Abstract
Sgs1 is a RecQ family DNA helicase required for genome stability in Saccharomyces cerevisiae whose human homologs BLM, WRN, and RECQL4 are mutated in Bloom's, Werner, and Rothmund Thomson syndromes, respectively. Sgs1 and mismatch repair (MMR) are inhibitors of recombination between similar but divergent (homeologous) DNA sequences. Here we show that SGS1, but not MMR, is critical for suppressing spontaneous, recurring translocations between diverged genes in cells with mutations in the genes encoding the checkpoint proteins Mec3, Rad24, Rad9, or Rfc5, the chromatin assembly factors Cac1 or Asf1, and the DNA helicase Rrm3. The S-phase checkpoint kinase and telomere maintenance factor Tel1, a homolog of the human ataxia telangiectasia (ATM) protein, prevents these translocations, whereas the checkpoint kinase Mec1, a homolog of the human ATM-related protein, and the Rad53 checkpoint kinase are not required. The translocation structures observed suggest involvement of a dicentric intermediate and break-induced replication with multiple cycles of DNA template switching.
RecQ-like DNA helicases play important roles in the maintenance of genome stability from bacteria to humans. The only member of the RecQ family of 3′ to 5′ DNA helicases in the yeast Saccharomyces cerevisiae is Sgs1. Sgs1 has been implicated in the coordination between DNA replication and recombination, in the regulation of homologous recombination (HR) and the suppression of crossover products during HR, and in S-phase checkpoint activation as well as in transcription (16, 26, 38, 49, 61, 96). As a consequence, cells that lack Sgs1 display a hyperrecombination phenotype, accumulate extrachromosomal ribosomal DNA (rDNA) circles, frequently missegregate chromosomes during mitosis and meiosis, have modestly increased rates of accumulating gross chromosomal rearrangements (GCRs), are sensitive to agents such as hydroxyurea and methyl-methanesulfonate, and show signs of premature aging (29, 58, 61, 86, 100, 101, 104).
To date, five human genes encoding RecQ-like (RECQL) proteins have been identified. Mutations in RECQL2 (WRN) (105), RECQL3 (BLM) (23), and RECQL4 (41, 42) cause three rare, cancer-prone disorders, Werner syndrome (WS), Bloom's syndrome (BS), and a subset of Rothmund Thomson syndrome (Type II RTS) (97), respectively, while defects in RECQL1 (73, 74) and RECQL5 (41) have not been linked to a disease. Besides short stature, early onset of diabetes mellitus, and immunodeficiency, BS is characterized by extreme cancer risk, which has been estimated to be 150 to 300 times higher than the risk of malignancy in the unaffected population; in 168 BS patients, 100 cancers of many types had arisen at a mean age of 24.7 years, with many of the patients suffering from multiple primary cancers (31). Although WS patients share some of these symptoms, including early onset of diabetes mellitus and increased cancer susceptibility, they also show numerous other signs of accelerated aging not typical for BS. RTS patients also show a high prevalence of cancers, especially osteosarcomas, and suffer from skeletal abnormalities and skin changes (95, 98).
WS, BS, and RTS cells exhibit a wide range of chromosomal aberrations. WS cells have an increased spontaneous mutation rate, mainly due to the accumulation of large deletions (>20 kb), but translocations and insertions have also been observed (27, 28, 79). Structural and numerical chromosome instability has been described for RTS cells (22, 51, 55, 67). Chromosome aberrations in BS cells include approximately 0.29 chromatid and chromosome breaks per cell, translocations, and ring chromosomes (34). The most striking feature of BS cells, however, is the increased rate of spontaneous reciprocal exchange of genetic material between sister chromatids (sister chromatid exchange [SCE]) as well as between chromatids of two different chromosomes leading to the appearance of, mostly, symmetric quadriradial (QR) chromosomes (12). While SCEs are not mutagenic per se, such hyperactivity of recombinogenic processes may result in mutations if it leads to recombination between homologs or sister chromatids at nonhomologous sites. Moreover, exchanges between ectopic homologous regions of single chromatids of two different chromosomes, whether they occur between homologous or nonhomologous chromosomes, as suggested by the formation of asymmetric QRs in BS cells, can lead to translocations as well as the formation of dicentric and acentric chromosomes which cannot be segregated properly. QRs are approximately 100-fold more frequent in BS cells than in normal cells, where they occur at a frequency of ∼1/1,000 (12, 91). The highly elevated rate of mitotic crossing over in BS cells between homologous chromosomes or regions of homology located on nonhomologous chromosomes, such as the rDNA regions in the satellite stalks of acrocentric chromosomes, has been shown to lead to a high degree of loss of heterozygosity (LOH) in BS cells and in BS mice, which may expose recessive mutations in tumor suppressor genes and has thus been suggested to be a source of tumorigenic chromosomal rearrangements in BS (52, 91).
Unlike most other DNA helicases, the 3′ to 5′ DNA helicases WRN, BLM, and Sgs1 have been shown to unwind a duplex DNA preferentially from an internal loop rather than from a blunt end or a 3′ overhang (6, 56). Moreover, WRN, BLM, and Sgs1 can unwind G-quadruplexes and Holliday junctions in vitro, while Sgs1 has also been shown to resolve three-way junctions (6, 37, 56). These structural substrate preferences suggest that these RecQ-like DNA helicases may be required for a variety of DNA metabolic processes during which such structures may arise, most prominently during HR and at stalled replication forks. In the absence of RecQ-like DNA helicases, recombinogenic lesions may instead be formed in an attempt to process anomalous replication forks or HR intermediates. The importance of RecQ family helicases for genome integrity is further supported by their physical interaction with proteins known to be involved in replicational and repair processes. For instance, Sgs1, BLM, and WRN interact with the single-stranded DNA binding protein RPA (11, 15, 17), Sgs1 and BLM interact with the strand exchange protein Rad51 (70, 103), RecQL4 colocalizes with Rad51 foci after induction of DNA damage (70), BLM and Sgs1 interact with topoisomerases (29, 101), and Sgs1, BLM, and WRN have been found in complexes with proteins that function in DNA damage response pathways (16, 20, 26, 47, 99). Moreover, genetic interactions have been demonstrated between sgs1 and mutations in DNA helicase genes RRM3 and SRS2 and the structure-specific endonuclease genes MUS81, MMS4, SLX1, and SLX4 (24, 30, 49, 59, 81, 93). Recently, additional interaction partners have been identified in large-scale genetic screens (66, 92). In contrast to Sgs1, BLM, and WRN helicases, recombinant RecQL4 purified from Escherichia coli cells lacks DNA helicase activity (53). Instead, Sangrithi et al. (78)identified a region at the N terminus of RecQL4 that shares homology with Sld2/Drc1, which is required for the establishment of replication forks in S. cerevisiae, thus suggesting a role of RecQL4 in the loading of replication factors at origins.
S. cerevisiae cells lacking Sgs1 have proven to be excellent model systems for some cellular phenotypes of the Bloom's and Werner syndromes, especially with respect to their hyperrecombination phenotype. Cells that lack Sgs1 display elevated rates (∼10-fold) of intrachromosomal HR between direct repeats and interchromosomal HR between homologous sequences or heteroalleles (100, 104). sgs1 mutations, including mutations that mimic two missense mutations found in BS, also cause an approximately fourfold elevated rate of SCE using an assay that measures reconstitution of a functional ADE3 gene from two nonfunctional ADE3 truncations containing a 305-bp overlap (40, 65). An increase in the frequency of LOH in diploid sgs1 mutants has also been reported and is mainly the result of chromosome loss and chromosome rearrangements in the form of ectopic interchromosomal rearrangements, such as translocations and unequal crossing over (3). In addition, sgs1 mutants exhibit an increased rate of recombination between similar, but nonidentical (homeologous), DNA sequences, leading to the conclusion that Sgs1 functions in the same pathway as the mismatch repair (MMR) proteins to suppress homeologous recombination (61, 87). Here we have evaluated the role of a wide range of DNA metabolic pathways in the suppression of spontaneous translocations between three highly diverged genes in sgs1 mutants of S. cerevisiae. We observed homology-driven translocations, which are suppressed by Sgs1 but not MMR, suggesting that a function of Sgs1 other than its regulation of homeologous recombination is responsible for its role in suppressing translocations between related genes. Based on our analysis of translocation structures, we propose a model for the formation of complex translocations by a single recombinational event that may be facilitated by the extraordinary relaxation of mitotic HR in the absence of Sgs1.
MATERIALS AND METHODS
Strains and general genetic methods.
S. cerevisiae strains used for determination of mutation rates are derivatives of S288C. Gene deletions in RDKY3615 (MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3), RDKY5027 (MATα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3), and their diploid derivatives were generated by HR-mediated integration of PCR fragments using standard methods. All haploid strains used for determination of mutation rates were obtained by sporulation of appropriate diploid strains. To minimize the emergence of suppressors, slow-growing haploid strains containing deletions of the DNA helicase genes SGS1 and RRM3 were freshly obtained by sporulation of the appropriate diploid strain for every experiment. S. cerevisiae strains were grown at 30°C. S. cerevisiae strains used in this study and their complete genotypes are listed in Table 1. Media for propagating S. cerevisiae strains have been described previously (14).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| RDKY3615 | MATaura3-5, trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 | 13 |
| RDKY3719 | RDKY3615 rad9::HIS3 | 61 |
| RDKY3731 | RDKY3615 tel1::HIS3 | 61 |
| RDKY3735 | RDKY3615 mec1::HIS3 sml1::TRP1 | 61 |
| RDKY3749 | RDKY3615 sml1::KAN rad53::HIS3 | 61 |
| RDKY4561 | RDKY3615 rfc5-1.TRP1 sgs1::HIS3 | 63 |
| RDKY4566 | RDKY3615 tel1::HIS3 sgs1::TRP1 | 63 |
| RDKY4588 | RDKY3615 sml1::KAN rad53::HIS3 sgs1::TRP1 | 63 |
| RDKY4753 | RDKY3615 cac1::HIS3 | 64 |
| RDKY4755 | RDKY3615 asf1::HIS3 | 64 |
| RDKY4765 | RDKY3615 sgs1::HIS3 cac1::TRP1 | 64 |
| RDKY4767 | RDKY3615 asf1::HIS, sgs1::TRP1 | 64 |
| RDKY5502 | RDKY3615 tsa1::KAN | 35 |
| RDKY5529 | RDKY3615 tsa1::KAN sgs1::HIS3 | 36 |
| RDKY5556 | RDKY3615 rrm3::TRP1 | This study |
| RDKY5558 | RDKY3615 sgs1::TRP1 | This study |
| RDKY5564 | RDKY3615 rad51::HIS3 sgs1::TRP1 rrm3::KAN | This study |
| RDKY5569 | RDKY3615 mec3::HIS3 | This study |
| RDKY5572 | RDKY3615 mec3::HIS3 sgs1::TRP1 | This study |
| RDKY5573 | RDKY3615 rad24::HIS3 | This study |
| RDKY5575 | RDKY3615 rad24::HIS3sgs1::TRP1 | This study |
| RDKY5577 | MATa/α ura3-52/ura3-52, trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2ΔBgl/lys2ΔBgl hom3-10/hom3-10 ade2Δ1/ade2Δ1 ade8/ade8 hxt13::URA3/hxt13::URA3 SGS1/sgs1::TRP1 RRM3/rrm3::KAN | This study |
| RDKY5579 | RDKY3615 mec3::HIS3 sgs1::TRP1 rrm3::KAN | This study |
| RDKY5772 | RDKY3615 mec3::KAN asf1::HIS3 sgs1::TRP1 | This study |
| RDKY5773 | RDKY3615 rad52::HIS3 | This study |
| RDKY5774 | RDKY3615 rad52::HIS3 sgs1::TRP1 | This study |
| RDKY5775 | RDKY3615 rad52::HIS3 sgs1::TRP1 mec3::KAN | This study |
| RDKY5776 | RDKY3615 sgs1::TRP1 rrm3::KAN rad51::HIS3 mec3::HIS3 | This study |
| RDKY5777 | RDKY3615 sgs1::HIS3 mec1::TRP1 sml1::KAN | This study |
| RDKY5778 | RDKY3615 rad9::HIS3 sgs1::TRP1 | This study |
| RDKY5779 | RDKY3615 msh2::KAN1 asf1::HIS3 | This study |
| RDKY5780 | RDKY3615 msh2::KAN1 cac1::HIS3 | This study |
| RDKY5781 | RDKY3615 msh2::KAN1 mec3::HIS3 | This study |
| RDKY5782 | RDKY3615 msh2::KAN1 rfc5-1.TRP1 | This study |
| RDKY5783 | RDKY3615 msh2::KAN1 tel1::HIS3 | This study |
| RDKY5784 | RDKY3615 msh2::KAN1 | This study |
| KHSY1357 | RDKY3615 sgs1::TRP1mec3::G418 rad51::HIS3 | This study |
| KHSY816 | MATa/α ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2ΔBgl/lys2ΔBgl hom3-10/hom3-10 ade2Δ1/ade2Δ1 ade8/ade8 hxt13::URA3/hxt13::URA3, SGS1/sgs1::TRP1 RRM3/rrm3::KANMEC3/mec3::HIS3 | This study |
| KHSY1448 | RDKY3615 msh6::TRP1 rad24::HIS3 | This study |
| KHSY1452 | RDKY3615 msh6::TRP1 mec3::HIS3 | This study |
GCR rates and rearrangement analysis.
GCR rates were determined independently by fluctuation analysis of at least 15 independent cultures from three different strain isolates, and the median rate is reported (13, 61). To minimize the emergence of suppressors in slow-growing S. cerevisiae strains, GCR rate measurements with rrm3 sgs1 and sgs1 rrm3 mec3 strains were carried out immediately after genotyping of freshly obtained, individual spore clones (i.e., day 7 after plating of spores on nonselective media). All of the cells originating from an individual spore clone were inoculated into 10 ml of yeast extract/peptone/glucose (YPD) medium, and the cultures were incubated for 2 to 3 days at 30°C with vigorous shaking and then plated on YPD and selective media to isolate GCRs. Rearrangements between the CAN1, ALP1, and LYP1 genes were identified by PCR using a primer pair that anneals to the 5′ end of CAN1 (5′-ATGACAAATTCAAAAGAAGACGCC-3′) and the 3′ end of ALP1 (5′-GAAAGGACATCCCAAACTCGTTGC-3′) or the 3′ end of LYP1 (5′-CAGCAGCCCAGAATTTCTCC-3′). PCR conditions were as follows: 5 min at 95°C and then 30 cycles of 30 s at 95°C, 30 s at 55°C, 4 min at 68°C, and 7 min at 68°C. PCR products were analyzed by 1% agarose gel electrophoresis. Prior to sequencing, PCR products were treated with shrimp alkaline phosphatase and exonuclease 1. The predicted orientation of the translocation target with respect to the centromere was determined by comparing the DNA sequences at the breakpoint to the Saccharomyces Genome Database. If the translocation target was predicted to contain a centromere, the translocation chromosome was classified as dicentric (e.g., CAN1-LYP1 translocation chromosomes). If the translocation target was not predicted to contain a centromere, then the translocation chromosome was classified as monocentric (e.g., CAN1-ALP1 and CAN1-LYP1-ALP1 translocation chromosomes).
RESULTS
Checkpoint proteins, chromatin assembly factors, and the DNA helicase Rrm3 suppress recurring translocations in the absence of Sgs1.
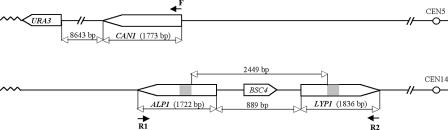
Cells lacking Sgs1 have a moderately increased rate of accumulating GCRs; these consist of broken chromosomes healed by de novo telomere addition (62%) and translocations with microhomology (25%) or without homology (13%) at the breakpoint (61). In contrast, rrm3Δ mutants do not have an increased GCR rate. sgs1Δ rrm3Δ double mutants have a severe growth defect that can be rescued by deletion of the HR genes RAD51, RAD55, and RAD57 (66, 81, 93). During a detailed investigation of checkpoint activation and genome instability in sgs1Δ rrm3Δ double mutants, we observed that sgs1Δ rrm3Δ double mutants have a synergistic increase in GCR rates that is reduced by mutations in HR genes and is increased by defects in checkpoint genes (K. H. Schmidt and R. Kolodner, unpublished data; also see Table 2). In this study, we repeatedly found translocations between the CAN1 gene on chromosome 5 and LYP1 or ALP1 on chromosome 14 in sgs1Δ rrm3Δ double mutants. CAN1, LYP1, and ALP1 are closely related genes, expressing basic amino acid transporters; the CAN1 gene is 64% identical to the LYP1 and ALP1 genes, while ALP1 and LYP1 show 59% sequence identity. This observation, taken together with the finding that, with the exception of a single CAN1-ALP1 translocation in a tlc1Δ rad51Δ mutant, sequencing of more than 358 translocation breakpoints in this laboratory has never identified translocations between CAN1 and any of its nine most closely related genes in any strain not carrying an sgs1Δ mutation (75), suggests that Sgs1 may be a regulator of translocations between highly diverged genes. To test this hypothesis, we combined an sgs1Δ mutation with mutations causing defects in cell cycle checkpoints (mec3Δ, rad24Δ, rad9Δ, rfc5-1, rad53Δ, mec1Δ, and tel1Δ), in HR (rad51Δ and rad52Δ), in the oxidative stress response (tsa1Δ), and in chromatin assembly (cac1Δ and asf1Δ), many of which are known to increase GCR rates, albeit through different defects, to determine the effect of the additional sgs1Δ mutation on the GCR rate and spectrum (36, 44). To facilitate this analysis, we designed a PCR assay to screen a large collection of independent GCRs isolated from these mutants for CAN1-ALP1 and CAN1-LYP1 translocations using two primer pairs that anneal to the 5′ end of CAN1, located on chromosome 5 (Fig. 1, primer F), and the 3′ end of either LYP1 or ALP1, located on chromosome 14 (Fig. 1, primers R1 and R2, respectively). The resulting PCR products were characterized by DNA sequencing, and the rate of each type of rearrangement was calculated.
TABLE 2.
Effect of defects in cell cycle checkpoints, chromatin assembly, homologous recombination, oxidative stress response, and DNA helicases on GCRs in sgs1 and mismatch repair mutants
| Relevant genotype | Total rate of GCRs (1010) | Total CAN1/LYP1/ALP1 translocations
|
Rates of individual CAN1/LYP1/ALP1 translocation classes (1010)b
|
||||
|---|---|---|---|---|---|---|---|
| Rate (1010)a | Frequency (%)b | CAN1-LYP1 | CAN1-ALP1 | CAN1-LYP1-ALP1 | CAN1-LYP1-ALP1- LYP1-ALP1 | ||
| Wild typec,d | 3.5 | ND (<0.2) | ND | ND | ND | ND | ND |
| sgs1 | 77 | <2.6 | <0.03 (0/30) | <3 (0/30) | <2.6 (0/30) | <2.6 (0/30) | <2.6 (0/30) |
| cac1e | 216 | <7.4 | <0.03 (0/29) | <7.4 (0/29) | <7.4 (0/29) | <7.4 (0/29) | <7.4 (0/29) |
| cac1 sgs1 | 355 | 60 | 17 (5/30) | <12 (0/30) | 24 (2/30) | 36 (3/30) | <12 (0/30) |
| asf1e | 250 | ND (<23) | ND | ND | ND | ND | ND |
| asf1 sgs1 | 318 | 64 | 23 (6/30) | <11 (0/30) | 21 (2/30) | 32 (3/30) | 11 (1/30) |
| rad24 | 23 | <0.8 | <0.03 (0/30) | <0.8 (0/30) | <0.8 (0/30) | <0.8 (0/30) | <0.8 (0/30) |
| rad24 sgs1 | 136 | 68 | 50 (15/30) | 4.5 (1/30) | 18 (4/30) | 41 (9/30) | 4.5 (1/30) |
| mec3c | 17 | <0.6 | <0.03 (0/30) | <0.6 (0/30) | <0.6 (0/30) | <0.6 (0/30) | <0.6 (0/30) |
| mec3 sgs1 | 339 | 79 | 23 (7/30) | 11 (1/30) | 45 (4/30) | 23 (2/30) | <11 (0/30) |
| mec3 sgs1 rad51 | 1,491 | 198 | 13 (4/30) | ND | ND | ND | ND |
| mec3 sgs1 asf1 | 2,588 | 431 | 17 (5/30) | 172 (2/30) | <86 (0/30) | 259 (3/30) | <86 (0/30) |
| rad52f | 161 | ND (<16) | ND | ND | ND | ND | ND |
| rad52 sgs1 | 125 | <4.3 | <0.03 (0/29) | <4.3 (0/29) | <4.3 (0/29) | <4.3 (0/29) | <4.3 (0/29) |
| rad52 sgs1 mec3 | 3,168 | <23 | <0.007 (0/136) | <40 (0/79) | <40 (0/79) | <40 (0/79) | <40 (0/79) |
| rad51 | <8 | ND | ND | ND | ND | ND | ND |
| rrm3 | 14 | <0.6 | <0.04 (0/24) | <0.6 (0/24) | <0.6 (0/24) | <0.6 (0/24) | <0.6 (0/24) |
| rrm3 sgs1 | 656 | 81 | 12 (7/57) | <12 (0/57) | 35 (3/57) | 46 (4/57) | <12 (0/57) |
| rrm3 sgs1 mec3 | 1,942 | 349 | 18 (7/39) | <50 (0/39) | 100 (2/39) | 249 (5/39) | <50 (0/39) |
| rrm3 sgs1 rad51 | 15 | 2.4 | 16 (3/19) | <0.8 (0/19) | <0.8 (0/19) | 2.4 (3/19) | <0.8 (0/19) |
| rrm3 sgs1 rad51 mec3 | 1,640 | 66 | 4 (2/50) | <33 (0/30) | 33 (1/30) | 33 (1/30) | <33 (0/30) |
| mec1 sml1c | 460 | ND (<51) | ND | ND | ND | ND | ND |
| mec1sml1 sgs1 | 1,930 | <10 | <0.005 (0/190) | <10 (0/190) | <10 (0/190) | <10 (0/190) | <10 (0/190) |
| tel1c | 2 | ND (<0.3) | ND | ND | ND | ND | ND |
| tel1 sgs1 | 126 | 21 | 17 (5/30) | <4 (0/30) | <4 (0/30) | 21 (5/30) | <4 (0/30) |
| rad53 sml1 | 95 | ND | ND | ND | ND | ND | ND |
| rad53 sml1 sgs1 | 879 | <29 | <0.03 (0/30) | <29 (0/30) | <29 (0/30) | <29 (0/30) | <29 (0/30) |
| rad9g | 20 | ND (<2) | ND | ND | ND | ND | ND |
| rad9 sgs1 | 748 | 25 | 3 (1/30) | <25 (0/30) | <25 (0/30) | 25 (1/30) | <25 (0/30) |
| rfc5-1c | 660 | ND (<66) | ND | ND | ND | ND | ND |
| rfc5-1 sgs1 | 631 | 42 | 7 (2/30) | <21 (0/30) | <21 (0/30) | 42 (2/30) | <21 (0/30) |
| tsa1h | 173 | ND (<17) | ND | ND | ND | ND | ND |
| tsa1 sgs1i | 1,139 | <38 | <0.03 (0/30) | <38 (0/30) | <38 (0/30) | <38 (0/30) | <38 (0/30) |
| msh2j | 5 | <0.3 | <0.07 (0/15) | <0.3 (0/15) | <0.3 (0/15) | <0.3 (0/15) | <0.3 (0/15) |
| msh2 asf1 | 100 | <3.7 | <0.04 (0/27) | <3.7 (0/27) | <3.7 (0/27) | <3.7 (0/27) | <3.7 (0/27) |
| msh2 cac1 | 296 | <11 | <0.04 (0/28) | <11 (0/28) | <11 (0/28) | <11 (0/28) | <11 (0/28) |
| msh2 mec3 | 32 | <1.5 | <0.05 (0/22) | <1.5 (0/22) | <1.5 (0/22) | <1.5 (0/22) | <1.5 (0/22) |
| msh2 rfc5-1 | 193 | <6.7 | <0.03 (0/29) | <6.7 (0/29) | <6.7 (0/29) | <6.7 (0/29) | <6.7 (0/29) |
| msh2 tel1 | 21 | <0.9 | <0.04 (0/23) | <0.9 (0/23) | <0.9 (0/23) | <0.9 (0/23) | <0.9 (0/23) |
| msh6 mec3 | 19 | <0.7 | <0.04 (0/27) | <0.7 (0/27) | <0.7 (0/27) | <0.7 (0/27) | <0.7 (0/27) |
| msh6 rad24 | 16 | <0.8 | <0.05 (0/20) | <0.8 (0/20) | <0.8 (0/20) | <0.8 (0/20) | <0.8 (0/20) |
ND, not determined; numbers in parentheses indicate an estimate of the CAN1/LYP1/ALP1 rate based on previous GCR breakpoint analyses as listed in footnotes c to h, in which no CAN1/LYP1/ALP1 rearrangements were found (75).
Numbers in parentheses indicate the number of CAN1/LYP1/ALP1 rearrangements detected among all GCR clones tested.
GCR rates and breakpoint analyses from reference 62. Breakpoint analysis results were the following: for rad53Δ sml1Δ, 5 telomere additions, 4 translocations with microhomology; rfc5-1, 10 telomere additions; mec1Δ sml1Δ, 9 telomere additions; tel1Δ, 6 translocations with microhomology; mec3Δ, 8 telomere additions, 2 translocations (0 nonhomology/2 microhomology/0 CAN1/ALP1/LYP1).
Breakpoint analysis for the wild type from references 61 and 69: 14 telomere additions, 3 translocation (1 nonhomology/2 microhomology/0 CAN1/ALP1/LYP1).
GCR rates and breakpoint analyses from reference 64. Breakpoint analysis results were the following: cac1Δ, 7 telomere additions, 3 translocations (2 nonhomology/1 microhomology/0 CAN1/ALP1/LYP1); asf1Δ, 9 telomere additions, 2 translocations (0 nonhomology/2 microhomology/0 CAN1/ALP1/LYP1).
Breakpoint analysis for rad52Δ from reference 60: 3 telomere additions, 7 translocations with microhomology.
GCR rate from reference 60. Breakpoint analysis for rad9Δ from reference 63: 8 telomere additions, 2 translocations with microhomology.
GCR rate and breakpoint analysis from reference 35. tsa1Δ, 8 telomere additions, 1 large deletion, 1 translocation (0 nonhomology/1 microhomology/0 CAN1/ALP1/LYP1).
GCR rate from reference 36.
Breakpoint analysis for msh2Δ from reference 61: 4 telomere additions, 2 independent base substitution mutations, 3 translocation (2 nonhomology/1 microhomology/0 CAN1/ALP1/LYP1).
FIG. 1.
Location of the CAN1, LYP1, and ALP1 genes in the yeast genome. CAN1 is located on chromosome 5, while ALP1 and LYP1 are located on the same arm of chromosome 14 in opposite orientations, separated by 889 bp that include a single 396-bp gene, BSC4. CAN1 and ALP1 are in the same orientation with respect to their centromeres, whereas LYP1 is in the opposite orientation. The lengths of the genes and distances between them are indicated in base pairs. The 174-bp regions of 93% sequence identity present in ALP1 and LYP1, indicated by a gray box, are 2,449 bp apart. The locations of the primers used to screen GCR clones for CAN1/LYP1/ALP1 rearrangements are indicated by F, R1, and R2 (see the text for details).
The analysis of GCRs described above in many cases yielded the expected PCR products for the CAN1-ALP1 translocations. Surprisingly, however, translocations that had been determined by other breakpoint mapping and sequencing methods (14) to contain CAN1-LYP1 breakpoints yielded CAN1-ALP1 PCR products instead of the expected CAN1-LYP1 PCR products. Sequencing of these PCR products revealed that these GCR breakpoint regions all contained a secondary LYP1-ALP1 breakpoint only a few hundred base pairs downstream of the identified primary CAN1-LYP1 breakpoint. Thus, these rearrangements were tripartite CAN1-LYP1-ALP1 translocations in which the region between LYP1 and ALP1 was deleted and both ALP1 and LYP1 sequences were now in the same orientation relative to each other, compared to being in inverted orientations on the normal chromosome 14. In total, PCR analysis of 880 chromosomal rearrangements isolated from 19 different sgs1 mutants revealed 41 tripartite CAN1-LYP1-ALP1 translocations, 18 CAN1-ALP1 translocations, 4 CAN1-LYP1 translocations without a secondary ALP1 breakpoint, and 2 CAN1 translocations with three breakpoints within LYP1 and ALP1, resulting in CAN1-LYP1-ALP1-LYP1-ALP1 rearrangements (Table 2).
None of these classes of translocations was identified in any single mutant analyzed, including the sgs1Δ single mutant. They were observed, however, if an sgs1Δ mutation was combined with rad24Δ, mec3Δ, asf1Δ, cac1Δ, or tel1Δ mutation and, at lower frequencies, with rad9Δ or rfc5-1 mutations. The synergistic increase in translocation rates in sgs1Δ mec3Δ and sgs1Δ asf1Δ double mutants and further increase in the translocation rate in the sgs1Δ asf1Δ mec3Δ triple mutant indicates that the formation of this class of translocations in sgs1Δ mutants is normally inhibited independently by DNA damage checkpoint sensors Mec3 and Rad24 and the chromatin assembly factors Cac1 and Asf1 (Table 2). A synergistic GCR rate increase was also observed when rrm3Δ and sgs1Δ mutations were combined, and introduction of a mec3Δ mutation into the sgs1Δ rrm3Δ double mutant led to a further GCR rate increase (Table 2). However, the doubling times of the rrm3Δ sgs1Δ and sgs1Δ rrmΔ3 mec3Δ strains are longer than those of HR-proficient sgs1Δ mutants with a functional Rrm3 helicase (e.g., Δrrm3 Δsgs1, 286 ± 32 min; sgs1Δ, 109 ± 3 min; rrm3Δ, 98 ± 1 min; wild type, 94 ± 2 min) (81); therefore, it is possible that their GCR rates may not be directly comparable to that of normally growing sgs1Δ mutants, even though all cultures were grown to the same final cell density during the fluctuation analysis. The critical role of the DNA damage checkpoint sensors Mec3 and Rad24 in suppressing these recurring translocations is further emphasized by the exceptionally high frequency of CAN-LYP1-ALP1 translocations, which make up 23% and 50% of all GCRs that were isolated from sgs1Δ mec3Δ and sgs1Δ rad24Δ double mutants, respectively. The weaker synergistic interactions and lower frequencies of CAN-LYP1-ALP1 translocations seen when rfc5-1 or rad9Δ mutation was combined with an sgs1Δ mutation indicate that the Rfc5 and Rad9 checkpoint proteins play a significant but lesser role in suppressing these translocations in sgs1Δ mutants than other checkpoint proteins. In contrast, we found no CAN1-LYP1-ALP1 translocations if the sgs1Δ mutation was combined with mutation in MEC1 or RAD53. This observation was surprising considering that Rad53, a central checkpoint kinase of the DNA damage checkpoint, is believed to act downstream of Rad24 and Mec3, while Mec1 has been implicated in virtually all DNA damage checkpoints in S. cerevisiae. Mec1 is known to phosphorylate Rad53, Rad9, and Ddc1, the latter of which is a subunit of the PCNA-like DNA damage-sensing complex that also contains Mec3 and interacts with Rad24, both of which we find are critical for the suppression of CAN1/LYP1/ALP1 translocations. In addition to its function in telomere maintenance, Tel1 forms a complex with Mre11 to establish a DNA damage checkpoint for double-strand breaks (19, 94). However, synthetic lethality between sgs1Δ and mre11Δ mutations prevented the further exploration of the role of MRE11 in the suppression of complex translocations in the absence of Sgs1. Altogether, these findings suggest that Rad24/Mec3-dependent processes effectively suppress translocations between related genes in the sgs1 mutant and that this suppression also requires the checkpoint kinase Tel1 but not the Mec1 kinase. Mutations that eliminated the HR pathway (rad52Δ) or the oxidative damage response system (tsa1Δ) did not lead to CAN1-LYP1-ALP1 translocations when combined with an sgs1Δ mutation; in fact, introduction of a rad52Δ mutation into the sgs1Δ mec3Δ double mutant eliminated the CAN1/LYP1/ALP1 translocations, demonstrating that HR is essential for the formation of these recurring translocations. However, introduction of a rad51Δ mutation, which causes a 4-fold reduction in mitotic recombination as opposed to the 3,000-fold reduction reported for the rad52Δ mutation (76), into the sgs1Δ mec3Δ double mutant did not reduce the rate of CAN1/LYP1/ALP1 translocations, suggesting that these translocations are formed by a Rad52-dependent, Rad51-independent recombination process.
Suppression of translocations between highly diverged genes depends on Sgs1 but not on Msh2 or Msh6.
MMR proteins have been shown to suppress recombination between homeologous DNA sequences (21, 83, 84, 88). To determine if the suppression of translocations between the highly diverged CAN1, LYP1, and ALP1 genes by Sgs1 is due to its role in the suppression of homeologous recombination, we analyzed GCRs isolated from MMR-defective mutants. We constructed msh2Δ mutants with an additional mutation in genes, such as CAC1, ASF1, MEC3, and TEL1, which had led to increased rates of CAN1/LYP1/ALP1 translocations in cells lacking Sgs1. All of these msh2Δ double mutants had GCR rates similar to those of the single mutants, and no rearrangements between CAN1 and ALP1 or LYP1 were detected (Table 2). However, in addition to its role in heteroduplex rejection, Msh2, together with the MMR protein Msh3, is also required for the removal of nonhomology from the ends of recombination intermediates (89), raising the possibility that the msh2Δ mutation may, in addition to inhibiting heteroduplex rejection, inhibit other early recombination steps that may prevent the formation of CAN1/LYP1/ALP1 translocations. We therefore combined an msh6 mutation, which inhibits heteroduplex rejection but not the removal of nonhomologous 3′ ends (89), with mec3Δ and rad24Δ mutations, which yield high frequencies of CAN1/LYP1/ALP1 translocations when combined with an sgs1Δ mutation (23% and 50% of total GCRs, respectively). We found that an msh6Δ mutation, like an msh2Δ mutation, did not lead to the formation of CAN1/LYP1/ALP1 translocations in the permissive mec3Δ and rad24Δ mutants, suggesting that relaxation of homeologous recombination is not sufficient for the formation of the complex translocations seen in sgs1Δ mutants, but that an Sgs1-specific function other than suppression of homeologous recombination normally prevents these rearrangements.
Structure of translocations between CAN1, ALP1, and LYP1.
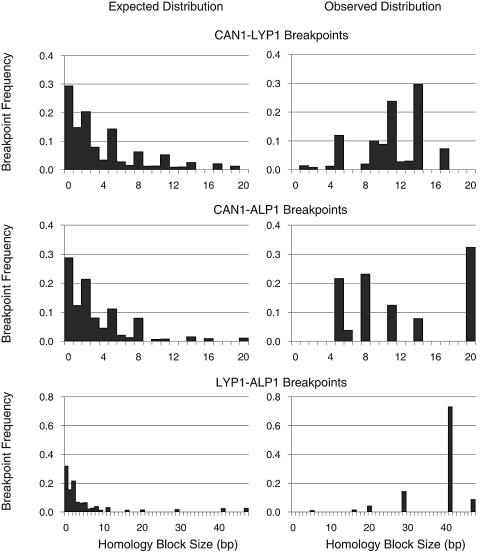
Sequence alignments between CAN1 and LYP1 as well as between CAN1 and ALP1 show 64% sequence identity, while ALP1 and LYP1 show 60% sequence identity. All of the 65 translocation breakpoints identified in this study were found within blocks of identical bases ranging from 1 to 17 bp in the CAN1-LYP1 alignment, from 5 to 20 bp in the CAN1-ALP1 alignment, and from 5 to 47 bp in the LYP1-ALP1 alignment (Fig. 2). Analysis of the frequency of breakpoints at specific sites revealed that longer homology blocks were utilized up to 31-fold more often than expected by chance, suggesting that CAN1/LYP1/ALP1 translocations are facilitated by the increasing length of homologous regions (Fig. 3). For instance, the 46 CAN1-LYP1 breakpoints were restricted to 26 homology blocks distributed over 717 bp near the 5′ end of CAN1 and LYP1 in a region of 74% sequence identity (Fig. 2A, breakpoints 1 to 26). The frequency of breakpoints in homology blocks of ≥9 bp was 2- to 10-fold higher than expected by chance, while those in homology blocks of <5 bp were underrepresented (Table 3; Fig. 3). The utilization of shorter than average homology blocks in the formation of CAN1-LYP1 translocations in mutants with rad51Δ or tel1Δ mutation (4.8 bp or 6 bp, respectively, versus the average of 8.6 bp) suggests that usage of long homology blocks for interchromosomal translocations between CAN1 and LYP1 depends on HR and may also depend on Tel1 (Table 4).
FIG. 2.
Location of translocation breakpoints in the CAN1, ALP1, and LYP1 genes. (A) The open reading frame (ORF) of CAN1 is shown. Regions of homology between the CAN1 and LYP1 genes that are longer than 4 bp are boxed with regions of homology that are unique to the CAN1-LYP1 alignment in uppercase letters and regions of homology that are identical in both CAN1-LYP1 and CAN1-ALP1 alignments in lowercase letters. The regions of homology associated with CAN1-LYP1 translocation breakpoints are numbered 1 to 26, with the most 5′ breakpoint having the lowest number; numbered red boxes indicate CAN1-LYP1 breakpoints, and numbered black boxes indicate CAN1-ALP1 breakpoints that occurred within a region of homology that is identical in CAN1-LYP1 and CAN1-ALP1 alignments. (B) The ORF of CAN1 is shown. Stretches of homology between the CAN1 and ALP1 genes that are longer than 4 bp are boxed with regions of homology that are unique to the CAN1-ALP1 alignment in uppercase letters and regions of homology that are identical in both CAN1-LYP1 and CAN1-ALP1 alignments in lowercase letters. Breakpoints are numbered 15 to 28, with the most 5′ breakpoint having the lowest number; numbered blue boxes indicate CAN1-ALP1 breakpoints, and numbered black boxes indicate CAN1-LYP1 breakpoints that occurred within a stretch of homology that is identical in CAN1-LYP1 and CAN1-ALP1 alignments. (C) The ORF of ALP1 is shown. Regions of homology between the ALP1 and LYP1 genes that are longer than 4 bp are boxed. Breakpoints are numbered 29 to 35, with the most 5′ breakpoint having the lowest number; numbered green boxes indicate LYP1-ALP1 breakpoints, and numbered dotted-line boxes indicate ALP1-LYP1 breakpoints that were part of CAN1-LYP1-ALP1-LYP1-ALP1 rearrangements. Breakpoint 34 was observed in LYP1-ALP1 rearrangements as well as in ALP1-LYP1 rearrangements.
FIG. 3.
Frequencies of CAN1-LYP1, CAN1-ALP1, and LYP1-ALP1 rearrangement breakpoints in homology blocks of varying lengths. The expected distribution assumes a random distribution of breakpoints.
TABLE 3.
Expected and observed frequencies of translocation breakpoints in homology blocks of various lengthsa
| Homology length (bp) | Breakpoint frequency for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
CAN1-LYP1
|
LYP1-ALP1
|
CAN1-ALP
|
|||||||
| Expected frequencyb | Observed frequencyc | Ratiod | Expected frequencyb | Observed frequencyc | Ratiod | Expected frequencyb | Observed frequencyc | Ratiod | |
| 1 | 0.146 | 0.064 (3) | 0.4 | 0.150 | 0 | 0 | 0.122 | 0 | 0 |
| 2 | 0.202 | 0.021 (1) | 0.1 | 0.210 | 0 | 0 | 0.211 | 0 | 0 |
| 4 | 0.031 | 0.021 (1) | 0.7 | 0.065 | 0 | 0 | 0.043 | 0 | 0 |
| 5 | 0.141 | 0.213 (10) | 1.5 | 0.061 | 0.064 (3) | 1.0 | 0.110 | 0.333 (6) | 3.0 |
| 6 | 0.026 | 0 | 0 | 0.016 | 0 | 0 | 0.019 | 0.056 (1) | 2.9 |
| 8 | 0.061 | 0.021 (1) | 0.3 | 0.033 | 0 | 0 | 0.078 | 0.278 (5) | 3.6 |
| 9 | 0.010 | 0.106 (5) | 10.2 | 0.009 | 0 | 0 | NA | NA | NA |
| 10 | 0.011 | 0.085 (4) | 7.4 | NA | NA | NA | 0.005 | 0 | 0 |
| 11 | 0.050 | 0.191 (9) | 3.8 | 0.028 | 0 | 0 | 0.005 | 0.111 | 20.2 |
| 12 | 0.007 | 0.021 (1) | 3.1 | NA | NA | NA | NA | NA | NA |
| 13 | 0.007 | 0.021 (1) | 2.9 | NA | NA | NA | NA | NA | NA |
| 14 | 0.023 | 0.191 (9) | 8.2 | NA | NA | NA | 0.014 | 0.056 | 4.0 |
| 16 | NA | NA | NA | 0.008 | 0.021 (1) | 2.7 | 0.008 | 0 | 0 |
| 17 | 0.019 | 0.043 (2) | 2.3 | NA | NA | NA | NA | NA | NA |
| 20 | NA | NA | NA | 0.010 | 0.064 (3) | 6.6 | 0.010 | 0.167 (3) | 17.3 |
| 22 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 29 | NA | NA | NA | 0.014 | 0.170 (8) | 12.2 | NA | NA | NA |
| 41 | NA | NA | NA | 0.019 | 0.596 (28) | 30.6 | NA | NA | NA |
| 47 | NA | NA | NA | 0.022 | 0.116 (4) | 3.8 | NA | NA | NA |
NA (not available) indicates homology blocks of certain lengths that were not found in the specified gene alignment.
The expected frequency of a breakpoint occurring in a homology block of a certain length was calculated by dividing the product of the homology block length and its number of occurrences within the alignment by the total length of the alignment. The total length of the alignment was defined as the sum of the products of homology block length and its number of occurrences.
The observed breakpoint frequency was calculated by dividing the number of observed breakpoints within a homology block of certain length by the total number of breakpoints observed between these two genes. Numbers in parentheses indicate the total number of breakpoints observed in a homology block of this length.
The ratio between the observed and expected number of breakpoints in a homology block of a certain length.
TABLE 4.
Lengths of homology blocks with breakpoint occurrences in sgs1 mutants with defects in cell cycle checkpoints, homologous recombination, chromatin assembly, or the DNA helicase Rrm3
| Relevant genotypea | Breakpoint(s) for:
|
|||||
|---|---|---|---|---|---|---|
|
CAN1-ALP1 (monocentric chromosome)
|
CAN1-LYP1 (dicentric intermediate)
|
LYP1-ALP1 (monocentric chromosome)
|
||||
| Homology block lengthb | Avg length | Homology block lengthb | Avg length | Homology block lengthb | Avg length | |
| sgs1Δ rad9Δc | NA | NA | 5 | 5.0 | 29 | 29.0 |
| sgs1Δ tel1Δd | NA | NA | 1, 1, 5, 9, 14 | 6.0 | 41, 41, 41, 41, 47 | 42.2 |
| sgs1Δ rad51Δe | 8 | 8.0 | 1, 4, 5, 9 | 4.8 | 20, 41, 41, 41 | 35.8 |
| sgs1Δ rrm3Δf | 5, 5, 8, 14, 20 | 10.4 | 5, 9, 9, 11, 11, 11, 14, 14, 14 | 10.9 | 16, 29, 29, 29, 29, 41, 41, 41, 41 | 33.8 |
| sgs1Δ mec3Δg | 5, 5, 5, 6, 11, 20 | 8.7 | 5, 5, 5, 9, 11, 11, 11, 14, 14, 14, 14, 14, 17 | 11.1 | 16, 29, 29, 41, 41, 41, 41, 41, 41, 41 | 36.1 |
| sgs1Δ asf1Δh | 5, 8 | 6.5 | 5, 11, 14, 14, 17, 5, 10, 12, 14 | 11.3 | 41, 41, 41, 41 | 41.0 |
| sgs1Δ cac1Δi | 8, 11 | 9.5 | 5, 11, 14 | 10.0 | 29, 41, 41 | 37.0 |
| sgs1Δ rad24Δj | 5, 8, 20, 20 | 13.3 | 2, 5, 5, 8, 9, 10, 10, 11, 11, 11, 17 | 9.0 | 5, 5, 20, 20, 41, 41, 41, 41, 41, 47 | 30.2 |
| sgs1Δ rfc5-1k | NA | NA | 10, 11 | 10.5 | 41, 47 | 44.0 |
| Avg lengthl | 9.4 | 8.6 | 36.6 | |||
For additional genotype information and strain numbers, see footnotes d to l. See Table 1 for complete genotypes of these strains.
The total length (in base pairs) of every homology block with an observed breakpoint is listed. NA (not available) indicates that this chromosomal rearrangement was not observed in this mutant.
CAN1/LYP1/ALP1 breakpoints were identified in the rad9Δ sgs1Δ (RDKY5778) mutant.
CAN1/LYP1/ALP1 breakpoints were identified in the tel1Δ sgs1Δ (RDKY4566) mutant.
CAN1/LYP1/ALP1 breakpoints were identified in rrm3Δ sgs1Δ rad51Δ (RDKY5564) and rrm3Δ sgs1Δ rad51Δ mec3Δ (RDKY5776) mutants.
CAN1/LYP1/ALP1 breakpoints were identified in rrm3Δ sgs1Δ (haploid strain freshly derived from RDKY5577) and rm3Δ sgs1Δ mec3Δ (RDKY5579) mutants.
CAN1/LYP1/ALP1 breakpoints listed were identified in sgs1Δ mec3Δ (RDKY5572), asf1Δ sgs1Δ mec3Δ (RDKY5772), and rrm3Δ sgs1Δ mec3Δ (RDKY5579) mutants.
CAN1/LYP1/ALP1 breakpoints were identified in asf1Δ sgs1Δ (RDKY4767) and asf1Δ sgs1Δ mec3Δ (RDKY5772) mutants.
CAN1/LYP1/ALP1 breakpoints were identified in the cac1Δ sgs1Δ (RDKY4765) mutant.
CAN1/LYP1/ALP1 breakpoints were identified in the rad24Δ sgs1Δ (RDKY5575) mutant.
CAN1/LYP1/ALP1 breakpoints were identified in the rfc5-1 sgs1Δ (RDKY4561) mutant.
The average length (in base pairs) of all homology blocks with a breakpoint occurrence is shown for each of the three rearrangements.
Of 47 CAN1-LYP1 translocations, we found only four that did not have a secondary LYP1-ALP1 breakpoint; these are currently being analyzed by array-based comparative genomic hybridization to detect potential secondary rearrangements. All LYP1-ALP1 breakpoints were confined to only seven homology blocks within a 173-bp stretch of 93% sequence identity in the center of the 1,722-bp LYP1-ALP1 alignment (Fig. 2C, homology blocks 29 to 35). Remarkably, two translocations were isolated from sgs1Δ rad24Δ and sgs1Δ asf1Δ double mutants that showed two additional breakpoints between LYP1 and ALP1, resulting in even more complex CAN1-LYP1-ALP1-LYP1-ALP1 translocations. These additional breakpoints were assigned on the basis of 1-bp discontinuities in the LYP1 or ALP1 alignment that corresponded to the ALP1 or LYP1 gene, respectively. All LYP1-ALP1 breakpoints occurred in homology blocks of at least 5 bp, and above this threshold longer homology blocks were favored over shorter homology blocks (Fig. 3; Table 3). In fact, 60% of the 47 breakpoints were located in the same 41-bp homology block (Fig. 2C, breakpoint 29), occurring with 31-fold higher frequency than expected by chance, while another 8 (17%) were located in the same 29-bp homology block (Fig. 2C, breakpoint 35), occurring with 12-fold higher frequency than expected. A rad51Δ mutation did not force rearrangements into shorter LYP1-ALP1 homology blocks or into regions without homology, as demonstrated by similar average homology block lengths in rad51Δ and RAD strains (35.8 bp and 36.7 bp, respectively). This disparity between the Rad51 independency of CAN1/LYP1/ALP1 translocation rates, Rad51 dependency of homology block length in CAN1-LYP1 translocations, and its lack with respect to LYP1-ALP1 rearrangements suggests that Rad51-independent mechanisms mediate the formation of secondary LYP1-ALP1 rearrangements (this pathway could involve Rad59, although we did not test this), whereas Rad51-dependent and Rad51-independent processes can mediate the formation of interchromosomal CAN1-LYP1 translocations with similar effectiveness but Rad51-dependent processes are used preferentially. The Rad52 dependency of translocations between CAN1 and LYP1 or ALP1, as indicated by the absence of CAN1/LYP1/ALP1 translocations in the sgs1Δ mec3Δ rad52Δ triple mutant, suggests that a Rad59-dependent HR pathway may be partially redundant with a Rad51-dependent HR pathway in promoting the initial HR event leading to the CAN1/LYP1/ALP1 translocations.
In addition to translocations between CAN1 and LYP1, translocations between CAN1 and ALP1 were observed in this study. All 18 CAN1-ALP1 breakpoints were confined to a 248-bp region of 76% sequence identity (Fig. 2B, breakpoints 15 to 28). Like CAN1-LYP1 and LYP1-ALP1 breakpoints, CAN1-ALP1 breakpoints occurred 3 to 20 times more frequently in larger homology blocks than expected, while no breakpoints were observed in regions with less than 4 bp of homology (Tables 3 and 4; Fig. 2B).
Note that we have excluded the possibility that CAN1-LYP1-ALP1 translocations may have been generated by translocation of CAN1 to a preexisting LYP1-ALP1 gene fusion on chromosome 14 (which could have resulted from unequal sister chromatid conversion or intrachromatid crossover between the two inverted homeologous genes) by confirming by PCR that the predicted LYP1-ALP1 rearrangements were not present in the clones containing GCRs and that intact ALP1 and LYP1 genes were present (data not shown). The presence of intact LYP1 and ALP1 genes in all GCR clones tested and the absence of ALP1-CAN1 rearrangements in all 12 tested GCR clones with CAN1-ALP1 translocations (data not shown) indicates that CAN1/LYP1/ALP1 rearrangements are nonreciprocal events, most likely generated by break-induced replication (BIR), rather than reciprocal events that occurred during G2/M. Since ALP1-CAN1 rearrangements are not selected against in our assay, reciprocal translocations in G2/M would predict an equal association of CAN1-ALP1 translocation chromosomes with a chromosome 14 that contains an intact ALP1 gene or an ALP1-CAN1 rearrangement, which we did not observe in this study.
The most overrepresented breakpoint location identified in this study is the 41-bp LYP1-ALP1 homology block. The observed 31-fold-higher-than-expected frequency of breakpoints in this location supports the correlation between longer regions of sequence identity and increased HR. However, the only threefold overrepresentation of the nearby 47-bp LYP1-ALP1 homology block shows that length of sequence identity is not the only determining factor for translocation target sites, but that structure and location of the homology block may also be important predictors of their suitability as an HR hotspot. In fact, the two most overrepresented LYP1-ALP1 breakpoints are not the longest blocks, but they are the first and the last homology block within the 173-bp homeologous region in the LYP1-ALP1 alignment (Fig. 2C, breakpoints 29 and 35), and they are preceded or followed by relatively long regions, 76 bp and 70 bp, respectively, that do not contain any homology blocks of at least 6 bp. This may suggest that the border between nonhomologous regions and regions of significant sequence identity may favor HR.
DISCUSSION
We have analyzed the formation of spontaneous translocations between three related but highly diverged genes (CAN1, ALP1, and LYP1) in their natural locations on two different chromosomes in S. cerevisiae and provided evidence for a central role of the DNA helicase Sgs1 in suppressing translocations between these related sequences. An sgs1Δ mutation caused a modest increase in the rate of translocations but did not result in the recovery of translocations between CAN1 and ALP1 or LYP1. However, combining an sgs1Δ mutation with additional defects in other DNA metabolic pathways often resulted in increased rates of translocations involving the divergent gene CAN1, ALP1, or LYP1. This was observed when an sgs1Δ mutation was combined with mutations causing defects in the DNA damage checkpoint sensors Mec3 and Rad24, the DNA damage checkpoint protein Rad9, the replication checkpoint protein Rfc5, the checkpoint kinase and telomere length maintenance factor Tel1, the DNA helicase Rrm3, and the chromatin assembly factors Cac1 and Asf1, but not the checkpoint kinases Mec1 and Rad53 and the Tsa1-dependent oxidative damage response pathway. A diversity of CAN1/LYP1/ALP1 translocations was observed, containing as many as four breakpoints that were preferentially located in regions of extended homology, suggesting these translocations were formed by HR. Where tested, these translocations were eliminated by a rad52Δ mutation but not by a rad51Δ mutation, indicating that the divergent sequence translocations were primarily formed by a Rad52-dependent HR pathway and that a Rad51-independent HR pathway could also promote translocations. An msh2Δ or an msh6Δ mutation in combination with mutations found to interact with an sgs1Δ mutation did not result in increased translocations between CAN1 and ALP1 or LYP1, indicating that Sgs1 plays a unique role in suppressing translocations between divergent sequences rather than simply acting in the MMR pathway that suppresses homeologous recombination.
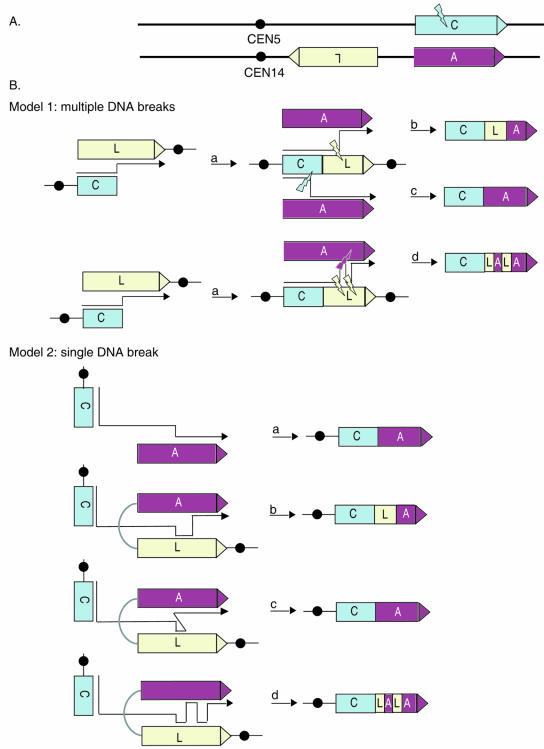
A striking feature of our results is the observation that 66% of the translocations involving CAN1, ALP1, and LYP1 involved two or more apparent translocation events. Assuming that these events are mediated by HR, there are two types of models that might explain these types of translocations (Fig. 4). All of the multiply translocated chromosomes appear to involve translocation from CAN1 to LYP1. Due to the opposite orientation of CAN1 and LYP1 relative to their respective centromeres, a translocation from CAN1 to LYP1 would yield a dicentric chromosome, which, as suggested by the first model, would then be predicted to break during cell division and undergo secondary rearrangements, yielding monocentric CAN1-ALP1, CAN1-LYP1-ALP1, or CAN1-LYP1-ALP1-LYP1-ALP1 translocation (Fig. 4, model 1). The CAN1-ALP1 translocations could also be generated by a single interchromosomal recombination event between CAN1 and ALP1. A general mechanism involving multiple, independent recombination events seems unlikely for several reasons: (i) the rate of a complex event would be expected to be the product of the rates of the individual steps, and given the rate of single translocation events is low, the observation of translocations involving two or more independent events seems unlikely; (ii) the formation of CAN1-ALP1 translocations needs to involve only one event, yet these translocations are less frequent (28%) than the CAN1-LYP1-ALP1 translocations (72%) that require two events, which is surprising unless there is selection against the CAN1-ALP1 translocations due to formation of active CAN1-ALP1 fusion genes (which we did not test), or ALP1 is a less favorable translocation target than LYP1 despite similar sequence identity and presence of comparable homology blocks; (iii) the CAN1-LYP1-ALP1-LYP1-ALP1 translocations, which would involve four events, would be predicted to be much more rare (square of the rate of two-event CAN1-LYP1-ALP1 translocations) than observed; and (iv) it is not clear what would select for CAN1-LYP1-ALP1-LYP1-ALP1 translocations, as all intermediate translocations containing ALP1 would no longer be dicentric and would not be subject to further breakage and translocation. An alternative model suggests that translocations may have formed as the result of repairing a single DNA break on chromosome 5 in CAN1 by BIR coupled with multiple DNA template switches (Fig. 4, model 2). This model suggests that all of the translocations are the product of a single, concerted series of events, which seems more consistent with the observed high frequency of translocation involving two or more rearrangements. A combination of factors, such as the location of ALP1 and LYP1 on the same chromosome arm, their close proximity (889 bp), and the presence of 173-bp regions of 93% sequence identity within LYP1 and ALP1 that are only separated by 2,530 bp, may have facilitated misannealing of the invading strand with the center of ALP1 after copying the 5′ end of LYP1 and dissociating from it, thereby facilitating the formation of complex translocations in our system. Since all sgs1 mutants with complex translocations between CAN1, LYP1, and/or ALP1 were mismatch repair proficient, it is possible that after the initial exchange between LYP1 and ALP1 occurred a LYP1-ALP1 heteroduplex intermediate was formed, and patchy rather than continuous mismatch correction, instead of multiple chromosome breakages or template switches, yielded the two CAN1-LYP1-ALP1-LYP1-ALP1 translocations rather than CAN1-LYP1-ALP1 translocations. Note that if this mechanism did occur, mismatch repair deficiency would only be expected to eliminate the CAN1-LYP1-ALP1-LYP1-ALP1 translocations and not the CAN1-LYP1-ALP1 translocations.
FIG. 4.
Models for the formation of complex translocations between CAN1, ALP1, and LYP1. (A) Location of CAN1 (“C”) on chromosome 5 and ALP1 (“A”) and LYP1 (“L”) on the same arm of chromosome 14, facing in opposite directions. (B) Model 1 shows formation of complex translocations as a result of multiple chromosome breaks. A single DNA break in chromosome 5 leads to invasion of the related LYP1 gene on chromosome 14, forming a D loop and initiating DNA synthesis to the end of chromosome 14 to yield a dicentric CAN1-LYP1 chromosome (a). A second independent DNA break similarly leads to recombination between LYP1 and ALP1, which is in the opposite orientation of LYP1, transforming the dicentric chromosome into a stable, monocentric CAN1-LYP1-ALP1 chromosome (b). Similarly, recombination between CAN1 and ALP1 upstream of the original CAN1-LYP1 breakpoint eliminates any LYP1 sequence from the translocation chromosome and transforms the CAN1-LYP1 chromosome into a CAN1-ALP1 chromosome (c). CAN1-LYP1-ALP1-LYP1-ALP1 translocations may have formed as a result of four independent break-mediated recombination events of the type described for step a above sequentially, leading to generation of a dicentric CAN1-LYP1 translocation, a CAN1-LYP1-ALP1 monocentric translocation, a CAN1-LYP1-ALP1-LYP1 dicentric translocation, and finally a CAN1-LYP1-ALP1-LYP1-ALP1 monocentric translocation. A related but alternative mechanism follows the two-break mechanism described for step a above, except that after initiating repair of the second DNA break in the CAN1-LYP1 chromosome by invading homologous sequences of ALP1 followed by short patch DNA synthesis, what occurs is dissociation from ALP1 and reinvasion of homologous sequences of LYP1, followed by another cycle of short patch DNA synthesis, dissociation, and reinvasion of homologous sequences of ALP1, after which DNA synthesis proceeds to the chromosome end (d). (B) Model 2 shows formation of complex translocations as a result of a single chromosome break in CAN1. Instead of forming a dicentric CAN1-LYP1 chromosome first, translocations may have formed by a single event, which may or may not have involved DNA template switching during DNA synthesis. CAN1-ALP1 translocations could be generated by repairing a DNA break in chromosome 5 through interchromosomal recombination between CAN1 and ALP1 (a). Discontinuous extension of the invading 3′ end could lead to the incorporation of multiple related DNA sequences: two cycles of strand invasion, DNA synthesis, and dissociation, first into LYP1 and then into the nearby ALP1, would lead to CAN1-LYP1-ALP1 translocations if dissociation and reannealing occur after LYP1-specific sequence has been copied (b); they would lead to CAN1-ALP1 translocations if dissociation from LYP1 and annealing to ALP1 occur prior to copying of LYP1-specific sequence at a homology block shared by all three genes, as is the case for 94% (17/18) of CAN1-ALP1 breakpoints (c); or four cycles of dissociation and reannealing would lead to CAN1-LYP1-ALP1-LYP1-ALP1 translocations (d). In all of the models discussed above, recombination most likely involves BIR, because the cells lose the region of chromosome 5 between CAN1 and the telomere but appear to retain an intact copy of chromosome 14 as evidenced by the presence of wild-type copies of LYP1 and ALP1.
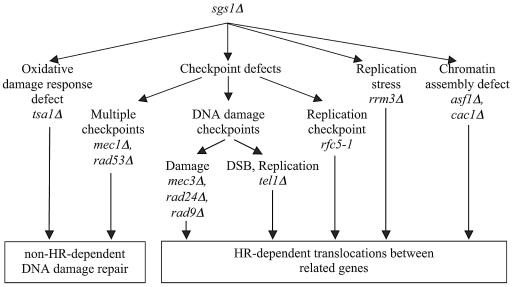
Defects in SGS1 appear to result in multiple defects that contribute to increased genome instability. These include defects in processing replication intermediates and defects in S-phase checkpoints (10, 24, 26, 30, 38, 49, 61, 81, 82, 93, 96) as well as defects in the suppression of aberrant recombination between divergent sequences documented here. However, the increased genome instability seen in sgs1Δ single mutants is modest and is most strikingly revealed when sgs1Δ mutations are combined with defects in other pathways that are important for maintaining genome stability. Here we observed three types of genetic interactions with sgs1Δ mutations: cases where no genetic interaction was observed; cases resulting in a synergistic increase in the GCR rate without the occurrence of translocations involving divergent sequences; and cases resulting in small to large synergistic increases in the GCR rate along with significantly increased rates of translocations involving divergent sequences. Combined with the results of previous studies, our results suggest two distinct scenarios occur. In some cases the synergistic interaction with sgs1Δ results in increased damage, but the lesions may not normally be substrates for HR and hence there is no effect of sgs1Δ on suppression of HR between divergent sequences (Fig. 5). Examples of this include the interaction between sgs1Δ and the mec1Δ and rad53Δ mutations that result in checkpoint defects or a tsa1Δ mutation that results in increased oxidative damage to DNA. In other cases the synergistic interaction with sgs1Δ usually, but possibly not always, results in increased damage yielding lesions that are substrates for HR, and hence there is an effect of sgs1Δ on suppression of HR between divergent sequences (Fig. 5). Examples of this include the interaction between sgs1Δ and the checkpoint-defective mutations rad9Δ, rad24Δ, mec3Δ, and rfc5-1, the checkpoint kinase-defective mutation tel1Δ, the rrm3Δ mutation that results in replication fork stalling, and the cac1Δ and asf1Δ mutations that result in defective chromatin assembly during DNA replication. Overall, these results raise the possibility that replication errors that can be processed by HR, as suggested by a number of studies (2, 8, 45, 46, 77), can be aberrantly acted on by HR to yield translocations, and that Tel1-dependent checkpoints may be critical to suppression of these replication errors or could act at later steps in the translocation suppression process (Fig. 5). The observation that a tel1Δ mutation did not cause as large a synergistic increase in divergent sequence translocations in combination with an sgs1Δ mutation as rad24Δ, mec3Δ, and rfc5-1 mutations did raises the possibility that Mec1 might be partially redundant with Tel1 (18, 57, 62, 80), although we did not test this.
FIG. 5.
Model for the suppression of homology-driven translocations in the absence of Sgs1 helicase. Replication stress as well as defects in DNA damage checkpoints, the DNA replication checkpoint, and chromatin assembly can lead to increased levels of HR-dependent translocations in sgs1Δ mutants, while lack of the checkpoint kinase Mec1 or Rad53 or a defective oxidative response preferentially leads to other GCR types, such as de novo telomere additions and translocations without homology at the breakpoint (see the text for details).
MMR proteins and Sgs1 function in the suppression of homeologous recombination in S. cerevisiae (61, 84, 87). That msh2Δ and msh6Δ mutations did not lead to CAN1/LYP1/ALP1 translocations in any of the permissive genetic backgrounds tested suggests that failure to suppress homeologous recombination was not the primary cause for the increased rate of translocations between divergent DNA sequences observed in sgs1Δ mutants. However, the difference between sgs1Δ and MMR mutations seen here could reflect a key mechanistic feature of the events studied. It is possible that MMR proteins might be more effective at suppressing the intrachromosomal recombination events measured in most previous studies compared to the interchromosomal recombination events assayed here. Additionally, the DNA sequences surrounding the homeologous genes may influence the activity of MMR proteins; for example, it was proposed that recombination between homeologous sequences may not be affected by MMR proteins if recombination is initiated within regions of nonhomology surrounding the homeologous sequences, whereas MMR proteins play an important role in the suppression of recombination between homeologous sequences embedded in homology (72). It is also possible that MMR, in contrast to Sgs1, cannot act to suppress translocations resulting from HR between sequences with the high levels of divergence studied here. In human tumors, MMR defects lead to mono- and dinucleotide repeat (microsatellites) instability (MIN) (25, 43, 48, 68), but MMR-defective tumors usually do not show chromosomal instability (CIN) (33, 43, 50). However, a subset of tumor cell lines is known to show both MIN and CIN (1, 32, 90). The results presented here are consistent with the observation that MMR-defective tumors do not usually show CIN and suggest that the reason for this is that suppression of homeologous recombination by MMR proteins may not be a major mechanism for the suppression of genome rearrangements. Our results also suggest that those MMR-defective tumor cell lines showing MIN and CIN (1, 32, 90) may contain an additional genetic defect inactivating a function that normally helps prevent genome instability.
Highly elevated levels of genetic exchange between identical sequences resulting in SCEs and symmetrical QRs are a hallmark of BS. Chromosomal aberrations including translocations have also been described in BS patients (4, 39, 71, 85, 102). Furthermore, the repeated observation of partial or complete loss of chromosome 7 in bone marrow cells of BS patients with acute lymphoblastic or myeloblastic leukemia may indeed suggest that lack of BLM can increase the frequency of specific, recurring chromosomal rearrangements (4, 39, 71, 85). However, translocation breakpoints from BS cells have not yet been cloned and sequenced, so little is known about the mechanisms that produce these translocations. Previous results showing that defects in the S. cerevisiae BLM homolog SGS1 result in increased recombination and altered control of crossing over and gene conversion (38, 100, 104) suggest how defects in BLM might result in increased SCEs and QRs. The results presented here showing that Sgs1 functions to prevent inappropriate recombination between highly diverged DNA sequences with minimal regions of identity, which would normally be excluded from HR during mitosis, suggest that such sequences may become effective target sites of chromosomal rearrangements in BS cells, leading to the translocations and other chromosome aberrations seen in BS cells. Moreover, HR between short interspersed elements, of which the 300-bp Alu element is the most abundant (∼106 copies/human genome), has been implicated as a major mutational mechanism in numerous common diseases with recurrent rearrangements (9). This raises the possibility that elevated short interspersed element-mediated recombination may also contribute to the increased formation of chromosomal rearrangements in BS. Similarly, elevation of nonallelic HR between region-specific, low-copy repeats, which are known mediators of recurrent rearrangements in the human genome, may contribute to chromosome rearrangements in BS. Recently the tumor suppressor protein p53 has also been implicated in the regulation of spontaneous HR (54; reviewed in reference 7), specifically in the suppression of DNA exchange between imperfectly homologous DNA sequences (5) and the suppression of HR induced by inhibition of replication (78), further highlighting the importance of stringent regulation of HR, like that mediated by Sgs1 in order to maintain stability of the repetitive human genome.
.
Acknowledgments
We thank Christopher Putnam, Vincent Pennaneach, Jorrit Enserinck, and Meng-Er Huang for helpful comments on the manuscript and Stephanie Ness for DNA sequencing.
This work was supported by NIH grants GM26017 and GM50006 to R.D.K.
REFERENCES
- 1.Abdel-Rahman, W. M., K. Katsura, W. Rens, P. A. Gorman, D. Sheer, D. Bicknell, W. F. Bodmer, M. J. Arends, A. H. Wyllie, and P. A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA 98:2538-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, A., S. Chavez, and F. Malagon. 2000. Mitotic recombination in yeast: elements controlling its incidence. Yeast 16:731-754. [DOI] [PubMed] [Google Scholar]
- 3.Ajima, J., K. Umezu, and H. Maki. 2002. Elevated incidence of loss of heterozygosity (LOH) in an sgs1 mutant of Saccharomyces cerevisiae: roles of yeast RecQ helicase in suppression of aneuploidy, interchromosomal rearrangement, and the simultaneous incidence of both events during mitotic growth. Mutat. Res. 504:157-172. [DOI] [PubMed] [Google Scholar]
- 4.Aktas, D., A. Koc, K. Boduroglu, G. Hicsonmez, and E. Tuncbilek. 2000. Myelodysplastic syndrome associated with monosomy 7 in a child with Bloom syndrome. Cancer Genet. Cytogenet. 116:44-46. [DOI] [PubMed] [Google Scholar]
- 5.Akyuz, N., G. S. Boehden, S. Susse, A. Rimek, U. Preuss, K. H. Scheidtmann, and L. Wiesmuller. 2002. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol. Cell. Biol. 22:6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, R. J., J. L. Keck, and J. C. Wang. 1999. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 289:235-248. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand, P., Y. Saintigny, and B. S. Lopez. 2004. p53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 20:235-243. [DOI] [PubMed] [Google Scholar]
- 8.Bierne, H., and B. Michel. 1994. When replication forks stop. Mol. Microbiol. 13:17-23. [DOI] [PubMed] [Google Scholar]
- 9.Bishop, A. J., and R. H. Schiestl. 2002. Homologous recombination and its role in carcinogenesis. J. Biomed. Biotechnol. 2:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjergbaek, L., J. A. Cobb, M. Tsai-Pflugfelder, and S. M. Gasser. 2005. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24:405-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brosh, R. M., Jr., J. L. Li, M. K. Kenny, J. K. Karow, M. P. Cooper, R. P. Kureekattil, I. D. Hickson, and V. A. Bohr. 2000. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275:23500-23508. [DOI] [PubMed] [Google Scholar]
- 12.Chaganti, R. S., S. Schonberg, and J. German. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71:4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23:81-85. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C., K. Umezu, and R. D. Kolodner. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2:9-22. [DOI] [PubMed] [Google Scholar]
- 15.Cobb, J. A., L. Bjergbaek, and S. M. Gasser. 2002. RecQ helicases: at the heart of genetic stability. FEBS Lett. 529:43-48. [DOI] [PubMed] [Google Scholar]
- 16.Cobb, J. A., L. Bjergbaek, K. Shimada, C. Frei, and S. M. Gasser. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Constantinou, A., M. Tarsounas, J. K. Karow, R. M. Brosh, V. A. Bohr, I. D. Hickson, and S. C. West. 2000. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craven, R. J., P. W. Greenwell, M. Dominska, and T. D. Petes. 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dasika, G. K., S. C. Lin, S. Zhao, P. Sung, A. Tomkinson, and E. Y. Lee. 1999. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 18:7883-7899. [DOI] [PubMed] [Google Scholar]
- 21.Datta, A., A. Adjiri, L. New, G. F. Crouse, and S. Jinks Robertson. 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Der Kaloustian, V. M., J. J. McGill, M. Vekemans, and H. R. Kopelman. 1990. Clonal lines of aneuploid cells in Rothmund-Thomson syndrome. Am. J. Med. Genet. 37:336-339. [DOI] [PubMed] [Google Scholar]
- 23.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 24.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fishel, R., M. K. Lescoe, M. R. Rao, N. G. Copeland, N. A. Jenkins, J. Garber, M. Kane, and R. Kolodner. 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75:1027-1038. [DOI] [PubMed] [Google Scholar]
- 26.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuchi, K., G. M. Martin, and R. J. Monnat, Jr. 1989. Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl. Acad. Sci. USA 86:5893-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuchi, K., K. Tanaka, J. Nakura, Y. Kumahara, T. Uchida, and Y. Okada. 1985. Elevated spontaneous mutation rate in SV40-transformed Werner syndrome fibroblast cell lines. Somat. Cell Mol. Genet. 11:303-308. [DOI] [PubMed] [Google Scholar]
- 29.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 31.German, J. 1997. Bloom's syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet. 93:100-106. [DOI] [PubMed] [Google Scholar]
- 32.Goel, A., C. N. Arnold, D. Niedzwiecki, D. K. Chang, L. Ricciardiello, J. M. Carethers, J. M. Dowell, L. Wasserman, C. Compton, R. J. Mayer, M. M. Bertagnolli, and C. R. Boland. 2003. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 63:1608-1614. [PubMed] [Google Scholar]
- 33.Grady, W. M. 2004. Genomic instability and colon cancer. Cancer Metastasis Rev. 23:11-27. [DOI] [PubMed] [Google Scholar]
- 34.Hojo, E. T., P. C. van Diemen, F. Darroudi, and A. T. Natarajan. 1995. Spontaneous chromosomal aberrations in Fanconi anaemia, ataxia telangiectasia fibroblast and Bloom's syndrome lymphoblastoid cell lines as detected by conventional cytogenetic analysis and fluorescence in situ hybridisation (FISH) technique. Mutat. Res. 334:59-69. [DOI] [PubMed] [Google Scholar]
- 35.Huang, M. E., and R. D. Kolodner. 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17:709-720. [DOI] [PubMed] [Google Scholar]
- 36.Huang, M. E., A. G. Rio, A. Nicolas, and R. D. Kolodner. 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100:11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber, M. D., D. C. Lee, and N. Maizels. 2002. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 30:3954-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwahara, Y., K. Ishii, S. Watanabe, H. Taguchi, H. Hara, and I. Miyoshi. 1993. Bloom's syndrome complicated by myelodysplastic syndrome and multiple neoplasia. Intern. Med. 32:399-402. [DOI] [PubMed] [Google Scholar]
- 40.Kadyk, L. C., and L. H. Hartwell. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132:387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitao, S., I. Ohsugi, K. Ichikawa, M. Goto, Y. Furuichi, and A. Shimamoto. 1998. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54:443-452. [DOI] [PubMed] [Google Scholar]
- 42.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 43.Kolodner, R. D., and G. T. Marsischky. 1999. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 9:89-96. [DOI] [PubMed] [Google Scholar]
- 44.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 45.Kraus, E., W. Y. Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lavin, M. F., and K. K. Khanna. 1999. ATM: the protein encoded by the gene mutated in the radiosensitive syndrome ataxia-telangiectasia. Int. J. Radiat. Biol. 75:1201-1214. [DOI] [PubMed] [Google Scholar]
- 48.Leach, F. S., N. C. Nicolaides, N. Papadopoulos, B. Liu, J. Jen, R. Parsons, P. Peltomaki, P. Sistonen, L. A. Aaltonen, M. Nystrom-Lahti, et al. 1993. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell 75:1215-1225. [DOI] [PubMed] [Google Scholar]
- 49.Lee, S. K., R. E. Johnson, S. L. Yu, L. Prakash, and S. Prakash. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339-2342. [DOI] [PubMed] [Google Scholar]
- 50.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1997. Genetic instability in colorectal cancers. Nature 386:623-627. [DOI] [PubMed] [Google Scholar]
- 51.Lindor, N. M., E. M. Devries, V. V. Michels, C. R. Schad, S. M. Jalal, K. M. Donovan, W. A. Smithson, L. K. Kvols, S. N. Thibodeau, and G. W. Dewald. 1996. Rothmund-Thomson syndrome in siblings: evidence for acquired in vivo mosaicism. Clin. Genet. 49:124-129. [DOI] [PubMed] [Google Scholar]
- 52.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancerpredisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 53.Macris, M. A., L. Krejci, W. Bussen, A. Shimamoto, and P. Sung. 2005. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Rep. (Amsterdam) 5:172-180. [DOI] [PubMed] [Google Scholar]
- 54.Mekeel, K. L., W. Tang, L. A. Kachnic, C. M. Luo, J. S. DeFrank, and S. N. Powell. 1997. Inactivation of p53 results in high rates of homologous recombination. Oncogene 14:1847-1857. [DOI] [PubMed] [Google Scholar]
- 55.Miozzo, M., P. Castorina, P. Riva, L. Dalpra, A. M. Fuhrman Conti, L. Volpi, T. S. Hoe, A. Khoo, J. Wiegant, C. Rosenberg, and L. Larizza. 1998. Chromosomal instability in fibroblasts and mesenchymal tumors from 2 sibs with Rothmund-Thomson syndrome. Int. J. Cancer 77:504-510. [DOI] [PubMed] [Google Scholar]
- 56.Mohaghegh, P., J. K. Karow, R. M. Brosh, Jr., V. A. Bohr, and I. D. Hickson. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter. 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82:831-840. [DOI] [PubMed] [Google Scholar]
- 58.Mullen, J. R., V. Kaliraman, and S. J. Brill. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073-1076. [DOI] [PubMed] [Google Scholar]
- 61.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 62.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 63.Myung, K., and R. D. Kolodner. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2000. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 459:203-209. [DOI] [PubMed] [Google Scholar]
- 66.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277-286. [DOI] [PubMed] [Google Scholar]
- 67.Orstavik, K. H., N. McFadden, J. Hagelsteen, E. Ormerod, and C. B. van der Hagen. 1994. Instability of lymphocyte chromosomes in a girl with Rothmund-Thomson syndrome. J. Med. Genet. 31:570-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parsons, R., G. M. Li, M. J. Longley, W. H. Fang, N. Papadopoulos, J. Jen, A. de la Chapelle, K. W. Kinzler, B. Vogelstein, and P. Modrich. 1993. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75:1227-1236. [DOI] [PubMed] [Google Scholar]
- 69.Pennaneach, V., and R. D. Kolodner. 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36:612-617. [DOI] [PubMed] [Google Scholar]
- 70.Petkovic, M., T. Dietschy, R. Freire, R. Jiao, and I. Stagljar. 2005. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 118:4261-4269. [DOI] [PubMed] [Google Scholar]
- 71.Poppe, B., H. Van Limbergen, N. Van Roy, E. Vandecruys, A. De Paepe, Y. Benoit, and F. Speleman. 2001. Chromosomal aberrations in Bloom syndrome patients with myeloid malignancies. Cancer Genet. Cytogenet. 128:39-42. [DOI] [PubMed] [Google Scholar]
- 72.Porter, G., J. Westmoreland, S. Priebe, and M. A. Resnick. 1996. Homologous and homeologous intermolecular gene conversion are not differentially affected by mutations in the DNA damage or the mismatch repair genes RAD1, RAD50, RAD51, RAD52, RAD54, PMS1 and MSH2. Genetics 143:755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puranam, K. L., and P. J. Blackshear. 1994. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem. 269:29838-29845. [PubMed] [Google Scholar]
- 74.Puranam, K. L., E. Kennington, S. N. Sait, T. B. Shows, J. M. Rochelle, M. F. Seldin, and P. J. Blackshear. 1995. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics 26:595-598. [DOI] [PubMed] [Google Scholar]
- 75.Putnam, C. D., V. Pennaneach, and R. D. Kolodner. 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol. Cell. Biol. 25:7226-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rattray, A. J., and L. S. Symington. 1994. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics 138:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 78.Saintigny, Y., D. Rouillard, B. Chaput, T. Soussi, and B. S. Lopez. 1999. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene 18:3553-3563. [DOI] [PubMed] [Google Scholar]
- 79.Salk, D., K. Au, H. Hoehn, and G. M. Martin. 1981. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet. Cell Genet. 30:92-107. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 80a.Sangrithi, M. N., J. A. Bernal, M. Madine, A. Philpott, J. Lee, W. G. Dunphy, and A. R. Venkitaraman. 2005. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell 121:887-898. [DOI] [PubMed] [Google Scholar]
- 81.Schmidt, K. H., and R. D. Kolodner. 2004. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 24:3213-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selva, E. M., A. B. Maderazo, and R. S. Lahue. 1997. Differential effects of the mismatch repair genes MSH2 and MSH3 on homeologous recombination in Saccharomyces cerevisiae. Mol. Gen. Genet. 257:71-82. [DOI] [PubMed] [Google Scholar]
- 84.Selva, E. M., L. New, G. F. Crouse, and R. S. Lahue. 1995. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics 139:1175-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shabtai, F., U. H. Lewinski, A. Meroz, D. Klar, M. Djaldetti, and I. Halbrecht. 1988. Non-random chromosomal aberrations in a complex leukaemic clone of a Bloom's syndrome patient. Hum. Genet. 80:311-314. [DOI] [PubMed] [Google Scholar]
- 86.Sinclair, D. A., and L. Guarente. 1997. Extrachromosomal rDNA circles-a cause of aging in yeast. Cell 91:1033-1042. [DOI] [PubMed] [Google Scholar]
- 87.Spell, R. M., and S. Jinks-Robertson. 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spell, R. M., and S. Jinks-Robertson. 2003. Role of mismatch repair in the fidelity of RAD51- and RAD59-dependent recombination in Saccharomyces cerevisiae. Genetics 165:1733-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tang, R., C. R. Changchien, M. C. Wu, C. W. Fan, K. W. Liu, J. S. Chen, H. T. Chien, and L. L. Hsieh. 2004. Colorectal cancer without high microsatellite instability and chromosomal instability-an alternative genetic pathway to human colorectal cancer. Carcinogenesis 25:841-846. [DOI] [PubMed] [Google Scholar]
- 91.Therman, E., and E. M. Kuhn. 1981. Mitotic crossing-over and segregation in man. Hum. Genet. 59:93-100. [DOI] [PubMed] [Google Scholar]
- 92.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 93.Torres, J. Z., S. L. Schnakenberg, and V. A. Zakian. 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24:3198-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 95.Vennos, E. M., M. Collins, and W. D. James. 1992. Rothmund-Thomson syndrome: review of the world literature. J. Am. Acad. Dermatol. 27:750-762. [DOI] [PubMed] [Google Scholar]
- 96.Versini, G., I. Comet, M. Wu, L. Hoopes, E. Schwob, and P. Pasero. 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22:1939-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang, L. L., A. Gannavarapu, C. A. Kozinetz, M. L. Levy, R. A. Lewis, M. M. Chintagumpala, R. Ruiz-Maldanado, J. Contreras-Ruiz, C. Cunniff, R. P. Erickson, D. Lev, M. Rogers, E. H. Zackai, and S. E. Plon. 2003. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J. Natl. Cancer Inst. 95:669-674. [DOI] [PubMed] [Google Scholar]
- 98.Wang, L. L., M. L. Levy, R. A. Lewis, M. M. Chintagumpala, D. Lev, M. Rogers, and S. E. Plon. 2001. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am. J. Med. Genet. 102:11-17. [DOI] [PubMed] [Google Scholar]
- 99.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 100.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 102.Werner-Favre, C., M. Wyss, C. Cabrol, F. Felix, R. Guenin, D. Laufer, and E. Engel. 1984. Cytogenetic study in a mentally retarded child with Bloom syndrome and acute lymphoblastic leukemia. Am. J. Med. Genet. 18:215-221. [DOI] [PubMed] [Google Scholar]
- 103.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 104.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]