Abstract
In this report, we show that the Saccharomyces cerevisiae protein Tpa1p (for termination and polyadenylation) influences translation termination efficiency, mRNA poly(A) tail length, and mRNA stability. Tpa1p is encoded by the previously uncharacterized open reading frame YER049W. Yeast strains carrying a deletion of the TPA1 gene (tpa1Δ) exhibited increased readthrough of stop codons, and coimmunoprecipitation assays revealed that Tpa1p interacts with the translation termination factors eRF1 and eRF3. In addition, the tpa1Δ mutation led to a 1.5- to 2-fold increase in the half-lives of mRNAs degraded by the general 5′→3′ pathway or the 3′→5′ nonstop decay pathway. In contrast, this mutation did not have any affect on the nonsense-mediated mRNA decay pathway. Examination of mRNA poly(A) tail length revealed that poly(A) tails are longer than normal in a tpa1Δ strain. Consistent with a potential role in regulating poly(A) tail length, Tpa1p was also found to coimmunoprecipitate with the yeast poly(A) binding protein Pab1p. These results suggest that Tpa1p is a component of a messenger ribonucleoprotein complex bound to the 3′ untranslated region of mRNAs that affects translation termination, deadenylation, and mRNA decay.
Eukaryotic translation termination is mediated by the release factors eRF1 and eRF3. eRF1 directly recognizes any of the three stop codons in the ribosomal A site and contains three functional domains (5, 41). Domain 1 mediates stop codon recognition and contains the highly conserved TASNIKS and YXCXXXF motifs (4, 15, 61). Domain 2 interacts with the peptidyl transferase center to facilitate release of the nascent polypeptide through the action of the conserved GGQ motif (27, 60). Finally, domain 3 mediates eRF1 binding to eRF3 (24, 50).
eRF3 is a GTPase that facilitates eRF1 stop codon recognition and enhances polypeptide chain release (26, 56). As with eRF1, three distinct functional domains in eRF3 have been identified. The N and M domains of Saccharomyces cerevisiae eRF3, comprising amino acids 1 to 253, are dispensable for both cell viability and translation termination (65). The N domain is capable of promoting the propagation of a prion form of yeast eRF3 known as [PSI+] (62). The N and M domains of eRF3 have also been shown to mediate an interaction between eRF3 and the mammalian (35) and yeast (42) forms of poly(A) binding protein (Pab1p). The C-terminal domain of eRF3 in all eukaryotes (comprising amino acids 254 to 479 in yeast) contains a conserved GTPase fold and is essential for cell viability (65). More recently, it was shown that the GTPase activity is the essential function of this domain, where it acts to modulate stop codon recognition by eRF1 (56). Consistent with the importance of GTP binding and hydrolysis by eRF3, it was recently shown that only the GTP-bound form of eRF3 can bind eRF1 (42).
Recent studies have revealed a connection between eukaryotic translation termination and mRNA turnover. For example, the nonsense-mediated mRNA decay (NMD) pathway degrades mRNAs that carry premature stop codons. Unlike the general yeast 5′→3′ mRNA turnover pathway that requires removal of the poly(A) tail prior to decapping and degradation by the Xrn1p exonuclease (20), the yeast NMD machinery facilitates decapping and 5′→3′ turnover of nonsense-containing mRNAs without prior removal of the poly(A) tail (3, 29). The NMD response requires the dedicated factors Upf1p, Upf2p, and Upf3p (44). While the loss of any one of these factors abrogates the NMD response, their absence also reduces the efficiency of translation termination (40, 45, 73, 74). In addition, each of the Upf factors has been shown to interact with the release factors, eRF1 and eRF3 (18). Although translation termination is required for the activation of NMD, polypeptide chain release alone is not sufficient to initiate the NMD process. Rather, eRF3 carries out additional interactions following polypeptide chain release that are required for the induction of NMD (42). This process is also dependent on the distance between the stop codon and the poly(A) tail of the mRNA (1).
Recently, yeast eRF3 was also shown to influence the half-lives of normal mRNAs through the interaction of its N and M domains with Pab1p (42). Interestingly, when the N and M domains of eRF3 bind to the C-terminal domain of Pab1p, the ability of Pab1p to oligomerize with other Pab1p molecules on the poly(A) tail of an mRNA is inhibited (35). This attenuation of Pab1p interactions may make the poly(A) tail transiently more accessible to the deadenylation machinery during each successive round of translation termination, leading to progressive shortening of the poly(A) tail. Ultimately, this process may shorten the poly(A) tail to a point at which Pab1p can no longer oligomerize efficiently, leading to the induction of mRNA turnover.
In the current study, we examine the function of a previously uncharacterized protein, Tpa1p. Tpa1p was found to interact with eRF1, eRF3, and Pab1p. Consistent with its association with components of the translation termination complex, the loss of Tpa1p increased readthrough of stop codons. A tpa1Δ mutation also caused an increase in the half-lives of both the ACT1 and CYH2 mRNAs and extended their poly(A) tail lengths in a manner similar to that observed in studies with eRF3 mutants that affected translation termination-coupled mRNA turnover (36, 42). Both the mRNA metabolism and translation termination defects observed in a tpa1Δ strain were abrogated by disruption of the PAN2 gene, which encodes the catalytic subunit of the poly(A) nuclease (PAN) deadenylation complex. Together, these findings suggest that the effects of Tpa1p on translation termination and mRNA stability are mediated by a messenger ribonucleoprotein (mRNP) complex bound to the 3′ untranslated region (UTR) of mRNAs that influences both translation termination and the deadenylation of mRNA poly(A) tails.
MATERIALS AND METHODS
Strains and growth conditions.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. Strains YDB415, YDB499, YDB635, and YDB646 were derived from strain YJW614. Strains YDB636 and YDB637 were derived from YJW615, while strain YDB644 was derived from PSY1209. All strain constructions were carried out using standard yeast genetic techniques. All gene knockouts were generated by deleting the entire open reading frame (14). Synthetic minimal dextrose medium is a supplemented minimal medium containing 2% glucose and other required nutritional supplements.
TABLE 1.
Strains used in this study
| Strain | Genotype |
|---|---|
| PSY1209 | MATaade2 his3 leu2 ura3 trp1 mex67::HIS3 [pRS316-URA3-MEX67] |
| PSY1799 | MATaade2 his3 leu2 ura3 trp1 mex67::HIS3 [pHT4667-URA3-mex67-5] |
| YJW614 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 [psi−] |
| YJW615 | MATα leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 tpa1::LEU2 [psi−] |
| YJW618 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 [PSI+] |
| YJW619 | MATα leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 tpa1::LEU2 [PSI+] |
| YDB415 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 upf1::TRP1 [psi−] |
| YDB499 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 ski7::TRP1 [psi−] |
| YDB635 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 pan2::TRP1 [psi−] |
| YDB636 | MATα leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 pan2::TRP1 tpa1::LEU2 [psi−] |
| YDB637 | MATα leu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 tpa1::LEU2 pop2::TRP1 [psi−] |
| YDB644 | MATaade2 his3 leu2 ura3 trp1 tpa1::LEU2 mex67::HIS3 [pRS316-URA3-MEX67] |
| YDB646 | MATaleu2-3,112 his3-11,15 trp1-1 ura3-1 ade1-14 pop2::TRP1 [psi−] |
Plasmids.
The plasmid used to express Tpa1p-HA (pDB1005) was made by PCR amplifying the TPA1 gene, including its promoter and transcription terminator regions, from yeast genomic DNA, using primers DB873 (5′-CCGGCTGCAGATCAAGAATGCTAATCAATTC-3′) and DB874 (5′-CCGGGGATCCAGTTAAACTTATATTCATTC-3′), and cloning the product into YCplac22 (with TRP1 as a selectable marker). A SalI restriction site was added on the 3′ end of the TPA1 gene by site-directed mutagenesis, using primers DB876 (5′-GGAAGATGAAGCGTCGACAATTAACCCGTC-3′) and DB877 (5′-GACGGGTTAATTGTCGACGCTTCATCTTCC-3′). A synthetic DNA fragment encoding a hemagglutinin (HA) epitope tag was then introduced into the SalI site, using the phosphorylated oligonucleotides DB2035 (5′-TCGACTACCCCTATGACGTCCCAGATTACGCATAAC-3′) and DB2036 (5′-TCGAGTTATGCGTAATCTGGGACGTCATAGGGGTAG-3′). A plasmid containing the CBP1 gene under the control of the GAL10 promoter (YEpI152-26 CBP1) was generously provided by Carol L. Dieckmann, University of Arizona (63). Finally, the HAC1 gene was PCR amplified from yeast genomic DNA, using primers DB2776 (5′-CTGCAGATGTTTAAGACG-3′) and DB2777 (5′-CCATCAGAGAACCACGAC-3′), and cloned into YCplac22 under the control of the CUP1 promoter to create pDB991.
Dual luciferase assays.
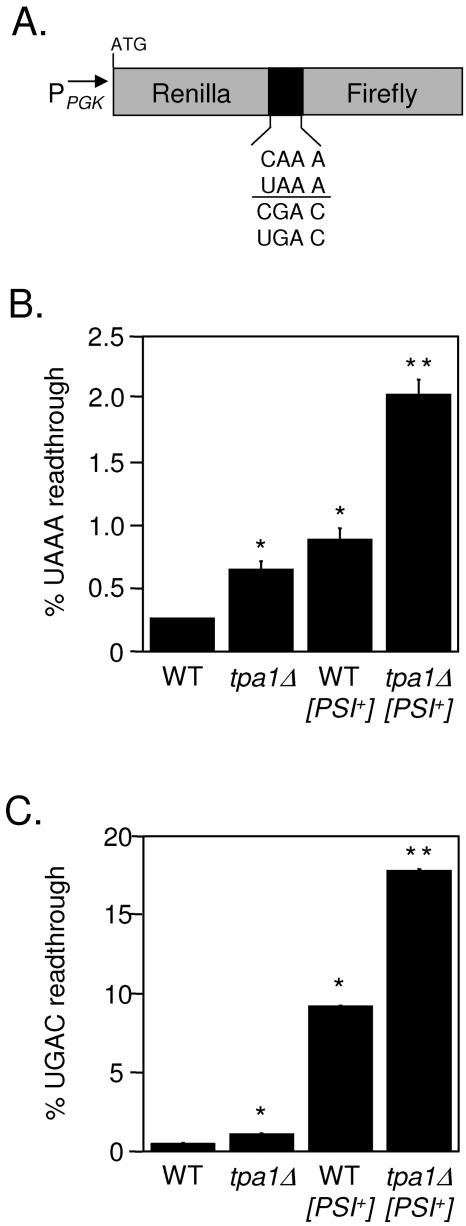
The dual luciferase reporters used to monitor readthrough of stop codons in yeast have been described previously (40). Briefly, this system utilizes tandem Renilla and firefly luciferase genes that are separated by a single in-frame stop codon or a corresponding sense codon control (Fig. 1A). The dual luciferase reporter plasmid pDB691 has a UGAC tetranucleotide termination signal between the Renilla and firefly genes, while pDB690 has the control CGAC sense sequence in the corresponding position. The plasmid pDB689 has a UAAA tetranucleotide termination signal between the Renilla and firefly genes, while pDB688 carries the control CAAA sense sequence in the corresponding position.
FIG. 1.
The efficiency of stop codon recognition is reduced in the tpa1Δ strain. (A) Diagram of dual luciferase readthrough reporter constructs. (B) Percent readthroughs of the UAAA tetranucleotide termination signal in WT, tpa1Δ, WT [PSI+], and tpa1Δ [PSI+] yeast strains. (C) Percent readthroughs of the UGAC tetranucleotide termination signal in WT, tpa1Δ, WT [PSI+], and tpa1Δ [PSI+] yeast strains. The percent readthrough values are expressed as means ± standard deviations. The presence of an asterisk (*) indicates a statistically significant increase in readthrough compared to the WT strain (P ≤ 0.002). The presence of a double asterisk (**) indicates a statistically significant increase in readthrough compared to the WT [PSI+] strain (P ≤ 0.002).
Yeast cells were transformed with the indicated dual luciferase reporter plasmids. Roughly 104 yeast cells were assayed for luminescence with a dual luciferase reporter assay system (Promega Corp.), using a Berthold Lumat LB9507 luminometer. The percent readthrough in each strain is expressed as the ratio of the firefly and Renilla luciferase activity units obtained from the nonsense construct divided by the ratio of the firefly and Renilla activity units produced by the sense construct and then multiplied by 100. Assays were done in quadruplicate, and the data are expressed as the means ± the standard deviations. Statistical significance was determined using the Mann-Whitney U test.
Coimmunoprecipitation experiments.
Yeast strains were grown to 0.5 A600 units/ml in synthetic minimal dextrose medium that maintained selection for plasmids in each strain. Ten A600 units of cells was harvested and washed once in ice-cold lysis buffer (20 mM HEPES buffer, pH 7.5, 10 mM potassium acetate, 20% glycerol, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 2 mg/ml pepstatin A, 2 mg/ml leupeptin, 2 mg/ml aprotinin) and then lysed in cold lysis buffer by mechanical disruption. The lysate was centrifuged in an Eppendorf microcentrifuge at 13,500 rpm at 4°C for 5 min to remove debris, and the supernatant was incubated with the appropriate antibody at 4°C for 2 h. The protein complexes were isolated using protein A-Sepharose beads (Sigma) and washed three times using cold lysis buffer. The samples were boiled in the presence of 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis buffer and then loaded onto an 8% SDS-polyacrylamide gel electrophoresis gel. After transfer of proteins to an Immobilon-P membrane (Millipore), using a semidry transfer apparatus (Bio-Rad Laboratories), the membrane was blotted with the appropriate antibody and visualized by incubation with 125I-protein A, followed by PhosphorImager analysis (GE Healthcare). Rabbit polyclonal antisera to eRF1 and eRF3 have previously been described (56), while a monoclonal antibody to F1β was a generous gift from Jeff Schatz. The mouse monoclonal antibody HA11 (Covance Research Products, Inc.) recognized the influenza HA epitope YPYDVPDYA. The rabbit polyclonal antibodies specific for yeast Pab1p were generously provided by David Mangus and Allan Jacobson, University of Massachusetts Medical School (46). To determine the dependence of these interactions on mRNA integrity, RNase A treatment of indicated samples was conducted by incubating the lysate for 2 h at 4°C with 1 mg/ml RNase A during the initial incubation with the primary antibody.
Northern blot and mRNA half-life analyses.
All yeast strains were grown to a cell density of 0.5 A600 units/ml prior to being harvested. To monitor the half-lives of mRNAs degraded by the 5′→3′ decay pathway, transcription was inhibited by the addition of 25 μg/ml thiolutin (CP-4092; Pfizer, Inc.). Aliquots of yeast cultures were collected at the indicated times after thiolutin addition. Total RNA was then isolated and analyzed by Northern blot analysis. To monitor the half-life of the CBP1 nonstop transcript degraded by the 3′→5′ turnover pathway, a protocol described previously (25) was utilized. Briefly, CBP1 transcription (under the control of the GAL10 promoter) was inhibited by a carbon source shift from 2% galactose to 2% glucose. Aliquots of cultures were taken at the indicated times after the shift, and RNA was isolated and subjected to Northern blot analysis. Total cellular RNA was isolated by SDS-phenol extraction (58). An equal amount of RNA (20 μg) from each strain was subjected to agarose gel electrophoresis in the presence of formaldehyde. Following transfer to nitrocellulose, the blots were incubated with probes labeled with [α-32P]dATP, using the random hexamer method. Radioactivity in specific hybrids was quantitated by PhosphorImager analysis (GE Healthcare). A probe for CYH2 analysis was PCR amplified from yeast genomic DNA, using the oligonucleotides DB585 (5′-CCGGTAAAGGTCGTATCGGT-3′) and DB586 (5′-GTTGATGCGCTTAAGCGATC-3′). A probe for ACT1 analysis was PCR amplified from yeast genomic DNA, using DB154 (5′-GCGCGGAATTCAACGTTCCAGCCTTCTACG-3′) and DB155 (5′-GGATGGAACAAAGCTTCTGG-3′). A probe for CBP1 analysis was PCR amplified from yeast genomic DNA, using DB2009 (5′-GCGACGAATCAACCACAGCA-3′) and DB2010 (5′-TGTGCCGATGGCATCATCAA-3′). The amount of RNA loaded on gels was normalized to the 25S rRNA level by quantitation using Quantity One Gel Doc software (Bio-Rad Laboratories, Inc.) after ethidium bromide staining.
Ligase-mediated poly(A) test.
The ligase-mediated poly(A) test (LMPAT) was carried out as previously described (57). Briefly, yeast cells were grown in yeast extract-peptone-dextrose medium to a cell density of 0.5 A600 units/ml, and total cellular RNA was isolated by the SDS-phenol method (58). Oligo(dT)18 (20 ng) was annealed to 250 ng of total RNA in the presence of T4 DNA ligase (Promega), followed by the addition of 200 ng of an anchored oligo(dT), DB2514 (5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′). The mRNA was then reverse transcribed using avian myeloblastosis virus reverse transcriptase (Promega Corp.) and PCR amplified using a 5′ primer specific to the 3′ end of either the CYH2 gene (DB585, 5′-CCGGTAAAGGTCGTATCGGT-3′) or the ACT1 gene (DB2223, 5′-CCGCTGCTCAATCTTCTTCA-3′) and the 3′ anchor primer (DB2295, 5′-GGCCACGCGTCGACTAGTAC-3′). Equal amounts of cDNA from each sample were visualized on a 6% native polyacrylamide gel, followed by ethidium bromide staining. DNA molecular weight markers were included on each gel as size standards.
RNase H/oligo(dT) Northern blot analysis.
Poly(A) tail lengths of the CYH2 transcript were also determined by RNase H endonuclease treatment, followed by Northern blot analysis (57). Total RNA (30 μg) was incubated with 25 pmol of the CYH2-specific oligonucleotide DB585 in the presence of RNase H buffer (40 mM HEPES, pH 7.9, 10 mM MgCl2, 60 mM KCl, 1 mM dithiothreitol [DTT]), 40 U RNasin (Promega), and 1 U RNase H (Promega) in a 20-μl reaction mixture with and without the addition of 500 pmol of oligo(dT)16 at 37°C for 30 min. The reaction was stopped by the addition of 1 μl of 0.5 M EDTA, and the RNA was ethanol precipitated and resuspended in 5 μl of gel tracking dye containing 8 M urea. The RNA samples were resolved on a 6% polyacrylamide gel containing 8 M urea for 4 h at 25 W. The RNA was transferred to Zetaprobe nylon (Bio-Rad), using a Bio-Rad Transblot SD semidry transfer cell for 30 min at 20 V. The membrane was baked at 80°C for 2 h, then incubated with a 300-bp CYH2 DNA probe that was PCR amplified from yeast genomic DNA, using the oligonucleotides DB585 (5′-CCGGTAAAGGTCGTATCGGT-3′) and DB2864 (5′-CCCTTACCCAAGATCTTACC-3′), and labeled with [α-32P]dCTP, using Ready-to-Go DNA labeling beads (Amersham). Radioactivity in specific hybrids was quantitated by PhosphorImager analysis (GE Healthcare). RNA molecular weight markers were included on each gel as size standards.
HAC1 mRNA export.
The HAC1 mRNA export assay was conducted as previously described (13) using isogenic strains that expressed either a wild-type or temperature-sensitive allele of the mRNA export receptor Mex67p (generously provided by Pam Silver, Harvard University). Both strains, PSY1209 and PSY1799, carried a deletion of the chromosomal MEX67 gene. PSY1209 carried a plasmid that expressed wild-type MEX67, while PSY1799 carried a plasmid that contained the temperature-sensitive mex67-5 mutant allele. The mutant Mex67p expressed in PSY1799 is unable to facilitate mRNA export at the restrictive temperature of 37°C (59).
Isogenic strains PSY1209 (wild type), PSY1799 (mex67-5), and YDB644 (tpa1Δ) were transformed with the HAC1 expression plasmid pDB991 and grown at 30°C (permissive temperature for mex67-5) in minimal selective media containing 2% glucose to 0.8 A600 units/ml. The cultures were shifted to 37°C for 30 min (nonpermissive temperature for mex67-5), and copper sulfate was added to the media at a final concentration of 0.5 mM to induce expression of HAC1 from the CUP1 promoter. The cells were cultured for an additional 10 min at 37°C, and 8 mM DTT was then added to induce the unfolded protein response (UPR) and splicing of the HAC1 transcript. Aliquots of the cultures were collected at the indicated times after DTT addition, and total RNA was extracted, analyzed by Northern blotting as described above, and quantitated by PhosphorImager analysis (GE Healthcare). A DNA probe for HAC1 mRNA was PCR amplified from yeast genomic DNA, using DB2776 (5′-CTGCAGATGTTTAAGACG-3′) and DB2777 (5′-CCATCAGAGAACCACGAC-3′). The percent spliced HAC1 mRNA was calculated by dividing the number of spliced HAC1 mRNAs by the number of spliced and unspliced HAC1 mRNAs and then multiplying by 100.
RESULTS
Loss of Tpa1p results in a readthrough phenotype.
The N-terminal domain of yeast eRF3 can induce the formation of a prion known as [PSI+] under certain conditions (62). In [PSI+] yeast strains, eRF3 molecules accumulate in prion aggregates that deplete the amount of functional eRF3 available for translation termination, resulting in a suppressor (or readthrough) phenotype. [PSI+] yeast cells can be reverted to [psi−] (also referred to as curing) by growth in the presence of guanidine hydrochloride (23, 53). Mutations in the TPA1 gene were originally identified in a Saccharomyces cerevisiae mutant screen designed to find “[PSI+] modifier” mutants. These mutants were selected based on their ability to block curing of [PSI+] by guanidine hydrochloride, as indicated by the persistence of a suppressor phenotype (L. Z. Osherovich and J. S. Weissman, unpublished data). Two possible mechanisms could account for the continued suppression phenotype in these potential “[PSI+] modifier” mutants: they could prevent curing of the [PSI+] phenotype, or they could simply reduce the efficiency of stop codon recognition in a [PSI+]-independent manner.
To distinguish between these possibilities for the tpa1 isolate, the TPA1 gene was deleted in [PSI+] and [psi−] backgrounds and stop codon readthrough was measured using dual luciferase readthrough reporter plasmids (40). The dual luciferase reporters contain an upstream Renilla luciferase gene and an in-frame downstream firefly luciferase gene that are separated by a readthrough cassette that contains either a stop codon or a sense codon (Fig. 1A). The efficiency of stop codon recognition is influenced primarily by the stop codon and the first downstream nucleotide, commonly referred to as the tetranucleotide termination signal (7, 11). Accordingly, each yeast strain was assayed using two sets of dual luciferase readthrough reporter plasmids. The first set contained a UAAA tetranucleotide termination signal, which was previously shown to be the most efficient termination signal. The second set contained a UGAC tetranucleotide termination signal, the least efficient termination signal (7, 40). In this system, the Renilla luciferase activity served as an internal control that provided a correction factor for any differences in mRNA stability or translation initiation rates between the different strains.
We measured a 2.5-fold increase in readthrough of the UAAA tetranucleotide termination signal in the tpa1Δ strain compared to that in the wild-type control. A 3.4-fold increase in readthrough was observed in the [PSI+] strain relative to that of the wild type, while the tpa1Δ [PSI+] strain showed an 8-fold increase in readthrough (Fig. 1B). A similar trend was observed at the UGAC tetranucleotide termination signal. A 1.8-fold increase in readthrough was measured for the tpa1Δ strain, a 9-fold increase in readthrough was measured for the [PSI+] strain, and an 18-fold increase in readthrough was measured for the tpa1Δ [PSI+] strain relative to that for the wild-type control (Fig. 1C). These results indicated that the tpa1Δ mutation caused a general defect in stop codon recognition in the absence of [PSI+]. Moreover, the increased readthrough associated with the tpa1Δ mutation could be superimposed on the readthrough associated with the [PSI+] state.
Tpa1p interacts with eRF1 and eRF3.
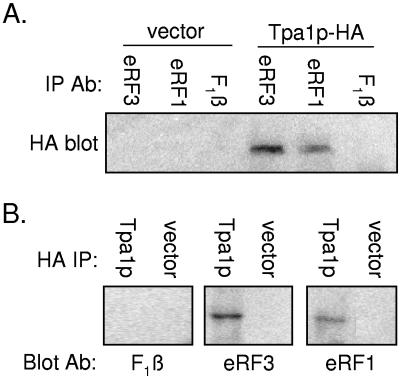
Since the tpa1Δ mutation caused a general increase in the readthrough of stop codons, we investigated whether Tpa1p interacts with the termination factors eRF1 and eRF3. To do this, a wild-type yeast strain was transformed with a low-copy-number plasmid expressing a version of Tpa1p containing an HA epitope tag (Tpa1p-HA) under the control of the TPA1 promoter. The plasmid expressing Tpa1p-HA was able to correct the readthrough phenotype associated with the tpa1Δ mutation (data not shown). To examine the association of Tpa1p with the termination factors, equal numbers of yeast cells carrying either the Tpa1-HA plasmid or vector alone were lysed and immunoprecipitated separately under nondenaturing conditions with antisera to eRF1 and eRF3 (56). As a negative control, the mitochondrial F1-ATPase β subunit (F1β) was also subjected to immunoprecipitation (Fig. 2A). The immunoprecipitated complexes were then subjected to a Western blot analysis with monoclonal antibodies to the HA epitope. We observed a band corresponding to the molecular mass of Tpa1p (74 kDa) in samples that expressed Tpa1-HA following immunoprecipitation with antiserum to eRF1 or eRF3 but not in samples that expressed vector alone. In addition, Tpa1-HA was not found to associate with F1β, demonstrating a specific Tpa1p interaction with each release factor. RNase A treatment of the cell lysates did not alter these interactions (data not shown), indicating that they were due to protein-protein interactions.
FIG. 2.
Tpa1p-HA interacts with eRF1 and eRF3. (A) The wild-type strain was transformed with vector alone or with a plasmid expressing a Tpa1p-HA construct. Cell lysates from these strains were subjected to immunoprecipitation (IP) under nondenaturing conditions, using antibodies (Ab) to the eRF1, eRF3, or F1β yeast protein. The protein complexes recovered were then subjected to Western blotting using a mouse monoclonal antibody to the HA epitope tag. (B) Yeast strains were transformed with vector alone or with a plasmid expressing Tpa1-HA. Cell lysates from these yeast strains were subjected to immunoprecipitation using a mouse monoclonal antibody to the HA epitope. The protein complexes recovered were then subjected to Western blotting using antiserum to the eRF1, eRF3, or F1β yeast protein.
The reciprocal immunoprecipitation experiment was also carried out. Lysates prepared from strains that carried the Tpa1p-HA plasmid or the vector alone were first immunoprecipitated with a monoclonal antibody to the HA epitope, and then a Western blot was probed with antiserum specific for eRF1, eRF3, or F1β (Fig. 2B). Again, an interaction was observed between Tpa1p and both eRF1 and eRF3 but not with F1β. When taken together, these results indicate that Tpa1p interacts with the eRF1 and eRF3 proteins. Currently, it is not known whether these represent direct interactions or whether other proteins provide a bridge between Tpa1p and the release factors.
Loss of Tpa1p does not affect NMD but prolongs the half-lives of mRNAs degraded by the general 5′→3′ decay pathway.
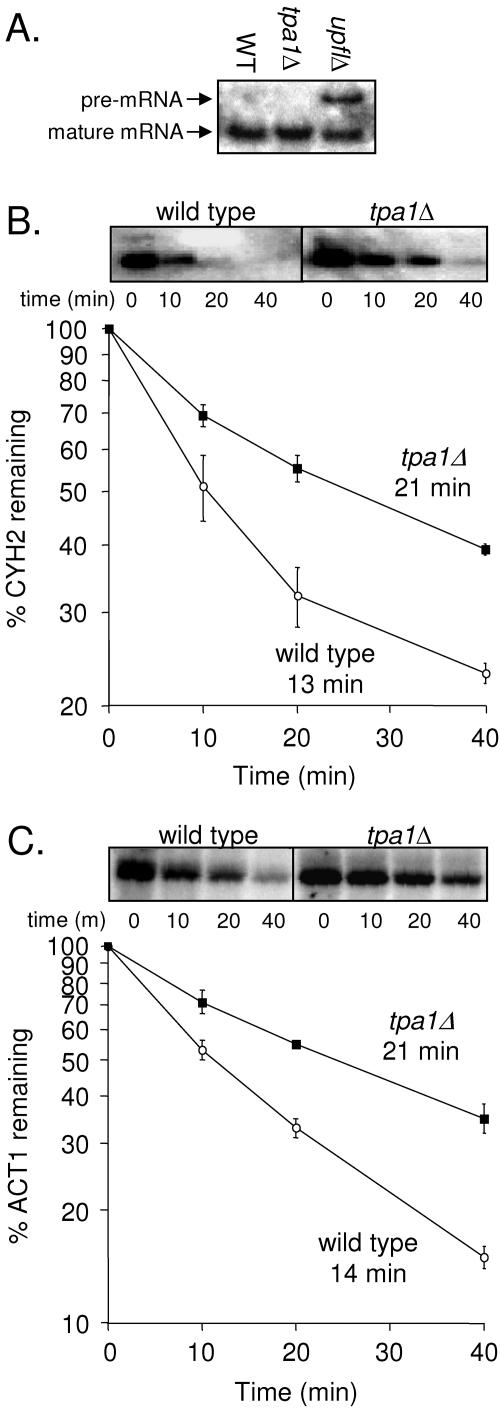
The NMD pathway degrades mRNAs that carry premature stop codons. Loss of one of the NMD factors such as Upf1p not only inhibits mRNA turnover by the NMD pathway but also causes readthrough of stop codons (40, 45, 72). To determine whether Tpa1p plays a role in the NMD pathway, we examined the efficiency of NMD in a tpa1Δ strain. In this experiment, wild-type and upf1Δ strains were included as controls. CYH2 transcripts are normally inefficiently spliced, resulting in the production of both unspliced and mature CYH2 mRNA species. Since the unspliced CYH2 mRNA contains an intron with an in-frame stop codon, it is a natural substrate for NMD and is present at very low levels in wild-type strains (31). However, the unspliced CYH2 mRNA is readily detected when a component of the NMD machinery such as Upf1p is missing or defective. When Northern blot analysis of total cellular RNA from the tpa1Δ strain was carried out using a probe that detects both CYH2 mRNA species, no accumulation of the CYH2 pre-mRNA was observed (Fig. 3A). These results suggest that Tpa1p does not play a significant role in mRNA turnover by the NMD pathway.
FIG. 3.
Substrates of the 5′→3′ mRNA decay pathway have longer half-lives in the tpa1Δ strain. (A) The effect of the tpa1Δ mutation on the NMD pathway was evaluated by monitoring the CYH2 pre-RNA by Northern blot analysis in WT, tpa1Δ, and upf1Δ strains. A representative blot is shown. (B) Quantitation of the amount of mature CYH2 mRNA remaining at specific times after transcription inhibition by thiolutin was monitored by Northern blot analysis. (C) Quantitation of the amount of mature ACT1 mRNA remaining at specific time points after transcription inhibition by thiolutin was monitored by Northern blot analysis. The abundance of each indicated mRNA was normalized to the level of 25S rRNA in each gel lane. The half-lives (in minutes) for the CYH2 and ACT1 mRNAs are indicated for the WT and tpa1Δ strains. Each time point shown is the mean ± standard deviation from three separate experiments.
We next asked whether Tpa1p plays a role in the turnover of normal cellular mRNAs, as indicated by the decay rates of the mature CYH2 and ACT1 mRNAs. To do this, cellular transcription was inhibited using the RNA polymerase inhibitor thiolutin (67). RNA was then isolated at the indicated times after transcription inhibition, and Northern blot analysis was carried out to determine the amount of RNA remaining at each time point, which allowed us to calculate the rate of mRNA turnover. We found that the half-lives of the mature CYH2 mRNA were 21 min in the tpa1Δ strain and 13 min in the wild-type strain (Fig. 3B), indicating that the loss of Tpa1p resulted in a 1.6-fold increase in the half-life of the CYH2 mRNA. Similarly, the half-lives of the ACT1 mRNA were 21 min in the tpa1Δ strain and 14 min in the wild-type strain, a 1.5-fold increase in mRNA half-life (Fig. 3C). Since most yeast mRNAs are degraded by the 5′→3′ turnover pathway (20, 52), these data suggest that the loss of Tpa1p results in a general decrease in the rate of mRNA turnover by this mechanism. Both the NMD and general 5′→3′ mRNA decay pathways are dependent upon the 5′→3′ exonuclease Xrn1p (51). Since the NMD pathway was unaffected by the loss of Tpa1p, Xrn1p activity does not appear to be inhibited by the loss of Tpa1p.
Loss of Tpa1p increases the half-life of an mRNA degraded by the nonstop 3′→5′ turnover pathway.
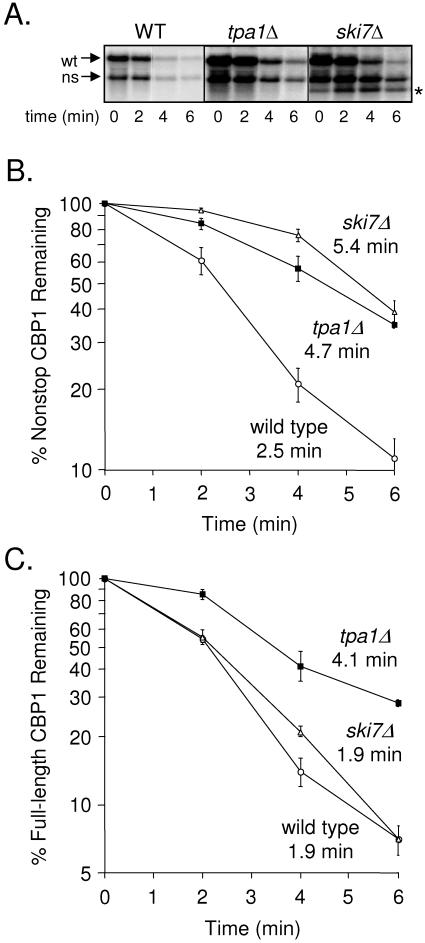
The nonstop mRNA decay pathway degrades transcripts that do not contain an in-frame stop codon (25, 71). Degradation of nonstop mRNAs is mediated by the 3′→5′ mRNA decay pathway that utilizes the cytoplasmic exosome, the SKI protein complex (composed of Ski2p, Ski3p, and Ski8p) (12), and Ski7p, which interacts with both the SKI complex and the exosome (2). To determine whether Tpa1p affects the half-life of mRNAs degraded by the 3′→5′ mRNA turnover pathway, we monitored the effect of the tpa1Δ mutation on the turnover of the CBP1 nonstop mRNA. Previous studies found that the CBP1 transcript carries a cryptic polyadenylation cleavage site within its coding sequence (49). Cleavage at this cryptic site leads to the production of an mRNA that is polyadenylated upstream of the translation termination signal, resulting in a truncated form of the CBP1 mRNA that lacks an in-frame stop codon. We expressed the CBP1 gene under the control of the GAL promoter in isogenic wild-type (WT), tpa1Δ, and ski7Δ yeast strains. After overnight growth in a medium containing galactose, the carbon source was shifted to glucose to shut off CBP1 transcription. The turnover rates of both nonstop and full-length CBP1 mRNAs in each strain were then monitored by Northern blot analysis (Fig. 4A). We found that the half-lives of the nonstop CBP1 transcript were 2.5 min in the wild-type strain and 4.7 min in the tpa1Δ strain (a 1.9-fold increase) (Fig. 4B). Similarly, the half-life of the nonstop CBP1 mRNA was extended to 5.4 min in the ski7Δ strain (a 2.2-fold increase).
FIG. 4.
A substrate of the 3′→5′ nonstop mRNA decay pathway has a longer half-life in the tpa1Δ strain. WT, tpa1Δ, and ski7Δ yeast strains were transformed with a plasmid expressing CBP1 under the control of the galactose-regulated GAL10 promoter. The amount of CBP1 mRNA remaining at specific times after CBP1 transcription was inhibited by a carbon source shift from galactose to glucose was monitored by Northern blot analysis. (A) Representative Northern blot of the normal (wt) and nonstop (ns) CBP1 mRNAs remaining at the indicated times after its transcription was inhibited by a carbon source shift from galactose to glucose. (B) Quantitation of the amount of nonstop CBP1 mRNA remaining at specific time points after its transcription was inhibited, as determined by Northern blot analysis. (C) Quantitation of the amount of full-length CBP1 mRNA remaining at specific time points after its transcription was inhibited, as determined by Northern blot analysis. The abundance of each indicated mRNA was normalized to the level of 25S rRNA in each gel lane. The half-lives (in minutes) of the nonstop and full-length CBP1 mRNAs are indicated for the WT, tpa1Δ, and ski7Δ strains. Each time point shown is the mean ± standard deviation from three separate experiments. The asterisk in panel A indicates a decay intermediate observed only in the ski7Δ strain.
The half-life of the full-length CBP1 mRNA was extended 2.2-fold by the tpa1Δ mutation, from 1.9 min to 4.1 min (Fig. 4C). In contrast, the half-life of this mRNA was not significantly altered in the ski7Δ strain. These results indicate that the full-length CBP1 mRNA is degraded primarily by the 5′→3′ mRNA decay pathway and not the 3′→5′ pathway. Since the tpa1Δ mutation resulted in prolonged half-lives of both the nonstop and full-length CBP1 mRNAs, it appears that the tpa1Δ mutation causes a more general defect in mRNA turnover than is observed with mutations that inactivate only the nonstop decay pathway. In addition, the presence of a prominent mRNA degradation product that accumulates in the ski7Δ strain (and presumably results from the block in the nonstop decay pathway) is not observed in the Northern blot from the tpa1Δ strain (Fig. 4A). Together, these findings suggest that the tpa1Δ mutation affects the 3′→5′ nonstop mRNA decay pathway in a manner that is distinct from that of the ski7Δ mutation.
Loss of Tpa1p leads to extended poly(A) tails.
The results presented above indicate that the tpa1Δ mutation increased the half-lives of mRNAs degraded by both the general 5′→3′ decay pathway and the 3′→5′ nonstop decay pathway but not those degraded by the NMD pathway. This led us to hypothesize that the length of poly(A) tails in the tpa1Δ strain may be increased, since both the 5′→3′ and 3′→5′ mRNA decay pathways are dependent upon the initial removal of the poly(A) tail while the yeast NMD pathway is not (3, 29). Two mRNA deadenylase complexes in yeast have been identified. The PAN complex trims poly(A) tails during processing of the 3′ ends of mRNAs (9, 10). Pan2p is the catalytic subunit of the PAN deadenylase complex, and the pan2Δ mutation leads to extended mRNA poly(A) tails. The second deadenylase, the CCR4 complex, is thought to mediate cytoplasmic poly(A) trimming (16, 19, 68, 69) and is composed of two subunits: the catalytic subunit Ccr4p and the Pop2p/Caf1p regulatory subunit that enhances CCR4 deadenylase activity. Loss of either subunit results in extended poly(A) tails on cellular mRNAs (19, 68, 69).
To examine the length of poly(A) tails in the tpa1Δ strain, we used two methods: an LMPAT and an RNase H/oligo(dT) Northern blot analysis. Total RNA was isolated from isogenic wild-type, tpa1Δ, pan2Δ, pop2Δ, tpa1Δ pan2Δ, and tpa1Δ pop2Δ yeast strains, and both assays were used on RNA from each strain to determine the length of the poly(A) tail on the CYH2 mRNA. We found that the poly(A) tail length of the CYH2 transcript was longer than normal in the tpa1Δ strain when assayed by the LMPAT (Fig. 5A) or the RNase H/oligo(dT) Northern blot analysis (Fig. 5B). As expected, both assays also indicated that the CYH2 poly(A) tail length was clearly longer in both the pan2Δ and pop2Δ strains than in the wild-type strain, and the poly(A) tail length in those strains also appears to be slightly longer than the CYH2 poly(A) tail length observed in the tpa1Δ strain. Interestingly, the CYH2 poly(A) tail length in the tpa1Δ pan2Δ double mutant was similar to that in the wild-type strain, indicating that the combination of these two mutations returned poly(A) tail length to normal. In contrast, the CYH2 poly(A) tail length in the tpa1Δ pop2Δ double mutant was longer than that in strains carrying either mutation alone. RNase H treatment in the presence of oligo(dT) indicated that each of the RNAs was of similar length, indicating that the site for cleavage and poly(A) addition was not altered in any of the mutants. Other LMPAT results indicated that the poly(A) tail length of the ACT1 transcript showed a trend similar to that observed for the CYH2 mRNA (data not shown). These results indicate that genetic interactions between tpa1Δ pan2Δ and tpa1Δ pop2Δ mutants influence poly(A) tail length on cellular mRNAs in distinct ways.
FIG. 5.
mRNAs have longer poly(A) tails in the tpa1Δ strain. Total RNA was isolated from wild-type, tpa1Δ, pan2Δ, pop2Δ, tpa1Δ pan2Δ, and tpa1Δ pop2Δ strains, and the lengths of poly(A) tails on the CYH2 mRNA were analyzed using an LMPAT or an RNase H/oligo(dT) Northern blot analysis. (A) LMPAT assay. The locations of primers used to PCR amplify the 3′ UTR of the CYH2 cDNA are indicated. In the LMPAT assay, RNA was reverse transcribed in the presence of oligo(dT), T4 DNA ligase, and an anchor primer that annealed preferentially to the end of the CYH2 poly(A) tail. The 3′ UTR region of the CYH2 transcript was PCR amplified using a 5′ primer that annealed 400 nucleotides upstream of the CYH2 stop codon and a 3′ anchor primer that annealed to the end of the poly(A) tail. Equal amounts of the PCR-amplified 3′ UTR region of the CYH2 cDNA obtained from RNA preparations isolated from each strain were resolved on a 6% native polyacrylamide gel and visualized by staining with ethidium bromide. (B) RNase H/oligo(dT) Northern blot analysis. The locations of the CYH2 primer and the oligo(dT) primer used to direct RNase H cleavage are indicated, as is the location of the CYH2 DNA probe. The CYH2 oligonucleotide annealed 400 nucleotides upstream of the stop codon. After RNase H digestion [in the presence or absence of oligo(dT)], the RNA was resolved on a denaturing 6% polyacrylamide gel and analyzed by Northern blotting.
mRNA export from the nucleus is not significantly altered in the tpa1Δ strain.
Maturation of the 3′ ends of mRNAs is a highly regulated process that occurs in a cotranscriptional manner. Normal 3′ end processing facilitates mRNA export from the nucleus, while faulty processing, such as hyperpolyadenylation, leads to retention of transcripts in the nucleus and degradation by the nuclear exosome (30, 32, 66). Previous studies have shown that Pab1p and the PAN complex are required for the efficient release of transcripts from their sites of transcription and subsequent nuclear export (13, 22). These findings predict that if hyperpolyadenylation occurs in the nucleus of the tpa1Δ strain, those transcripts should be retained in the nucleus and a defect in mRNA export should be observed.
To test whether the longer poly(A) tails in the tpa1Δ mutant caused a defect in mRNA export from the nucleus, we examined export of the HAC1 mRNA as previously described (13). Hac1p is a transcription factor whose expression is dependent upon a novel cytoplasmic splicing reaction. Splicing of the HAC1 mRNA is mediated by the endoplasmic reticulum-associated endonuclease Ire1p, which is activated only upon induction of the UPR (39, 55). This system provides a direct means of monitoring the rates of HAC1 mRNA export from the nucleus in wild-type and tpa1Δ strains by monitoring the appearance of the spliced species as a function of time following induction of the UPR.
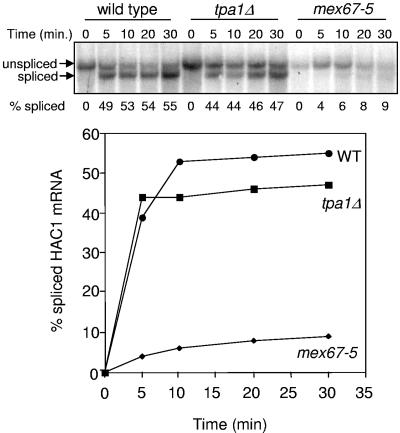
Our experiment utilized a wild-type strain as a positive control and a mex67-5 strain as a negative control. Mex67p is an essential mRNA export receptor, and the mex67-5 mutation imposes a strong block on mRNA export at 37°C (59). Expression of the HAC1 mRNA was induced from the CUP1 promoter, and the UPR was induced by the addition of DTT to the growth medium. Samples were then harvested at various times for Northern blot analysis to monitor splicing of the HAC1 mRNA (Fig. 6). We found that roughly 45 to 50% of the HAC1 mRNA was spliced in both the wild-type and tpa1Δ strains by 5 min after induction of the UPR. The fraction of spliced HAC1 mRNA gradually rose to 55% in the wild-type strain and 47% in the tpa1Δ strain by 30 min after induction. In contrast, only 4% of the HAC1 mRNA was spliced in the mex67-5 mutant by 5 min after induction of the UPR at the nonpermissive temperature, and the level of spliced mRNA rose to only 9% by 30 min after induction. When taken together, these results indicate that the tpa1Δ mutation does not cause a significant defect in mRNA export from the nucleus.
FIG. 6.
The tpa1Δ mutation does not significantly inhibit mRNA export from the nucleus. The indicated strains were grown at 37°C, HAC1 gene expression was induced from the CUP1 promoter, and DTT was added to induce the UPR. Samples were harvested at the indicated times after induction of the UPR to prepare RNA for Northern blot analysis using a HAC1-specific probe. (Upper panel) Northern blot showing HAC1 mRNA splicing in wild-type, tpa1Δ, and mex67-5 strains following induction of the UPR. (Lower panel) Graph showing the percentage of total HAC1 mRNA spliced as a function of time after UPR induction.
Tpa1p interacts with Pab1p.
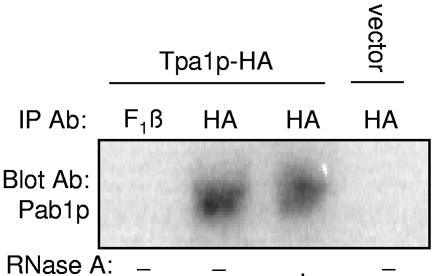
The results presented above indicate that Tpa1p influences the efficiency of translation termination and associates with both eRF1 and eRF3. eRF3 has previously been shown to interact with Pab1p in a manner that is important for both poly(A) tail length and mRNA stability (34, 42, 70). Since our results indicate that Tpa1p also affects poly(A) tail length and mRNA stability, we next investigated whether Tpa1p interacts with Pab1p. Lysates derived from a wild-type strain carrying either the Tpa1p-HA plasmid or the vector alone were immunoprecipitated using an antibody to the HA epitope. The precipitated complexes were then subjected to Western blot analysis using antiserum specific for Pab1p (46) or F1β as a negative control. As shown in Fig. 7, no interaction was detected either in samples expressing the vector alone or in samples probed with F1β antibodies. However, we found a specific Pab1p band in the Western blot of the Tpa1p immunoprecipitation, indicating an association between Tpa1p-HA and Pab1p. Moreover, RNase A treatment did not alter this result, suggesting that this association is due to protein-protein interactions. It has not yet been determined whether this represents a direct binding between Tpa1p and Pab1p or whether these two proteins reside together in a larger protein complex.
FIG. 7.
Tpa1p-HA interacts with Pab1p. The wild-type yeast strain was transformed with vector alone or with a plasmid expressing Tpa1p-HA. Cell lysates from these yeast strains were subjected to immunoprecipitation (IP) using a mouse monoclonal antibody (Ab) to the HA epitope or rabbit polyclonal antiserum to the yeast F1β protein with or without the addition of RNase A treatment. The protein complexes were then blotted using a rabbit polyclonal antibody to the yeast Pab1p protein.
Other factors that alter poly(A) tail length can also affect termination efficiency.
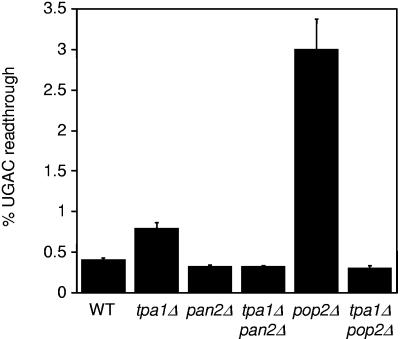
It was recently shown that eRF3 interacts with Pab1p [the yeast poly(A) binding protein] and influences both Pab1p oligomerization and the rate of mRNA turnover (36, 42, 70). It is therefore possible that other factors that bind to the 3′ UTR of a transcript and affect poly(A) tail length might also influence eRF3 function during translation termination. This potential effect on eRF3's ability to participate in the termination process could occur by various means, such as by limiting the number of eRF3 molecules available to engage in the termination complex and by altering the GTPase activity or conformational state of eRF3. To test this possibility, we used the dual luciferase readthrough reporter system to compare the efficiency of stop codon recognition in the tpa1Δ strain with those of the the pan2Δ and pop2Δ strains that also have extended poly(A) tails. As described before, we observed a twofold increase in readthrough of the UGAC tetranucleotide termination signal in the tpa1Δ strain, but we did not find any increase in readthrough at the same termination signal in the pan2Δ strain (Fig. 8). In contrast, we found that the pop2Δ mutation resulted in a 4.3-fold increase in readthrough of the UGAC termination signal compared to the wild-type strain.
FIG. 8.
Effect of other mutations that alter poly(A) tail length on translation termination efficiency. Readthrough of the UGAC tetranucleotide termination signal in the indicated strains was measured using dual luciferase readthrough reporter plasmids. The percent UGAC readthrough is expressed as the mean ± standard deviation.
We also examined the effects of combining the tpa1Δ mutation with the pan2Δ or pop2Δ mutation. We found that the level of readthrough in the tpa1Δ pan2Δ strain was normal, indicating that the pan2Δ mutation is epistatic to the tpa1Δ mutation with regard to translation termination. Similarly, we found that readthrough was also completely normal in the tpa1Δ pop2Δ strain (even though the presence of either mutation alone led to a readthrough phenotype). Together, these results indicate that genetic interactions between tpa1Δ pan2Δ and tpa1Δ pop2Δ strains influence the efficiency of translation termination in a complex manner.
DISCUSSION
The length of the poly(A) tail plays an essential role in mRNA function, subcellular localization, and half-life. mRNA 3′ end formation occurs in a cotranscriptional manner via a coupled, three-step reaction that includes endonucleolytic cleavage of the nascent transcript, the addition of the poly(A) tract by poly(A) polymerase (Pap1p), and poly(A) trimming (8, 33, 48). The PAN complex (Pan2p and Pan3p) trims nascent poly(A) tails in a Pab1p-dependent manner in the nucleus (6, 9, 10). Abnormally long poly(A) tails on nuclear mRNAs can result from either excessive poly(A) addition or defective poly(A) trimming. The presence of an extended poly(A) tail has been shown to block mRNA export from the nucleus, leading to its rapid degradation by the nuclear exosome (22, 32).
Longer poly(A) tails can also result from a defect in cytoplasmic deadenylation. The CCR4 complex (Ccr4p and Pop2p) has been reported to function as the major cytoplasmic deadenylation in yeast (16, 19, 43, 68, 69). However, several lines of evidence suggest that the PAN complex may also play a role in cytoplasmic deadenylation. First, the yeast PAN complex interacts with yeast Pab1p (48), which is bound to mRNAs in both the nucleus and cytoplasm (13, 22). Second, both Pan2p and Pan3p are located primarily in the cytoplasm (21, 37). Third, the loss of either Ccr4p or Pop2p does not cause a complete loss of cytoplasmic deadenylase activity. It was suggested that this residual activity is due to the PAN complex functioning as a cytoplasmic deadenylase, since all residual deadenylase activity was lost in a ccr4Δ pan2Δ double mutant (69). Finally, a recent study provided evidence that the PAN and CCR4 complexes together carry out the cytoplasmic deadenylation of mammalian mRNAs in a sequential manner (75).
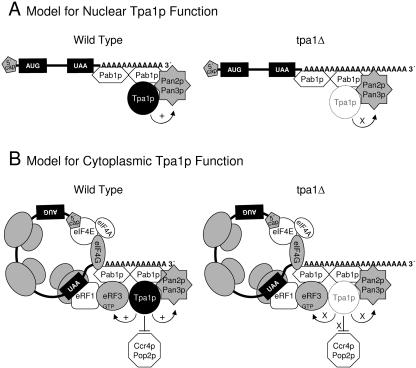
Our results indicate that the tpa1Δ mutation results in extended poly(A) tails. One possibility is that Tpa1p may alter polyadenylation or poly(A) trimming in the nucleus. Prior studies have shown that Tpa1p is located primarily in the nucleus, although some cytoplasmic staining was also observed (37, 43). Consistent with its nuclear location, Tpa1p has also been copurified with both the yeast nuclear pore complex (54) and the NuA3 histone acetyltransferase complex (38). These data suggest that Tpa1p may bind to mRNAs while still in the nucleus through its interactions with Pab1p (or possibly the PAN complex). It is possible that the absence of Tpa1p could influence the poly(A) tail length of mRNAs while still in the nucleus, or its absence could lead to the assembly of an aberrant mRNP complex in the nucleus that is subsequently transported into the cytoplasm, where defects in translation termination and deadenylation could occur (Fig. 9A).
FIG. 9.
Two models for Tpa1p function. (A) Tpa1p influences poly(A) tail length and mRNP assembly on poly(A) tails in the nucleus and may indirectly influence translation termination following export of the aberrant mRNP to the cytoplasm. (B) Tpa1p is a component of mRNP complexes bound to the 3′ UTR of cytoplasmic mRNAs, where it acts directly to couple translation termination and mRNA deadenylation. Tpa1p denoted by the dashed lines on the right (tpa1Δ) side of the figure indicates the absence of this factor.
However, other data are more consistent with a model in which Tpa1p plays a more direct role in cytoplasmic translation termination and deadenylation. First, the tpa1Δ mutation did not cause a significant defect in the rate of HAC1 mRNA export from the nucleus, which frequently occurs when nuclear 3′ end formation is compromised (22, 32). This suggests that extended poly(A) tails observed in the tpa1Δ strain may not be formed in the nucleus. Second, Tpa1p was found to associate with the cytosolic translation factors eRF1 and eRF3, and the tpa1Δ mutation also altered the efficiency of translation termination. This also suggests that some Tpa1p functions are carried out in the cytoplasm. Third, we found that poly(A) tails are extended in the tpa1Δ, pop2Δ, and pan2Δ strains (6, 9, 10, 69), while poly(A) tail length was normal in a tpa1Δ pan2Δ double mutant. The epistasis of the pan2Δ mutation over the tpa1Δ mutation suggests that the PAN complex functions upstream in the same pathway as Tpa1p. In contrast, poly(A) tails in the tpa1Δ pop2Δ strain were longer than those in either the tpa1Δ or the pop2Δ strain.
We also found that the tpa1Δ and the pop2Δ strains exhibited a readthrough phenotype, while the pan2Δ strain showed normal stop codon recognition. The tpa1Δ pan2Δ double mutant also did not have a readthrough phenotype, again indicating that the pan2Δ mutation is epistatic to the tpa1Δ mutation. This indicates that Pan2p functions upstream of Tpa1p, which is consistent with the initial function of Pan2p being poly(A) trimming in the nucleus, while Tpa1p functions later (possibly after mRNAs are exported to the cytoplasm). The pop2Δ mutation exhibited an even stronger readthrough phenotype than the tpa1Δ mutation. This effect is also consistent with its assigned role in cytoplasmic deadenylation.
When taken together, these data are consistent with a model in which Tpa1p is a direct component of the mRNP complex on mRNA poly(A) tails that functions to modulate cytoplasmic deadenylation (Fig. 9B). In this model, Tpa1p serves as a positive regulator of the PAN complex and a negative regulator of the CCR4 complex during cytoplasmic mRNA deadenylation, as suggested by the genetic interaction data. The idea that Tpa1p evokes opposite effects on these deadenylase complexes is consistent with previous observations that Pab1p stimulates the deadenylase activity of the PAN complex (47) and inhibits the deadenylase activity of the CCR4 complex (68). Since Tpa1p binds Pab1p and appears to stimulate PAN deadenylase activity, our data are most consistent with the PAN complex carrying out the initial phase of cytoplasmic mRNA deadenylation. As the poly(A) tail length is reduced, both the PAN complex and Tpa1p may dissociate, thereby allowing the CCR4 complex to gain access to the poly(A) tail to complete the deadenylation process. Furthermore, this sequential deadenylation facilitated first by the PAN complex and then by the CCR4 complex is consistent with the recently proposed model for the cytoplasmic deadenylation of mammalian mRNAs (75). Clearly, further studies are required to determine whether one (or both) of these models of Tpa1p function is correct.
Previous studies reported that an interaction between eRF3 and Pab1p stimulates deadenylation and subsequent mRNA turnover (35, 42). The following evidence from our study suggests that Tpa1p may also be involved in termination-coupled mRNA deadenylation. First, Tpa1p was found in protein complexes with the cytoplasmic translation termination factors eRF1 and eRF3 as well as Pab1p. An interaction with all three proteins is not surprising, since both eRF1 (64, 76) and Pab1p (17, 35) are known to interact with eRF3. Second, the tpa1Δ mutation resulted in longer poly(A) tails and an increase in the half-lives of the CYH2 and ACT1 mRNAs. These defects associated with the tpa1Δ mutation were strikingly similar to the phenotypes caused by mutations in the GTPase domain of eRF3 or by deletion of the N and M domains of eRF3 that mediate its interaction with Pab1p (35, 42). These data suggest that eRF3 and Tpa1p may act together to couple translation termination and mRNA deadenylation, leading to the observed changes in mRNA turnover. Technical difficulties precluded us from directly testing whether Tpa1p is required for the interaction between eRF3 and Pab1p. However, it has previously been shown that purified mammalian PABP (the homologue of yeast Pab1p) and eRF3 can interact in the absence of Tpa1p (35).
Our finding that the tpa1Δ mutation increases readthrough of stop codons suggests that Tpa1p may normally stimulate some aspect of release factor function during the termination process. One way Tpa1p could mediate this effect is by stimulating the GTP hydrolysis by eRF3. This mechanism would be consistent with the previous finding that mutations in the GTPase domain of eRF3 result in extended poly(A) tails and longer mRNA half-lives (42). However, mutations that influence the GTPase activity of eRF3 have also been associated with an increase in the half-lives of mRNA substrates degraded by the NMD pathway (42), while we did not observe an NMD defect associated with the tpa1Δ mutation. Interestingly, protein abundance measurements available at the Yeast GFP Fusion Localization Database indicate that Tpa1p is roughly twofold less abundant than eRF1 and ninefold less abundant than eRF3 (28). This suggests that Tpa1p may associate only with a subset of termination complexes in the cell and may not join those that form at premature stop codons. Additional experiments will be required to determine how Tpa1p influences only a subset of the mRNA turnover pathways influenced by eRF3.
Finally, database searches indicate that homologues of the TPA1 gene are present in other fungal species as well as in higher eukaryotes such as Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, Mus musculus, and Homo sapiens. The ubiquitous presence of Tpa1p throughout the eukaryotic kingdom suggests that it may influence translation termination, mRNA deadenylation, and mRNA turnover in all eukaryotic species.
Acknowledgments
We thank Carol L. Dieckmann, Roy Parker, David Mangus, Jeff Schatz, Pam Silver, and Allan Jacobson for reagents and Sunnie Thompson for critically reading the manuscript.
This work was supported by NIH grant RO1 GM 68854 (D.M.B.).
REFERENCES
- 1.Amrani, N., R. Ganesan, S. Kervestin, D. A. Mangus, S. Ghosh, and A. Jacobson. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112-118. [DOI] [PubMed] [Google Scholar]
- 2.Araki, Y., S. Takahashi, T. Kobayashi, H. Kajiho, S. Hoshino, and T. Katada. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20:4684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 4.Bertram, G., H. A. Bell, D. W. Ritchie, G. Fullerton, and I. Stansfield. 2000. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA 6:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertram, G., S. Innes, O. Minella, J. Richardson, and I. Stansfield. 2001. Endless possibilities: translation termination and stop codon recognition. Microbiology 147:255-269. [DOI] [PubMed] [Google Scholar]
- 6.Boeck, R., S. Tarun, Jr., M. Rieger, J. A. Deardorff, S. Muller-Auer, and A. B. Sachs. 1996. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271:432-438. [DOI] [PubMed] [Google Scholar]
- 7.Bonetti, B., L. Fu, J. Moon, and D. M. Bedwell. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251:334-345. [DOI] [PubMed] [Google Scholar]
- 8.Brodsky, A. S., and P. A. Silver. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. E., and A. B. Sachs. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18:6548-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, C. E., S. Z. Tarun, Jr., R. Boeck, and A. B. Sachs. 1996. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5744-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, C. M., M. E. Dalphin, P. A. Stockwell, and W. P. Tate. 1993. The translational termination signal database. Nucleic Acids Res. 21:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, J. T., X. Bai, and A. W. Johnson. 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brune, C., S. E. Munchel, N. Fischer, A. V. Podtelejnikov, and K. Weis. 2005. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11:517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke, D., D. C. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Chavatte, L., A. Seit-Nebi, V. Dubovaya, and A. Favre. 2002. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 21:5302-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J., Y. C. Chiang, and C. L. Denis. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosson, B., A. Couturier, S. Chabelskaya, D. Kiktev, S. Inge-Vechtomov, M. Philippe, and G. Zhouravleva. 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI+] propagation. Mol. Cell. Biol. 22:3301-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czaplinski, K., M. J. Ruiz-Echevarria, S. V. Paushkin, X. Han, Y. Weng, H. A. Perlick, H. C. Dietz, M. D. Ter-Avanesyan, and S. W. Peltz. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daugeron, M. C., F. Mauxion, and B. Seraphin. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 21.Dheur, S., K. R. Nykamp, N. Viphakone, M. S. Swanson, and L. Minvielle-Sebastia. 2005. Yeast mRNA poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent poly(A) nuclease activity. J. Biol. Chem. 280:24532-24538. [DOI] [PubMed] [Google Scholar]
- 22.Dunn, E. F., C. M. Hammell, C. A. Hodge, and C. N. Cole. 2005. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Dev. 19:90-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eaglestone, S. S., L. W. Ruddock, B. S. Cox, and M. F. Tuite. 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eurwilaichitr, L., F. M. Graves, I. Stansfield, and M. F. Tuite. 1999. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol. 32:485-496. [DOI] [PubMed] [Google Scholar]
- 25.Frischmeyer, P. A., A. van Hoof, K. O'Donnell, A. L. Guerrerio, R. Parker, and H. C. Dietz. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258-2261. [DOI] [PubMed] [Google Scholar]
- 26.Frolova, L., X. Le Goff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334-341. [PMC free article] [PubMed] [Google Scholar]
- 27.Frolova, L. Y., R. Y. Tsivkovskii, G. F. Sivolobova, N. Y. Oparina, O. I. Serpinsky, V. M. Blinov, S. I. Tatkov, and L. L. Kisselev. 1999. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 29.Hagan, K. W., M. J. Ruiz-Echevarria, Y. Quan, and S. W. Peltz. 1995. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 15:809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammell, C. M., S. Gross, D. Zenklusen, C. V. Heath, F. Stutz, C. Moore, and C. N. Cole. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22:6441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, F., X. Li, P. Spatrick, R. Casillo, S. Dong, and A. Jacobson. 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12:1439-1452. [DOI] [PubMed] [Google Scholar]
- 32.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 33.Hilleren, P., and R. Parker. 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshino, S., N. Hosoda, Y. Araki, T. Kobayashi, N. Uchida, Y. Funakoshi, and T. Katada. 1999. Novel function of the eukaryotic polypeptide-chain releasing factor 3 (eRF3/GSPT) in the mRNA degradation pathway. Biochemistry (Moscow) 64:1367-1372. [PubMed] [Google Scholar]
- 35.Hoshino, S., M. Imai, T. Kobayashi, N. Uchida, and T. Katada. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274:16677-16680. [DOI] [PubMed] [Google Scholar]
- 36.Hosoda, N., T. Kobayashi, N. Uchida, Y. Funakoshi, Y. Kikuchi, S. Hoshino, and T. Katada. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278:38287-38291. [DOI] [PubMed] [Google Scholar]
- 37.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 38.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 39.Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8:1845-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeling, K. M., J. Lanier, M. Du, J. Salas-Marco, L. Gao, A. Kaenjak-Angeletti, and D. M. Bedwell. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kisselev, L., M. Ehrenberg, and L. Frolova. 2003. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 22:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi, T., Y. Funakoshi, S. Hoshino, and T. Katada. 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 279:45693-45700. [DOI] [PubMed] [Google Scholar]
- 43.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maderazo, A. B., F. He, D. A. Mangus, and A. Jacobson. 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangus, D. A., N. Amrani, and A. Jacobson. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18:7383-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangus, D. A., M. C. Evans, N. S. Agrin, M. Smith, P. Gongidi, and A. Jacobson. 2004. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 24:5521-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangus, D. A., M. M. Smith, J. M. McSweeney, and A. Jacobson. 2004. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 24:4196-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer, S. A., and C. L. Dieckmann. 1989. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3′-end formation. Mol. Cell. Biol. 9:4161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merkulova, T. I., L. Y. Frolova, M. Lazar, J. Camonis, and L. L. Kisselev. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443:41-47. [DOI] [PubMed] [Google Scholar]
- 51.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 52.Muhlrad, D., C. J. Decker, and R. Parker. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15:2145-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ness, F., P. Ferreira, B. S. Cox, and M. F. Tuite. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22:5593-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 55.Ruegsegger, U., J. H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107:103-114. [DOI] [PubMed] [Google Scholar]
- 56.Salas-Marco, J., and D. M. Bedwell. 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24:7769-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salles, F. J., W. G. Richards, and S. Strickland. 1999. Assaying the polyadenylation state of mRNAs. Methods 17:38-45. [DOI] [PubMed] [Google Scholar]
- 58.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seit-Nebi, A., L. Frolova, J. Justesen, and L. Kisselev. 2001. Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res. 29:3982-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seit-Nebi, A., L. Frolova, and L. Kisselev. 2002. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 3:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Serio, T. R., and S. L. Lindquist. 1999. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu. Rev. Cell Dev. Biol. 15:661-703. [DOI] [PubMed] [Google Scholar]
- 63.Sparks, K. A., S. A. Mayer, and C. L. Dieckmann. 1997. Premature 3′-end formation of CBP1 mRNA results in the downregulation of cytochrome b mRNA during the induction of respiration in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4199-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ter-Avanesyan, M. D., V. V. Kushnirov, A. R. Dagkesamanskaya, S. A. Didichenko, Y. O. Chernoff, S. G. Inge-Vechtomov, and V. N. Smirnov. 1993. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 7:683-692. [DOI] [PubMed] [Google Scholar]
- 66.Thomsen, R., D. Libri, J. Boulay, M. Rosbash, and T. H. Jensen. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9:1049-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tipper, D. J. 1973. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J. Bacteriol. 116:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad, and R. Parker. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 70.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 71.van Hoof, A., P. A. Frischmeyer, H. C. Dietz, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262-2264. [DOI] [PubMed] [Google Scholar]
- 72.Wang, W., K. Czaplinski, Y. Rao, and S. W. Peltz. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20:880-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weng, Y., K. Czaplinski, and S. W. Peltz. 1996. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol. Cell. Biol. 16:5477-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weng, Y., K. Czaplinski, and S. W. Peltz. 1996. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol. Cell. Biol. 16:5491-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamashita, A., T. C. Chang, Y. Yamashita, W. Zhu, Z. Zhong, C. Y. Chen, and A. B. Shyu. 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 12:1054-1063. [DOI] [PubMed] [Google Scholar]
- 76.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]