Abstract
Eukaryotic RNA polymerases are large complexes, 12 subunits of which are structurally or functionally homologous across the three polymerase classes. Each class has a set of specific subunits, likely targets of their cognate transcription factors. We have identified and characterized a human RNA polymerase I (Pol I)-specific subunit, previously identified as ASE-1 (antisense of ERCC1) and as CD3ɛ-associated signal transducer (CAST), and here termed CAST or human Pol I-associated factor of 49 kDa (hPAF49), after mouse orthologue PAF49. We provide evidence for growth-regulated Tyr phosphorylation of CAST/hPAF49, specifically in initiation-competent Pol Iβ complexes in HeLa cells, at a conserved residue also known to be important for signaling during T-cell activation. CAST/hPAF49 can interact with activator upstream binding factor (UBF) and, weakly, with selectivity factor 1 (SL1) at the rDNA (ribosomal DNA repeat sequence encoding the 18S, 5.8S, and 28S rRNA genes) promoter. CAST/hPAF49-specific antibodies and excess CAST/hPAF49 protein, which have no effect on basal Pol I transcription, inhibit UBF-activated transcription following functional SL1-Pol I-rDNA complex assembly and disrupt the interaction of UBF with CAST/hPAF49, suggesting that interaction of this Pol I-specific subunit with UBF is crucial for activation. Drawing on parallels between mammalian and Saccharomyces cerevisiae Pol I transcription machineries, we advance one model for CAST/hPAF49 function in which the network of interactions of Pol I-specific subunits with UBF facilitates conformational changes of the polymerase, leading to stabilization of the Pol I-template complex and, thereby, activation of transcription.
In eukaryotes, the three nuclear DNA-dependent RNA polymerases share a similar structural layout and catalytic activity but fulfill different specialist functions in cells: RNA polymerase I (Pol I) synthesizes rRNA precursors, Pol II synthesizes the pre-messenger RNAs, and Pol III synthesizes tRNAs and 5S rRNA, among other small RNAs. The α2ββ′ω subunit composition of a prokaryotic core RNA polymerase is conserved for all three nuclear RNA polymerases from yeast to humans (reviewed in reference 8). The largest and second largest subunits of eukaryotic RNA polymerases share substantial homology with prokaryotic β′ and β subunits, respectively, and possess most of the enzymatic functions. Heterodimers AC40-AC19 of Pol I and Pol III and RPB3-RPB11 of Pol II are functional homologues of the prokaryotic α2 dimer, and the RPB6 (ABC23) subunit, shared between all three polymerases, is a structural and functional homologue of the bacterial ω subunit (Table 1).
TABLE 1.
Human orthologues of 13 of the 14 S. cerevisiae RNA polymerase I subunits
| Yeast subunit | Human Pol I subunit orthologue
|
|||
|---|---|---|---|---|
| Pol I subunit (synonym)a | Unique or shared in (Pol[s]): | Name (synonym[s])d | Accession no.e | Interaction(s) in PICf |
| RPA190 (A190) | I | hRPA190 (A190 or A194) | U33460 | |
| RPA135 (A135) | Ι | hRPA135 (A127) | NM_019014 | |
| RPA49 (A49)b | I | hRPA49 (hPAF53) | NM_022490 | UBF |
| RPA43 (A43) | I | hRPA43 (A43/TWIST neighbor) | EAL24277 | hRRN3 |
| RPA40 (AC40) | Ι, ΙΙΙ | hRPA40 (AC40/hRPA5) | NM_203290 | |
| RPA34.5 (A34.5)c | I | CAST (ASE-1/hPAF49/hRPA34.5) | NM_012099 | UBF, SL1 |
| RPB5 (ABC27) | I, II, III | hRPB5 | NM_002695 | |
| RPB6 (ABC23) | Ι, ΙΙ, ΙΙΙ | hRPB6 | NM_021974 | |
| RPA19 (AC19) | Ι, ΙΙΙ | hRPA19 (AC19) | NM_015972 | |
| RPB8 (ABC14.5) | I, II, III | hRPB8 | NM_006232 | |
| RPA14 (A14)c | I | ND | ||
| RPA12.2 (A12.2)b | I | hRPA12.2 | AF230338 | |
| RPB10 (ABC10β) | I, II, III | hRPB10 | U37690 | |
| RPB12 (ABC10α) | I, II, III | hRPB12 | NM_005034 | |
Most Pol I subunits are essential for viability in yeast (as determined by null mutant analysis).
Conditional mutants.
Subunits not essential for viability.
Names (proposed) of the human Pol I subunits (following primarily the yeast nomenclature); some synonyms are given. ND, not determined.
Database accession numbers of the nucleotide sequences of the human Pol I subunit orthologues.
Interactions of Pol I subunits with other factors in the human PIC.
Eukaryotic RNA polymerases are more complex than their bacterial counterpart and require an additional four subunits to synthesize RNA from a nonspecific DNA template, which are shared between all three polymerases (RPB5, RPB8, RPB10, and RPB12). The unique subunits A12.2, RPB9, and C11 for yeast Pols I, II, and III, respectively, complete the 10 “core” RNA polymerase complex subunits (Table 1), the structure of which for Saccharomyces cerevisiae class I and/or II enzymes has been studied extensively by electron microscopy and by crystallography (2, 9, 15, 20). In addition to the core complex subunits, polymerases I, II, and III each contain a heterodimer (RPB4/RPB7, A14/A43, and C17/C25, respectively) with shared genetic, biochemical, and structural characteristics and sequence homology between RPB7, A43, and C25 (22, 27, 34). There are two additional subunits specific to Pol I (A49 and A34.5) and five specific to Pol III (C82, C53, C37, C34, and C31) (4, 6, 14). Given the unique tasks of the eukaryotic polymerases in transcribing subsets of genes, the polymerase-specific subunits are likely targets of their cognate transcription factors.
We are particularly interested in the human Pol I enzyme complex, which transcribes the rDNA (ribosomal DNA repeat sequence encoding the 18S, 5.8S, and 28S rRNA genes) to produce the major ribosomal RNAs, a process inextricably linked to ribosome biogenesis and cell growth (reviewed in reference 29). Detailed information of the composition of the Pol I complex has come from genetic and biochemical studies in yeast (4). Yeast Pol I is a complex of ∼600 kDa, comprised of 14 subunits, 10 core subunits of which are conserved in metazoa, including humans (Table 1) (33). A mammalian orthologue has yet to be identified for the Pol I-specific subunit A14, but the A43 subunit is evolutionarily conserved (5, 35). The mammalian Pol I-specific A43 subunit interacts with human RRN3 (TIF-IA in rodents) and thereby contributes to formation of a productive preinitiation complex (PIC) at the rDNA promoter via interactions of RRN3 with basal transcription factor SL1 (selectivity factor 1) (3, 23, 24, 28, 39). The A49 subunit (21) is also evolutionarily conserved. Mammalian A49, also known as Pol I-associated factor of 53 kDa (PAF53), has been reported to interact with the upstream activator of Pol I transcription, upstream binding factor (UBF) (17, 32).
Here, we have identified a putative human orthologue of the yeast Pol I-specific A34.5 subunit by using mass spectrometry of polypeptides in highly purified Pol I preparations. This 72-kDa protein was initially discovered as a human autoantigen (encoded antisense in the region of the ERCC1 gene, or ASE-1) and was shown to interact with UBF in vitro and to colocalize with UBF in nucleoli, suggesting that it might have a role in Pol I transcription, although its function was unknown (37). Later, the same protein was rediscovered as one associated with the CD3ɛ-signaling module of the T-cell receptor, named CAST (CD3ɛ-associated signal transducer), and was shown to be important in signaling and gene expression following T-cell activation (40). A mouse homologue of ASE-1/CAST, PAF of 49 kDa (PAF49), was reported while this paper was in preparation (38). Mouse PAF49 can interact in solution with one of the SL1 subunits (TAFI48) (38).
We have shown previously that the Pol I enzyme in human cells is found in at least two distinct complexes of over 1 MDa (Pol Iα and Pol Iβ), each with discrete functions (23), and we demonstrate here that ASE-1/CAST, hereafter referred to as CAST/hPAF49, is associated with both of the human Pol Iα and Pol Iβ complexes. Phosphorylation of the unique tyrosine residue of CAST/hPAF49 is detectable in the initiation-competent Pol Iβ complex but not in Pol Iα. The phosphorylation of Tyr82 of CAST/hPAF49 in Pol Iβ is growth regulated. We show that CAST/hPAF49 can interact with UBF, in contrast to mouse PAF49 (38), and weakly with SL1 at the rDNA promoter. We provide novel insights into the role of this Pol I-specific subunit. We present evidence to suggest that the interaction of CAST/hPAF49 with UBF is important for UBF-dependent transactivation of rDNA transcription in vitro. Based on our results and the structural data available for yeast Pol I, we propose one model for the role of Pol I-specific subunit CAST/hPAF49 (and hPAF53) within the Pol I complex.
MATERIALS AND METHODS
Antibodies, immunoblotting, and immunoprecipitation.
Primary antibodies, specific for the following, were used: CAST (40), hRPA19 (hAC19) and peptide affinity-purified hA190 (sheep polyclonals) (23), mouse monoclonal antibodies for PAF53 (Transduction Laboratories), phosphotyrosine (clone 4G10 [Upstate]; PY-20 [Santa Cruz Biotechnology]), affinity-purified rabbit polyclonals for CAST/hPAF49 (40), and rabbit polyclonals for hA127 (also known as A135; Santa Cruz Biotechnology). Appropriate secondary antibodies conjugated to horseradish peroxidase (Jackson Immuno Research) were used to detect immunocomplexes on the blots by chemiluminescence (ECL Plus; Amersham Biosciences).
All buffers used for preparation of nuclear extracts and for immunoprecipitation contained EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (set 2; Calbiochem). For immunoprecipitation of Flag-CAST/hPAF49 (wild type [WT] or Y82F mutant), HeLa cells were first transfected, using the Effectene method (QIAGEN), with pcFCAST/hPAF49 expression vectors, which are a fusion of Flag-peptide coding sequence (Sigma) and the full-length cDNA of CAST/hPAF49 (WT or Y82F mutant [40] subcloned into pcDNA3.2/V5-DEST vector; Invitrogen). Then, 0.4 mg of nuclear extract from these cells, precleared for 30 min with 10 μl of protein A Dynabeads (Dynal), was incubated with 25 μl of anti-Flag M2 affinity beads (Sigma) in TM10 (50 mM Tris-HCl, pH 7.9, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 1 mM sodium metabisulfite) buffer-0.2 M KCl and 0.015% NP-40 for 2 h at 4°C. Beads were washed four times in 1 ml of TM10-0.25 M KCl buffer and once with 100 μl of TM10-0.05 M KCl and 0.015% NP-40, and precipitants were eluted with an excess of Flag peptide (Sigma) as follows: 25 μl of washed beads was incubated with 25 μl of 200 ng/μl Flag peptide in TM10-0.05 M KCl and 0.015% NP-40 for 10 min on ice. The supernatant was removed, and the beads were incubated for another 10 min with 25 μl of fresh Flag peptide elution buffer on ice. The supernatants were pooled and analyzed by immunoblotting or used in S1 nuclease protection and in nonspecific transcription assays.
Pol I transcription components and mass spectroscopy.
Pol I and SL1 were purified from HeLa cell nuclear extracts as previously described (23). Human UBF (hUBF or UBF1) and Flag-hUBF were expressed in insect cells and purified to apparent homogeneity as previously described (11, 23). Human PAF53 (UltimatORF clone IOH27877; Invitrogen) and human CAST/hPAF49 were cloned in pBAD-DEST49 Gateway vector (Invitrogen). Proteins were expressed in Escherichia coli TOP10 cells (Invitrogen) according to the manufacturer's instructions (a 0.02% final concentration of arabinose was used for induction). Proteins were purified on a HisTrap column and a MonoS (GE Health-Amersham Bioscience) column to near homogeneity. Matrix-assisted laser desorption ionization-time of flight mass spectrometry and identification of polypeptides in Pol I were performed as previously described (36).
Nonspecific transcription assay.
In nonspecific (promoter independent and randomly initiated) transcription assays, samples (up to 5 μl) were tested in a 25-μl reaction mixture with 2.5 μg of sheared calf thymus DNA, which yields, on average, transcripts of over 500 nucleotides long (data not shown), 500 μM ATP, 500 μM GTP, 500 μM UTP, 10 μM CTP, 2.5 μCi of [α-32P]CTP (ICN), 0.1 mg/ml α-amanitin (Sigma), 1.5 mM MnCl2, and 0.015% NP-40 in TM10-0.05 M KCl for 45 min at 30°C. The reaction was stopped by the addition of 100 μl of 50 mM sodium pyrophosphate, 50 mM EDTA, and 1 mg/ml calf thymus DNA, and then nucleic acid was precipitated with 100 μl of 20% ice-cold trichloroacetic acid for at least 1 h on ice. Precipitated nucleic acids were recovered on Whatman GF/C filters, which were then washed with 10 ml of ice-cold 0.1 M sodium pyrophosphate and 1 mM HCl, followed by a rinse in 100% ethanol. Filters were air dried, and radiolabel incorporation was determined by Cerenkov counting.
In vitro transcription and immobilized template assays.
In vitro transcription reactions with immobilized linear rDNA promoter fragment (Fr4) were performed and analyzed by an S1 nuclease protection assay in which synthesis of the first 40 nucleotides of the pre-rRNA is measured (26). Signals were quantitated using a Fuji Phosphorimager and Aida software.
In vitro binding assays.
Specific interactions between CAST/hPAF49, UBF, SL1, and hPAF53 were analyzed using in vitro translated 35S-labeled CAST/hPAF49, Flag-CAST/hPAF49, and hPAF53. In vitro translation of a full-length cDNA of CAST/hPAF49, Flag-CAST/hPAF49, and hPAF53 subcloned into pcDNA3.2/V5-DEST vector (Invitrogen) was performed using a TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. Highly purified and transcriptionally active Flag-hUBF (from recombinant baculovirus-infected insect cells, purified as previously described [11]) or in vitro translated 35S-labeled Flag-CAST/hPAF49 was immobilized on anti-Flag M2 affinity beads (Sigma) according to the manufacturer's instructions. Beads were extensively washed with 0.8 M KCl in TM10, incubated with bovine serum albumin (0.01 mg/ml), and equilibrated in 75 mM KCl in TM10 buffer. Immobilized template (IT) DNA (biotinylated Fr4 rDNA promoter fragment bound to streptavidin-coated paramagnetic beads from Dynal) was prepared as previously described (26). SL1 and/or UBF proteins were bound to IT promoter DNA by incubation at 75 mM KCl in TM10 buffer plus 0.015% NP-40 for 20 min at 0°C, and the beads were then washed with binding buffer. In vitro binding assays were performed in 75 mM KCl in TM10 buffer plus 0.015% NP-40 for 30 min at 0°C. After binding, beads were extensively washed with binding buffer.
RESULTS
CAST/ASE-1 is a subunit of the human Pol Iα complex.
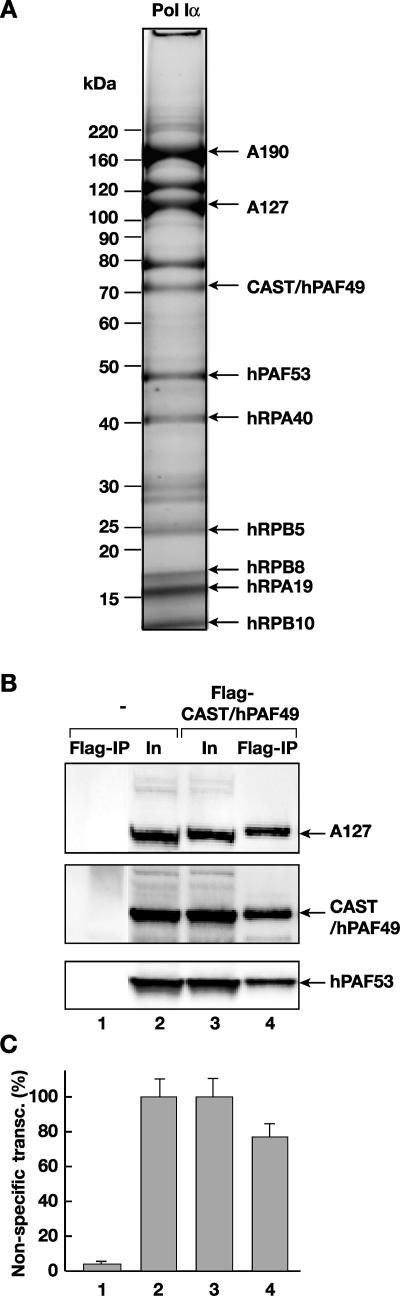
Human Pol Iα was purified from HeLa nuclear extract as previously described (23) and analyzed by mass spectrometry. A polypeptide of 72 kDa (Fig. 1A) was identified as CAST (40), also known as ASE-1 (37), by mass fingerprinting; matched peptides covered 63% of the protein (data not shown). CAST/ASE-1 is referred to here as CAST/hPAF49, after the mouse homologue mPAF49 (38). CAST/hPAF49 is likely to be present within the Pol I complex in stoichiometric amounts, given the relative intensity of Sypro Ruby staining of CAST/hPAF49 on the gel compared to the intensities of the bands of known Pol I subunits (A190, A127, hPAF53, hRPA40, and hRPB5) (Fig. 1A).
FIG. 1.
CAST is a subunit of Pol I. (A) The subunit structure of highly purified Pol Iα was analyzed on a gradient (4 to 12%) of sodium dodecyl sulfate-polyacrylamide gel stained with Sypro Ruby. The po sitions of known Pol I subunits and associated factors are shown (arrows). A polypeptide with an apparent molecular mass of 72 kDa was identified as CAST/ASE-1 by mass fingerprinting (CAST/hPAF49). Molecular mass standard positions are indicated. (B) Pol I subunits coimmunoprecipitate with Flag-CAST/hPAF49. Nuclear extracts from HeLa cells, untransfected (lanes 1 and 2) or transfected with an expression construct for Flag-tagged CAST/hPAF49 (lanes 3 and 4), were immunoprecipitated with Flag-specific antibodies, and immunocomplexes were eluted using Flag-peptide and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Flag-IP; lanes 1 and 4). Control lanes 2 and 3 contain 40 μg of input (In) nuclear extracts (10% of the total protein subjected to immunoprecipitation). Proteins were immunoblotted and probed with antibodies specific for human A127, CAST/hPAF49, and PAF53. (C) Pol I activity coimmunoprecipitates with Flag-CAST/hPAF49. Immunocomplexes from panel B were washed extensively before being tested for nonspecific transcription activity (lanes 1 and 4). Forty micrograms of the input protein (10% of total protein subjected to immunoprecipitation) was tested for nonspecific transcription (transc) activity as controls (lanes 2 and 3). The activity associated with the immunocomplexes was expressed as a percentage of the activity detected in the control samples, set at 100%, and experimental error bars are included. The data are from one representative experiment (for which the immunoblot is shown in B), which was repeated three times.
To establish whether the CAST/hPAF49 polypeptide is part of the human Pol I complex, we determined whether Pol I proteins coimmunoprecipitate with CAST/hPAF49. Immunoprecipitation was performed using Flag-specific antibodies and nuclear extracts from HeLa cells transfected with Flag-CAST/hPAF49 expression vector (Fig. 1B, lane 4) or nontransfected cells (Fig. 1B, lane 1). Pol I subunits, A127 and hPAF53, coimmunoprecipitated with Flag-CAST/hPAF49 (Fig. 1B, lane 4). Furthermore, Pol I transcription activity was detected in the Flag-CAST/hPAF49 immunoprecipitate (Fig. 1C, lane 4) and not in the control sample (Fig. 1C, lane 1). The same results were obtained using hemagglutinin-specific antibodies and expression vector hemagglutinin-CAST/hPAF49 (data not shown). These results suggest that CAST/hPAF49, previously reported to be involved in T-cell signaling, is an integral component of the Pol I enzyme complex in HeLa cells.
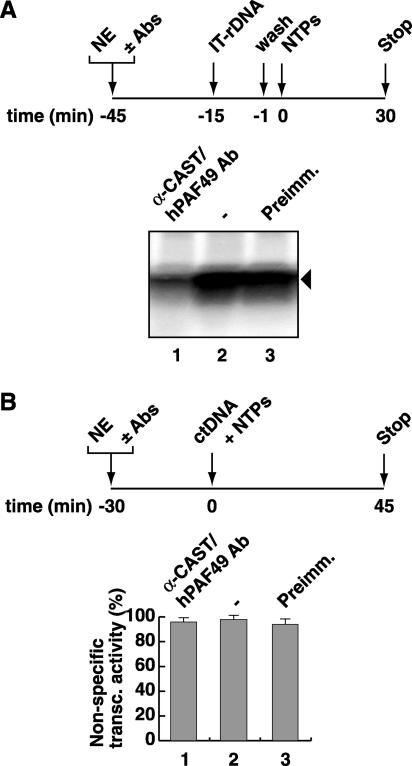
To determine the requirement for CAST/hPAF49 function in promoter-specific transcription, we looked for an effect of CAST/hPAF49-specific antibodies on transcription from the rDNA promoter in the immobilized template assay outlined in Fig. 2A. Additionally, the effect of these antibodies on Pol I was tested in a nonspecific (random) transcription assay, as outlined in Fig. 2B. CAST/hPAF49-specific antibodies were found to repress promoter-specific transcription (Fig. 2A, lane 1) but not random transcription (Fig. 2B, 1), implying that elongation of transcription is not affected by the CAST/hPAF49-specific antibodies. In the mouse system, the Pol I transcriptional capacity of nuclear extracts was also repressed in the presence of mPAF49-specific antibodies (38), although the effect on nonspecific transcription was not reported.
FIG. 2.
CAST/hPAF49-specific antibodies repress rDNA promoter-directed Pol I transcription by UBF but have no effect on nonspecific Pol I transcription. (A) CAST/hPAF49-specific antibodies repress Pol I promoter-specific transcription. HeLa nuclear extracts (NE; 50 mM KCl) were incubated with CAST/hPAF49-specific antibodies (Ab; lane 1), buffer (lane 2), or preimmune sera (Preimm.; lane 3) for 30 min on ice. Immobilized template DNA (IT-rDNA) was added and, following incubation for a further 14 min, protein-IT-rDNA complexes were washed in TM10-0.05 M KCl. Transcription was initiated with the addition of NTPs, and the reaction was allowed to proceed for 30 min at 30°C. Transcript synthesis was analyzed by S1 nuclease protection assays and autoradiography. (B) CAST/hPAF49-specific antibodies do not affect nonspecific (promoter-independent, randomly initiated) transcription. HeLa nuclear extracts (NE; 50 mM KCl) were incubated with CAST/hPAF49-specific antibodies (Ab; lane 1), buffer (2), or preimmune (Preimm.) sera (3) for 30 min on ice. Nonspecific transcription was initiated by addition of sheared calf thymus DNA (ctDNA) as template and NTPs, and reactions were allowed to proceed for 45 min at 30°C. The efficiency of nonspecific transcription (transc) was determined in five independent experiments by measuring the radioactivity incorporated into the acid insoluble fraction and is expressed relative to that measured in the reaction with buffer alone, set at 100%. Standard deviations are indicated.
Therefore, our data suggest a role for CAST/hPAF49 in the early events of transcription from rDNA promoters, that is, in PIC formation, initiation, and/or promoter escape.
CAST/hPAF49 is a subunit of both Pol Iα and Pol Iβ, tyrosine phosphorylated only in Pol Iβ.
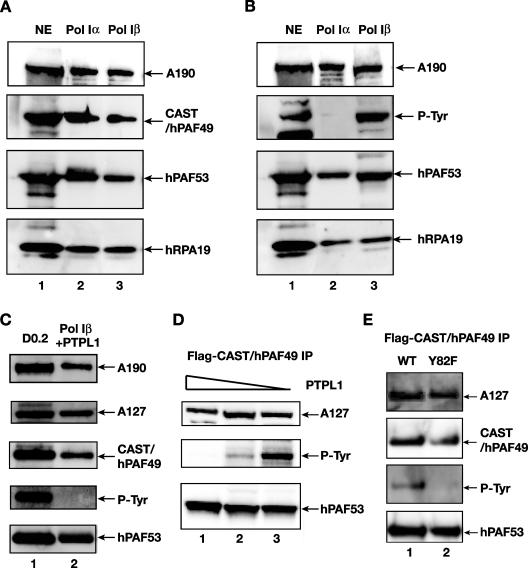
Human Pol I is found in two forms in HeLa cell nuclear extracts, the abundant Pol Iα complexes, which support only random RNA synthesis, and the less abundant Pol Iβ complexes, which are competent for specific initiation of transcription at the rDNA promoters (23). We asked whether CAST/hPAF49 is present in both of these Pol I complexes. Highly purified Pol Iα and Pol Iβ were analyzed by immunoblotting with antibodies specific for Pol I subunits A190, PAF53, or hRPA19 or for CAST/hPAF49. The data indicate that CAST/hPAF49 is present in both Pol Iα and initiation-competent Pol Iβ (Fig. 3A, lanes 2 and 3). Interestingly, in mouse Pol I, two forms of Pol I (A and B) have been identified, separable by chromatography over a CM-Sepharose column, and PAF49 copurifies with Pol I-associated protein PAF53 in Pol IB, which can support promoter-specific transcription, whereas mPAF49 and PAF53 are absent from mouse Pol IA (38). The mouse Pol IA and Pol IB isoforms are therefore not the equivalents of the human Pol Iα and Pol Iβ complexes.
FIG. 3.
CAST/hPAF49 is a subunit of both Pol Iα and Pol Iβ, but the unique tyrosine residue (Tyr82) in CAST/hPAF49 is detectably phosphorylated in only initiation-competent Pol Iβ and not in Pol Iα. (A) CAST/hPAF49 is present in Pol Iα and Pol Iβ. HeLa nuclear extract (NE; lane 1), Pol Iα (lane 2), and Pol Iβ (lane 3) were immunoblotted using antibodies specific for human A190, CAST/hPAF49, PAF53, and hRPA19. (B) Phosphotyrosine-specific antibody recognizes a polypeptide of the same electrophoretic mobility as CAST/hPAF49 (∼72 kDa) in Pol Iβ but not in Pol Iα. HeLa nuclear extract (NE; lane 1), Pol Iα (lane 2), and Pol Iβ (lane 3) were immunoblotted using antibodies specific for human A190, phosphotyrosine, PAF53, and hRPA19. (C) Phosphatase treatment of Pol Iβ leads to disappearance of the 72-kDa band which cross-reacts with phosphotyrosine-specific antibody 4G10. Pol Iβ was incubated with 10 μg of protein tyrosine phosphatase PTPL1 for 15 min at 37°C. The phosphatase-treated Pol Iβ was immunoblotted using antibodies specific for human A190, A127, CAST/hPAF49, phosphotyrosine, and PAF53 (lane 2). Lane 1 is a marker for the Pol I subunits (DEAE 0.2 M fraction [D0.2]). (D) CAST/hPAF49 immunoprecipitated from cells cross-reacts with phosphotyrosine-specific antibody, and this signal decreases with phosphatase treatment. Nuclear extracts from HeLa cells transfected with an expression construct for Flag-tagged CAST/hPAF49 were immunoprecipitated with Flag-specific antibodies, and immunocomplexes were extensively washed and eluted by Flag peptide. The immunocomplexes were treated with PTPL1 phosphatase (10, 5, and 1 μg in lanes 1, 2, and 3, respectively) prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using antibodies specific for human A127, phosphotyrosine (P-Tyr), and PAF53. (E) CAST/hPAF49 with a Y82F mutation did not react with phosphotyrosine-specific antibodies. Nuclear extracts from actively growing HeLa cells transfected with an expression construct for Flag-tagged CAST/hPAF49 (WT or Y82F mutant in lanes 1 and 2, respectively) were immunoprecipitated with Flag-specific antibodies, and immunocomplexes were extensively washed and eluted by Flag peptide. Proteins were immunoblotted and probed with antibodies specific for human A127, CAST/hPAF49, phosphotyrosine (P-Tyr), and PAF53.
Human CAST/hPAF49 contains a single tyrosine residue at position 82 (Tyr82), which is phosphorylated upon stimulation of the T-cell receptor (40). We have analyzed the Tyr82 phosphorylation status of CAST/hPAF49 in highly purified Pol Iα and Pol Iβ by immunoblotting using antibodies specific for phosphotyrosine residues and Pol I subunits (A190, PAF53, and hRPA19). Phosphotyrosine-specific antibody 4G10 cross-reacted with a polypeptide with the same apparent molecular mass (72 kDa) as CAST/hPAF49 in Pol Iβ (Fig. 3B, lane 3). By contrast, antibody 4G10 did not cross-react with any polypeptides between 60 and 100 kDa in Pol Iα (Fig. 3B, lane 2). The same results were obtained using phosphotyrosine antibody PY-20 (data not shown). Treatment of Pol Iβ with tyrosine phosphatase PTPL1 (30) led to disappearance of the 72-kDa band (Fig. 3C, lane 2). We then sought to establish that the 72-kDa polypeptide in Pol Iβ cross-reacting with phosphotyrosine-specific antibodies was CAST/hPAF49. Phosphotyrosine-specific antibodies recognized a protein with the same mobility as CAST/hPAF49 in Pol I complexes immunoprecipitated, using Flag-specific antibodies, from nuclear extracts of Flag-CAST/hPAF49-transfected actively growing HeLa cells (Fig. 3D, lane 3). Treatment of immunoprecipitated Flag-CAST/hPAF49 with tyrosine phosphatase PTPL1 led to disappearance of the 72-kDa band (Fig. 3D, lanes 1 and 2). Moreover, no band between 60 and 100 kDa was detected by phosphotyrosine antibodies in immunoprecipitates from nuclear extracts of HeLa cells transfected with a Flag-CAST/hPAF49-Y82F mutant construct (Fig. 3E, lane 2).
Taken together, the data indicate that Tyr82 phosphorylation of CAST/hPAF49 occurs in the context of the Pol I complex in HeLa cells, as well as in the context of the T-cell receptor complex in T cells upon T-cell activation. Furthermore, phosphorylation of CAST/hPAF49 at Tyr82 is specifically associated with initiation-competent Pol Iβ and not with Pol Iα.
Tyrosine phosphorylation of CAST/hPAF49 is decreased in serum-starved cells.
CAST/hPAF49 can interact with the CD3ɛ-signaling module of the T-cell receptor, and phosphorylation of the unique Tyr82 residue of CAST/hPAF49, which occurs upon T-cell receptor stimulation, is necessary to transduce an activation signal downstream, leading, for example, to NFAT-mediated gene activation (40). We therefore considered the possibility that CAST/hPAF49, as a downstream effector of a signaling pathway, might be involved in the regulation of Pol I transcription via phosphorylation of residue Tyr82. As we have shown, the tyrosine-phosphorylated form of CAST/hPAF49 is associated with initiation-competent Pol Iβ. Therefore, we considered the possibility that tyrosine phosphorylation of CAST/hPAF49 would be downregulated under conditions in which Pol Iβ activity is downregulated. To test this, we compared Pol I activities and Tyr82 phosphorylation of CAST/hPAF49 in actively growing and serum-starved cells.
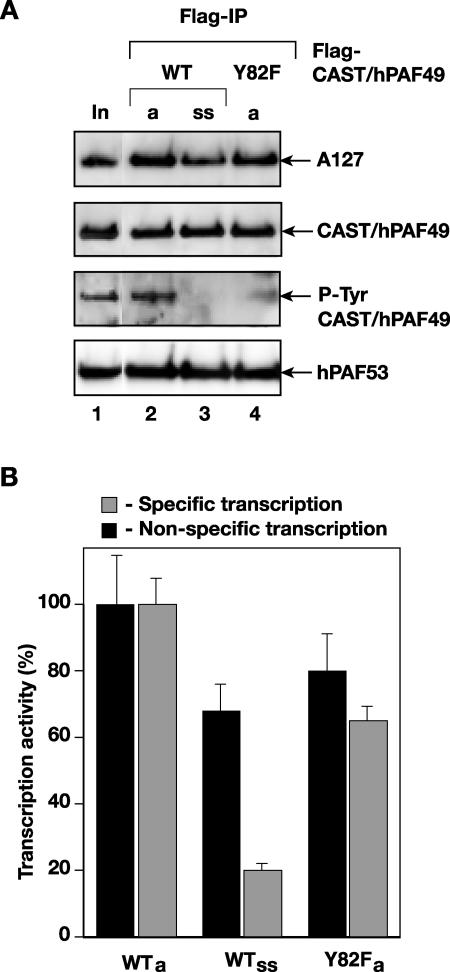
Pol I was immunoprecipitated, using Flag antibodies, from nuclear extracts of actively growing and serum-starved cells in which Flag-CAST/hPAF49 had been overexpressed. Similar amounts of Flag-CAST/hPAF49 were immunoprecipitated from actively growing and serum-starved cells (Fig. 4A, lanes 2 and 3). However, the extent of tyrosine phosphorylation of CAST/hPAF49 was drastically reduced in the immunoprecipitate from serum-starved cells compared to that from actively growing cells (Fig. 4A, P-Tyr, lane 3 compared to lane 2).
FIG. 4.
CAST/hPAF49 in Pol I is Tyr-phosphorylated in actively growing cells, whereas in serum-starved cells, in which promoter-specific Pol I transcription is downregulated, CAST/hPAF49 in Pol I is not detectably Tyr phosphorylated. (A) CAST/hPAF49 in Pol I is Tyr-phosphorylated in actively growing cells but not detectably in serum-starved cells. Nuclear extracts from actively growing (lanes a) or serum-starved (lane ss) HeLa cells (no serum for 24 h) transfected with an expression construct for Flag-tagged CAST/hPAF49 (WT or Y82F mutant) were immunoprecipitated with Flag-specific antibodies (Flag-IP), and immunocomplexes were extensively washed and eluted by Flag peptide. Proteins were immunoblotted and probed with antibodies specific for human A127, CAST/hPAF49, phosphotyrosine (P-Tyr), and PAF53. Lane 1 (In, input) is a marker for the Pol I subunits (DEAE 0.2 M fraction). (B) Downregulation of promoter-specific Pol I transcription in serum-starved cells. Immunoprecipitated and Flag peptide-eluted Pol I complexes from panel A (actively growing cells, WTa; serum-starved cells, WTss; and Y82F) were tested for nonspecific transcription activity (black) or for promoter-dependent transcription activity in a reconstituted transcription reaction with purified SL1 and recombinant UBF (gray). The data sets of panels A and B are of a representative experiment, which has been repeated twice. The activity associated with immunocomplexes was expressed as a percentage of that derived from actively growing cells (set at 100%), and experimental error bars have been included.
Pre-rRNA synthesis by Pol I is down-regulated in serum-starved cells. We found that random RNA synthesis activity of Flag-CAST/hPAF49-immunoprecipitated Pol I from serum-starved cells was reduced to ∼70% of that of actively growing cells (Fig. 4B, compare black bars in lanes ss and a). This could perhaps be explained at least in part by reduced incorporation of Flag-CAST/hPAF49 into the Pol I complex in serum-starved compared to actively growing cells (Fig. 4A, compare CAST/hPAF49 and Pol I subunit levels in lanes ss and a). The ability of the Flag-CAST/hPAF49-immunoprecipitated Pol I from serum-starved cells to support promoter-specific transcription was reduced to 20% of that from actively growing cells (Fig. 4B, compare gray bars in lanes ss and a). The reduced nonspecific RNA synthesis activity of immunoprecipitated Pol I from serum-starved cells cannot solely account for this reduction, indicating that Pol Iβ activity was downregulated in these cells, consistent with inactivation of the Pol Iβ-associated factor TIF-IA/hRRN3 in serum-starved cells (reviewed in reference 29). The reduction in Pol Iβ activity correlates with loss of Tyr82 phosphorylation of CAST/hPAF49 in Pol Iβ in serum-starved cells. This raises the possibility that CAST/hPAF49 Tyr82 phosphorylation might be linked to growth factor signaling pathways' affecting Pol I transcription (19), and we are currently pursuing this hypothesis.
Pol I immunoprecipitated from nuclear extracts of cells transfected with the Flag-CAST/hPAF49-Y82F mutant construct was able to support both nonspecific (Fig. 4B, Y82F, black) and promoter-driven transcription (Fig. 4B, Y82F, gray), though at a somewhat reduced level compared to that from cells transfected with the WT Flag-CAST/hPAF49 construct. This suggests that Tyr82 phosphorylation of CAST/PAF49 is not essential for Pol I complex assembly and Pol I transcription activity. However, it does not rule out a role for Tyr82 phosphorylation of CAST/PAF49 in the upregulation of Pol I transcription in cells.
CAST/hPAF49 interacts with UBF.
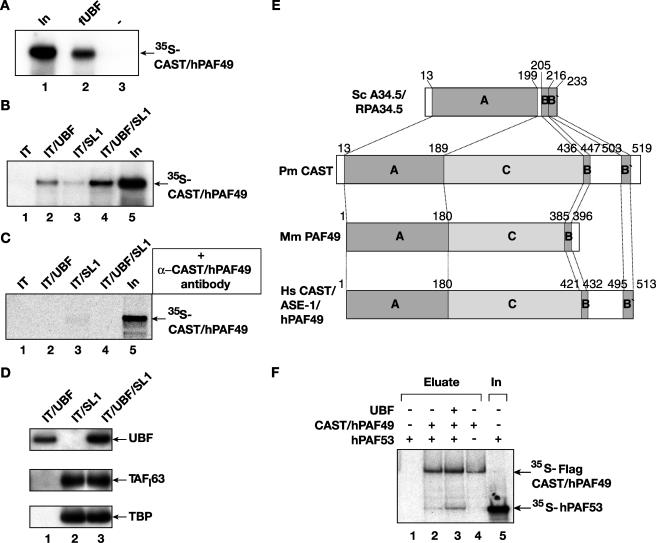
Given the evidence that CAST/hPAF49 functions as a Pol I subunit in rDNA transcription, we were interested in determining its interactions with other components of the Pol I transcription machinery. We demonstrate that recombinant CAST/hPAF49 (in vitro translated) binds to recombinant Flag-tagged UBF (baculovirus and insect cell expressed and purified) in an in vitro binding assay (Fig. 5A, lane 2), in agreement with previous results obtained with the human ASE-1 protein (37). Intriguingly, this is in contrast to findings in the mouse system, where mPAF49 (glutathione transferase fusion) does not interact detectably with UBF (in vitro translated) (38), and it will be interesting to determine whether this is due to differences between the mouse and human proteins. Here, we have extended the analysis to test for interactions between CAST/hPAF49 and UBF or SL1 at the rDNA promoter at salt concentrations compatible with transcription. The data show that CAST/hPAF49 interacts with promoter-bound UBF under these conditions (Fig. 5B, lane 2). A weak interaction of CAST/hPAF49 with promoter-bound SL1 was also detected (Fig. 5B, lane 3), in accord with an observed interaction in solution of mouse PAF49 with the TAFI48 subunit of mouse TIF-IB/SL1 (38). When SL1 and UBF were both present at the rDNA promoter, the CAST/hPAF49 interaction signal was significantly greater than the sum of the signals for CAST/hPAF49 interactions with UBF and SL1 separately (Fig. 5B, lane 4 compared to lanes 2 and 3). This can be accounted for by an increase in the amount of UBF associated with the rDNA in the presence of SL1 (Fig. 5D, compare lanes 1 and 3), as a result of stabilization of UBF by SL1 at the rDNA promoter through a reduction in the dissociation rate of UBF (11). The interaction of CAST/hPAF49 with transcription activator UBF is precluded by preincubation of CAST/hPAF49 with CAST/hPAF49-specific affinity-purified antibodies in the in vitro binding assay (Fig. 5C, lanes 2 and 4). Collectively, the results support the conclusion that CAST/hPAF49 can interact with UBF at the rDNA promoter under conditions compatible with transcription. In an attempt to map a domain of CAST/hPAF49 responsible for its interaction with UBF and also to analyze the C terminus of CAST/hPAF49 in particular, which is shorter in mPAF49, we generated truncated versions of CAST/hPAF49 approximately following the homology domains A, B, B′, and C (Fig. 5E). However, whereas the full-length CAST/hPAF49 protein interacted with UBF, none of these truncated versions interacted with UBF in our assay (data not shown), perhaps suggesting that interaction occurs through multiple domains.
FIG. 5.
CAST/hPAF49 interacts with UBF and SL1 at the rDNA promoter. (A) CAST/hPAF49 interacts with UBF in solution. 35S-labeled in vitro translated CAST/hPAF49 was incubated with highly purified and transcriptionally active Flag-tagged UBF immobilized on Flag-specific antibody beads (lane 2) or beads alone (lane 3) at 75 mM KCl for 30 min on ice. Following washes, bound protein was detected by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Twenty-five percent of CAST/hPAF49 input was loaded on the gel (lane 1). (B) CAST/hPAF49 interacts with UBF and SL1 at the rDNA promoter. Immobilized promoter fragment (IT) was incubated with UBF and/or with SL1 (lanes 2 to 4) or without transcription factors (lane 1) at 75 mM KCl for 20 min on ice and then washed to remove unbound protein. Immobilized template-protein complexes were incubated with 35S-labeled CAST/hPAF49 at 75 mM KCl for 30 min on ice, and, following washes, bound CAST/hPAF49 was detected by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Twenty-five percent of CAST/hPAF49 input was loaded on the gel (lane 5). (C) CAST/hPAF49-specific antibodies block CAST/hPAF49-UBF interactions. Immobilized promoter fragment (IT) was incubated with UBF and/or with SL1 as in panel B. 35S-labeled CAST/hPAF49 was preincubated with affinity-purified CAST/hPAF49-specific antibodies for 1 h at 4°C. The CAST/hPAF49-antibody complex was incubated with the immobilized template-protein complexes for 30 min on ice, and, following washes, bound CAST/hPAF49 was detected by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Twenty percent of CAST/hPAF49 input was loaded on the gel (lane 5). (D) The amount of UBF bound to immobilized template DNA is higher in the presence of SL1. Immobilized promoter fragment (IT) was incubated with UBF and/or with SL1 as in panel B. Bound proteins were detected by immunoblotting using antibodies specific for UBF, TAFI63, and TBP. (E) Yeast (Sc) A34.5, lamprey fish (Pm) CAST, mouse (Mm) PAF49, and human (Hs) CAST/ASE-1/hPAF49 proteins are represented, with shaded boxes indicating regions of homology. The identity and similarity, respectively, for each region of human CAST (hPAF49) with the orthologues were determined: for domain A, yeast Rpa34 and human CAST, 19% and 33%; lamprey fish CAST and human CAST, 34% and 48%; mouse PAF49 and human CAST 71% and 72; for domain B, yeast Rpa34 and human CAST, 67% and 83%; lamprey fish CAST and human CAST, 33% and 83%; mouse PAF49 and human CAST, 38% and 54%; for domain B′, yeast Rpa34 and human CAST, 27% and 55%; lamprey fish CAST and human CAST, 28% and 40%; for domain C, lamprey fish CAST and human CAST 11% and 21%; mouse PAF49 and human CAST, 38% and 45%. Truncated mutant versions of CAST/hPAF49 (nucleotides 1 to 180,180 to 421, 421 to 513, and 432 to 513) were used in experiments to map the regions of CAST/hPAF49 interaction with UBF. None of the truncated mutant proteins interacted with UBF (data not shown). (F) CAST/hPAF49 interacts with hPAF53 in the absence or presence of UBF. 35S-labeled in vitro translated hPAF53 was incubated with 35S-labeled in vitro translated Flag-CAST/hPAF49 immobilized on Flag-specific antibody beads (lanes 2 and 3) or beads alone (lane 1) in the absence (lane 2) or in the presence (lane 3) of recombinant UBF at 75 mM KCl for 30 min on ice. Following washes, bound proteins were eluted by an excess of Flag peptide and were detected by autoradiography after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Control lane 4 contains no hPAF53. Lane 5 contains 25% of hPAF53 input. In, input.
We considered the possibility that the phosphorylation of Tyr82 might be required for the interaction between CAST/hPAF49 and UBF and thereby upregulate Pol I transcription. However, a CAST/hPAF49-Y82F mutant interacted with UBF as efficiently as wild-type CAST/hPAF49 (data not shown), suggesting that phosphorylation of Tyr82 is not essential for the interaction between CAST and UBF.
UBF interacts with mammalian PAF53. Mouse PAF49 has also been reported to interact with mouse PAF53 (38). Our analysis of CAST/hPAF49 in in vitro binding assays (Fig. 5F) demonstrates conservation of this interaction as human CAST/hPAF49, the orthologue of yeast RPA34.5 (see Table 1), interacted with human PAF53, the orthologue of yeast RPA49 (Fig. 5F, lane 2); UBF does not interfere with this interaction (Fig. 5F, compare lanes 3 and 2), despite being able to interact with both these Pol I subunits (17, 37; the present study). Thus, UBF can interact with human Pol I via interaction with Pol I subunit CAST/hPAF49 as well as hPAF53, and these Pol I subunits also interact with each other.
CAST/hPAF49-specific antibodies and excess CAST/hPAF49 and/or hPAF53 proteins block UBF-dependent activation of Pol I transcription.
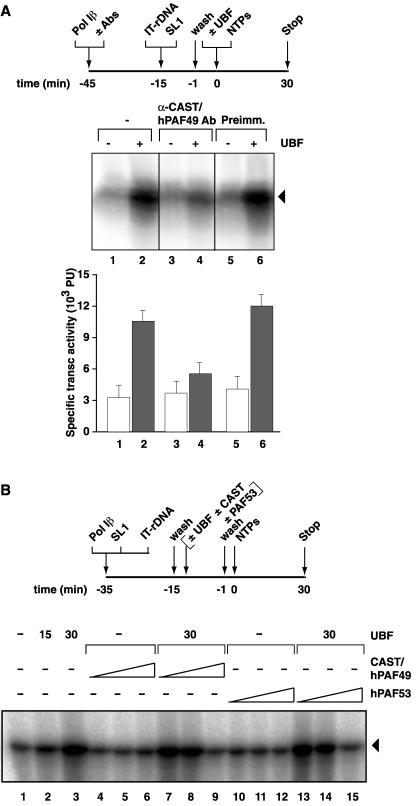
We reasoned that, since CAST/hPAF49-specific antibodies repressed specific transcription but had no effect on random transcription (Fig. 2), the interaction of Pol I with one of the general transcription factors UBF or SL1 might be perturbed. Therefore, we extended our studies to examine the functional significance of interaction of CAST/hPAF49 with UBF during transcription, using CAST/hPAF49-specific antibodies in a highly reconstituted transcription system and the immobilized template assay outlined in Fig. 6. Pol Iβ was preincubated with or without CAST/hPAF49-specific antibodies and then added to SL1-rDNA promoter complexes. Following a further incubation, the templates were washed to remove any unbound factors, and transcription was initiated with nucleoside triphosphates (NTPs) and with or without UBF. Basal transcription (SL1-Pol I-directed transcription) was not affected by the incubation of Pol I with CAST/hPAF49-specific antibodies (Fig. 6A, compare lanes 1, 3, and 5), suggesting that functional SL1-Pol I-rDNA PICs can assemble unimpeded in the presence of CAST/hPAF49-specific antibodies. Activation of transcription by UBF from the preassembled PICs was significantly reduced by the CAST/hPAF49-specific antibodies (Fig. 6A, compare lanes 2 and 6 with 4) that disrupted the interaction between CAST/hPAF49 and UBF (Fig. 5C). The results suggest that the interaction of CAST/hPAF49 with UBF is important for UBF-activated transcription by Pol I at a step following functional SL1-Pol I-rDNA PIC formation.
FIG. 6.
CAST/hPAF49-specific antibodies and preincubation of UBF with CAST/hPAF49 or hPAF53 block activation of Pol I transcription by UBF but have no effect on basal Pol I transcription. (A) CAST/hPAF49-specific antibodies (Abs) repress activation of Pol I transcription by UBF but do not affect basal transcription. Pol Iβ at 75 mM KCl was incubated with buffer (lanes 1 and 2), with CAST/hPAF49-specific affinity-purified antibodies (lanes 3 and 4) or with preimmune serum (lanes 5 and 6) for 30 min on ice. Immobilized template DNA (IT-rDNA) and SL1 were added to each reaction. Following incubation for another 14 min, protein-IT-rDNA complexes were washed in TM10-0.05 M KCl. To gauge the effect of CAST antibodies on activation of transcription by UBF, recombinant human UBF was added to reactions in lanes 2, 4, and 6. Transcription was initiated upon the addition of NTPs, and the reaction was allowed to proceed for 30 min at 30°C. Transcript synthesis was analyzed by S1 nuclease protection assay. The bar graph (in arbitrary phosphorimager units [PU]) represents the efficiency of transcription (transc) from two independent experiments. White bars represent basal transcription levels (SL1-Pol I-rDNA); gray bars represent UBF-activated transcription levels (SL1-UBF-Pol I-rDNA). (B) Preincubation of UBF with CAST/hPAF49 or hPAF53 represses activation of Pol I transcription by UBF but does not affect basal transcription. Pol Iβ was incubated with SL1 and IT-rDNA (immobilized promoter template) for 30 min on ice. The SL1-Pol I-rDNA complexes were washed, and then UBF (15 or 30 ng; lanes 2, 3, 7 to 9, and 13 to 15), preincubated with CAST/hPAF49 (60, 250, or 500 ng; lanes 4 to 6 and 7 to 9) or hPAF53 (60, 250, or 500 ng; lanes 10 to 12 and 13 to 15) for 25 min on ice, was added. Following incubation for a further 14 min, beads and supernatant were separated, and beads were washed in TM10-0.05 M KCl. To gauge the effect of CAST/hPAF49 or hPAF53 on activation of transcription by UBF, transcription was initiated upon the addition of NTPs, and the reaction was allowed to proceed for 30 min at 30°C. Transcript synthesis was analyzed by S1 nuclease protection assay. In lane 1 the products of a basal transcription reaction were loaded. The experiment shown is representative of three independent experiments.
We reasoned that if CAST/hPAF49 was involved specifically in UBF-activated transcription, the presence of excess CAST/hPAF49 might squelch UBF-activated transcription, perhaps via its interaction with UBF. Similarly, PAF53 might squelch UBF-activated transcription via its interaction with UBF. Therefore, UBF preincubated with excess CAST/hPAF49 or hPAF53 was added to Pol Iβ and SL1 in the immobilized template assay. Basal transcription was not affected by excess CAST/hPAF49 or hPAF53 (Fig. 6B, compare lane 1 with lanes 4 to 6 and 10 to 12, respectively), but UBF-activated transcription was reduced significantly in a dose-dependent manner, down to basal levels of transcription (Fig. 6B, compare lane 3 with lanes 7 to 9 and 13 to 15, respectively).
Taken together, our data support the idea that the interaction between CAST/hPAF49 and UBF is functionally significant for UBF-activated transcription.
DISCUSSION
In this study, we provide evidence that human CAST/hPAF49 is an integral component of both human Pol I isoforms, initiation-competent Pol Iβ and the abundant Pol Iα, which is catalytically active but does not support rDNA promoter-directed transcription. Intriguingly, phosphorylation of the unique Tyr82 residue of CAST/hPAF49 (a residue conserved from yeast to human), previously demonstrated to be essential for its role in T-cell activation (40), is detectable only in the initiation-competent Pol Iβ complex. Furthermore, we demonstrate a correlation between Tyr82 phosphorylation of CAST/hPAF49 and Pol I transcription activity in serum-starved and actively growing HeLa cells, which could imply a link between Tyr82 phosphorylation of CAST/hPAF49 and growth factor signaling pathways influencing Pol I transcription. Additionally, our data suggest that CAST/hPAF49 is an important target in Pol I for UBF in the activation of rDNA transcription.
CAST/hPAF49 in UBF-dependent activation of Pol I transcription.
CAST/hPAF49/ASE-1 interacts with UBF in solution (reference 37 and this paper), and we have also demonstrated an interaction between UBF and CAST/hPAF49 at the rDNA promoter, at which UBF likely resides in a higher-order nucleoprotein structure (1). Intriguingly, no interaction between mouse PAF49 and UBF has been detected (in a glutathione transferase pull-down assay) (38), which could reflect differences in the properties of CAST/hPAF49 and mouse PAF49 proteins or the experimental conditions. The functional significance of the interaction between UBF and CAST/hPAF49 was established by blockage of the interaction by two experimental strategies, which abrogated activation of transcription by UBF. CAST/hPAF49-specific antibodies blocked protein-protein interaction between UBF and CAST/hPAF49, and, crucially, we detected an inhibitory effect of these antibodies on UBF-dependent activation of transcription. This was a specific effect upon UBF activation, not due to inactivation or loss of Pol I, because basal transcription directed by SL1 and Pol I at the rDNA promoter was not affected by the antibodies. Furthermore, we provide evidence that the activation of transcription by UBF can be abrogated following functional SL1-Pol I-rDNA PIC formation, in agreement with our studies which demonstrate that UBF can activate transcription postrecruitment of SL1 and Pol I (26a). We suggest that the interaction between UBF and CAST/hPAF49 in Pol I is not essential for elongation of transcription per se, because the CAST/hPAF49-specific antibodies did not block random RNA synthesis by Pol I and, therefore, that this interaction might be critical at the early stages of transcription. The inhibition of UBF-activated, but not basal, transcription by excess recombinant CAST/hPAF49 or hPAF53 protein (also known to interact with UBF [17]) further substantiates the importance of the activator-Pol I interaction in activated transcription.
Sequence analyses have revealed that CAST/hPAF49 and mPAF49 have homology to the yeast (S. cerevisiae) A34.5 subunit of Pol I (13). Genetic depletion of A34.5 results in a cryo-sensitive but viable yeast strain (13, 21). We have used a yeast strain lacking the A34.5 gene in various genetic backgrounds to establish whether or not CAST/hPAF49 could complement the absence of the A34.5 subunit. We were unable to detect any effect of CAST/hPAF49 expression, either toxic or rescuing (data not shown); however, it is possible that CAST/hPAF49 cannot function properly in yeast Pol I, perhaps because it is twice the size of A34.5.
Interaction of yeast A34.5 with the core enzyme is dependent upon the presence of Pol I subunit A49 and vice versa (13, 18, 21). The importance of this interaction is suggested by its evolutionary conservation, since the putative A34.5 mouse and human orthologues, mPAF49 and CAST/hPAF49, interact with PAF53, the mammalian orthologue of yeast Pol I subunit A49 (17, 38) (Fig. 5F). The CAST/hPAF49-hPAF53 complex was maintained in the presence of UBF, suggesting that the interactions of these subunits with UBF and with each other are unlikely to be mutually exclusive in the PIC. The mouse Pol I subunits PAF53 and PAF49 can be separated from the Pol I complex under certain chromatographic conditions, to yield a complex Pol IA that has random RNA synthesis activity but cannot initiate rDNA promoter-directed transcription (17). There are interesting parallels to be drawn with the yeast Pol I subunits. The A34.5 and A49 subunits of purified yeast Pol I can also be separated from Pol I (in high salt), and, importantly, the polymerase lacking these subunits, PolA*, displays a reduced specific activity in RNA synthesis from calf thymus DNA and a higher sensitivity to α-amanitin, suggesting a role for these subunits in elongation or enzyme processivity (18).
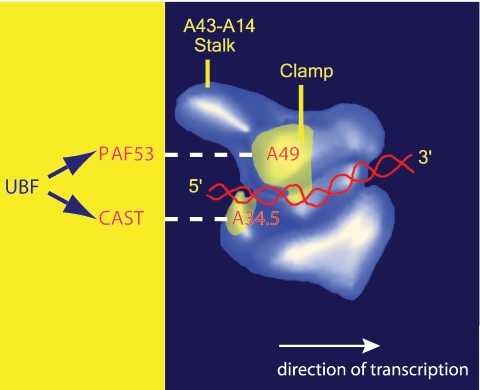
Strikingly, as with human CAST/hPAF49, the A34.5 subunit in yeast Pol I can interact with UBF (31) and with yeast protein Hmo1, a high-mobility-group box protein which might be functionally equivalent to UBF (12). Electron microscopy studies of yeast Pol I have located the A34.5 subunit at the entry to the cleft (2), which has been shown to bind the DNA template (15), adjacent to the second largest subunit, A127. The A34.5 subunit is proposed to stabilize the interaction of the DNA template with the core enzyme via its C-terminal domain (2). Thus, the location of A34.5 close to the upstream edge of the DNA binding groove (cleft) and, by inference, the location of CAST/hPAF49 in Pol I are consistent with its interaction with activating factors bound to the upstream promoter DNA (Fig. 7). PAF53 (mammalian A49) can also interact with UBF (17), and yeast A49 has been shown to act synergistically with Hmo1 (12). Cryo-electron microscopy studies have located the yeast A49 subunit on the head region of the clamp formed by A190/RPA190, the largest subunit of Pol I (2, 10), which forms one side of the cleft (Fig. 7). It was suggested that A49 could affect the conformation of the clamp (2), moving it inwards to hold downstream DNA more firmly, or that it could interact directly with the DNA and so increase the processivity of the enzyme (9, 15). Indeed, conformational subpopulations of the yeast Pol I enzyme have been described (10), and this is consistent with conformational flexibility in other RNA polymerases, where an open conformation of the clamp might reflect the initiation-competent form of the polymerase enzyme, whereas the clamp adopts a closed conformation during elongation (8, 10).
FIG. 7.
The inferred positions of CAST and PAF53 in Pol I and their interaction surfaces for the upstream activator of rDNA transcription, UBF. Based on yeast Pol I data, the relative positions of CAST and PAF53, the mammalian orthologues of yeast A34.5 and A49, respectively, and their interaction with UBF are indicated. Yeast A49 and A34.5 subunit-specific antibody binding sites are indicated (in yellow) on the surface of the electron microscopy-derived three-dimensional model of yeast RNA Pol I, and superimposed upon this is the approximate trajectory of the DNA template (red) to illustrate the direction of transcription (white arrow) (image kindly provided by Patrick Schultz).
Therefore, given our evidence that CAST/hPAF49 and hPAF53 function in UBF-activated transcription at a step following the assembly of SL1-Pol I-rDNA PICs, one possibility is that, in the course of initiation of transcription, targeting of these Pol I subunits by UBF might facilitate conformational changes in Pol I and influence the interaction of Pol I with the DNA, perhaps leading to closure of the cleft, “locking” Pol I onto the template. For now, however, biochemical probing of conformational changes in the mammalian Pol I enzyme complex, facilitated by the UBF activator of transcription, might be beyond that which is technically feasible, perhaps requiring single molecule Pol I transcription analyses with recombinant Pol I enzyme complexes.
CAST/hPAF49 and the regulation of Pol I transcription.
CAST/hPAF49 can interact with the CD3ɛ-signaling module of the T-cell receptor, and phosphorylation of the unique Tyr82 residue of CAST/hPAF49, which occurs upon T-cell receptor stimulation, is necessary to transduce an activation signal downstream, leading, for example, to NFAT-mediated gene activation (40). We present several lines of evidence that collectively suggest that Tyr82 of CAST/hPAF49 in Pol Iβ is phosphorylated in actively growing HeLa cells: phosphotyrosine-specific antibodies cross-react with a protein of the same electrophoretic mobility as CAST/hPAF49 (72 kDa) in Pol Iβ but not in Pol Iα; this phosphotyrosine signal disappears upon protein tyrosine-phosphatase treatment of Pol Iβ; phosphotyrosine-specific antibodies cross-react with Flag-tagged CAST/hPAF49 immunoprecipitated from cells; this phosphotyrosine signal disappears upon protein tyrosine-phosphatase treatment of immunoprecipitated Flag-tagged CAST/hPAF49; and no tyrosine phosphorylation of proteins of ∼72 kDa was detectable using phosphotyrosine-specific antibodies in the Flag-antibody immunoprecipitate from cells transfected with a Flag-tagged CAST/hPAF49 mutant in which Tyr82 had been replaced by phenylalanine.
Intriguingly, we observed a correlation between the tyrosine phosphorylation status of CAST/hPAF49 and Pol I activity in serum-starved and actively growing cells, suggesting a positive role for Tyr82 phosphorylation of CAST/hPAF49 in Pol I transcription. Preliminary studies suggest that the interaction between CAST/hPAF49 and UBF is not dependent upon Tyr82 phosphorylation (data not shown). Furthermore, we have shown that the Tyr phosphorylation of CAST/hPAF49 is not essential for transcription, as the Flag-tagged CAST/hPAF49 Y82F mutant protein was incorporated into Pol I complexes that displayed specific transcription initiation activity, albeit at a reduced level. Significantly, phosphorylation at Tyr82 of CAST/hPAF49 is detectable in only initiation-competent Pol Iβ, leading us to speculate that phosphorylation of this unique tyrosine residue in human CAST/hPAF49, rather than regulating the catalytic activity of the Pol I enzyme per se, could trigger a chain of events to rapidly generate Pol Iβ in response to extracellular signals, such as growth factors, thereby leading to an increase in Pol I transcription necessary to support accelerated cell growth. Intriguingly, mPAF49 (and mPAF53) were found to relocalize following serum starvation (17, 38); although this has not always been seen for mPAF53 (32), it might suggest the possibility that the availability in cells of PAF49 for incorporation into initiation-competent Pol I complexes affects the level of Pol I activity. It will therefore be interesting to determine the localization and mobility of Tyr82-phosphorylated and nonphosphorylated CAST/hPAF49 in cells under different growth conditions.
The presence of tyrosine-phosphorylated CAST/hPAF49 exclusively in the initiation-competent form of Pol I and the requirement for tyrosine phosphorylation of CAST/hPAF49 in T-cell activation suggest a signaling pathway that links the CAST/hPAF49 phosphorylation to an increase in the level of initiation-competent Pol I. There are data to suggest that the levels of initiation-competent Pol I can also vary according to the growth status of cells (reviewed in references 16, 25, and 29). We consider that a regulatory pathway involving CAST/hPAF49 might not be exclusive to T cells but might also be activated and contribute to regulation of rDNA transcription under other circumstances in which a rapid increase in Pol I transcription is required.
RNA polymerase subunits and transcription regulation.
There is evidence to support the prediction that transactivators which upregulate transcription by RNA polymerases I, II, and III interact specifically with shared polymerase subunits (7). Here, on the other hand, we have identified a Pol I-specific subunit with a selective role in Pol I-specific activation of transcription. We propose that the direct interaction of the activator UBF with the Pol I-specific subunit CAST/hPAF49 is crucial for UBF-dependent activation of Pol I transcription. The dual roles for CAST/hPAF49 in Pol I transcription in the nucleolus and as a component of the membrane-bound T-cell receptor complex in T-cell activation, together with the observation that the protein C17/CGRP-RCP functions as a Pol III subunit as well as a hormone receptor component of a signal transduction cascade related to membrane-bound G proteins (34), leads us to suggest that direct regulation of RNA polymerase subunits might be a common denominator in the control of transcription in response to external cellular signals. Currently, we are exploring this possibility through analysis of the role(s) of CAST/hPAF49 in human cells (small interfering RNA) and in mouse knockout model systems.
. . . . . . .
Acknowledgments
We thank P. Schultz (Ecole Superieure de Biotechnologie de Strasbourg, France) for the yeast Pol I image, G. Kular for PTPase (MRC Protein phosphorylation Unit, Dundee), D. Lamont and K. Beattie in the Proteomics Facility (School of Life Sciences, University of Dundee) for peptide mass fingerprinting by matrix-assisted laser desorption ionization-time of flight mass spectrometry, and the National Cell Culture Center (Minneapolis, MN) for supplying HeLa cell nuclei. We thank our colleagues in the Zomerdijk laboratory for advice and critical reading of the manuscript.
T.B.P. received a BBSRC Ph.D. studentship. J.C.B.M.Z. is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
REFERENCES
- 1.Bazett Jones, D. P., B. Leblanc, M. Herfort, and T. Moss. 1994. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264:1134-1137. [DOI] [PubMed] [Google Scholar]
- 2.Bischler, N., L. Brino, C. Carles, M. Riva, H. Tschochner, V. Mallouh, and P. Schultz. 2002. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 21:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf1, H. Delius1, M. Vingron, and I. Grummt. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carles, C., and M. Riva. 1998. Yeast RNA polymerase I subunits and genes, p. 9-38. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.
- 5.Cavanaugh, A. H., I. Hirschler-Laszkiewicz, Q. Hu, M. Dundr, T. Smink, T. Misteli, and L. I. Rothblum. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277:27423-27432. [DOI] [PubMed] [Google Scholar]
- 6.Chedin, S., M. L. Ferri, G. Peyroche, J. C. Andrau, S. Jourdain, O. Lefebvre, M. Werner, C. Carles, and A. Sentenac. 1998. The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harbor Symp. Quant. Biol. 63:381-389. [DOI] [PubMed] [Google Scholar]
- 7.Cheong, J. H., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer, P. 2002. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 12:89-97. [DOI] [PubMed] [Google Scholar]
- 9.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876.11313498 [Google Scholar]
- 10.De Carlo, S., C. Carles, M. Riva, and P. Schultz. 2003. Cryo-negative staining reveals conformational flexibility within yeast RNA polymerase I. J. Mol. Biol. 329:891-902. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich, J. K., K. I. Panov, P. Cabart, J. Russell, and J. C. Zomerdijk. 2005. TBP-TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J. Biol. Chem. 280:29551-29558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadal, O., S. MariotteLabarre, S. Chedin, E. Quemeneur, C. Carles, A. Sentenac, and P. Thuriaux. 1997. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell. Biol. 17:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310:1-26. [DOI] [PubMed] [Google Scholar]
- 15.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876-1882.11313499 [Google Scholar]
- 16.Grummt, I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691-1702. [DOI] [PubMed] [Google Scholar]
- 17.Hanada, K., C. Z. Song, K. Yamamoto, K. Yano, Y. Maeda, K. Yamaguchi, and M. Muramatsu. 1996. RNA polymerase I associated factor 53 binds to the nucleolar transcription factor UBF and functions in specific rDNA transcription. EMBO J. 15:2217-2226. [PMC free article] [PubMed] [Google Scholar]
- 18.Huet, J., J. M. Buhler, A. Sentenac, and P. Fromageot. 1975. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl. Acad. Sci. USA 72:3034-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, M. J., and J. C. Zomerdijk. 2004. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 279:8911-8918. [DOI] [PubMed] [Google Scholar]
- 20.Kettenberger, H., K. J. Armache, and P. Cramer. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16:955-965. [DOI] [PubMed] [Google Scholar]
- 21.Liljelund, P., S. Mariotte, J. M. Buhler, and A. Sentenac. 1992. Characterization and mutagenesis of the gene encoding the A49 subunit of RNA polymerase A in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:9302-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meka, H., G. Daoust, K. B. Arnvig, F. Werner, P. Brick, and S. Onesti. 2003. Structural and functional homology between the RNAP(I) subunits A14/A43 and the archaeal RNAP subunits E/F. Nucleic Acids Res. 31:4391-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, G., K. I. Panov, J. K. Friedrich, L. Trinkle-Mulcahy, A. I. Lamond, and J. C. Zomerdijk. 2001. hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J. 20:1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moorefield, B., E. A. Greene, and R. H. Reeder. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. USA 97:4724-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moss, T. 2004. At the crossroads of growth control: making ribosomal RNA. Curr. Opin. Genet. Dev. 14:210-217. [DOI] [PubMed] [Google Scholar]
- 26.Panov, K. I., J. K. Friedrich, and J. C. Zomerdijk. 2001. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol. Cell. Biol. 21:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Panov, K. I., J. K. Friedrich, J. Russell, and J. C. B. M. Zomerdijk. UBF activates RNA polymerase transcription by stimulating promoter escape. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 27.Peyroche, G., E. Levillain, M. Siaut, I. Callebaut, P. Schultz, A. Sentenac, M. Riva, and C. Carles. 2002. The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl. Acad. Sci. USA 99:14670-14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peyroche, G., P. Milkereit, N. Bischler, H. Tschochner, P. Schultz, A. Sentenac, C. Carles, and M. Riva. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19:5473-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, J., and J. C. Zomerdijk. 2005. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 30:87-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saras, J., L. Claesson-Welsh, C. H. Heldin, and L. J. Gonez. 1994. Cloning and characterization of PTPL1, a protein tyrosine phosphatase with similarities to cytoskeletal-associated proteins. J. Biol. Chem. 269:24082-24089. [PubMed] [Google Scholar]
- 31.Schnapp, G., F. Santori, C. Carles, M. Riva, and I. Grummt. 1994. The HMG box-containing nucleolar transcription factor UBF interacts with a specific subunit of RNA polymerase I. EMBO J. 13:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seither, P., O. Zatsepina, M. Hoffmann, and I. Grummt. 1997. Constitutive and strong association of PAF53 with RNA polymerase I. Chromosoma 106:216-225. [DOI] [PubMed] [Google Scholar]
- 33.Shematorova, E. K., and G. V. Shpakovskii. 2002. Structure and function of eukaryotic nuclear DNA-dependent RNA polymerase I. Mol. Biol. (Moscow) 36:3-26. (In Russian.) [PubMed] [Google Scholar]
- 34.Siaut, M., C. Zaros, E. Levivier, M. L. Ferri, M. Court, M. Werner, I. Callebaut, P. Thuriaux, A. Sentenac, and C. Conesa. 2003. An Rpb4/Rpb7-like complex in yeast RNA polymerase III contains the orthologue of mammalian CGRP-RCP. Mol. Cell. Biol. 23:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thuriaux, P., S. Mariotte, J. M. Buhler, A. Sentenac, L. Vu, B. S. Lee, and M. Nomura. 1995. Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J. Biol. Chem. 270:24252-24257. [DOI] [PubMed] [Google Scholar]
- 36.Walsh, E. P., D. J. Lamont, K. A. Beattie, and M. J. Stark. 2002. Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 41:2409-2420. [DOI] [PubMed] [Google Scholar]
- 37.Whitehead, C. M., R. J. Winkfein, M. J. Fritzler, and J. B. Rattner. 1997. ASE-1: a novel protein of the fibrillar centres of the nucleolus and nucleolus organizer region of mitotic chromosomes. Chromosoma 106:493-502. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, K., M. Yamamoto, K. Hanada, Y. Nogi, T. Matsuyama, and M. Muramatsu. 2004. Multiple protein-protein interactions by RNA polymerase I-associated factor PAF49 and role of PAF49 in rRNA transcription. Mol. Cell. Biol. 24:6338-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, R. T., Y. Nogi, J. A. Dodd, and M. Nomura. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J. 15:3964-3973. [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki, T., Y. Hamano, H. Tashiro, K. Itoh, H. Nakano, S. Miyatake, and T. Saito. 1999. CAST, a novel CD3ɛ-binding protein transducing activation signal for interleukin-2 production in T cells. J. Biol. Chem. 274:18173-18180. [DOI] [PubMed] [Google Scholar]