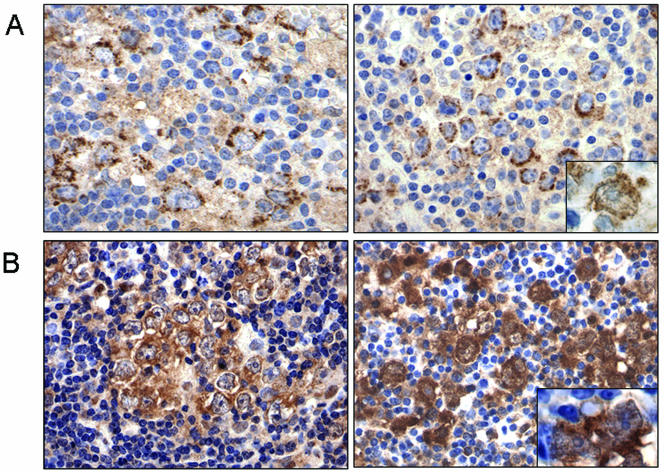
Given the coregulation of BAL1 and BBAP by a single IFN-γ-responsive, bidirectional promoter, the demonstrated role of BAL1 as a modulator of transcription, and the increased abundance of BAL1 and BBAP transcripts in HR tumors, we next evaluated the expression of these proteins in HR DLBCLs by immunohistochemistry. As expected, BAL1 and BBAP were primarily expressed by the malignant B cells rather than the infiltrating host immune/inflammatory cells in HR tumors. Unexpectedly, however, the immunohistochemistry analysis revealed significant levels of BAL1 and BBAP protein in the cytoplasm of these cells, with detectable, but less prominent, nuclear localization (Fig. 5A and B).
FIG. 5.
Subcellular localization of BAL1 and BBAP in primary HR DLBCLs. (A) Cytoplasmic and nuclear (inset, right panel) staining of BAL1 in the tumor cells of two primary HR DLBCLs (left and right panels). (B) Cytoplasmic and nuclear (inset, right panel) staining of BBAP in tumor cells of the same two primary HR DLBCLs. Magnification, ×400; inset, ×1,000.
Since our previous analyses of BAL1 nuclear immunolocalization were with transfected fibroblasts without detectable BBAP expression (4), we reasoned that concurrent BBAP expression might alter the subcellular localization of BAL1. To assess this possibility, COS-7 cells were transfected with eGFP-tagged BAL1, full-length FLAG-tagged BBAP, or a combination of eGFP-BAL1 and full-length BBAP and analyzed by immunofluorescence microscopy. Consistent with previous reports (4), cells transfected with eGFP-tagged BAL1 alone exhibited prominent nuclear fluorescence (Fig. 6A, upper panel). In contrast, full-length BBAP transfectants had predominant cytosolic fluorescence (Fig. 6A, bottom panel) whereas cells cotransfected with eGFP-tagged BAL1 and FLAG-tagged BBAP had abundant cytoplasmic and less prominent nuclear expression of the binding partners (Fig. 6B). Similar results were obtained when eGFP-BAL1 was cotransfected with a BBAP N-terminal construct (amino acids [aa] 1 to 560) that includes the BAL1 binding domain (34) (data not shown).
Taken together, the data suggested that BBAP might regulate the subcellular localization of BAL1. Although the BAL1 sequence includes a putative nuclear localization signal (RKKK, aa 650 to 653), BBAP contains both strong predicted nuclear localization signals (PRVRRKL [aa 20 to 26] and RKHLHQTKFADDFRKRH [aa 462 to 478]) and an export signal (LNHQFTKLLI [aa 325 to 334]). To assess the possibility that BBAP protein might shuttle between the nuclear and cytoplasmic compartments, we transfected full-length or N-terminally FLAG-tagged BBAP into COS-7 cells in the presence or absence of the nuclear export inhibitor LMB. Following LMB treatment, full-length and N-terminal BBAP (aa 1 to 560) were retained primarily in the nucleus (Fig. 6C, upper and bottom panels, respectively). When BAL1 and N-terminal BBAP were cotransfected in the presence of LMB, both proteins were trapped in the nucleus (Fig. 6D). Taken together, these data suggest that BBAP regulates BAL1 subcellular localization, facilitating nuclear export and shuttling from the nucleus to the cytosol (Fig. 6D).
To further define the role of BBAP in regulating BAL1 subcellular localization, we transduced a cell line that expressed high levels of endogenous BAL1 and BBAP (HeLa) with a retrovirus encoding a BBAP-specific short hairpin RNA (shRNA), an empty virus or a virus expressing a scrambled control sequence. After confirmation of the efficacy of BBAP knockdown (Fig. 6E, left panel), nuclear and cytosolic fractions were analyzed for BAL1 protein by immunoblotting and scanning densitometry (Fig. 6E, right panel). In cells transduced with either control, BAL1 was detected in both the cytosol and nucleus; in marked contrast, BAL1 was localized primarily to the nucleus in cells with RNA interference-mediated knockdown of BBAP (Fig. 6E, right panel). Therefore, the subcellular localization of BAL1 is determined by its coordinately regulated binding partner.
DISCUSSION
Herein, we show that BAL1 and BBAP are induced by IFN-γ and overexpressed in a subset of DLBCLs with a brisk, but ineffective, host inflammatory response (HR tumors) and IFN-γ signature. The basis for the coregulated, IFN-γ-induced expression of BAL1 and BBAP is a shared, bidirectional promoter containing a phylogenetically conserved IFN-γ response element. Coregulated expression of the BAL1 and BBAP binding partners results in the formation of a biological complex that shuttles between the cytoplasm and the nucleus.
The BAL1 and BBAP genes are located in a “head-to-head” orientation on chromosome 3q21. The human genome contains numerous pairs of genes with similar “head-to-head” orientations and transcription start sites separated by less than 1 kb (36). Several of these gene pairs are known to be regulated by a single bidirectional promoter (36). Bidirectional promoters typically have high GC contents, frequently lack TATA boxes, and often are conserved among mouse orthologs (36). All of these structural features are present in the bidirectional human, mouse, and rat BAL1/BBAP promoters.
Bidirectional promoters typically ensure the coordinate expression of genes with complementary roles (14, 36). For example, bidirectional promoters (i) maintain stoichiometric quantities of transcribed genes (5), (ii) drive the transcription of genes involved in the same cellular pathway or/and activated by the same cellular signal (14), and/or (iii) regulate the expression of genes that need to be sequentially activated (21). In this regard, the bidirectional BAL1/BBAP promoter ensures tightly coordinated transcriptional regulation of the BAL1/BBAP complex in response to IFN-γ. In addition, BBAP regulates the subcellular localization of BAL1 by a dynamic shuttling mechanism, highlighting the functional requirement for coordinated BBAP and BAL1 expression.
There are multiple examples of binding partners regulating the nucleocytoplasmic trafficking of transcription factors (2, 7, 18-20). Numerous cell cycle regulators and/or tumor suppressors shuttle between nucleus and cytoplasm in a tightly regulated fashion, where binding either impedes export and leads to nuclear retention of the complex or facilitates export of the binding partners (7, 17, 20). BAL1/BBAP binding facilitates the export of the BAL1/BBAP complex, likely due to the BBAP nuclear export signal.
BAL1 and BBAP are most abundant in HR DLBCLs that have a prominent immune/inflammatory infiltrate that includes interdigitating dendritic cells and CD2+/CD3+ T cells, a likely source of IFN-γ (27). However, the IFN-γ-inducible BAL1 and BBAP proteins are primarily expressed by tumor cells, underscoring the dynamic interaction between HR tumor cells and their microenvironment. In this context, IFN-γ-induced BAL1 expression and BAL1-induced IFN-stimulated genes directly place BAL1 in the IFN signaling pathway.
The unique clinical and biological features of HR tumors, including fewer known genetic abnormalities and younger ages of patients at presentation than with non-HR tumors, prompt speculation regarding the cause and nature of an associated, ineffective host immune response. Previous studies suggest a dual role of IFN-γ in tumor immunity, initially protecting the host from tumor formation and development but subsequently immunoediting established tumors to resist attack. For example, IFN-γ delays the growth and increases the rejection of carcinogen-induced tumors in mice (13, 23, 32). However, IFN-γ also induces the expression NK/T-cell inhibitory molecules, making IFN-γ-treated tumor cell lines more resistant to NK cell- or lymphokine activated killer-mediated lysis in vitro (11, 35). Given the IFN-γ inducibility of BAL1, the modulation of IFN-stimulated genes by BAL1, and the adverse effect of BAL1 expression on DLBCL outcome, it is possible that BAL1/BBAP may play a role in editing or inhibiting the host immune response against lymphoma. The current studies defining the regulation of BAL1 expression in HR DLBCLs begin to address this important issue.
REFERENCES
- 1.Abramson, J. S., and M. A. Shipp. 2005. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood 106:1164-1174. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Shaar, M., H. D. Ryoo, and R. S. Mann. 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguiar, R. C., K. Takeyama, C. He, K. Kreinbrink, and M. A. Shipp. 2005. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280:33756-33765. [DOI] [PubMed] [Google Scholar]
- 4.Aguiar, R. C., Y. Yakushijin, S. Kharbanda, R. Salgia, J. A. Fletcher, and M. A. Shipp. 2000. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96:4328-4334. [PubMed] [Google Scholar]
- 5.Albig, W., P. Kioschis, A. Poustka, K. Meergans, and D. Doenecke. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314-322. [DOI] [PubMed] [Google Scholar]
- 6.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, S. D., K. Y. Tsai, and T. Jacks. 2000. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat. Cell Biol. 2:563-568. [DOI] [PubMed] [Google Scholar]
- 8.Chow, W. A., J. J. Fang, and J. K. Yee. 2000. The IFN regulatory factor family participates in regulation of Fas ligand gene expression in T cells. J. Immunol. 164:3512-3518. [DOI] [PubMed] [Google Scholar]
- 9.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 10.Davuluri, R. V., I. Grosse, and M. Q. Zhang. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29:412-417. [DOI] [PubMed] [Google Scholar]
- 11.de Fries, R. U., and S. H. Golub. 1988. Characteristics and mechanism of IFN-gamma-induced protection of human tumor cells from lysis by lymphokine-activated killer cells. J. Immunol. 140:3686-3693. [PubMed] [Google Scholar]
- 12.De Vos, J., M. Jourdan, K. Tarte, C. Jasmin, and B. Klein. 2000. JAK2 tyrosine kinase inhibitor tyrphostin AG490 downregulates the mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription (STAT) pathways and induces apoptosis in myeloma cells. Br. J. Haematol. 109:823-828. [DOI] [PubMed] [Google Scholar]
- 13.Dighe, A. S., E. Richards, L. J. Old, and R. D. Schreiber. 1994. Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity 1:447-456. [DOI] [PubMed] [Google Scholar]
- 14.Dovhey, S. E., N. S. Ghosh, and K. L. Wright. 2000. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 60:5789-5796. [PubMed] [Google Scholar]
- 15.Dunn, G. P., A. T. Bruce, K. C. Sheehan, V. Shankaran, R. Uppaluri, J. D. Bui, M. S. Diamond, C. M. Koebel, C. Arthur, J. M. White, and R. D. Schreiber. 2005. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 6:722-729. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, G. P., L. J. Old, and R. D. Schreiber. 2004. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 21:137-148. [DOI] [PubMed] [Google Scholar]
- 17.Fabbro, M., and B. R. Henderson. 2003. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 282:59-69. [DOI] [PubMed] [Google Scholar]
- 18.Fabbro, M., S. Schuechner, W. W. Au, and B. R. Henderson. 2004. BARD1 regulates BRCA1 apoptotic function by a mechanism involving nuclear retention. Exp. Cell Res. 298:661-673. [DOI] [PubMed] [Google Scholar]
- 19.Galea, M. A., A. Eleftheriou, and B. R. Henderson. 2001. ARM domain-dependent nuclear import of adenomatous polyposis coli protein is stimulated by the B56 alpha subunit of protein phosphatase 2A. J. Biol. Chem. 276:45833-45839. [DOI] [PubMed] [Google Scholar]
- 20.Geyer, R. K., Z. K. Yu, and C. G. Maki. 2000. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2:569-573. [DOI] [PubMed] [Google Scholar]
- 21.Guarguaglini, G., A. Battistoni, C. Pittoggi, G. Di Matteo, B. Di Fiore, and P. Lavia. 1997. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem. J. 325:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, X. L., R. Pawliczak, X. L. Yao, M. J. Cowan, M. T. Gladwin, M. J. Walter, M. J. Holtzman, P. Madara, C. Logun, and J. H. Shelhamer. 2003. Interferon-gamma induces p11 gene and protein expression in human epithelial cells through interferon-gamma-activated sequences in the p11 promoter. J. Biol. Chem. 278:9298-9308. [DOI] [PubMed] [Google Scholar]
- 23.Kaplan, D. H., V. Shankaran, A. S. Dighe, E. Stockert, M. Aguet, L. J. Old, and R. D. Schreiber. 1998. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA 95:7556-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loots, G. G., and I. Ovcharenko. 2004. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32:W217-W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malmberg, K. J., V. Levitsky, H. Norell, C. T. de Matos, M. Carlsten, K. Schedvins, H. Rabbani, A. Moretta, K. Soderstrom, J. Levitskaya, and R. Kiessling. 2002. IFN-gamma protects short-term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J. Clin. Investig. 110:1515-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monti, S., K. J. Savage, J. L. Kutok, F. Feuerhake, P. Kurtin, M. Mihm, B. Wu, L. Pasqualucci, D. Neuberg, R. C. Aguiar, P. Dal Cin, C. Ladd, G. S. Pinkus, G. Salles, N. L. Harris, R. Dalla-Favera, T. M. Habermann, J. C. Aster, T. R. Golub, and M. A. Shipp. 2005. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 105:1851-1861. [DOI] [PubMed] [Google Scholar]
- 28.Perche, P. Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531-1534. [DOI] [PubMed] [Google Scholar]
- 29.Sandelin, A., W. W. Wasserman, and B. Lenhard. 2004. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 32:W249-W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherf, M., A. Klingenhoff, and T. Werner. 2000. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol. 297:599-606. [DOI] [PubMed] [Google Scholar]
- 31.Shankaran, V., H. Ikeda, A. T. Bruce, J. M. White, P. E. Swanson, L. J. Old, and R. D. Schreiber. 2001. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410:1107-1111. [DOI] [PubMed] [Google Scholar]
- 32.Street, S. E. A., E. Cretney, and M. J. Smyth. 2001. Perforin and interferon-γ activities independently control tumor initiation, growth, and metastasis. Blood 97:192-197. [DOI] [PubMed] [Google Scholar]
- 33.Street, S. E. A., J. A. Trapani, D. MacGregor, and M. J. Smyth. 2002. Suppression of lymphoma and epithelial malignancies effected by interferon γ. J. Exp. Med. 196:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeyama, K., R. C. Aguiar, L. Gu, C. He, G. J. Freeman, J. L. Kutok, J. C. Aster, and M. A. Shipp. 2003. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278:21930-21937. [DOI] [PubMed] [Google Scholar]
- 35.Tomita, Y., H. Watanabe, H. Kobayashi, T. Nishiyama, S. Tsuji, M. Fujiwara, and S. Sato. 1992. Interferon gamma but not tumor necrosis factor alpha decreases susceptibility of human renal cell cancer cell lines to lymphokine-activated killer cells. Cancer Immunol. Immunother. 35:381-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trinklein, N. D., S. F. Aldred, S. J. Hartman, D. I. Schroeder, R. P. Otillar, and R. M. Myers. 2004. An abundance of bidirectional promoters in the human genome. Genome Res. 14:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright, G., B. Tan, A. Rosenwald, E. H. Hurt, A. Wiestner, and L. M. Staudt. 2003. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 100:9991-9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y., and E. C. Uberbacher. 1997. Automated gene identification in large-scale genomic sequences. J. Comput. Biol. 4:325-338. [DOI] [PubMed] [Google Scholar]
- 39.Yuan, B., R. Latek, M. Hossbach, T. Tuschl, and F. Lewitter. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32:W130-W134. [DOI] [PMC free article] [PubMed] [Google Scholar]