Abstract
Zac is a C2H2 zinc finger protein that regulates apoptosis and cell cycle arrest through DNA binding and transactivation. The coactivator proteins p300/CBP enhance transactivation through their histone acetyltransferase (HAT) activity by modulating chromatin structure. Here, we show that p300 increases Zac transactivation in a strictly HAT-dependent manner. Whereas the classic recruitment model proposes that coactivation simply depends on the capacity of the activator to recruit the coactivator, we demonstrate that coordinated binding of Zac zinc fingers and C terminus to p300 regulates HAT function by increasing histone and acetyl coenzyme A affinities and catalytic activity. This concerted regulation of HAT function is mediated via the KIX and CH3 domains of p300 in an interdependent manner. Interestingly, Zac zinc fingers 6 and 7 simultaneously play key roles in DNA binding and p300 regulation. Our findings demonstrate, for the first time, that C2H2 zinc fingers can link DNA binding to HAT signaling and suggest a dynamic role for DNA-binding proteins in the enzymatic control of transcription.
Zac is a zinc finger (ZF) protein which potently induces apoptosis and cell cycle arrest and prevents tumor formation in nude mice (44, 52). Expression of Lot1, the rat orthologue of Zac, is lost during spontaneous transformation of ovary surface epithelial cells in vitro (1), and the human orthologue ZAC/LOT1/PLAGL1, which is widely expressed in normal tissues, is frequently down-regulated in a methylation-sensitive manner in various tumors (4, 5).
Evidence that Zac expression during embryogenesis in mesenchymal and neural stem/progenitor cells is tightly regulated in a spatiotemporal fashion and evidence that Zac is upregulated following seizures and transient focal cerebral ischemia in mice suggest that the protein may have additional roles, e.g., in neural differentiation and plasticity (2, 15, 48-50). Indeed, Zac was recently recloned in a subtractive hybridization screen for genes involved in neuronal cell fate specification (31). Moreover, Zac expression can be influenced by hormonal and epigenetic signals during regeneration, differentiation, and age-related degenerative processes, although its exact role in these conditions requires further studies (9, 13, 46, 57).
In adults, Zac is highly expressed in most steroid-responsive tissues (35, 36, 48, 52). Interestingly, Zac potently coactivates or corepresses the hormone-dependent activity of nuclear receptors, including that of androgen, estrogen, glucocorticoid, and thyroid hormone receptors (20); all of these receptors are key regulators of cell growth, differentiation, homeostasis, and development and act in a cell-specific manner. Furthermore, recent studies disclosed the importance of Zac's imprinting status in the etiology of transient neonatal diabetes mellitus, an uncommon form of childhood diabetes (NCBI entry OMIM *601410 [http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=601410]) (28, 51).
Our earlier studies revealed that Zac can act as a transcription factor through its monomeric or dimeric binding either to a GC-rich palindromic DNA element or to GC-rich direct and reverse repeat elements, respectively (6, 18, 52). The two closely related members of the Zac family Plag1 and Plagl2 share at their N termini virtually identical DNA-binding domains consisting of canonical zinc fingers of the C2H2 type (52). In line with this observation, Zac and Plag target genes identified so far (18, 54, 58) contain closely related GC-rich sequences in their promoter regions. Zac family members show, however, marked differences in their expression patterns in several tissues, indicating different biological roles. Moreover, proteins of the Zac family strongly diverge in their C termini, which could determine specific interactions in transcriptional regulation. In fact, only Zac has been reisolated in a yeast screen for mammalian proteins that bind to the C-terminal activation domain of the nuclear receptor coactivator SRC-1 (GRIP1) (20). Moreover, Zac binds to the C-terminal domain of the homologous coactivators p300 and CREB-binding protein (CBP) and strongly coactivates or corepresses nuclear receptors and p53 in a context-dependent manner (19, 20, 39).
The transcriptional coactivator proteins p300/CBP exert key roles in cellular differentiation, growth control, and homeostasis (16). In response to diverse physiological cues, these proteins coordinate and integrate multiple signal-dependent events at the transcriptional level by virtue of their histone acetyltransferase (HAT) activity. This allows them to influence chromatin activity by modulating nucleosomal histones, modify transcription factors, and influence DNA recognition, protein-protein interactions, and protein stability (25). The HAT activity of these coactivators is markedly increased upon their interaction with specific transcription factors (24, 43), although the underlying mechanisms of this switch in transcriptional activity are poorly understood. Whereas the classic recruitment model proposes that coactivation simply reflects the capacity of the activator to recruit the coactivator, we demonstrate here that HAT activity is controlled by the coordinated binding of Zac to p300. Furthermore, our experiments reveal a new function of C2H2 zinc fingers in the regulation of HAT activity, suggesting a dynamic role of DNA-binding proteins in the enzymatic control of transcription.
MATERIALS AND METHODS
Cell culture, plasmids, and transfections.
LLC-PK1 (porcine kidney epithelial [ATCC CL-101]), 293T (human embryonic kidney epithelial [ATCC CRL-11268]), PA-TU-8988T (human pancreatic adenoma carcinoma epithelial [DSMZ, Germany]) (14), and HeLa (human adenocarcinoma cervix epithelial [ATCC CCL-2.1]) cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Trypsinized cells were centrifuged, resuspended in EP1x buffer (50 mM K2HPO4, 20 mM CH3CO2K, 20 mM KOH, 26.7 mM MgSO4, pH 7.4, with CH3CO2H) and mixed with plasmid DNA or small interfering RNA (siRNA) oligonucleotides. After 5 min incubation at room temperature, cell suspensions (150 μl) were pulsed by using an electroporation apparatus (BTX 600).
Zac constructs and reporter plasmids were described previously (18). Coactivator cDNAs used were pCMV-p300-HA (11), pCMV-p300(DY-HAT)-myc (21), pcDNA3-CBP, pcDNA3-CBP(HAT−)L(1690)K, C(1691)L (42), pCX.Flag-PCAF (7), and pCR3.1.SRC1a (34). Single or combined deletion mutants of the p300 KIX (aa 566 to 650) and CH3 (aa 1673 to 1966) domains were generated by quick-change site-directed mutagenesis (Stratagene). p300 (10), CBP (29), or control siRNA oligonucleotides were purchased from MWG, Germany. Luciferase values were standardized by cotransfection of a β-galactosidase expression vector (0.1 μg of pRK7β-Gal). Results represent means and standard deviations from at least two experiments done in triplicate.
GST pulldown assays and in vitro translations.
The CH1 (amino acids [aa] 302 to 528), KIX (aa 566 to 829), HAT (aa 1197 to 1673), and CH3 (aa 1673 to 1966) domains were amplified by PCR, cloned into the BamHI site of pGEX-2TK (Amersham Biosciences) and pRK7Flag, and the sequence was verified. Glutathione S-transferase (GST) fusion proteins of p300 and Zac were grown in DH5α, purified on glutathione Sepharose 4B (product code 17-0756-01; Amersham Biosciences), and checked by Coomassie blue staining. Detailed information on the generation of p300 constructs used for in vitro translation is available upon request. In vitro translations were done with the TNT kit (Promega). The figures show representative autoradiograms and results are means and standard deviations from at least three independent experiments. Signals were quantified in a liquid scintillation counter (Beckman LS 6000IC).
For in vitro competition experiments, hemagglutinin (HA)-tagged Zac zinc fingers (aa 1 to 208), C terminus (aa 382 to 667), or luciferase was in vitro translated in the presence or absence of [35S]methionine. Relative concentrations were determined by use of anti-HA antibody. Results represent means and standard deviations from four independent experiments.
Immunoprecipitations and immunoblots.
Monoclonal anti-Flag (product code F3165; Sigma), anti-HA (product code 12CA5; Roche), anti-p300 (sc-584), anti-CBP (sc-369), anti-SRC (sc-8995), and antiactin (sc-7210) antibodies (all from Santa Cruz), and anti-Flag M2 affinity gel (product code A-2220; Sigma) were used according to the manufacturers' recommendations. Cells were lysed in 1 ml of lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Extracts were incubated for 1 h on ice and then centrifuged for 5 min at 18,000 relative centrifugal force. Following a preclearing step, soluble extracts were incubated at 4°C on a rocking platform with either anti-Flag M2-agarose affinity gel (A-2220; Sigma) or anti-p300 (sc-584). Immunoprecipitates were washed three times with 1 × TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl), eluted with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer and immunoblotted.
Immunoblots were performed on total cell lysates (5 to 50 μg protein) and peroxidase-linked anti-rabbit or anti-mouse immunoglobulin G (product code A-0545 or A-6782; Sigma).
A p300 cDNA fragment encoding part of the N-terminal region (aa 1 to 347) or part of the C-terminal region (aa 2042 to 2157) was subcloned into pGEX-2TK (Amersham Biosciences). Rabbits were immunized four times with 100 μg of the fusion protein at 10-day intervals, and antisera were collected after 75 days. The specificity of the antibody (1:500 to 1:1,000) was tested by immunoblotting of mock- or p300-transfected 293T cells. Cellular lysates (50 μg) were blotted and probed with preimmune sera, antibody anti-p300-N (1:1,000) or anti-p300-C (1:500), or p300 antibodies preabsorbed on GST-, GST-p300-N-glutathione-Sepharose, or GST-p300-C-glutathione-Sepharose, respectively. Signals were specific for p300-transfected cells. No signals were detected with either the preimmune or preabsorbed p300 sera.
Electrophoretic mobility shift assays.
Nuclear extracts were checked with anti-Flag, anti-HA, and anti-p300-N antibodies for equivalent levels of Zac or ZacΔC1 expression relative to p300 and derivatives. Then, 10 μg of nuclear cell extract was used for binding reactions as described previously (18).
In vitro HAT assays.
Acetylation reactions were performed essentially as described previously (27). GST-p300-HAT (aa 1197 to 1673) (25 nM) was incubated at 30°C for 30 min with GST proteins (GST-Zac, GST-Zac-ZF, GST-TFIIE, or GST-TFIIF, 1 μM each) as substrates in the presence of 3.5 μl [3H]acetyl coenzyme A ([3H]acetyl-CoA) in 1 × HAT buffer (50 mM Tris-HCl, pH 8.0, 10% glycerol, 1 mM EDTA, 10 mM phenylmethylsulfonyl fluoride) in a total volume of 40 μl. Reactions were fractionated on a 10% SDS-polyacrylamide gel. Gels were treated with 2,5-diphenyloxazol (56) and exposed to Hyperfilm MP (product code RPN6K; Amersham Biosciences) for 1 to 3 days. The experiment was done three times, and all experiments yielded similar results.
In vitro-translated p300 proteins (p300, p300ΔKIX, and p300ΔCH3) were preincubated with an equimolar dose of GST-Zac proteins (GST-Zac, GST-ZacΔC1, and GST-ZacΔZF6, 1 nM each) for 10 min at room temperature in 1 × HAT buffer in a total volume of 45 μl. Different amounts of GST fusion proteins were used for Fig. 10C. The reactions for kinetic analysis and for HAT progression curves were initiated by the addition of 300 μM biotinylated N-terminal H4 (aa 2 to 24) peptide (product code 12-372; Upstate Biotechnology) and 0.75 μCi [3H]acetyl-CoA (5.3 Ci mmol−1) (product code TRK688; Amersham Biosciences) in a volume of 5 μl, and the mixtures were incubated for 1 to 15 min at 30°C. Saturating (300 μM) or differing concentrations of core histones (product code H9250; Sigma) or H4 peptide were assayed in parallel for Fig. 10A and B. Saturation analysis of H4 peptide and acetyl-CoA binding reactions were initiated likewise, and the mixtures were incubated for 2 min at 30°C. After the addition of 500 μl 1 × HAT buffer with 30 μl of 50% slurry of streptavidine agarose (product code 69203-3; Novagen), the samples were incubated for 20 min at 4°C on a rotating wheel. The beads were washed twice with radioimmunprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 0.1% phenylmethylsulfonyl fluoride) and counted in a liquid scintillation counter. All reactions were conducted in the presence of saturating amounts of the direct repeat (DR) element.
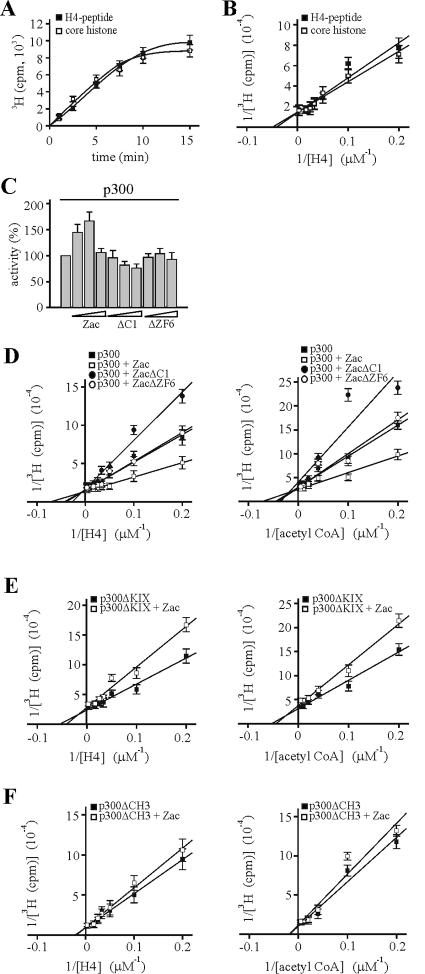
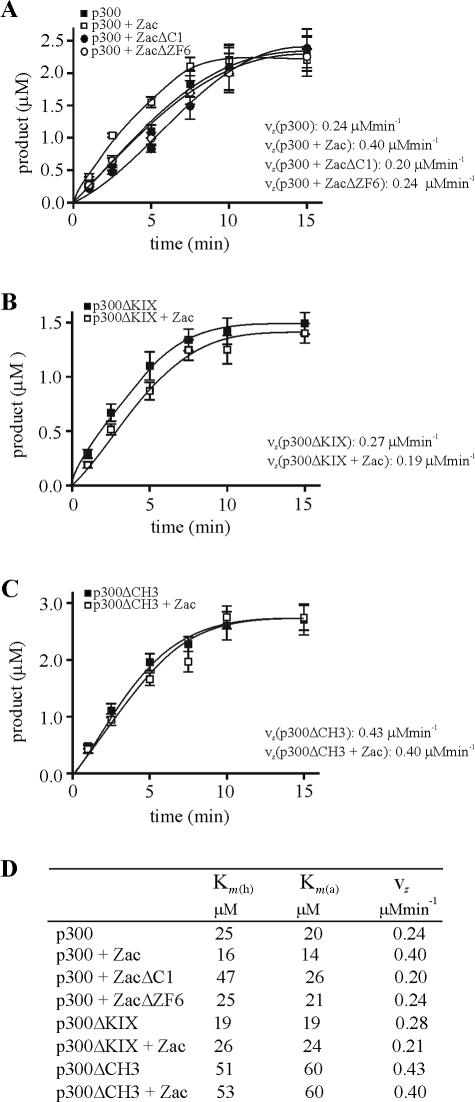
FIG. 10.
Coordinated Zac binding regulates in vitro p300-HAT substrate affinities. Data represent means and standard deviations from at least three experiments done in duplicate. (A) Kinetic analysis of acetylation reaction. Saturating amounts of core histones, H4 peptide, and [3H]acetyl-CoA were incubated with in vitro-translated p300. 3H acetylation was measured for the indicated times. (B) Various concentrations of core histones and H4 peptide were assayed with in vitro-translated p300 and saturating concentrations of [3H]acetyl-CoA. (C) Zac or derivatives dose-dependently regulate p300-HAT activity. p300 (1 nM) was incubated for 2 min with saturating amounts of substrates and with differing concentrations of recombinant Zac, ZacΔC1, or ZacΔZF6 at a ratio of 3:1, 1:1, or 1:3, respectively. (D) Saturation analysis. p300 (1 nM) was incubated for 2 min with differing concentrations of H4 peptide (left) and acetyl-CoA (right) in the absence or presence of equimolar amounts of recombinant Zac, ZacΔC1, or ZacΔZF6. All reactions contained the DR element. Acetylation products and substrate concentrations were plotted in a double-reciprocal graph. (E and F) p300ΔKIX and p300ΔCH3 (1 nM each) were tested as detailed for panel D in the absence or presence of equimolar amounts of Zac.
For HAT reaction progress curves, the specific activity of acetyl-CoA was determined by dividing the total cpm from the unwashed sample by the total concentration of acetyl-CoA in the assay (cpm/μM). For each sample, the cpm observed was directly related to the concentration (μM) of acetylated protein by the specific activity (cpm/μM) of acetyl-CoA. After the background cpm was subtracted from the enzyme reaction cpm, the resulting cpm value was converted to concentration of product formed by dividing by the specific activity of acetyl-CoA. Data are plotted as amounts of product formed versus time. Linear least-squares fit was performed over the linear portion of the data. If the concentration of substrates was saturating (generally 10 times the Km), the slope of the line was taken to represent the maximum rate of 3H acetylation (vz) for this concentration of enzyme.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (43). Primers used for PCR were from the direct repeat reporter (18) flanking the Zac response elements (5′ primer, 5′-TATCTTATGGTACTGTAACTGAGC-3′; 3′ primer, 5′-CTAGAGGATAGAATGGCGCCGGGC-3′). Antibodies against acetyl-histone H4 were from Upstate Biotechnology (product code 06-598). Levels of expression of HA-tagged Zac, Flag-tagged p300, and their derivatives were monitored by immunoblotting with anti-HA and anti-Flag antibodies. In all cases, aliquots of PCR carried out for 25 or 35 cycles were analyzed to ensure that amplification was maintained in the linear range.
RESULTS
p300 mediates Zac transactivation.
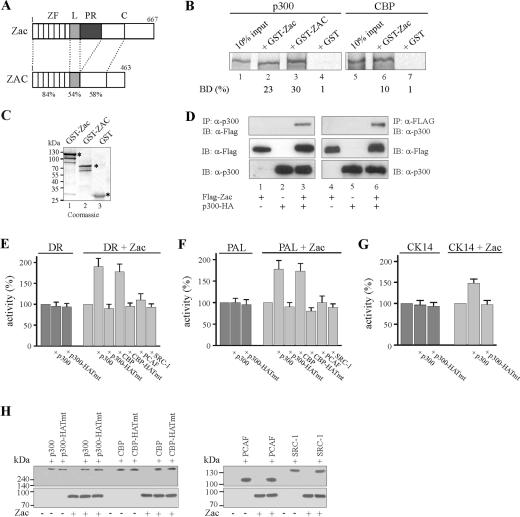
Mouse and human Zac proteins contain virtually identical N-terminal DNA-binding domains and show high conservation for the N-terminal parts of their C termini but differ by the presence of a central proline-rich transactivation domain specific to mice (Fig. 1A) (18, 44, 52). To study Zac interaction with p300/CBP, we conducted pulldown assays with in vitro-translated p300 and CBP proteins and mouse and human Zac GST fusion proteins. Both fusion proteins (Fig. 1B, lanes 2, 3, and 6) but not GST alone (lanes 4 and 7) efficiently retained p300 or CBP. Equivalent amounts of GST fusion proteins were used in these experiments (Fig. 1C).
FIG. 1.
p300/CBP enhance Zac transactivation. (A) Scheme of Zac proteins. Numbers denote amino acids, and domains are boxed. Mouse Zac and human ZAC protein contain virtually identical zinc finger domains (ZF) and a strongly conserved linker (L) and N-terminal region within the C terminus (C), whereas the central proline-rich transactivation domain (PR) is specific to mice. Identity (%) is indicated. (B) Pulldown assay. Equal amounts of in vitro-translated p300 or CBP were incubated with adjusted amounts of GST-Zac, GST-ZAC, or GST alone. The fraction of the input (100%) bound by each GST protein [BD (%)] is indicated. (C) The Coomassie blue stain shows adjusted amounts of GST-Zac, GST-ZAC, or GST used in pulldown assays. Asterisks mark bands of predicted molecular mass (kDa). (D) Coimmunoprecipitation experiment. Zac (0.5 μg of pRK7Flag-Zac) and p300 (3 μg of pCMV-p300-HA) were cotransfected in 293T cells as indicated in the text. Immunoprecipitation (IP) and immunoblotting (IB) were done with anti-Flag and anti-p300 antibodies. (E to G) Reporter assays. Zac (0.1 μg of pRK7Flag-Zac) was cotransfected with the DR element, PAL element, and cytokeratin 14 promoter (CK14) (2 μg each) in the absence (dark gray) or presence (light gray) of Zac. Activity of the reporter alone was set to 100% and compared to cotransfection of p300 or the p300 HAT-deficient mutant (p300-HATmt). Activity of the reporter in the presence of Zac was set to 100% and compared to the activity in the presence of wild-type p300, CBP, PCAF, SRC-1, or HAT-deficient mutants (1 μg each). (H) Immunoblot. Expression of Zac (0.1 μg of pRK7Flag-Zac) in the presence or absence of coactivators (1 μg each) as indicated. Coexpressions did not alter Zac or coactivator protein levels. Antibodies were anti-p300 (sc-584), anti-CBP (sc-369), anti-SRC (sc-8995), anti-Flag (F3165), and anti-Zac.
Zac bound p300 consistently stronger than CBP, corroborating earlier experiments in which, however, solely the C termini of p300 (aa 1571 to 2414) or CBP (aa 1594 to 2441) were used (20). To assess whether this interaction also occurred in vivo we carried out coimmunoprecipitation experiments using epitope-tagged proteins. We detected Zac solely in the presence of p300-HA and vice versa (Fig. 1D). Moreover, Zac interacted with CBP although to a lesser degree than with p300 (data not shown).
Given this interaction in vitro and in vivo, we investigated its relevance for Zac transactivation. Apoptotic cell death is known to cause global nuclear condensation, probably leading to broad transcriptional repression (37), confounding coactivator or transcriptional assays, and to potentially target p300/CBP for degradation (38). Therefore, we performed reporter assays with LLC-PK1 cells, which fairly resist Zac-induced cell death at 24 h following transfection (44, 52). We cotransfected Zac together with a reporter plasmid harboring two DR elements or four palindromic elements (PAL) in the absence or presence of additional amounts of p300 or CBP. Coexpression of wild-type p300 or of a HAT-defective version failed to enhance basal activities of the DR or PAL reporters, which were set to 100% (Fig. 1 E and F). In contrast, Zac-induced DR or PAL reporter activities set to 100% (Fig. 1 E and F) were about twofold induced by coexpression of p300 or CBP, while the HAT-deficient versions were completely ineffective in this respect. Similarly, coexpression of p300 but not of its HAT-deficient version specifically increased Zac-dependent transactivation of the CK14 promoter (Fig. 1G), a previously identified Zac target gene (18). In contrast, coexpression of broadly differing doses of the unrelated coactivators PCAF and SRC-1 (GRIP1) failed to stimulate Zac-dependent transactivation of the DR and PAL reporter plasmids (Fig. 1E and F and data not shown) although both PCAF (data not shown) and SRC-1 readily bound to Zac (20; data not shown). To rule out the possibility that these discrepancies simply resulted from differences in Zac expression in the presence of individual coactivators, we carried out immunoblotting. As shown in Fig. 1H, cotransfection of p300 or p300-HATmt did not influence Zac protein levels and vice versa. This was also the case for CBP, CBP-HATmut, PCAF, and SRC-1. Together, these data show that solely p300 and CBP support Zac transactivation in a strictly HAT-dependent manner.
In a complementary approach to these experiments, we depleted HeLa cells of endogenous p300 or CBP by RNA interference. HeLa cells were chosen at this step because they could be efficiently transfected by electroporation (>70% β-galactosidase-positive cells following transfection of 100 ng of pRK7-β-galactorisdase expression vector). p300 or control siRNA oligonucleotides (Fig. 2A, columns 2 and 3) failed to affect basal DR reporter activity, which was set to 100% (column 1). In contrast, Zac-dependent reporter activity (similarly set to 100% [Fig. 2A, column 4]) was diminished by p300 siRNA at the highest dose by fivefold (column 7), whereas control siRNA oligonucleotides of the same dose were largely ineffective (column 8). Immunoblotting evidenced that p300 siRNA reduced p300 protein in a dose-dependent manner, which correlated well with reductions in Zac-dependent transactivation (Fig. 2A, bottom panel). Additionally, to exclude the possibility that p300 or control siRNA affected Zac DNA binding, we carried out electrophoretic mobility shift assays (EMSA). These experiments displayed no differences between samples from either treatment or control conditions, attesting to the specificity of the results (data not shown). Similar to how we tested p300, we further tested the role of endogenous CBP to enhance Zac transactivation. CBP siRNA did not reduce basal DR reporter activity and impaired only weakly Zac-dependent activity at the highest dose tested (Fig. 2B). Immunoblotting revealed, however, efficient reduction of CBP protein levels, comparable to the results obtained for p300. In addition, CBP siRNA did not change Zac DNA binding (data not shown). Therefore, the strong decline in Zac transactivation upon p300 knockdown compared to the weak effect of CBP knockdown indicated a preferential interaction of Zac with endogenous p300 to enhance transactivation.
FIG. 2.
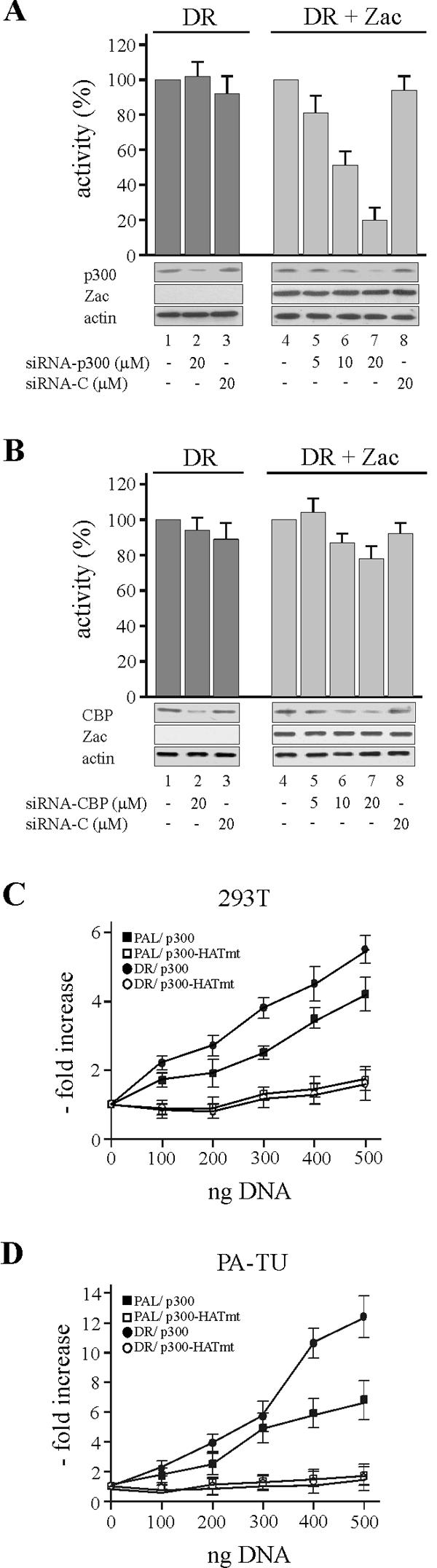
p300 preferentially mediates Zac coactivation. (A) RNA interference. The DR reporter (2 μg) was cotransfected in the absence (dark gray) or in the presence (light gray) of Flag-Zac (0.1 μg of pRK7Flag-Zac) with p300 or control siRNA into HeLa cells. Activity of the reporter alone or of the reporter plus Zac in the absence of siRNA was each set to 100% and compared to conditions containing siRNA. Columns represent means and standard deviations from three experiments. Representative immunoblots of p300, Zac and actin proteins are shown. (B) CBP or control siRNA was tested as described in the text. (C and D) Cotransfection of 293T and PA-TU cells. Given amounts of Zac (0.1 μg of pRK7Flag-Zac) were cotransfected with the DR or PAL reporters (2 μg each) in the presence of increasing doses of wild-type or HAT-deficient p300 as indicated.
Given these findings, we considered two additional cellular models. First, we considered the human embryonic kidney cell line 293T, which contains functionally impaired p300 and CBP due to high levels of adenovirus 5 and simian virus 40 large-T-antigen proteins. Second, we considered the pancreatic adenocarcinoma cell line PA-TU-8988T, which harbors solely one copy of a hypomorph p300 allele (14). In the absence of ectopic p300, we measured weak Zac-mediated transactivation of the DR and PAL reporter plasmids in both cell lines (data not shown). However, cotransfection of wild-type but not of HAT-deficient p300 markedly restored transactivation (Fig. 2C and D). So far, our results reveal an interaction between Zac and p300/CBP in vitro and in vivo and show that Zac transactivation is preferentially enhanced by p300 in a strictly HAT-dependent manner.
Mapping Zac and p300 interactions.
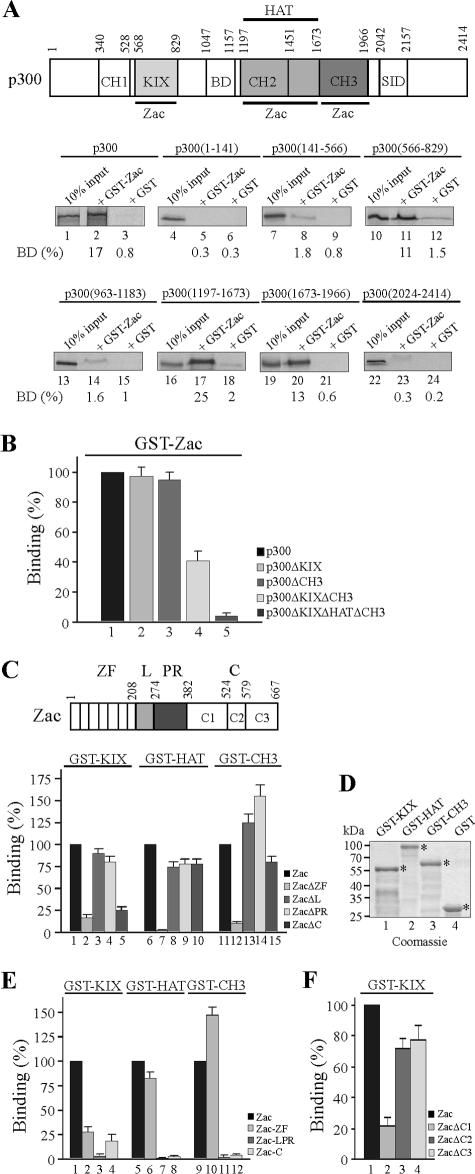
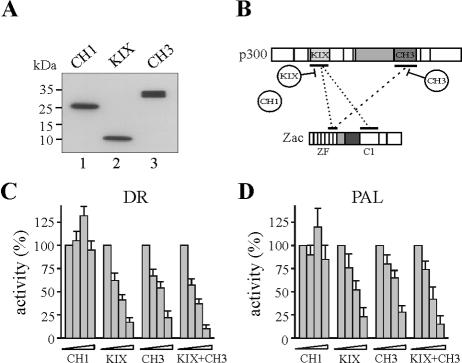
To delineate the domains of Zac and p300 interacting with each other, we in vitro translated successive segments of p300, schematically depicted at the top of Fig. 3A. Pulldown assays evidenced that the N-terminal KIX domain (aa 566 to 829), the central HAT domain (aa 1197 to 1673), and the C-terminal CH3 domain (aa 1673 to 1966) bound with similar affinities to GST-Zac protein. No binding occurred in the case of the CH1 domain, bromodomain, and steroid receptor interacting domain. To follow up the contribution of the KIX and CH3 domains to overall binding, we deleted them singly or together (Fig. 3B). Zac binding to singly deleted p300 derivatives persisted (columns 2 and 3), whereas combined deletion reduced binding by 60% (column 4). As expected, absence of either Zac binding site abolished p300 binding (column 5).
FIG. 3.
p300-Zac interactions. (A) Mapping p300 domains. A scheme of p300 is shown. Numbers denote amino acids, and domains are boxed. Abbreviations: CH, cystidine-histidine-rich region; BD, bromodomain; HAT, histone acetyltransferase domain; SID, steroid receptor interacting domain. Zac binding sites are underlined. Equal amounts of in vitro-translated p300 segments were incubated with adjusted amounts of GST-Zac or GST alone; representative autoradiograms are shown. The fraction of the input (100%) bound by each GST protein is given as [BD (%)]. (B) p300 derivatives singly deleted (p300ΔKIX and p300ΔCH3) bound indistinguishably from p300 to GST-Zac, while combined deletion (p300ΔKIXΔCH3) reduced binding by 60%. The absence of either Zac binding site (p300ΔKIXΔHATΔCH3) abolished the interaction. p300 binding was set to 100%. (C) Mapping Zac domains. A scheme of Zac is shown. Abbreviations: ZF, zinc finger domain; L, linker domain; PR, proline repeat domain; C, C terminus, further subdivided in C1 to C3. Equal amounts of in vitro-translated Zac segments were incubated with adjusted amounts of GST-KIX, -HAT, -CH3, and GST alone. Fraction of the input (100%) bound by each GST protein is given [BD (%)], with the binding of Zac set to 100%. (D) The Coomassie blue stain shows adjusted amounts of GST-KIX, GST-HAT, GST-CH3, and GST used in pulldown assays. Asterisks mark bands of predicted molecular mass (kDa). (E) Equal amounts of in vitro-translated Zac, zinc fingers, linker-proline domain, or C terminus were incubated with GST-KIX, -HAT, -CH3, or GST alone. The binding of Zac was set to 100%. (F) The conserved C1 region binds to GST-KIX. Equal amounts of in vitro-translated Zac, ZacΔC1, ZacΔC2, and ZacΔC3 were incubated with GST-KIX or GST alone. Zac binding was set to 100%.
While these findings showed that either the KIX or CH3 domain in conjunction with the HAT domain enabled efficient Zac binding, they left unanswered which parts of Zac underlie this activity. Therefore, we evaluated Zac (a scheme is shown at the top of Fig. 3C) and the following derivatives: ZacΔZF, shortened by the N-terminal zinc finger domain; ZacΔL and ZacΔPR, with the N-terminal or C-terminal part of the central transactivation domain deleted; and ZacΔC, shortened by the C terminus. Interestingly, the absence of the zinc fingers strongly reduced binding to the KIX, HAT, or CH3 domains (Fig. 3C, columns 2, 7, and 12). In contrast, the deletion of the central transactivation domains showed no effect (columns 3, 4, 8, 9, 13, and 14), whereas deletion of the C terminus selectively impeded binding to the KIX domain (compare column 5 with columns 10 and 15). Equivalent amounts of GST fusion proteins or of GST alone were used in these experiments (Fig. 3D).
In line with these results, the isolated zinc fingers efficiently bound to the HAT and the CH3 domains (Fig. 3E, columns 6 and 10), whereas the central transactivation domain failed to interact with any of the p300 domains (columns 3, 7, and 11). Moreover, both the zinc fingers and the C terminus alone weakly bound to the KIX domain (columns 2 and 4), suggesting that they cooperated in binding. Irrespective of this, Zac and ZacΔC indistinguishably bound to p300 (as opposed to the behavior of isolated domains), suggesting that overall binding reflected multipoint interactions (data not shown).
Mouse and human Zac proteins share high homology in the N-terminal part of the C terminus (labeled C1) but not in the central and C-terminal parts (labeled C2 and C3, respectively), which are specific to mice (Fig. 1A and 3C, top) (18, 44, 52). Therefore, we determined the region of mouse Zac responsible for binding to the KIX domain. In fact, the C1 region accounted for the interaction with p300 (Fig. 3F, column 2). This suggested a conserved function to this part of human ZAC, which actually bound with affinity similar to that of the mouse one to the KIX domain (data not shown). Our previous work evidenced this region of human ZAC to contain high transactivation potency as opposed to the complete absence of transactivation in the corresponding part in mice (18, 52). As a result, transactivation potency and p300 binding are strictly separated in the case of the mouse C terminus but coexist in the case of the human one. Conclusively, Zac interacts with the p300-KIX, -HAT, and -CH3 domains through the zinc fingers and the C1 region. The zinc fingers are necessary and sufficient for binding to the HAT and CH3 domains, while binding of the C1 region is restricted to the KIX domain, possibly in cooperation with the zinc fingers.
Zinc fingers interact cooperatively and selectively with p300.
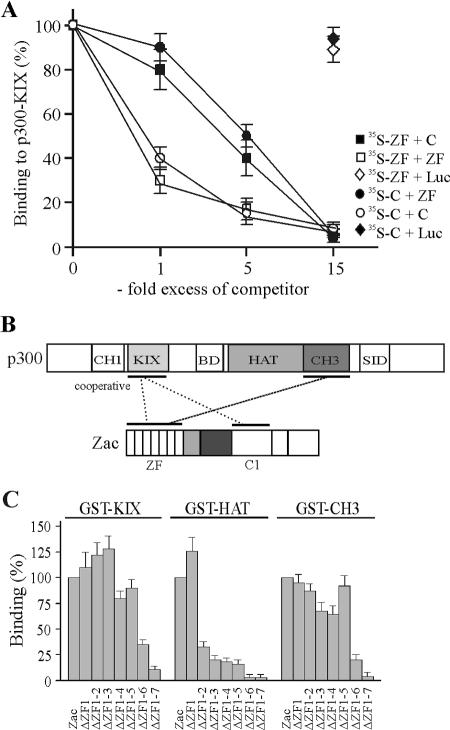
To establish more firmly whether the decline in binding of the isolated C terminus and the zinc fingers to the KIX domain stemmed from cooperative binding, we conducted in vitro competition experiments. Results shown in Fig. 3E showed that the isolated zinc fingers and the C terminus bound with similar affinities to the KIX domain. Therefore, we competed binding of the C terminus to limiting concentrations of the KIX domain with increasing amounts of the zinc fingers and vice versa (Fig. 4A). Equimolar amounts of the unlabeled zinc fingers caused a twofold decline in self-competition but weakly decreased binding of the C terminus. We obtained similar results for the reverse experiment, in which equimolar amounts of the unlabeled C terminus weakly reduced binding of the zinc fingers yet led to a twofold decline in self-competition. In further support of these findings, a fivefold excess of the zinc fingers only halved binding of the C terminus and vice versa but generated a sixfold decrease in self-competition. Finally, a 15-fold excess of competitor completely abolished binding either of the zinc fingers or of the C terminus to the KIX domain, suggesting that they shared overlapping sites. In contrast, competition of the zinc fingers or of the C terminus with a 15-fold excess of in vitro-translated luciferase protein failed to reduce significantly their binding to the KIX domain, attesting to the specificity of these results. Together, these data support a cooperative binding of the zinc fingers and of the C terminus to the KIX domain at possibly overlapping sites. On the basis of these experiments, we surmised the following model for the interaction between Zac and p300 (Fig. 4B): (i) the zinc fingers and the C1 region cooperatively bind to the KIX domain, and (ii) the zinc fingers additionally interact with the CH3 domain. In this regard, numerous transcription factors are known to bind to p300/CBP at multiple sites, although the functional implications of such multipoint interactions remain poorly understood (16, 17). Therefore, Zac binding to the KIX and CH3 domains raised the question of whether their binding is functionally coordinated or whether the KIX and CH3 domains operate independently of each other to mediate Zac coactivation.
FIG. 4.
Zinc fingers interact cooperatively and selectively with p300. (A) In vitro competition experiment. Binding of the in vitro-translated zinc fingers to GST-KIX (aa 566 to 650) protein was competed by the indicated amounts of in vitro-translated C terminus (C) or by itself. Similarly, binding of the in vitro-translated C terminus was competed by the indicated amounts of in vitro-translated zinc fingers or by itself. In control experiments, a 15-fold excess of in vitro-translated luciferase (Luc) protein failed to compete binding of the zinc fingers (open diamonds) or of the C terminus (filled diamonds) to the KIX domain. (B) Model of Zac-p300 interaction. Zinc fingers and C1 region cooperatively bind to the KIX domain. The zinc fingers additionally bind to the CH3 domain. SID, steroid receptor interacting domain; BD, bromodomain. (C) Pulldown assays. Zac and successive deletion mutants of the zinc fingers were in vitro translated and tested for binding to GST fusions of the KIX, HAT, or CH3 domains. Zac binding was set to 100%.
Among the seven canonical C2H2 zinc fingers present in the Zac DNA-binding domain, ZF2, ZF3, ZF4, ZF6, and ZF7 control binding to imperfect and perfect DR elements, while ZF6 and ZF7 control binding to the PAL element (18). This prompted us to examine in more detail whether zinc fingers engaged in DNA binding additionally recruit p300 or whether these functions are mutually exclusive. We successively deleted single zinc fingers of Zac and tested the corresponding in vitro translation products for binding to GST fusions of the isolated KIX, HAT, and CH3 domains (Fig. 4C). In the case of the KIX domain, binding strongly declined following the deletion of ZF6 and ZF7. In contrast, binding to the HAT domain sharply decreased subsequently to deletion of ZF2, followed by a gradual decline for further deletions. Conversely, the binding pattern of the CH3 domain closely mirrored the one of the KIX domain, with ZF6 and ZF7 mainly contributing. While ZF1 and ZF5 are not involved in DNA binding, and while ZF2, ZF3, and ZF4 are only necessary for binding to the DR element, ZF7 is strictly required for DNA binding to both the DR and PAL elements (18). Moreover, ZF6 is indispensable for Zac binding to the imperfect DR elements and the PAL element. Therefore, these findings evidence a new role of ZF6 and ZF7 in binding to the KIX and CH3 domains in addition to their previously described core function in DNA binding (18).
Zinc fingers and p300 interact in a DNA-bound complex.
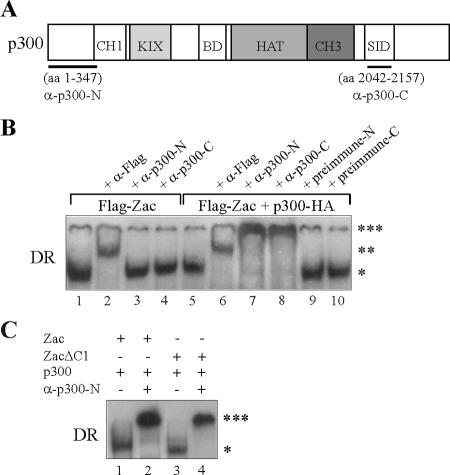
To test whether DNA-bound Zac stably complexes with p300 and, if so, whether ZF6 and ZF7 actually participate in p300 binding, we firstly established EMSA with extracts prepared from PA-TU cells expressing ectopic Zac protein tagged with the Flag epitope. Control experiments confirmed that DNA complex formation was Zac dependent and sequence specific (data not shown). The addition of Flag antibody to the reaction mix supershifted Zac complexes on the DR and PAL elements in a dose-dependent manner (Fig. 5B, lane 2; data not shown). While we detected no significant differences in the affinity of Zac-DNA complexes in the absence or presence of ectopic p300, prolonged electrophoresis disclosed a tiny but consistent decrease in mobility (data not shown). Based on this initial evidence, we raised polyclonal p300 antibodies directed against N- or C-terminal regions of p300 as outlined in the legend to Fig. 5A. Both antibodies failed to affect Zac-DNA complexes in the absence of ectopic p300 (Fig. 5B, lanes 3 and 4). However, following cotransfection of p300, they caused dose-dependent supershifts (lanes 7 and 8), while the respective preimmune sera were ineffective (lanes 9 and 10). EMSA with the PAL element showed similar results under these conditions (data not shown). Supershifted complexes migrated at largely reduced mobility and demonstrated that nearly all of Zac was complexed with p300. These findings begged the question of whether ZF6 and ZF7 in a DNA-bound state participated in p300 binding or whether, alternatively, this function solely reflected the presence of the C1 region. To evaluate the contribution of the C1 region to Zac-p300-DNA complex formation, we compared Zac and ZacΔC1 in EMSA. Both Zac proteins were strongly supershifted on the DR and PAL elements in the presence of p300 (Fig. 5C; data not shown). Together with results from Fig. 4C, showing that ZF2, ZF6, and ZF7 underlie p300 binding, Zac interaction with p300 despite the absence of the C1 region strongly suggests that DNA-bound ZF6 and ZF7 participate in p300 binding.
FIG. 5.
Zac zinc fingers and p300 form a DNA-bound complex. (A) Scheme of p300. Fragments used for immunization are underlined; numbers denote amino acids. The corresponding antibodies are labeled α-p300-N and α-p300-C. SID, steroid receptor interacting domain; BD, bromodomain. (B) EMSA. PA-TU cells were transfected with Zac (0.2 μg of pRK7Flag-Zac) in the absence or presence of p300-HA (1.0 μg of pCMV-p300-HA). Zac-DNA complexes on the DR element ( ) are supershifted by anti-Flag antibodies (
) are supershifted by anti-Flag antibodies (
 ). Anti-p300 antibodies (α-p300-N and α-p300-C) were ineffective in the absence of ectopic p300 (lanes 3 and 4) but strongly supershifted Zac-DNA complexes (
). Anti-p300 antibodies (α-p300-N and α-p300-C) were ineffective in the absence of ectopic p300 (lanes 3 and 4) but strongly supershifted Zac-DNA complexes (

 ) upon p300 cotransfection (lanes 7 and 8). Preimmune sera were ineffective (lanes 9 and 10). (C) EMSA. Adjusted amounts of Zac (0.2 μg of pRK7Flag-Zac) and ZacΔC1 (0.04 μg of pRK7Flag-ZacΔC1) were transfected in PA-TU cells and tested as described above.
) upon p300 cotransfection (lanes 7 and 8). Preimmune sera were ineffective (lanes 9 and 10). (C) EMSA. Adjusted amounts of Zac (0.2 μg of pRK7Flag-Zac) and ZacΔC1 (0.04 μg of pRK7Flag-ZacΔC1) were transfected in PA-TU cells and tested as described above.
C1 region mediates coactivation via p300.
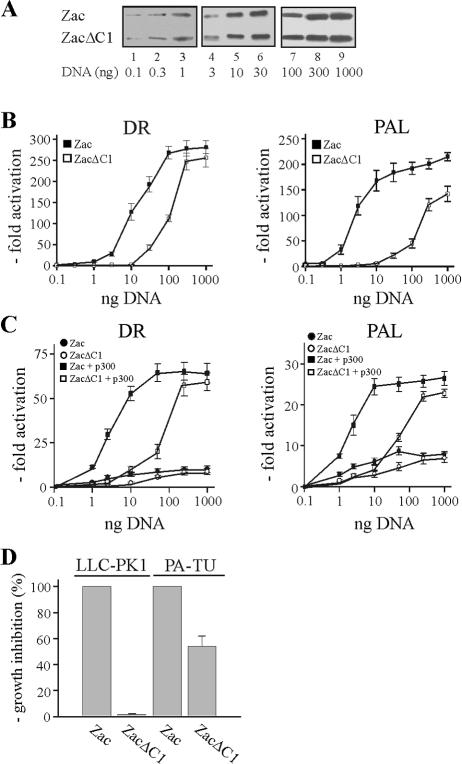
Above experiments showed that DNA-bound Zac formed a stable complex with p300 even in the absence of the C1 region. Then, what is the function of the C1 region in p300 binding? To answer this question, we cotransfected Zac and ZacΔC1 into LLC-PK1 cells at low doses to account for rate-limiting concentrations of endogenous p300. Immunoblots of adjusted amounts of the respective expression vectors demonstrated equal levels of Zac proteins (Fig. 6A). Strikingly, we measured a dramatic difference in transactivation efficiency between these two Zac proteins on the DR and PAL elements (Fig. 6B). These differences were strongest at low doses of Zac (>100-fold) and steadily declined at higher ones, to almost disappear for maximal transactivation. We further cotransfected Zac or ZacΔC1 into p300-negative PA-TU cells and measured a largely blunted twofold difference in transactivation efficiency (Fig. 6C). Interestingly, cotransfection of a given amount of p300 strongly restored Zac but not ZacΔC1 activity at low concentrations on the DR and PAL DNA elements (Fig. 6C). Together, these data suggested a role for cooperative binding of the C1 region and of the zinc fingers to the KIX domain to mediate coactivation via p300. If so, high doses of ZacΔC1 could overcome impaired binding of the zinc fingers to the KIX domain, restoring similar levels of maximal transactivation. Moreover, the almost complete loss in transactivation at low doses of ZacΔC1 indicates that coordinated binding to the KIX and CH3 domains controls Zac coactivation by p300.
FIG. 6.
The C1 region mediates coactivation via p300. (A) Zac or ZacΔC1 were transfected into LLC-PK1 cells, and different amounts of cell extracts (left lanes, 50 μg; middle lanes, 20 μg; and right lanes, 5 μg) were immunoblotted with α-Zac. Indicated amounts of DNA refer to Zac with ZacΔC1 adjusted appropriately. (B) The indicated doses of Zac or adjusted amounts of ZacΔC1 were cotransfected with the DR or PAL reporter plasmids (2 μg each) into LLC-PK1 cells. (C) Zac or ZacΔC1 was cotransfected with the indicated reporters in the absence or presence of a given amount of p300 (0.5 μg of pCMV-p300-HA) into PA-TU cells. (D) Colony formation assay. Zac (0.2 μg of pRK7Flag-Zac) or ZacΔC1 (0.04 μg of pRK7Flag-ZacΔC1) were cotransfected with a puromycin resistance vector (pRK7Pur) at a ratio of 3:1 into LLC-PK1 and PA-TU cells. Selection (puromycin, 2 μg/ml) began on day 2, and medium was renewed every third day. Colonies were stained with MTT (methylthiazolyldiphenyl-tetrazolium bromide; 1 mg/ml) on day 10 and counted. Growth inhibition by Zac was set to 100%.
The fact that the C1 region and zinc fingers together seemed to regulate coactivation by p300 preferentially at low doses appears pertinent to Zac's biological function regarding rather low protein levels in vivo (4, 35, 36, 49, 50). Since Zac-dependent regulation of target genes and antiproliferation demands transactivation (18), we carried out colony formation assays to test whether the differences noted above correlate with Zac's biological activity. While Zac potently repressed cell proliferation in LLC-PK1 cells, we noted a >10-fold decline in PA-TU cells (data not shown). More importantly, there was a >50-fold difference between Zac and ZacΔC1 in growth inhibition of LLC-PK1 cells, while the difference between Zac and ZacΔC1 in growth inhibition of PA-TU cells was only twofold (Fig. 6D). Thus, coactivation by p300 appears to correlate well with Zac antiproliferation in vivo.
KIX and CH3 domains interdependently regulate Zac coactivation.
We conducted in vivo competition experiments for p300 binding to infer the function of the KIX and CH3 domains in Zac coactivation (diagrammed in Fig. 7B). We cotransfected Zac with adjusted amounts of the isolated CH1, KIX, and CH3 domains into LLC-PK1 cells (Fig. 7A). Interestingly, increasing amounts of either the KIX or CH3 domain severely impaired Zac transactivation on the DR and PAL elements (Fig. 7C and D). Overexpression of the KIX domain in conjunction with the CH3 domain, which most likely disrupted the interaction between Zac and p300, was only slightly more effective than overexpression of either domain alone. Because overexpression of the CH1 domain hardly affected Zac transactivation, transcriptional squelching appeared rather unlikely. In support of this idea, p53 displayed a differential response (data not shown) with the CH1 and CH3 domains, producing moderate and potent inhibition, respectively (17). Conceptually, competition between ectopically expressed p300 domains and endogenous p300 for Zac binding indicated that coactivation required both the KIX and CH3 domains. Additionally, overexpression of the CH3 domain, which competed solely the interaction of the zinc fingers with the CH3 domain of endogenous p300, pointed to a regulatory role of the zinc fingers in coactivation via p300.
FIG. 7.
KIX and CH3 domains are necessary for Zac coactivation. (A) Immunoblot of LLC-PK1 cells expressing adjusted amounts of single p300 domains (0.5 μg of pRK7Flag-CH1 [aa 302 to 528], 1.0 μg of pRK7Flag-KIX [aa 566 to 650], and 3.0 μg of pRK7Flag-CH3 [aa 1197 to 1673]) as detected by α-Flag. (B) Competition scheme. Broken lines show binding of Zac ZF6 and ZF7 and of the C1 region to the KIX and CH3 domains of endogenous p300. Intact lines symbolize competition by overexpression of the isolated KIX and CH3 domains (circles). (C and D) In vivo competition experiment. Zac and DR or PAL reporters were cotransfected with increasing amounts of the CH1, KIX, and CH3 domains singly or together into LLC-PK1 cells.
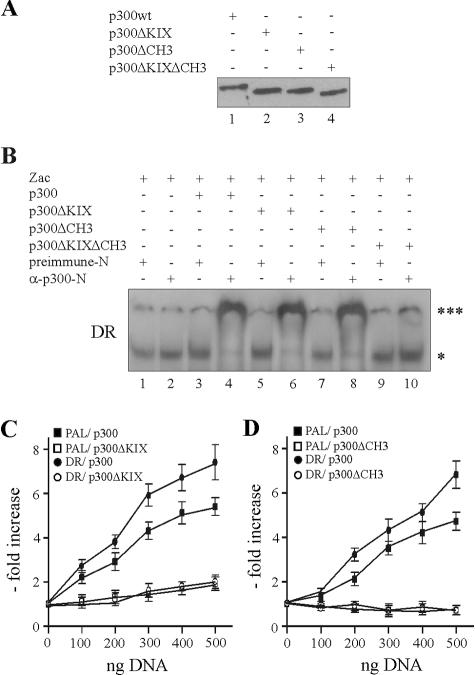
We further studied this concept by expressing p300 and derivatives in PA-TU cells. To investigate the stability of complexes formed by DNA-bound Zac and p300 derivatives, we adjusted the concentrations of the expression vectors to achieve similar protein levels for p300, p300ΔKIX, p300ΔCH3, and p300ΔKIXΔCH3 as shown in Fig. 8A. Subsequent EMSA with the DR element evidenced indistinguishable supershifts between p300 (Fig. 8B, lane 4) and derivatives in which the KIX or CH3 domains were singly deleted (lanes 6 and 8), whereas absence of both domains completely prevented the formation of Zac-p300 complexes (lane 10). Similar results were obtained for the PAL element (data not shown). Reporter assays conducted in PA-TU cells showed that cotransfection of p300ΔKIX failed to enhance Zac transactivation on the DR or PAL elements (Fig. 8C), while p300ΔCH3 even inhibited residual Zac activity (Fig. 8D). As expected, p300ΔKIXΔCH3 failed to affect transactivation in agreement with the absence of stable Zac-p300 complexes (data not shown). These experiments strengthened the concept that coordinated p300 binding via the KIX and CH3 domains mediates Zac coactivation.
FIG. 8.
KIX and CH3 domains interdependently regulate Zac coactivation. (A) Immunoblot of PA-TU cells expressing adjusted amounts of p300 or derivatives as detected by α-Flag (10 μg of pCI.Flag-p300, 5 μg of pCI.Flag-p300ΔKIX, 3 μg of pCI.Flag-p300ΔCH3, or 1 μg of pCI.Flag-p300ΔKIX ΔCH3). (B) EMSA with DR element. PA-TU cells were transfected with Zac in the absence or presence of adjusted amounts of p300, p300ΔKIX, p300ΔCH3, or p300ΔKIXΔCH3. Zac-DNA complexes ( ) were unaffected by preimmune-N or α-p300-N sera in the absence of p300 (lanes 1 and 2). They were indistinguishably supershifted (
) were unaffected by preimmune-N or α-p300-N sera in the absence of p300 (lanes 1 and 2). They were indistinguishably supershifted (

 ) by α-p300-N but not by preimmune-N serum in the presence of p300, p300ΔKIX, or p300ΔCH3 (lanes 3 to 8), while p300ΔKIXΔCH3 was ineffective (lanes 9 and 10). (C and D) Zac (0.1 μg of pRK7Flag-Zac) and DR or PAL reporters (2 μg each) were cotransfected with increasing amounts of p300, p300ΔKIX, and p300ΔCH3 into PA-TU cells. Indicated amounts of DNA refer to p300 and appropriately adjusted amounts of the derivatives.
) by α-p300-N but not by preimmune-N serum in the presence of p300, p300ΔKIX, or p300ΔCH3 (lanes 3 to 8), while p300ΔKIXΔCH3 was ineffective (lanes 9 and 10). (C and D) Zac (0.1 μg of pRK7Flag-Zac) and DR or PAL reporters (2 μg each) were cotransfected with increasing amounts of p300, p300ΔKIX, and p300ΔCH3 into PA-TU cells. Indicated amounts of DNA refer to p300 and appropriately adjusted amounts of the derivatives.
p300 does not acetylate Zac.
Only few reports have described an interaction between p300/CBP and canonical C2H2 zinc fingers controlling DNA binding; these examples include the transcription factors Sp1, KLF5, YY1, and WT1 (33, 45, 55, 59). Although the functional significance of these interactions with p300 remains elusive, increasing evidence has accumulated for a role of p300/CBP in regulating transcription factor activity through acetylation (25). Indeed, p300/CBP HAT-mediated acetylation of KLF5 and YY1 zinc fingers modifies their DNA-binding affinities (33, 59). Given these precedents, we investigated whether recombinant p300-HAT acetylates Zac or its isolated zinc finger domain. Results in Fig. 9 gave no evidence for either case, whereas the general transcription factors TFIIF (74 kDa) and TFIIE (30 kDa), which we included as positive controls, as well as p300-HAT itself, were potently acetylated. Even extended exposure for one week revealed no acetylation of Zac.
FIG. 9.
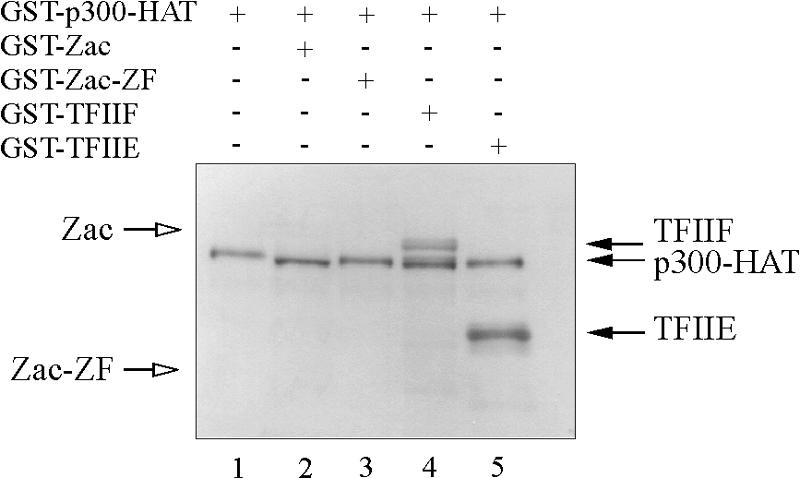
p300 does not acetylate Zac. Acetylation assay. GST-p300-HAT (25 nM) was incubated with GST fusions of Zac, Zac zinc fingers, or the general transcription factors TFIIF or TFIIE (1 μM each). Reaction mixtures were fractionated on a 10% SDS-polyacrylamide gel. A representative autoradiogram following 2,5-diphenyloxazol treatment and exposure for 2 days is shown. Open arrows indicate predicted positions of Zac and zinc fingers; filled arrows mark positions of acetylated TFIIF, TFIIE, and autoacetylated p300-HAT.
Coordinated Zac binding regulates in vitro p300-HAT activity.
Our model of Zac's interaction with p300 suggested that rather than recruitment per se coordinated binding of ZF6 and ZF7 together with the C1 region to the CH3 and KIX domains regulated HAT-dependent coactivation.
To exclude any other potential effectors that could assist binding to p300 and modify HAT activity in vivo, we performed in vitro HAT assays with recombinant Zac in the presence of its DNA elements and in vitro-translated p300 protein. Recent reports suggest that small peptides are suitable substrates for kinetic or mechanistic studies of p300-HAT activity (8, 47). Therefore, we compared firstly core histones to a peptide (corresponding to the first 24 amino acids of histone H4) that is coupled to biotin to allow rapid quantification of acetylation products. A kinetic analysis with saturating amounts of core histones or H4 peptide and [3H]acetyl-CoA revealed closely related acetylation reactions with a linear range between 1 and 7 min (Fig. 10A). We additionally tested various concentrations of core histones and H4 peptide, using as the shortest possible period an incubation time of 2 min, and plotted the incorporated radioactivity (3H acetylation) in a double-reciprocal graph. The deduced Km values for core histones [Km(ch), 21 μM] and H4 peptide [Km(h), 25 μM] were strongly related (Fig. 10B), confirming the validity of H4 peptides as substrates for p300-mediated acetylation.
The addition of Zac dose-dependently enhanced HAT activity, reaching a maximum at equimolar ratios of Zac and p300 (Fig. 10C). Higher Zac doses, however, caused a decline, probably due to competition for limited amounts of available p300 binding sites, leading to one-sided contacts blocking p300 activation. In contrast to the biphasic response for increasing Zac concentrations, we observed a steady decline for ZacΔC1, probably due to binding solely to the CH3 domain. We conducted this experiment also in the presence of ZacΔZF6, which binds to perfect DR elements (18) and p300 indistinguishably from wild-type Zac (data not shown). However, ZacΔZF6 did not significantly regulate HAT activity, strengthening the regulatory role of the zinc fingers (Fig. 10C).
Mutational and crystallographic studies suggest that nuclear acetyltransferases share a structurally conserved central core domain, which mediates acetyl-CoA binding and catalysis, whereas the sequence variability within a structurally related framework of the N- and C-terminal domains determines substrate binding specificity. A recent report, however, demonstrated that amino acid residues in the middle of the p300-HAT domain, which is the evolutionarily best-conserved subregion, are intimately involved in both acetyl-CoA and histone binding (8). Given this functional entity, we studied in parallel the influences of Zac on p300 histone and acetyl-CoA binding with differing concentrations of H4 peptide or [3H]acetyl-CoA and an excess of the Zac DR element. We plotted the incorporated radioactivity (3H acetylation) in a double-reciprocal graph to deduce Km values for histone [Km(h)] and acetyl-CoA binding [Km(a)]. We conducted these experiments in the absence or presence of Zac, ZacΔC1, or ZacΔZF6. Interestingly, the addition of Zac enhanced histone and acetyl-CoA binding [Km(h), 16 μM; Km(a), 14 μM] compared to p300 alone [Km(h), 25 μM; Km(a), 20 μM] (Fig. 10D). In contrast, ZacΔC1 reduced [Km(h), 47 μM; Km(a), 26 μM], while ZacΔZF6 failed to regulate p300-HAT activity [Km(h), 25 μM; Km(a), 21 μM]. Results for Zac and ZacΔC1 in the presence of the DR element were identical to those measured in the presence of the PAL element (data not shown). Since ZacΔZF6 fails to bind the PAL element (18), we tested regulation of HAT activity in this case solely in the presence of the DR element. Moreover, Zac but not ZacΔC1 or ZacΔZF6 seemed to increase 3H-acetylation rates (see below).
Together, our data indicate (i) that equimolar ratios of Zac and p300 most efficiently induce HAT activity, (ii) that coordinated Zac binding similarly increases p300 histone and acetyl-CoA binding, (iii) that ZF6 and ZF7 exert an important regulatory function, since binding to the CH3 domain alone inverts this enhancement, and (iv) that the zinc fingers are indispensable to HAT regulation.
To validate this concept, we further studied the regulation of p300ΔKIX and p300ΔCH3. Kinetic analysis displayed robust HAT activity similar to p300 (see below). The apparent Km values of p300ΔKIX [Km(h), 19 μM; Km(a), 19 μM] mirrored the ones of p300 [Km(h), 25 μM; Km(a), 20 μM], while they clearly increased by two- and threefold, respectively [Km(h), 51 μM; Km(a), 60 μM] in the case of p300ΔCH3 (Fig. 10E and F; for a survey, see Fig. 11D). This suggested a critical role of the CH3 domain in modulating the affinity of p300 substrate binding. In accord with results given above, binding of Zac solely to the CH3 domain of p300ΔKIX relatively impaired the affinities [Km(h), 26 μM; Km(a), 24 μM], whereas binding to the KIX domain of p300ΔCH3 was largely ineffective. These results evidenced the differential but interdependent functions of the KIX and CH3 domains in Zac-mediated regulation of p300 histone and acetyl-CoA binding.
FIG. 11.
Coordinated Zac binding regulates in vitro the catalytic activity of p300-HAT. (A) p300 (1 nM) was incubated in the absence or presence of equimolar amounts of Zac, ZacΔC1, or ZacΔZF6 with saturating amounts of H4 peptide and [3H]acetyl-CoA for the indicated times. All reactions contained the DR element. Linear least-squares fit was used to calculate maximal vz. (B and C) vz of p300ΔKIX or of p300ΔCH3 (1 nM each) in the absence or presence of equimolar amounts of Zac was determined as described above. (A through C) Data represent means and standard deviations from at least three experiments done in duplicate. (D) Survey of Km(h), Km(a), and vz values for p300 and derivatives in the absence or presence of Zac, ZacΔC1, and ZacΔZF6.
As Zac apparently enhanced p300 catalytic activity, we measured HAT reaction progress curves in the absence or presence of Zac, ZacΔC1, or ZacΔZF6. Linear least-squares fit was performed over the linear portion of data to deduce maximal rates of vz. Interestingly, equimolar amounts of Zac robustly increased vz by 67%, whereas ZacΔC1 impeded vz by 17% (Fig. 11A). Again, ZacΔZF6 failed to regulate HAT activity, reinforcing the essential role of the zinc fingers in this process. We additionally studied Zac regulatory effects on p300ΔKIX and p300ΔCH3 (Fig. 11B and C). p300ΔKIX resembled wild-type p300, while the vz of p300ΔCH3 paradoxically increased by 79%. This could indicate a more dynamic role of the CH3 domain in p300-HAT function by its activating substrate binding at low concentrations and by its inhibiting catalytic activity at high ones. Irrespective of this topic, the addition of Zac reduced the vz of p300ΔKIX by 25% and of p300ΔCH3 by 7%. Hence, Zac binding to either the KIX or CH3 domain alone failed to enhance or even reduced p300 acetylation rates. Therefore, in accord with Zac-mediated enhancement of p300 substrate binding, regulation of the catalytic activity strictly required coordinated binding of the C1 region and of the zinc fingers to p300.
Coordinated Zac binding regulates in vivo p300-HAT activity.
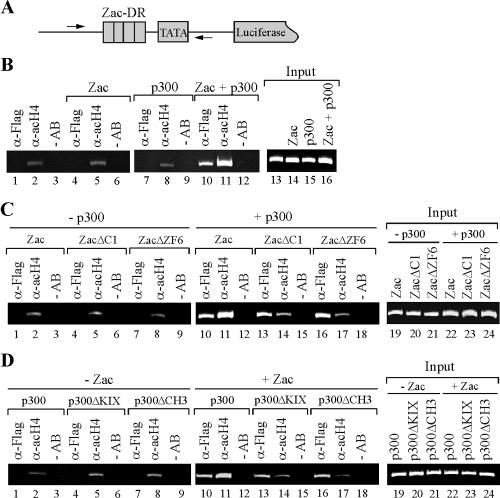
To validate whether Zac regulation of in vitro HAT activity mirrored regulation of p300-HAT activity in vivo, we cotransfected the DR element and HA-tagged Zac or derivatives (18) in the absence or presence of Flag-tagged p300 or derivatives into PA-TU cells. Following cultivation for 24 h, we carried out ChIP experiments with an antibody directed against acetylated histone H4. While H4 acetylation of the reporter (Fig. 12B, lane 2) increased weakly or not at all following cotransfection of either Zac or p300 alone (lanes 5 and 8), we detected strikingly increased H4 acetylation following cotransfection of Zac and p300 together (lane 11). Moreover, p300 recruitment to the reporter plasmid strictly correlated with the presence of Zac (compare lanes 7 and 10).
FIG. 12.
Coordinated Zac binding regulates in vivo p300-HAT activity. (A) Scheme of Zac reporter plasmid. Arrows indicate positions of PCR primers next to the direct repeat DNA elements (DR). (B) Zac-dependent p300-mediated histone H4 acetylation. p300-negative PA-TU cells transfected with Zac (20 ng of pRK7.HA-Zac) or p300 (0.5 μg of pCI.Flag-p300) alone or together were subjected to ChIP analysis with antibodies against p300 (α-Flag) or acetylated histone H4 (α-acH4) or without antibodies (−AB). PCR analysis with input chromatin confirmed that equal amounts were used for all reactions. (C) Coordinated Zac binding to p300 regulates H4 acetylation. p300 was transfected singly or together with adjusted amounts of Zac, ZacΔC1, or ZacΔZF6 (20 ng of pRK7.HA-Zac, 4 ng of pRK7.HA-ZacΔC1, or 20 ng of pRK7.HA-ZacΔZF6). ChIP analysis was done as described above. (D) KIX and CH3 domains interdependently regulate Zac-induced acetylation. Adjusted amounts of p300, p300ΔKIX, or p300ΔCH3 (0.5 μg of pCI.Flag-p300, 0.25 μg of pCI.Flag-p300 ΔKIX, or 0.15 μg of pCI.Flag-p300 ΔCH3) were transfected singly or together with Zac (20 ng of pRK7.HA-Zac). ChIP analysis was done as described above. (B through D) Results are representative of four independent experiments.
Having established conditions to detect Zac-controlled p300-mediated H4 acetylation, we evaluated Zac, ZacΔC1, and ZacΔZF6 in the absence or presence of ectopic p300 for their capability to support acetylation. In the absence of p300, they uniformly showed low levels of H4 acetylation (Fig. 12C, lanes 2, 5, and 8). In contrast, Zac but not ZacΔC1 or ZacΔZF6 strongly enhanced H4 acetylation in the presence of p300 (Fig. 12C, compare lane 11 with lanes 14 and 17). Importantly, all Zac constructs recruited p300 similarly (lanes 10, 13, and 16), demonstrating that coordinated Zac binding determined regulation of p300-HAT activity. To corroborate further this idea, we investigated H4 acetylation by p300 and its derivatives in the absence or presence of Zac. Neither p300 nor any of its derivatives enhanced H4 acetylation in the absence of Zac (Fig. 12D, lanes 2, 5, and 8). In contrast, solely p300 (lane 11) but neither p300ΔKIX nor p300ΔCH3 (lanes 14 and 17) conferred potent H4 acetylation in the presence of Zac. In sum, these data faithfully reflect the specific mode of Zac-p300 interaction deduced from the preceding sections and closely recapitulate the conclusions from the in vitro HAT assays.
DISCUSSION
The major conclusion from this work is that coordinated binding of Zac to p300, rather than recruitment per se, regulates HAT activity by simultaneously increasing substrate affinities and catalytic activity. The C2H2 zinc finger domain of Zac, which links DNA binding to HAT signaling, is a key element for this regulation. Thus, our results define a new function for this classic DNA recognition motif and demonstrate a molecular mechanism of potentially general importance to the direct enzymatic regulation of transcription by other DNA recognition motifs.
Coordinated Zac binding regulates p300 enzymatic activity.
The typical activator operates by binding to specific sites on DNA and contacting with its activating region the multiprotein machinery that directs transcription. According to this model, the specificity and magnitude of activation depend largely on the DNA-binding address of the activator and its capacity to recruit the transcriptional machinery (23). In continuation of this concept, recruitment of p300/CBP is a key event in triggering transcriptional activation through these coactivators. Consequently, the level of transcription and/or HAT activity induced by coactivator-dependent transcriptional factors is postulated to correlate with the strength of the transcription factor-coactivator interaction (22, 40).
In contrast, we show here that coordinated binding of Zac to p300 regulates HAT activity. In support of our concept, dominant negative mutants of the hepatocyte nuclear factor 1α paradoxically exhibit even stronger interaction with either CBP or PCAF than the wild-type protein; however, CBP and PCAF recruited by these mutants lack HAT activity (43). This suggests that the specific mode of interaction between a transcription factor and a coactivator regulates HAT activity, possibly by allosteric mechanisms. In the present study, we developed a comprehensive model for Zac binding to p300 by assigning possible interactions from a functional perspective based on complementary in vitro and in vivo experiments. Our model proposes that the Zac C1 region and the zinc fingers function as possible allosteric effectors of HAT activity by coordinated binding to the KIX and CH3 domains. Mechanistically, Zac increases the affinity for histone and acetyl-CoA binding and the catalytic activity. Since Zac binding to either the KIX or the CH3 domain alone confers no or even negative regulation, respectively, the KIX and CH3 domains must operate in a differential but interdependent all-or-nothing manner to mediate Zac coactivation. Our results represent the first demonstration of how a transcription factor can coordinately modulate various enzymatic properties of a HAT. Further, they raise the possibility that allosteric regulation of enzyme activity might extend to transcription factor-coactivator interactions, leading us to speculate that this type of regulation may be of general importance, potentially opening new opportunities for more selective targeting of aberrant HAT activity in disease.
Multitasking C2H2 zinc fingers link Zac DNA binding to the regulation of HAT activity.
While several proteins, including Zac, contain multiple C2H2 fingers which typically comprise the DNA recognition motif, other classes of zinc fingers act exclusively in protein-only or protein-lipid interactions. Among the latter is the RING family of zinc fingers that is involved in ubiquitination processes, phosphate inositol signaling (FYVE and PHD domains), and the assembly of large protein complexes (LIM, TAZ, and PHD domains) (32). A remarkably small number of DNA-bound C2H2 zinc fingers interacts with other proteins (30); for those that do, recent reports emphasized the importance of posttranslational modifications including acetylation (33, 59).
Zinc finger domains are common, relatively small protein motifs that fold around one or more zinc ions. Classic C2H2 zinc fingers conform to the well-characterized ββα-fold, comprising a short β-hairpin followed by a 10- to 11-residue α-helix. Residues near the N-terminal half of the α-helix are primarily responsible for contacts to nucleotides, binding in an antiparallel manner to DNA, with the N terminus directed into the major groove and the C terminus directed towards the surrounding environment. Therefore, DNA recognition and p300 binding most likely localize to separate surfaces of Zac ZF6 and ZF7 and illustrate that domains as small as 30 to 40 residues may represent two (or perhaps more) separable functional surfaces in transcriptional regulation. If so, what is the advantage of such a functionally linked entity? Most likely, coupling DNA binding to regulation of HAT activity would provide a potent link to transcription. While a large group of transcription factors may conform to the classic recruitment model, in which HAT activity is rather passively tethered, a subgroup, as exemplified by Zac, may regulate HAT activity more dynamically. Kinetic profiles of p300 occupancy in vivo predict that sustained association marks genes containing CpG islands, where local methylation and histone acetylation exert significant regulatory influence (41). Zac DNA-binding sites are GC rich, a feature typical of CpG islands (3). A restraint chromatin structure at Zac- or other DNA-binding sites may require potent HAT activity to promote transcription. Moreover, biochemical studies implicate the amino-terminal tails of histones both in folding of nucleosomes into higher order chromatin (12) and in directly controlling transcription factor binding to nucleosomal DNA (26, 53). Histone acetylation stabilizes the binding of transacting factors and might occur either before or concurrently with the interactions of transcription factors with their binding sites in chromatin (26, 53). From this perspective, the link between Zac DNA binding to HAT regulation would provide additional stability to the binding of Zac to nucleosomal DNA.
Based on the results presented here, we propose that this new mechanism, linking DNA binding to enzymatic regulation of transcription, may also apply to other activators which comprise C2H2 or other DNA recognition motifs and could be of general importance.
. . . . . . . . .
Acknowledgments
We gratefully acknowledge the gifts of p300, CBP, and PCAF plasmids by J. Boyes, R. Eckner, T. Kouzarides, and I. Talianidis. We are thankful to O. X. Almeida for critical reading of the manuscript.
D.S. was supported by the DFG.
REFERENCES
- 1.Abdollahi, A., A. K. Godwin, P. D. Miller, L. A. Getts, D. C. Schultz, T. Taguchi, J. R. Testa, and T. C. Hamilton. 1997. Identification of a gene containing zinc-finger motifs based on lost expression in malignantly transformed rat ovarian surface epithelial cells. Cancer Res. 57:2029-2034. [PubMed] [Google Scholar]
- 2.Alam, S., D. Zinyk, L. Ma, and C. Schuurmans. 2005. Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dyn. 234:772-782. [DOI] [PubMed] [Google Scholar]
- 3.Arima, T., T. Kamikihara, T. Hayashida, K. Kato, T. Inoue, Y. Shirayoshi, M. Oshimura, H. Soejima, T. Mukai, and N. Wake. 2005. Zac, Lit1 (Kcnq1Ot1) and P57(Kip2) (Cdkn1C) are in an imprinted gene network that may play a role in Beckwith-Wiedemann syndrome. Nucleic Acids Res. 33:2650-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basyuk, E., V. Coulon, D. A. Le, M. Coisy-Quivy, J. P. Moles, A. Gandarillas, and L. Journot. 2005. The candidate tumor suppressor gene ZAC is involved in keratinocyte differentiation and its expression is lost in basal cell carcinomas. Mol. Cancer Res. 3:483-492. [DOI] [PubMed] [Google Scholar]
- 5.Bilanges, B., A. Varrault, E. Basyuk, C. Rodriguez, A. Mazumdar, C. Pantaloni, J. Bockaert, C. Theillet, D. Spengler, and L. Journot. 1999. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene 18:3979-3988. [DOI] [PubMed] [Google Scholar]
- 6.Bilanges, B., A. Varrault, A. Mazumdar, C. Pantaloni, A. Hoffmann, J. Bockaert, D. Spengler, and L. Journot. 2001. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene 20:1246-1253. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, J. C., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, Y. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bordoli, L., S. Husser, U. Luthi, M. Netsch, H. Osmani, and R. Eckner. 2001. Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 29:4462-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciani, E., A. Hoffmann, P. Schmidt, L. Journot, and D. Spengler. 1999. Induction of the PAC1-R (PACAP-type I receptor) gene by p53 and Zac. Mol. Brain Res. 69:290-294. [DOI] [PubMed] [Google Scholar]
- 10.Debes, J. D., L. J. Schmidt, H. Huang, and D. J. Tindall. 2002. p300 mediates androgen-independent transactivation of the androgen receptor by interleukin 6. Cancer Res. 62:5632-5636. [PubMed] [Google Scholar]
- 11.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 12.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 13.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 14.Gayther, S. A., S. J. Batley, L. Linger, A. Bannister, K. Thorpe, S. F. Chin, Y. Daigo, P. Russell, A. Wilson, H. M. Sowter, J. D. Delhanty, B. A. Ponder, T. Kouzarides, and C. Caldas. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24:300-303. [DOI] [PubMed] [Google Scholar]
- 15.Gillardon, F., R. Hata, and K. A. Hossmann. 1998. Delayed up-regulation of Zac1 and PACAP type I receptor after transient focal cerebral ischemia in mice. Mol. Brain Res. 61:207-210. [DOI] [PubMed] [Google Scholar]
- 16.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 17.Grossman, S. R. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 268:2773-2778. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, A., E. Ciani, J. Boeckardt, F. Holsboer, L. Journot, and D. Spengler. 2003. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol. Cell. Biol. 23:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, S. M., A. H. Schonthal, and M. R. Stallcup. 2001. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene 20:2134-2143. [DOI] [PubMed] [Google Scholar]
- 20.Huang, S. M., and M. R. Stallcup. 2000. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol. Cell. Biol. 20:1855-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janknecht, R., and T. Hunter. 1996. Transcription. A growing coactivator network. Nature 383:22-23. [DOI] [PubMed] [Google Scholar]
- 23.Kadonaga, J. T. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116:247-257. [DOI] [PubMed] [Google Scholar]
- 24.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 27.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, D., J. P. H. Shield, W. Dean, I. Leclerc, C. Knauf, R. Burcelin, G. A. Rutter, and G. Kelsey. 2004. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J. Clin. Investig. 114:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, H., C. Nguyen, K. S. Lee, and M. Kahn. 2005. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene 24:3619-3631. [DOI] [PubMed] [Google Scholar]
- 30.Mackay, J. P., and M. Crossley. 1998. Zinc fingers are sticking together. Trends Biochem. Sci. 23:1-4. [DOI] [PubMed] [Google Scholar]
- 31.Mattar, P., O. Britz, C. Johannes, M. Nieto, L. Ma, A. Rebeyka, N. Klenin, F. Polleux, F. Guillemot, and C. Schuurmans. 2004. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 273:373-389. [DOI] [PubMed] [Google Scholar]
- 32.Matthews, J. M., and M. Sunde. 2002. Zinc fingers—folds for many occasions. IUBMB Life 54:351-355. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto, S., T. Suzuki, S. Muto, K. Aizawa, A. Kimura, Y. Mizuno, T. Nagino, Y. Imai, N. Adachi, M. Horikoshi, and R. Nagai. 2003. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 23:8528-8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 35.Pagotto, U., T. Arzberger, E. Ciani, F. Lezoualc'h, C. Pilon, L. Journot, D. Spengler, and G. K. Stalla. 1999. Inhibition of Zac1, a new gene differentially expressed in the anterior pituitary, increases cell proliferation. Endocrinology 140:987-996. [DOI] [PubMed] [Google Scholar]
- 36.Pagotto, U., T. Arzberger, M. Theodoropoulou, Y. Grubler, C. Pantaloni, W. Saeger, M. Losa, L. Journot, G. K. Stalla, and D. Spengler. 2000. The expression of the antiproliferative gene ZAC is lost or highly reduced in nonfunctioning pituitary adenomas. Cancer Res. 60:6794-6799. [PubMed] [Google Scholar]
- 37.Robertson, J. D., S. Orrenius, and B. Zhivotovsky. 2000. Review: nuclear events in apoptosis. J. Struct. Biol. 129:346-358. [DOI] [PubMed] [Google Scholar]
- 38.Rouaux, C., N. Jokic, C. Mbebi, S. Boutillier, J. P. Loeffler, and A. L. Boutillier. 2003. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 22:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozenfeld-Granot, G., J. Krishnamurthy, K. Kannan, A. Toren, N. Amariglio, D. Givol, and G. Rechavi. 2002. A positive feedback mechanism in the transcriptional activation of Apaf-1 by p53 and the coactivator Zac-1. Oncogene 21:1469-1476. [DOI] [PubMed] [Google Scholar]
- 40.Shaywitz, A. J., S. L. Dove, J. M. Kornhauser, A. Hochschild, and M. E. Greenberg. 2000. Magnitude of the CREB-dependent transcriptional response is determined by the strength of the interaction between the kinase-inducible domain of CREB and the KIX domain of CREB-binding protein. Mol. Cell. Biol. 20:9409-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, J. L., W. J. Freebern, I. Collins, A. De Siervi, I. Montano, C. M. Haggerty, M. C. McNutt, W. G. Butscher, I. Dzekunova, D. W. Petersen, E. Kawasaki, J. L. Merchant, and K. Gardner. 2004. Kinetic profiles of p300 occupancy in vivo predict common features of promoter structure and coactivator recruitment. Proc. Natl. Acad. Sci. USA 101:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soutoglou, E., G. Papafotiou, N. Katrakili, and I. Talianidis. 2000. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem. 275:12515-12520. [DOI] [PubMed] [Google Scholar]
- 43.Soutoglou, E., B. Viollet, M. Vaxillaire, M. Yaniv, M. Pontoglio, and I. Talianidis. 2001. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 20:1984-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spengler, D., M. Villalba, A. Hoffmann, C. Pantaloni, S. Houssami, J. Bockaert, and L. Journot. 1997. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 16:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 5:29-41. [DOI] [PubMed] [Google Scholar]
- 46.Theodoropoulou, M., J. Zhang, S. Laupheimer, M. Paez-Pereda, C. Erneux, T. Florio, U. Pagotto, and G. K. Stalla. 2006. Octreotide, a somatostatin analogue, mediates its antiproliferative action in pituitary tumor cells by altering phosphatidylinositol 3-kinase signaling and inducing zac1 expression. Cancer Res. 66:1576-1582. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, P. R., D. Wang, L. Wang, M. Fulco, N. Pediconi, D. Zhang, W. An, Q. Ge, R. G. Roeder, J. Wong, M. Levrero, V. Sartorelli, R. J. Cotter, and P. A. Cole. 2004. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11:308-315. [DOI] [PubMed] [Google Scholar]
- 48.Valente, T., and C. Auladell. 2001. Expression pattern of Zac1 mouse gene, a new zinc-finger protein that regulates apoptosis and cellular cycle arrest, in both adult brain and along development. Mech. Dev. 108:207-211. [DOI] [PubMed] [Google Scholar]
- 49.Valente, T., M. I. Dominguez, A. Bellmann, L. Journot, I. Ferrer, and C. Auladell. 2004. Zac1 is up-regulated in neural cells of the limbic system of mouse brain following seizures that provoke strong cell activation. Neuroscience 128:323-336. [DOI] [PubMed] [Google Scholar]
- 50.Valente, T., F. Junyent, and C. Auladell. 2005. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev. Dyn. 233:667-679. [DOI] [PubMed] [Google Scholar]
- 51.Varrault, A., B. Bilanges, D. J. G. Mackay, E. Basyuk, B. Ahr, C. Fernandez, D. O. Robinson, J. Bockaert, and L. Journot. 2001. Characterization of the methylation-sensitive promoter of the imprinted ZAC gene supports its role in transient neonatal diabetes mellitus. J. Biol. Chem. 276:18653-18656. [DOI] [PubMed] [Google Scholar]
- 52.Varrault, A., E. Ciani, F. Apiou, B. Bilanges, A. Hoffmann, C. Pantaloni, J. Bockaert, D. Spengler, and L. Journot. 1998. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc. Natl. Acad. Sci. USA 95:8835-8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508-2518. [PMC free article] [PubMed] [Google Scholar]
- 54.Voz, M. L., J. Mathys, K. Hensen, H. Pendeville, V. Van, I., H. C. Van, M. Chavez, D. B. Van, M. B. De, Y. Moreau, and d. Van, V. 2004. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene 23:179-191. [DOI] [PubMed] [Google Scholar]
- 55.Wang, W., S. B. Lee, R. Palmer, L. W. Ellisen, and D. A. Haber. 2001. A functional interaction with CBP contributes to transcriptional activation by the Wilms tumor suppressor WT1. J. Biol. Chem. 276:16810-16816. [DOI] [PubMed] [Google Scholar]
- 56.Waterborg, J. H., and H. R. Matthews. 1994. Fluorography of polyacrylamide gels containing tritium. Methods Mol. Biol. 32:163-167. [DOI] [PubMed] [Google Scholar]
- 57.Yan, Z., S. Choi, X. Liu, M. Zhang, J. J. Schageman, S. Y. Lee, R. Hart, L. Lin, F. A. Thurmond, and R. S. Williams. 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 278:8826-8836. [DOI] [PubMed] [Google Scholar]
- 58.Yang, M. C., J. C. Weissler, L. S. Terada, F. Deng, and Y. S. Yang. 2005. Pleiomorphic adenoma gene-like-2, a zinc finger protein, transactivates the surfactant protein-C promoter. Am. J. Respir. Cell Mol. Biol. 32:35-43. [DOI] [PubMed] [Google Scholar]
- 59.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]