Abstract
The spliceosomal B complex is the substrate that undergoes catalytic activation leading to catalysis of pre-mRNA splicing. Previous characterization of this complex was performed in the presence of heparin, which dissociates less stably associated components. To obtain a more comprehensive inventory of the B complex proteome, we isolated this complex under low-stringency conditions using two independent methods. MS2 affinity-selected B complexes supported splicing when incubated in nuclear extract depleted of snRNPs. Mass spectrometry identified over 110 proteins in both independently purified B complex preparations, including ∼50 non-snRNP proteins not previously found in the spliceosomal A complex. Unexpectedly, the heteromeric hPrp19/CDC5 complex and 10 additional hPrp19/CDC5-related proteins were detected, indicating that they are recruited prior to spliceosome activation. Electron microscopy studies revealed that MS2 affinity-selected B complexes exhibit a rhombic shape with a maximum dimension of 420 Å and are structurally more homogeneous than B complexes treated with heparin. These data provide novel insights into the composition and structure of the spliceosome just prior to its catalytic activation and suggest a potential role in activation for proteins recruited at this stage. Furthermore, the spliceosomal complexes isolated here are well suited for complementation studies with purified proteins to dissect factor requirements for spliceosome activation and splicing catalysis.
Pre-mRNA splicing is catalyzed by a large RNP molecular machine, termed the spliceosome, which consists of the U1, U2, U4/U6, and U5 snRNPs and a multitude of non-snRNP proteins (reviewed in reference 48). The active site(s) responsible for the catalysis of pre-mRNA splicing is not preformed but, rather, is created anew during the highly dynamic process of spliceosome assembly. The latter is an ordered process during which several intermediates, termed E, A, B, and B*, can be detected in vitro (reviewed in reference 48). Assembly is initiated by the ATP-independent interaction of the U1 snRNP with the conserved 5′ splice site of the pre-mRNA, forming the E complex. At this stage, the U2 snRNP is loosely associated with the pre-mRNA (11). In a subsequent step requiring ATP, the U2 snRNP stably interacts with the pre-mRNA's branch site, leading to formation of the A complex (also called the prespliceosome). Spliceosome assembly culminates with the formation of the spliceosomal B complex, during which the preformed U4/U6.U5 tri-snRNP particle interacts with the A complex. The B complex thus contains a full set of U snRNAs in a precatalytic state. It subsequently undergoes major rearrangements, including destabilization or loss of the U1 and U4 snRNPs, leading to catalytic activation and the formation of the so-called activated spliceosome (B* complex).
Splicing catalysis subsequently proceeds by a two-step mechanism (reviewed in reference 29). During the first step the 5′ splice site is cleaved, and the 5′ end of the intron is covalently linked to the branch site forming a lariat structure. This step of splicing is accompanied by the formation of the spliceosomal C complex. During the second step of splicing, the 3′ splice site is cleaved, releasing the intron, and the 5′ and 3′ exons are ligated to form the mRNA. Upon dissociation of the spliceosome, both pre-mRNA splicing products are ultimately released, and the individual subunits of the spliceosome take part in subsequent rounds of splicing. Within the spliceosome a complex, highly dynamic RNA-RNA network is formed (reviewed in references 31 and 48). During the catalytic activation of the spliceosome, rearrangements in this RNA-RNA network bring the reactive groups of the pre-mRNA (i.e., the 5′ splice site and branch site) into close proximity, and the RNAs forming this network (primarily U2 and U6) are thought to form the core of the active center that catalyzes the first step of splicing.
In addition to the snRNAs, proteins play an essential role in splicing. Indeed the spliceosome is an extremely protein-rich molecular machine (reviewed in reference 48). Each snRNP particle consists of a set of particle-specific proteins, plus seven Sm proteins (B/B′, D1, D2, D3, E, F, and G) found in all of the spliceosomal snRNPs. Over 50 unique proteins are recruited to the spliceosome as stable components of the snRNPs. In addition, a large number of non-snRNP proteins are also associated with the spliceosome, as evidenced by several recent studies in which spliceosomes were affinity-selected and their protein composition determined by mass spectrometry (MS). MS studies of a mixture of spliceosomal complexes (including the mRNP) that were isolated under mild conditions (60 to 100 mM salt) indicated that the number of proteins that associate with spliceosomes assembled in HeLa nuclear extract under splicing conditions lies between ∼170 (including hnRNP proteins) (52) and ∼260 (excluding keratins and ribosomal proteins) (33). This apparently large discrepancy in the total number of spliceosome-associated proteins likely reflects the fact that in these studies: (i) different isolation protocols were used (MS2 affinity selection versus streptavidin-biotin affinity selection) and (ii) different criteria were used to decide whether a given protein is a bona fide spliceosomal protein. For example, in the first study, only those proteins found associated with both of two different substrates analyzed were designated spliceosome associated (52).
To learn more about the dynamics of the spliceosome's proteome, MS studies of spliceosomal complexes isolated at defined assembly or functional stages have also been performed. Complexes analyzed to date include the A, BΔU1, B*, and C complexes (19, 21, 26, 27). The latter three complexes were isolated under stringent conditions (i.e., in the presence of heparin), and thus only those proteins that are stably associated at these stages could be determined. The similar isolation conditions used allowed meaningful comparisons to be made between the proteomes of the BΔU1, B*, and C complexes; these studies (i) confirmed the dynamic nature of the spliceosome's protein composition, (ii) revealed stabilization/destabilization events involving spliceosomal proteins during catalytic activation, and (iii) provided information about a given protein's potential window of function during splicing.
However, due to the highly stringent conditions employed to isolate the BΔU1, B*, and C complexes, information about the recruitment of proteins that were not stably incorporated into the spliceosome at these stages of assembly/function—but are nonetheless functionally important—could not be obtained. That is, an unknown number of proteins were likely stripped away in the presence of heparin. Indeed, the previously analyzed “B-like” BΔU1 complex lacked the U1 snRNP (and thus, presumably, other proteins), which was apparently lost due to treatment with heparin (27). The stringent nature of the isolation protocol previously used to isolate B complexes has also prevented any meaningful comparison of the proteome of the BΔU1 complex with that of the A complex, which was isolated in the absence of heparin (19). Thus, the isolation of spliceosomes at defined stages under more native conditions may provide important information about the complete set of proteins associated with a particular spliceosome assembly or functional intermediate and thus indicate when spliceosomal proteins could potentially act during the splicing cycle and at what stage they are initially recruited or lost.
Of particular importance are those protein recruitment events that are a prerequisite and/or that accompany the catalytic activation of the spliceosome. A number of recent studies have greatly expanded our understanding of the role of proteins at this critical stage of spliceosome maturation. Prp19 and a number of proteins that associate with it play a key role in the catalytic activation of the spliceosome (27, 39, 40). In humans, Prp19 is stably associated with seven to nine proteins, including CDC5 (1, 27). This hPrp19/CDC5 complex appears to be involved in the catalytic activation of the spliceosome, as its depletion from HeLa nuclear extract blocks pre-mRNA splicing prior to the first catalytic step but at a stage after U4/U6.U5 tri-snRNP association (27). In Saccharomyces cerevisiae, Prp19 is present in the heteromeric NTC (for nineteen complex) protein complex that consists of at least eight proteins (9), only three of which have homologues that are present in the human hPrp19/CDC5 complex. Recent studies have revealed that the NTC acts subsequent to U4 dissociation, apparently stabilizing the association of U5 and U6 with the activated spliceosome (8). The human hPrp19/CDC5 complex and its associated proteins appear to be stably integrated into the spliceosome (i.e., they remain bound in the presence of heparin) at the time of its activation (27), but whether it is recruited at an earlier stage is not clear. In yeast, immunoprecipitation studies suggested that Prp19 first associates after B complex formation (39). However, more recent data suggest that at least one component of the NTC complex, Clf1p, functions during tri-snRNP integration and thus may be recruited at an earlier stage of spliceosome assembly (10, 46).
The ability to affinity purify spliceosomal complexes has not only allowed the analysis of their protein composition using MS but also enabled the first views of the three-dimensional structure of the spliceosome to be generated by electron microscopy (EM) studies. Higher-order structures at a resolution of 30 to 40 Å were obtained for the human spliceosomal BΔU1 and C complexes by using single-particle electron cryo-microscopy (7, 22). These complexes were isolated after performing in vitro splicing in HeLa nuclear extract, followed by affinity selection in the presence of heparin; thus several less tightly associated proteins may have been lost during purification. The BΔU1 spliceosome, which has not yet undergone catalytic activation and contains the U2, U4/U6, and U5 snRNPs, has a triangular body and an additional upper globular domain and is 370 Å long with a maximum width of 260 Å (7). The C complex, on the other hand, which has undergone step I of splicing and contains the U2, U5, and U6 snRNPs, exhibits an asymmetrical shape with three major domains and overall dimensions of 270 by 220 by 240 Å (22). Thus, it appears to be considerably smaller than the B complex. More recently, the three-dimensional structure of a single subunit of large 200S spliceosomal complexes (referred to as supraspliceosomes) was determined by cryo-EM (2). This complex, which is thought to be a single spliceosome, was isolated by biochemical means from HeLa nuclear extract, but its precise assembly stage(s) and protein composition are presently not known. In contrast to the structures described above, it exhibits a globular shape with a maximum height of 280 Å and consists of two distinct subunits. Thus, the three-dimensional structures currently available from three different spliceosomal preparations differ considerably in their sizes and shapes. Some of these apparent discrepancies are likely due to the fact that (i) different assembly/functional stages of the spliceosome were analyzed and (ii) that different purification methods were employed. Therefore, additional EM analyses are clearly needed to resolve current questions that remain regarding the structure of the human spliceosome.
To learn more about the catalytic activation of the spliceosome, as well as the dynamics of spliceosomal protein recruitment, we have isolated the human spliceosomal B complex, which is the spliceosome assembly intermediate that is structurally remodeled during catalytic activation. B complexes were assembled in HeLa nuclear extract and purified under native, low-stringency conditions using two independent purification methods: (i) glycerol gradient centrifugation followed by MS2-based affinity selection and (ii) tobramycin affinity selection coupled with anti-61K/hPrp31 immunoaffinity purification. In both cases purified complexes contained stoichiometric amounts of unspliced pre-mRNA and the U1, U2, U4, U5, and U6 snRNAs. MS2 affinity-selected B complexes were functionally committed for subsequent catalytic activation and splicing catalysis as evidenced by in vitro splicing assays with nuclear extract depleted of snRNPs. MS analyses revealed more than 110 proteins common to both affinity-purified human B complexes. Furthermore, they indicated that, in addition to tri-snRNP proteins, a large number of non-snRNP proteins, including the hPrp19/CDC5 complex and related proteins, are recruited to the pre-mRNA prior to catalytic activation. Finally, negative-stain EM was performed to elucidate the two-dimensional structure of the human B complex isolated under physiological conditions. These data provide important information about proteins associated with the precatalytic spliceosome, as well as its structure, just prior to the functionally decisive, spliceosome activation step. The ability to purify native, precatalytic spliceosomal complexes paves the way for complementation studies with purified proteins to dissect factor requirements for the subsequent activation and catalytic steps of splicing.
MATERIALS AND METHODS
Generation of aptamer-tagged pre-mRNA.
A transcription template for the MINX pre-mRNA was generated from pMINX plasmid (54) by PCR. A PCR product containing three MS2 coat protein RNA binding sites was generated using pAdML-M3 (52), which was a kind gift from R. Reed. A transcription template for MINX pre-mRNA tagged with three MS2 RNA aptamers (MS2-tagged MINX) was generated by overlapping PCR of the MINX and MS2 PCR products. MINX pre-mRNA tagged with the J6f1 tobramycin RNA aptamer was generated as described previously (19). Uniformly 32P-labeled, m7G(5′)ppp(5′)G-capped pre-mRNA was synthesized in vitro by T7 runoff transcription.
MS2 affinity selection of spliceosomal B complexes.
HeLa nuclear extract was prepared according to Dignam et al. (13). To isolate spliceosomal B complexes under native conditions, we incubated MS2-tagged MINX pre-mRNA with a 20-fold molar excess of purified MS2-MBP fusion protein (21) at 4°C for 30 min prior to splicing in 20 mM HEPES-KOH (pH 7.9)-60 mM KCl. To affinity purify preparative amounts of the B complex, a reaction was carried out with a 12-ml splicing mixture containing 15 nM 32P-labeled MS2-tagged MINX pre-mRNA (specific activity, 8,000 cpm/pmol). The splicing reaction was supplemented with 25 mM KCl, 3.0 mM MgCl2, 20 mM creatine phosphate, 2 mM ATP, and 40% HeLa cell nuclear extract in buffer D (20 mM HEPES-KOH, pH 7.9, 100 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, pH 8.0, 10% [vol/vol] glycerol, 0.5 mM dithioerythritol, and 0.5 mM phenylmethylsulfonyl fluoride). B complexes were assembled by incubating at 30°C for 8 min. Two-milliliter aliquots of the splicing reaction were loaded onto six 14-ml linear 10 to 30% (vol/vol) glycerol gradients containing G buffer (20 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 150 mM NaCl). Gradients were centrifuged for 16 h at 80,000 × g in a Sorvall TST 41.14 rotor and harvested manually in 500-μl fractions from the top. Peak fractions containing the B complex were pooled, divided into two fractions, and loaded onto two separate columns containing 250 μl of amylose beads (New England Biolabs) equilibrated with G buffer. The matrix was washed 6× with 1 ml of G buffer, and spliceosomal complexes were eluted dropwise with 900 μl of elution buffer (G buffer containing 12 mM maltose). Protein and RNA were recovered and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10/13% polyacrylamide gel or by denaturing PAGE on an 8.3 M urea-9.6% polyacrylamide gel, respectively, and visualized by staining with silver and/or by autoradiography. For functional and EM studies, 600 μl of the eluate were loaded onto a 3.8-ml linear (10 to 30%, vol/vol) glycerol gradient containing G buffer (EM studies) or G buffer with only 75 mM KCl (functional assays). Gradients were centrifuged for 107 min at 374,000 × g in a Sorvall TH660 rotor, and 175-μl fractions were harvested from the top (functional studies) or bottom (EM studies) of the gradient.
In vitro splicing assays with MS2 affinity-selected B complexes.
HeLa nuclear extract was depleted of U2 or U4/U6 snRNPs by streptavidin agarose affinity selection using a biotinylated 2′-O-methyl (2′-OMe) RNA oligonucleotide complementary to the U2 snRNA or the 3′ end of the U6 snRNA, respectively, as previously described (6, 34). HeLa nuclear extract was treated with micrococcal nuclease as described previously (26). In vitro splicing (15-μl reaction volume) was performed essentially as described above for the indicated times with 30% U2-depleted, 30% U4/U6-depleted, 30% mock-depleted, or 35% micrococcal nuclease (MN)-treated HeLa nuclear extract and 5 μl of the peak gradient fraction containing MS2 affinity-selected B complexes (∼13 fmoles). In parallel, an equimolar amount of 32P-labeled MS2-tagged MINX was incubated under the same conditions. To assay whether the B complex is formed de novo during splicing complementation, in vitro splicing was performed with 32P-labeled MS2-tagged MINX pre-mRNA or equimolar amounts of affinity-selected spliceosomal B complexes (5 μl of peak gradient fraction) (specific activity of tagged pre-mRNA, 250,000 cpm/pmol) in the presence of an equimolar amount of radioactive, untagged MINX pre-mRNA (specific activity, 500,000 cpm/pmol) for 0 to 90 min. In each case, RNA was isolated, fractionated on an 8.3 M urea-9.6% polyacrylamide gel, and visualized by autoradiography.
Tobramycin affinity selection of spliceosomal B complexes.
A mixture of A and B complexes was purified by affinity selection with a tobramycin aptamer-tagged MINX pre-mRNA essentially as described previously (19). B complex was subsequently immunoaffinity purified from this mixture with anti-peptide antibodies directed against amino acids 484 to 497 of the U4/U6-61K (hPrp31) protein as previously described for the BΔU1 complex, except that heparin was omitted (27). Briefly, 24 15-μl aliquots of tobramycin matrix bound with a tobramycin aptamer-tagged MINX pre-mRNA were prepared, and 750 μl of a standard splicing reaction was added per 15 μl of matrix and incubated for 60 min. After bound complexes were washed and eluted with an excess of tobramycin, glycerol was added to the eluate to a final concentration of 3% (vol/vol). The eluate was diluted fivefold with immunoprecipitation (IP) buffer (20 mM HEPES-KOH, pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol) containing 0.05% NP-40 and incubated for 3 h at 4°C with 250 μl of protein A Sepharose bound with affinity-purified anti-61K/hPrp31 antibodies (approximately 350 μg) that had been preblocked with 0.5 mg/ml bovine serum albumin and 0.05 mg/ml yeast tRNA. Beads were washed four times with IP buffer lacking NP-40, and bound material was eluted by incubating 1 h with IP buffer containing 3% glycerol (vol/vol) and 0.6 mg/ml cognate peptide. Protein and RNA were recovered and analyzed as described for the MS2 affinity selection of the B complex.
MS.
MS analyses were performed on two individual preparations of both the MS2 (no. 1 and 2) and tobramycin affinity-selected complexes (no. 1 and 2). Proteins recovered from MS2 or tobramycin affinity-selected B complexes were separated by 10/13% SDS-PAGE and stained with Coomassie blue. An entire lane of the Coomassie blue-stained gel was cut into ∼60 slices, and proteins were digested in gel with trypsin and extracted according to Shevchenko et al. (35). The extracted peptides from the tobramycin (no. 1 and 2) and MS2 (no. 1) samples were analyzed in a liquid chromatography-coupled electrospray ionization quadrupole time of flight (Q-ToF Ultima; Waters) mass spectrometer, and MS2 (no. 2) was analyzed in a linear ion trap (4000 QTrap; Applied Biosystems) mass spectrometer, both under standard conditions. Proteins were identified by searching fragment spectra of sequenced peptides against the NCBI nonredundant database using Mascot as search engine.
EM.
Negative staining was carried out by the double-carbon-film method (23) after fixing RNP complexes with glutaraldehyde. Briefly, complexes in solution were allowed to adsorb to a thin carbon film. The immobilized complexes were then transferred to a copper grid covered with a perforated carbon support film and stained for 2 min in a 2% (wt/vol) uranyl formate solution. Subsequently, they were overlaid with a second carbon film. Images of the sandwiched B complex were recorded with a Philips CM 200 FEG electron microscope (Philips, Eindhoven, Netherlands) at a magnification of ×122,000 with a TVIPS 4,000- by 4,000-pixel charge-coupled-device (CCD) camera (TemCam-F415; Gauting, Germany) using a Philips holder. The BΔU1 complex was isolated as previously described (7) and imaged with a TVIPS 1,000- by 1,000-pixel camera at a magnification of ×27,000. For the B complex, the camera was used with a twofold coarsening of the pixels. Images were taken under low-dose conditions at a 1.5- to 3-μm defocus and were computationally coarsened to a final pixel size corresponding to 9.8 Å on the specimen scale. Imagic-5 (Image Science, Berlin, Germany) was used for image processing of 7,500 individual molecular images (43). Briefly, after a “reference-free” alignment procedure (alignment by classification) (15), images were subjected to multivariate statistical analysis (MSA) (42) and classification (41). Based on the classification, 15 to 20 individual molecular images were averaged. The resulting class averages were used as reference images in subsequent rounds of alignment until the computed class averages were stable.
RESULTS
MS2 affinity selection of spliceosomal B complex.
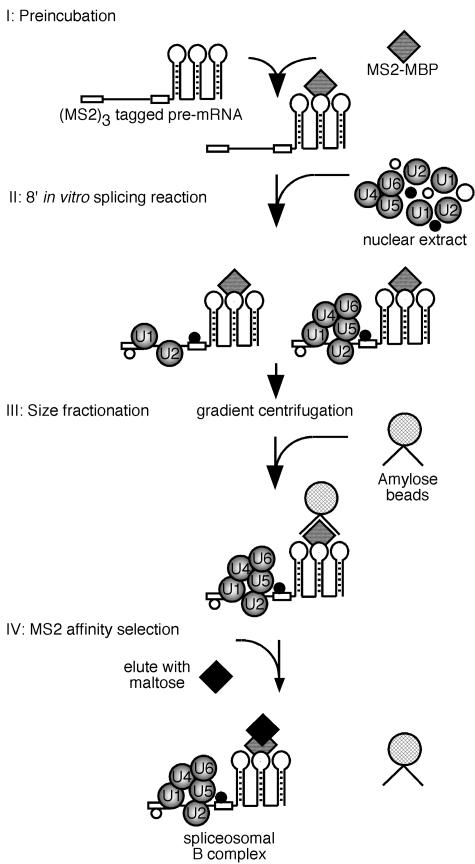
To isolate native B complexes under mild, low-stringency conditions, we first employed an MS2 affinity selection method previously used by other groups to isolate spliceosomal complexes (11, 21). We introduced three bacteriophage MS2 coat protein binding sites (i.e., RNA aptamers) at the 3′ end of exon 2 of the MINX pre-mRNA substrate (Fig. 1). Prior to incubating with HeLa nuclear extract under splicing conditions, 32P-labeled pre-mRNA was incubated with a fusion protein consisting of the MS2 coat protein and maltose binding protein (MBP) (Fig. 1, step I). Addition of the MS2 RNA aptamers to the splicing substrate had no effect on the kinetics of the in vitro splicing reaction; splicing intermediates were first observed after 10 min of incubation with both MINX and MS2-tagged MINX pre-mRNA (Fig. 2A and data not shown). Analysis of splicing complex formation by native gel electrophoresis revealed that the B complex first appeared after 4 min of splicing and reached its maximal level by 8 min (data not shown). Thus, to isolate complexes that are in a precatalytic state, splicing was carried out with the MS2-tagged MINX pre-mRNA for 8 min under splicing conditions in HeLa nuclear extract (Fig. 1, step II).
FIG. 1.
Schematic diagram of the MS2 affinity purification strategy used to isolate human spliceosomal B complexes. MINX pre-mRNA tagged at its 3′ end with three RNA binding sites of the MS2 coat protein was preincubated with a fusion protein of the MS2 coat protein and MBP (step I). Subsequently, nuclear extract was added and spliceosomes were allowed to form for 8 min (step II). Complexes were then fractionated by size on a linear 10 to 30% glycerol gradient (step III). Spliceosomal B complexes from the 40S peak fractions were affinity selected using amylose beads and subsequently eluted with maltose (step IV).
FIG. 2.
Characterization of MS2 affinity-selected B complexes. (A) In vitro splicing kinetics of 32P-labeled MS2-tagged MINX pre-mRNA. Splicing was performed in HeLa cell nuclear extract for 0 to 20 min as indicated above each lane. RNA was recovered and separated on an 8.3 M urea-10% polyacrylamide gel. The 32P-labeled pre-mRNA and splicing intermediates or products were detected by autoradiography, and their positions are indicated on the right; note that the straight line represents debranched, excised intron. The lengths (in base pairs) of a size marker (3′ end-labeled MspI cleaved pBR322) (NEB) are indicated on the left. (B) RNA composition of MS2 affinity-purified B complexes. Spliceosomal complexes were isolated as described in Fig. 1, and RNA was recovered from 20% of the MS2 eluate (lanes 2 and 4). For comparison, RNA was also isolated from 0.2% of the input splicing reaction (lanes 1 and 3). RNA was separated on an 8.3 M urea-10% polyacrylamide gel and visualized by staining with silver (lanes 1 and 2) or by autoradiography (lanes 3 and 4). The positions of the pre-mRNA, splicing intermediates or products, and/or snRNAs are indicated on the right and left. The band indicated by an asterisk is a pre-mRNA degradation product.
The splicing reaction was then subjected to glycerol gradient centrifugation in order to separate the B complex from other RNP complexes such as the spliceosomal A complex (Fig. 1, step III). Analysis of the distribution of radioactivity across the gradient revealed that the majority of complexes peaked in either the 30S or the 40S region of the gradient (data not shown). Characterization of the pre-mRNA and snRNA content of these peaks revealed that the 30S peak contains a mixture of H and A complexes, whereas the 40S peak contains the B complex (data not shown). To further purify the B complex, affinity-selection with amylose beads was performed with the 40S gradient fractions, and bound spliceosomal complexes were eluted under native conditions by incubating with an excess of maltose (Fig. 1, step IV).
RNA was subsequently recovered from the eluate and analyzed by denaturing-PAGE, followed by silver staining. The eluate contained equimolar amounts of uncleaved pre-mRNA, U1, U2, U4, U5, and U6 snRNA (Fig. 2B, lane 2), confirming that the vast majority of complexes present are B complexes and have not undergone catalytic activation (i.e., release of U1 and U4). Only minor amounts of high-molecular-weight RNA were also visible on the gel. Aside from unspliced pre-mRNA, only faint amounts of the intron-lariat intermediate (but no splicing products) were detected in the eluate, as evidenced by autoradiography (Fig. 2B, lane 4), confirming that essentially pure precatalytic spliceosomal complexes had been isolated.
Prior to functional and structural studies using electron microscopy (see below), the eluate was fractionated on a second 10 to 30% glycerol gradient in order to remove the excess of maltose that was used to elute amylose-bound complexes. The majority of the eluted complexes peaked in the 40S region of the gradient (data not shown), suggesting that the purified B complexes are relatively stable. However, analysis of the RNA content of the 40S peak fraction revealed that, whereas nearly equimolar amounts of pre-mRNA, U2, U4, U5, and U6 were present, U1 was somewhat underrepresented (data not shown).
MS2 affinity-selected B complexes are functionally committed for subsequent activation and splicing catalysis.
To assay whether or not MS2 affinity-purified B complexes are functionally committed to splicing, we first depleted HeLa nuclear extract either specifically of U2 snRNPs or U4/U6 snRNPs using biotinylated 2′-OMe oligonucleotides complementary to U2 or U6, respectively, followed by streptavidin-agarose affinity selection, or of all endogenous snRNPs by treating with MN. Analysis of the RNA content of these extracts revealed that, in each case, quantitative digestion or depletion of the targeted snRNPs was achieved (data not shown). Significantly, in the presence of U2- or U4/U6-depleted nuclear extract, purified B complexes catalyzed both steps of splicing (Fig. 3A and B), whereas no splicing was observed even after 90 min when an equimolar amount of naked, 32P-labeled MS2-tagged MINX pre-mRNA was added (data not shown). Likewise, purified B complexes, but not naked MINX pre-mRNA, underwent splicing in MN-digested extract, which is devoid of all snRNPs (Fig. 3C). To rule out the possibility that the affinity-selected B complexes dissociate under splicing conditions and new splicing-active B complexes are subsequently formed, we performed splicing in MN-treated nuclear extract in the presence of an equimolar amount of 32P-labeled MINX pre-mRNA lacking a tag and MS2 affinity-purified B complexes. As a control, splicing was also performed with an equimolar amount of MS2-tagged and untagged MINX pre-mRNA. As expected, neither naked MS2-tagged pre-mRNA nor untagged pre-mRNA was spliced in the MN-treated extract (Fig. 3D, lanes 1 to 6). Significantly, although the tagged substrate within the purified B complexes underwent splicing, splicing of the untagged pre-mRNA in the same reaction was not detected (Fig. 3D, lanes 7 to 12). Thus, splicing-active B complexes are not formed de novo under these conditions; identical results were also obtained with the MS2 eluate that was not subjected to a second gradient centrifugation step (data not shown).
FIG. 3.
MS2 affinity-selected spliceosomal B complexes catalyze splicing in nuclear extracts depleted of snRNPs. MS2 affinity-selected spliceosomal B complex was incubated for the indicated times (0 to 90 min) in HeLa nuclear extract from which U2 snRNPs (A) or U4/U6 snRNPs (B) were depleted using biotinylated 2′-OMe oligonucleotides complementary to the U2 or U6 snRNA. (C) Naked 32P-labeled MS2-tagged MINX pre-mRNA (MS2-pre-mRNA) (lanes 1 to 6) or MS2 affinity-selected spliceosomal B complex (B complex) (lanes 7 to 12) was incubated from 0 to 90 min under splicing conditions in the presence of MN-treated HeLa nuclear extract. (D) Naked 32P-labeled MS2-tagged MINX pre-mRNA (MS2-pre-mRNA) (lanes 1 to 6) or MS2 affinity-selected spliceosomal B complex (B complex) (lanes 7 to 12) was incubated for 0 to 90 min (as indicated above each lane) under splicing conditions together with an equimolar amount of untagged, naked 32P-labeled MINX pre-mRNA in the presence of MN-nuclear extract. In each case, RNA was recovered and analyzed as described in the legend of Fig. 2A. The positions of the intron 3′ exon intermediate, pre-mRNA, spliced out intron, spliced mRNA, and 5′ exon intermediate are indicated on the right. NX, nuclear extract.
Taken together, these experiments demonstrate that MS2 affinity-selected B complexes do not require complementation with any of the spliceosomal snRNPs for their activity but, rather, that they are functionally committed for the subsequent steps of splicing (as opposed to being “dead-end” complexes). However, incubation of purified B complexes (subjected to either one or two rounds of glycerol gradient centrifugation) under splicing conditions but in the absence of extract did not result in splicing (data not shown). Thus, these purified complexes appear either to lack one or more factors required for subsequent spliceosome activation and/or catalysis or potentially to contain a full protein complement but lack functionally important posttranslational protein modifications (see Discussion).
Tobramycin affinity selection of spliceosomal B complex.
For MS studies, the human B complex was additionally purified by an independent, low-stringency method, involving tobramycin affinity selection followed by immunoaffinity selection. Tobramycin RNA aptamer-tagged MINX pre-mRNA was first immobilized on a tobramycin-derivatized Sepharose matrix (Fig. 4A, step I). The matrix-bound pre-mRNA substrate was incubated for 60 min under splicing conditions (Fig. 4A, step II). The kinetics of the solid phase splicing reaction are much slower than that observed in solution, with a very low level of splicing intermediates (but no products) appearing first after 60 min (19; data not shown). Thus, after 60 min, essentially only precatalytic spliceosomal complexes have formed. Spliceosomal complexes were then eluted with an excess of tobramycin (Fig. 4A, step III). The resultant eluate contained a mixture of predominantly A and B complexes (19; data not shown).
FIG. 4.
Purification strategy and RNA composition of B complexes purified by tobramycin affinity selection coupled with anti-61K immunoaffinity purification. (A) Schematic diagram of the two-step tobramycin-immunoaffinity purification strategy used to isolate human spliceosomal B complexes. Pre-mRNA tagged at its 3′ end with the tobramycin RNA aptamer was bound to a tobramycin matrix (step I). Spliceosomal complexes were allowed to form for 60 min by incubating under splicing conditions in the presence of nuclear extract (step II). After elution with tobramycin (step III), precatalytic spliceosomal B complexes were isolated by immunoaffinity selection using anti-peptide antibodies directed against the U4/U6-61K (hPrp31) protein (step IV). Spliceosomal B complexes were subsequently eluted with cognate peptide (step V). (B) RNA composition of tobramycin-immunoaffinity purified B complexes. RNA was recovered from 12% of the anti-61K/hPrp31 eluate (lanes 2 and 4) and also isolated from 0.2% of the input material used for binding to the tobramycin matrix (lanes 1 and 3). RNA was separated on an 8.3 M urea-10% polyacrylamide gel and visualized by staining with silver (lanes 1 and 2) or by autoradiography (autorad; lanes 3 and 4). The positions of the pre-mRNA, splicing intermediates or products, and/or snRNAs are indicated on the right and left. The band indicated by an asterisk is a pre-mRNA degradation product. Tob, tobramycin; NHS, N-hydroxysuccinimide.
To purify the B complex from this mixture, immunoaffinity selection was subsequently performed with anti-peptide antibodies directed against the U4/U6-61K (hPrp31) protein that had been immobilized on protein A Sepharose (Fig. 4A, step IV). Anti-61K/hPrp31 antibodies, which specifically react with this U4/U6 protein in immunoblots, were previously used to isolate BΔU1 spliceosomes, albeit in the presence of heparin (27). Analytical immunoprecipitation studies indicated that B complexes could also be efficiently precipitated by these antibodies even in the absence of heparin (data not shown). Bound material was subsequently eluted under native conditions with an excess of 61K (hPrp31) peptide (Fig. 4A, step V), and the RNA content of the eluate was analyzed by denaturing PAGE. As shown in Fig. 4B, lane 4, the eluate contained almost exclusively unspliced tobramycin-tagged pre-mRNA and only faint amounts of lariat intermediate but no product. Thus, essentially only precatalytic spliceosomal complexes had been isolated. As the tri-snRNP is first incorporated during B complex formation, the eluate should also be free of H/A complexes which do not contain the U4/U6-61K (hPrp31) protein. Indeed, complexes in the eluate contained the full set of snRNAs in nearly stoichiometric amounts, indicating that predominantly B complexes had been isolated (Fig. 4B, lane 2). As with the MS2 approach, minor amounts of high-molecular-weight RNA were also visible on the gel. Nonetheless, our data indicate that by using a combination of tobramycin and immunoaffinity selection, highly pure human B complexes can be obtained.
MS of the MS2 and tobramycin affinity-selected B complexes.
To determine the proteome of the human spliceosomal B complex isolated under mild conditions, proteins were recovered from two independent preparations of both the MS2 and tobramycin affinity-selected B complexes and separated by SDS-PAGE. Proteins were subsequently identified by liquid chromatography-tandem MS, and their presence in each of the four B complex preparations (indicated by the absolute number of peptides sequenced in each preparation) is summarized in Tables 1 and 2. Most proteins identified were consistently identified in all B complex preparations (i.e., 4 out of 4 isolates), and thus they clearly represent B complex components. A few additional proteins were found in both MS2 and tobramycin affinity-purified complexes but were detected in only two or three out of four preparations, suggesting that they are less stably associated B complex components that are more readily lost during purification. Finally, 35 proteins were detected in only one out of four preparations. Thus, they likely represent non-B complex components that are associated with other spliceosomal complexes (i.e., A, B*, and C complexes) that may be present in low amounts in our B complex preparations (Table 2). Indeed, based on the absolute number of peptides identified for these proteins, most of them appear to be present in very low amounts.
TABLE 1.
Protein composition of the human spliceosomal B complex isolated under physiological conditionsa
| Proteinb | Mol mass (kDa) | GenBank accession no. | No. of peptides sequenced in B complex preparationc
|
Present in A complexd | Saccharomyces cerevisiae genee | Schizosaccharomyces pombe genee | |||
|---|---|---|---|---|---|---|---|---|---|
| TOB no. 1 | TOB no. 2 | MS2 no. 1 | MS2 no. 2 | ||||||
| Sm proteins | |||||||||
| B | 24.6 | gi 4507125 | 17 | 39 | 84 | 74 | + | SMB1 | smb1 |
| D1 | 13.3 | gi 5902102 | 11 | 18 | 48 | + | SMD1 | smd1 | |
| D2 | 13.5 | gi 29294624 | 10 | 46 | 32 | 103 | + | SMD2 | cwf9/smd2 |
| D3 | 13.9 | gi 4759160 | 5 | 23 | 65 | 45 | + | SMD3 | smd3 |
| E | 10.8 | gi 4507129 | 1 | 22 | 45 | 28 | + | SME1 | sme1 |
| F | 9.7 | gi 4507131 | 3 | 11 | 5 | + | SMX3 | smf1 | |
| G | 8.5 | gi 4507133 | 11 | 28 | 11 | + | SMX2 | smg1 | |
| U1 snRNP | |||||||||
| U1-70K | 51.6 | gi 29568103 | 10 | 38 | 10 | 14 | + | SNP1 | U1-70k/SPAC19A8.13 |
| U1-A | 31.3 | gi 4759156 | 8 | 33 | 12 | 27 | + | MUD1 | U1-A/SPBC4B4.07c |
| U1-C | 17.4 | gi 4507127 | 1 | 1 | 1 | 2 | + | YHC1 | SPBP35G2.09 |
| U2 snRNP | |||||||||
| U2A′ | 28.4 | gi 50593002 | 13 | 45 | 41 | 67 | + | LEA1 | U2-A/SPBC1861.08c |
| U2B" | 25.4 | gi 4507123 | 17 | 8 | 31 | 28 | + | MSL1 | U2-B/SPBC8D2.09c |
| SF3a120 | 88.9 | gi 5032087 | 58 | 71 | 71 | 43 | + | PRP21 | sap114 |
| SF3a66 | 49.3 | gi 21361376 | 13 | 13 | 24 | 56 | + | PRP11 | sap62 |
| SF3a60 | 58.5 | gi 5803167 | 27 | 41 | 52 | 7 | + | PRP9 | sap61 |
| SF3b155 | 145.8 | gi 54112117 | 29 | 59 | 111 | 109 | + | HSH155 | prp10/sap155 |
| SF3b145 | 100.2 | gi 55749531 | 46 | 44 | 70 | 69 | + | CUS1 | sap145 |
| SF3b130 | 135.5 | gi 54112121 | 11 | 121 | 252 | 93 | + | RSE1 | prp12/sap130 |
| SF3b49 | 44.4 | gi 5032069 | 9 | 13 | 9 | 10 | + | HSH49 | sap49 |
| SF3b14a/p14 | 14.6 | gi 7706326 | 7 | 12 | 20 | 30 | + | SPBC29A3.07c | |
| SF3b14b | 12.4 | gi 14249398 | 6 | 5 | 12 | 12 | + | RDS3 | ini1 |
| SF3b10 | 10.1 | gi 13775200 | 3 | 6 | 2 | + | YNL138W-A/RCP10/YSF3 | SPBC211.05 | |
| U2-related | |||||||||
| hPRP43 | 90.9 | gi 68509926 | 2 | 14 | 29 | 28 | + | PRP43 | prp43 |
| SPF45 | 45.0 | gi 14249678 | 1 | 2 | 5 | + | |||
| SPF30 | 26.7 | gi 5032113 | 4 | 7 | 8 | 18 | + | spf30 | |
| U2AF65 | 53.5 | gi 6005926 | 2 | 5 | 18 | 11 | + | MUD2 | prp2/mis11/U2AF59 |
| U2AF35 | 27.9 | gi 5803207 | 2 | 7 | 26 | + | U2AF23/SPAP8A3.06 | ||
| Non-snRNP proteins* | |||||||||
| RNPC2 (CC1.3/CAPER/fSAP59) | 59.4 | gi 4757926 | 5 | 11 | 46 | 33 | + | rsd1 | |
| ELAV (HuR) | 36.1 | gi 38201714 | 1 | 5 | 3 | 10 | + | ||
| YB-1 | 35.9 | gi 34098946 | 3 | 6 | 17 | 25 | + | ||
| ASR2B | 100.0 | gi 58331218 | 81 | 69 | 8 | 47 | + | SPBC725.08 | |
| p68 (DDX5) | 69.2 | gi 4758138 | 8 | 19 | 5 | + | DBP2 | dbp2 | |
| DNaJ homolog (DNAJC6) | 100.0 | gi 7662146 | 1 | 1 | + | SWA2 | ucp11 | ||
| SF4 (F23858) | 72.5 | gi 33469964 | 3 | 1 | + | YNL224C (?) | SPAC2G11.04 (?) | ||
| NFAR | 95.4 | gi 24234750 | 11 | 3 | + | ||||
| TLS/FUS | 53.4 | gi 544357 | 4 | 1 | + | ||||
| SR proteins | |||||||||
| SRp20 | 19.4 | gi 4506901 | 10 | 19 | + | ||||
| 9G8 | 27.4 | gi 72534660 | 17 | 24 | 13 | 22 | + | ||
| SRp30c | 25.5 | gi 4506903 | 8 | 12 | 4 | + | |||
| SRp40 | 31.3 | gi 3929378 | 11 | 16 | 9 | 4 | + | ||
| SRp55 | 39.6 | gi 20127499 | 11 | 27 | 6 | 2 | + | ||
| SRp75 | 56.8 | gi 21361282 | 6 | 7 | 2 | + | |||
| SF2/ASF | 27.8 | gi 5902076 | 55 | 68 | 18 | 37 | + | ||
| SC35 (SFRS2) | 25.5 | gi 47271443 | 8 | 2 | + | ||||
| hTra-2 alpha | 32.7 | gi 9558733 | 3 | 3 | |||||
| hTra-2 beta/SFRS10 | 33.7 | gi 4759098 | 3 | 4 | 6 | ||||
| SR-related proteins | |||||||||
| FLJ10154 | 32.9 | gi 48675817 | 9 | 11 | 2 | ||||
| SRm160 | 102.5 | gi 3005587 | 2 | 4 | SPCC825.05c | ||||
| SRm300 | 300.0 | gi 19923466 | 14 | 71 | CWC21 (?) | cwf21 (?) | |||
| hnRNP | |||||||||
| hnRNP A0 | 30.9 | gi 5803036 | 16 | 10 | |||||
| hnRNP A1 | 38.7 | gi 4504445 | 16 | 24 | 38 | 27 | + | ||
| hnRNP A3 | 39.6 | gi 34740329 | 1 | 4 | 2 | + | |||
| hnRNP A/B | 36.0 | gi 12803583 | 3 | 1 | + | ||||
| hnRNP A2/B1 | 37.4 | gi 14043072 | 13 | 12 | 9 | 4 | + | ||
| hnRNP C1/C2 | 33.3 | gi 4758544 | 13 | 34 | 11 | 5 | + | ||
| hnRNP G | 42.4 | gi 56699409 | 7 | 4 | 8 | + | |||
| hnRNP M | 77.5 | gi 14141152 | 6 | 12 | |||||
| PCBP1 | 37.5 | gi 5453854 | 6 | 7 | 7 | 14 | + | HEK2 | |
| RALY | 32.5 | gi 8051631 | 3 | 2 | + | ||||
| Cap binding complex | |||||||||
| CBP20 | 18.0 | gi 19923387 | 6 | 19 | 4 | 16 | + | CBC2 | SPBC13A2.01c |
| CBP80 | 91.8 | gi 4505343 | 12 | 34 | 2 | 45 | + | STO1 | SPAC6G10.07 |
| U5 snRNP | |||||||||
| 220K | 273.7 | gi 3661610 | 2 | 91 | 184 | 306 | + | PRP8 | spp42/cwf6/prp15 |
| 200K | 244.5 | gi 45861372 | 17 | 124 | 95 | 267 | + | BRR2 | brr2 |
| 116K | 109.4 | gi 41152056 | 30 | 51 | 153 | 76 | + | SNU114 | cwf10/spef2/snu114 |
| 40K | 39.3 | gi 4758560 | 7 | 19 | 28 | 34 | spf38/cwf17 | ||
| 102K | 106.9 | gi 40807485 | 38 | 83 | 78 | 99 | + | PRP6 | prp1 |
| 15K | 16.8 | gi 5729802 | 4 | 1 | 5 | 7 | DIB1 | dim1 | |
| 100K | 95.6 | gi 41327771 | 9 | 23 | 33 | 48 | + | PRP28 | prp28 |
| 52K | 37.6 | gi 5174409 | 4 | 5 | 5 | + | LIN1 (SNU40) | SPBC83.09c | |
| Lsm proteins | |||||||||
| Lsm2 | 10.8 | gi 10863977 | 2 | 17 | 14 | LSM2 | lsm2 | ||
| Lsm3 | 11.8 | gi 7657315 | 3 | 8 | 3 | 5 | LSM3 | lsm3 | |
| Lsm4 | 15.4 | gi 6912486 | 10 | 23 | 21 | LSM4 | lsm4 | ||
| Lsm5 | 9.9 | gi 6912488 | LSM5 | lsm5 | |||||
| Lsm6 | 9.1 | gi 5919153 | 6 | 8 | 20 | LSM6 | lsm6 | ||
| Lsm7 | 11.6 | gi 7706423 | 3 | 6 | 4 | LSM7 | lsm7 | ||
| Lsm8 | 10.4 | gi 7706425 | 2 | 4 | 1 | 7 | LSM8 | lsm8 | |
| U4/U6 snRNP | |||||||||
| 90K | 77.6 | gi 4758556 | 20 | 43 | 82 | 85 | PRP3 | SPAC29E6.02 | |
| 60K | 58.4 | gi 45861374 | 27 | 22 | 49 | 41 | PRP4 | SPAC227.12 | |
| 20K | 20.0 | gi 5454154 | 3 | 15 | 22 | 46 | CPR6 or CPR3 (?) | cyp3 | |
| 61K | 55.4 | gi 40254869 | 12 | 39 | 62 | 53 | PRP31 | prp31/spp13 | |
| 15.5K | 14.2 | gi 4826860 | 1 | 4 | 2 | 8 | SNU13 | snu13/rph1 | |
| U4/U6.U5 tri-snRNP | |||||||||
| 110K | 90.2 | gi 13926068 | 33 | 34 | 29 | 64 | + | SNU66 | snu66 |
| 65K | 65.4 | gi 56550051 | 8 | 20 | 15 | 35 | SAD1 | ubp10 | |
| 27K | 18.9 | gi 24307919 | SPCC162.01c | ||||||
| hSnu23 | 23.6 | gi 21389511 | 2 | 8 | 3 | 4 | SNU23 | SPAC22F3.11c | |
| hPRP38 | 37.5 | gi 24762236 | 3 | 11 | 24 | PRP38 | SPBC19C2.08 | ||
| TFIP11 | 96.8 | gi 8393259 | 2 | 6 | 1 | 14 | SPP382/YLR424W | SPAC1486.03c | |
| hPRP19/CDC5L complex | |||||||||
| hPRP19 | 55.2 | gi 7657381 | 40 | 55 | 46 | 50 | + | PRP19 | cwf8/prp19 |
| CDC5L | 92.2 | gi 11067747 | 33 | 48 | 43 | 50 | CEF1 | cdc5 | |
| SPF27 | 21.5 | gi 5031653 | 6 | 18 | 21 | 20 | SNT309 | cwf7/spf27 | |
| PRL1 | 57.2 | gi 4505895 | 2 | 13 | 19 | 20 | PRP46 | cwf1/prp5/prl1/pi024 | |
| CCAP1 (hsp73) | 70.4 | gi 5729877 | 2 | 5 | SSA1 | ssa1 | |||
| CCAP2 (hspc148/AD-002) | 26.6 | gi 7705475 | 6 | 12 | 7 | 13 | CWC15 | cwf15 | |
| Catenin, β-like 1 (CTNNBL1/NAP) | 65.1 | gi 18644734 | 5 | 9 | 11 | SPAC1952.06c | |||
| Npw38BP | 70.0 | gi 7706501 | 22 | 38 | 25 | 42 | |||
| Npw38 | 30.5 | gi 5031957 | 8 | 28 | 19 | 17 | |||
| hPRP19/CDC5L-related | |||||||||
| PRCC | 52.4 | gi 40807447 | 1 | 1 | 1 | 4 | |||
| RBM22 (fSAP47) | 46.9 | gi 8922328 | 8 | 15 | 8 | 1 | ECM2/SLT11 | cwf5/ecm2 | |
| hSYF1 (XAB2) | 100.0 | gi 55770906 | 2 | 5 | 15 | 45 | SYF1 | cwf3/syf1 | |
| CRNKL1/hSYF3 | 100.6 | gi 30795220 | 5 | 14 | 16 | 44 | CLF1 | cwf4/syf3 | |
| hIsy1 (fSAP133) | 33.0 | gi 20149304 | 1 | 7 | 3 | 12 | ISY1 | isy1/cwf12 | |
| SKIP | 51.1 | gi 6912676 | 26 | 30 | 28 | 41 | PRP45 | prp45/snw1/cwf13 | |
| Cyp-E | 33.4 | gi 5174637 | 3 | 3 | 8 | 11 | |||
| PPIase-like 1 (PPIL1) | 18.2 | gi 7706339 | 1 | 8 | 8 | 14 | |||
| KIAA0560 (fSAP164) | 171.3 | gi 38788372 | 8 | 39 | cwf11 | ||||
| G10 (fSAP17) | 17.0 | gi 32171175 | 2 | 2 | 8 | 20 | BUD31/CWC14 | cwf14 | |
| RES complex | |||||||||
| SNIP1 | 45.8 | gi 21314720 | 2 | 5 | 3 | YLR016C/PML1 | |||
| MGC13125 (fSAP71) | 70.5 | gi 14249338 | 3 | 6 | 13 | BUD13 | cwf26 | ||
| CGI-79 | 39.7 | gi 4929627 | 1 | 1 | IST3/SNU17 | cwf29 | |||
| Miscellaneous | |||||||||
| FBP21 | 42.5 | gi 6005948 | 3 | 8 | 4 | 8 | SPBC18H10.07 | ||
| MFAP1 | 51.9 | gi 50726968 | 18 | 19 | 23 | 23 | SPAC1782.03 | ||
| RED | 65.6 | gi 10835234 | 30 | 38 | 31 | 35 | SPBC1539.02 | ||
| hSmu-1 (fSAP57) | 57.5 | gi 8922679 | 30 | 38 | 26 | 40 | |||
| LOC124245 | 104.0 | gi 31377595 | 12 | 22 | 5 | 53 | |||
| TCERG1 (CA150) | 123.9 | gi 21327715 | 24 | 20 | 3 | 14 | YPR152C | dre4 | |
| hsp27 | 22.8 | gi 4504517 | 7 | 11 | 2 | 4 | |||
| LOC84081/DKFZp434k1421 | 66.4 | gi 14149807 | 1 | 1 | 1 | 5 | SPAC29A4.06c | ||
| PPP1CA | 38.6 | gi 4506003 | 6 | 3 | 4 | GLC7 | dis2/bws1/sds1 | ||
| UBL5 | 8.5 | gi 13236510 | 2 | 6 | 7 | HUB1 | ubl4/hub1 | ||
| MGC23918 | 19.2 | gi 21389497 | 2 | 4 | 6 | cwf18 | |||
| MGC20398 | 42.0 | gi 49472814 | 3 | 3 | |||||
| matrin 3 | 94.6 | gi 21626466 | 3 | 4 | 4 | ECM13 | |||
| PPIL2/Cyp-60 | 59.5 | gi 7657473 | 2 | 2 | 1 | 27 | |||
| NF45 | 43.0 | gi 24234747 | 13 | 6 | |||||
| hPRP2 | 119.2 | gi 4503293 | 3 | 8 | PRP2 | cdc28/prp8 | |||
| DDX3 | 73.3 | gi 87196351 | 2 | 6 | DED1 | sum3 | |||
| HSP70 | 70.0 | gi 5123454 | 20 | 30 | SSA2 | ssa2 | |||
| FBP11 | 48.5 | gi 88953744 | 17 | 24 | PRP40 | prp40 | |||
| S164 (fSAP94) | 100.1 | gi 4050087 | 12 | 18 | SPBC839.10 | ||||
| p72/DDX17 | 80.5 | gi 3122595 | 1 | 3 | (DBP2) | (dbp2) | |||
| RBM4 | 40.9 | gi 4506445 | 1 | 4 | |||||
| RBM7 | 30.5 | gi 5070698 | 2 | 6 | |||||
| NRIP2 | 31.3 | gi 13899327 | 1 | 9 | |||||
| NIPP1 | 38.5 | gi 13699256 | 6 | 4 | |||||
| Proteins not previously found in spliceosomes | |||||||||
| MEP50 | 36.7 | gi 13129110 | 7 | 8 | |||||
| PRMT5 (SKB1) | 72.7 | gi 20070220 | 11 | 25 | HSL7 | skb1 | |||
| PPIL4 | 57.2 | gi 20911035 | 1 | 3 | |||||
| B*/C complex and step II proteins | |||||||||
| hPRP17 | 65.5 | gi 7706657 | 1 | 8 | CDC40 | prp17 | |||
| hSLU7 | 68.4 | gi 27477111 | 1 | 1 | SLU7 | slu7 | |||
| EJC protein, Y14 | 19.9 | gi 4826972 | 1 | 3 | SPAC23A1.09 | ||||
Proteins were identified by liquid chromatography-tandem mass spectrometry after separation by SDS-PAGE.
Asterisk, non-snRNP proteins that were found previously in the A complex and also were detected more than once in B complex preparations.
The presence of a protein (indicated on the left) in tobramycin and MS2 B complex preparations is indicated by a number which represents the absolute number of peptides sequenced for that protein. Note that a number of proteins were detected in multiple bands; also because the tandem MS analysis of MS2 no. 2 was performed in a linear ion trap, the overall number of sequenced peptides per protein is higher (on average), compared to the other B complex preparations (which were analyzed in a quadropole-time of flight MS instrument). TOB 1 and 2, B complexes purified by tobramycin affinity selection followed by immunoaffinity purification with antibodies against the U4/U6-61K (hPrp31) protein; MS2 1 and 2, B complexes purified by glycerol gradient centrifugation followed by MS2 affinity selection.
Proteins identified previously in tobramycin affinity-selected A complexes (19) are indicated by a plus sign. Several ribosomal proteins and translation elongation factors (not shown) were also detected but, based on the low number of peptides identified, appear to be present in substoichiometric amounts and likely represent contaminants.
The gene names of the S. cerevisae and S. pombe homologues of the indicated human protein are given. A question mark indicates that it is currently not clear whether the indicated yeast protein is a true functional homolog. BLAST alignments of both p68 (DDX5) and p72 (DDX17) revealed significant homology to S. cerevisiae DBP2 and S. pombe dbp2 (between 56 and 59% sequence identity, respectively). Since true homology remains to be verified, DBP2 and dbp2 are both enclosed in parentheses.
TABLE 2.
Proteins detected by MS in only one of four preparations of the human spliceosomal B complexa
| Protein | Mol mass (kDa) | GenBank accession no. | No. of peptides sequenced in B complex preparationb
|
Present in A complexc | Saccharomyces cerevisiae gene | Schizosaccharomyces pombe gene | |||
|---|---|---|---|---|---|---|---|---|---|
| TOB no. 1 | TOB no. 2 | MS2 no. 1 | MS2 no. 2 | ||||||
| SPF31 | 31.0 | gi 7657611 | 3 | + | spf31 | ||||
| SR140 (fSAPa) | 118.2 | gi 51492636 | 8 | + | SPBC11C11.01 | ||||
| CHERP | 100.0 | gi 21359884 | 3 | + | |||||
| PUF60 | 59.9 | gi 17298690 | 5 | ||||||
| SF3b125 | 103.0 | gi 45446743 | 1 | ||||||
| CDC2L2 | 91.0 | gi 16357494 | 4 | SPBC18H10.15 | |||||
| RBM10 | 103.5 | gi 12644371 | 2 | SPAC17H9.04c (?) | |||||
| tat SF1 | 85.7 | gi 21361437 | 2 | CUS2 | uap2 | ||||
| KIAA0073 (CyP64/PPWD1) | 73.6 | gi 24308049 | 4 | ||||||
| KIAA1604 (fSAPb) | 105.5 | gi 55749769 | 6 | CWC22 | cwf22 | ||||
| SRp38 | 31.3 | gi 5730079 | 2 | ||||||
| hnRNP F | 45.7 | gi 4826760 | 3 | ||||||
| hnRNP H2 | 49.3 | gi 9624998 | 1 | ||||||
| hnRNP Q3 | 69.6 | gi 23397427 | 4 | + | |||||
| hnRNP R | 70.9 | gi 5031755 | 17 | + | |||||
| hPRP4-Kinase | 117.1 | gi 17999535 | 1 | YAK1 (?) | prp4 | ||||
| Abstrakt | 69.8 | gi 21071032 | 2 | ||||||
| DBPA | 40.1 | gi 14602477 | 1 | ||||||
| GCIP p29 (fSAP29) | 28.7 | gi 7661636 | 5 | SYF2 | SPBC3E7.13c | ||||
| DDX9 | 142.0 | gi 4503297 | 5 | + | |||||
| Magoh | 17.2 | gi 4505087 | 1 | SPBC3B9.08c | |||||
| eIF4A3 | 46.9 | gi 7661920 | 8 | FAL1 (?) | tif412 | ||||
| THOC4 | 26.9 | gi 55770864 | + | YRA1 | mlo3/rai1 | ||||
| UAP56 | 49.1 | gi 21040371 | 3 | SUB2 | uap56 | ||||
| Pinin | 81.6 | gi 33356174 | 1 | ||||||
| Acinus (fSAP152) | 151.8 | gi 7662238 | 3 | ||||||
| HCNGP | 33.9 | gi 9994179 | 12 | SPAC3H1.03 | |||||
| KIAA1627 (MT-A70) | 52.1 | gi 24308265 | 2 | KAR4 | |||||
| Nm23 | 19.6 | gi 38045913 | 1 | YNK1 | ndk1 | ||||
| NPM1 (NUMATRIN) | 32.6 | gi 10835063 | 21 | ||||||
| PCBP2 | 38.6 | gi 14141168 | 1 | PBP2 | rnc1 | ||||
| PABP | 70.7 | gi 46367787 | 4 | PAB1 | pabp/pab1 | ||||
| HsKin17 | 45.4 | gi 13124883 | 11 | RTS2 | SPBC365.09c | ||||
| LOC51325 (GCFC/fSAP105) | 104.8 | gi 22035565 | 11 | ||||||
| NY-CO-10 | 53.8 | gi 64276486 | 6 | CWC27 | cyp7/cwf27 | ||||
Column designations and abbreviations are as described in the notes to Table 1.
The presence of a protein (indicated on the left) in tobramycin (TOB) and MS2 B complex preparations is indicated by a number which represents the absolute number of peptides sequenced for that protein.
Proteins identified previously in tobramycin affinity-selected A complexes (19) are indicated by a plus sign.
A total of 118 proteins were identified at least once in both B complex preparations (Table 1), whereas 11 proteins were found solely in the MS2 affinity-selected B complex, and 11 were found solely in the tobramycin affinity-selected B complex. These differences in protein composition are likely due to differences in the two purification methods employed, which may lead to the loss of a specific set of proteins in each case. Alternatively, as they were not observed in both affinity-selected B complex preparations, they might represent contaminants. However, all but 3 of the proteins identified solely in the MS2 or tobramycin affinity-selected B complexes (i.e., PPIL4, PRMT5, and MEP50) have been shown previously (i) to participate in pre-mRNA splicing and/or (ii) to be associated with spliceosomal complexes, arguing that most are bona fide spliceosomal proteins.
As in the human A complex, which was isolated previously under similar conditions (19), all U1 snRNP-specific (i.e., C, A, and 70K) and 17S U2 snRNP-specific proteins (i.e., U2-A′/B" and all SF3a and SF3b subunits) were detected in both affinity-selected B complex preparations (Table 1). Several U2-associated proteins (e.g., SPF45, SPF30, and hPrp43p) were also found in both preparations. In addition to numerous SR and hnRNP proteins, the cap binding proteins CBP20 and CBP80, and the early factors U2AF65 and U2AF35, many of the other non-snRNP proteins previously detected in tobramycin affinity-purified A complexes were also found in the B complex (Table 1). However, five non-snRNP proteins previously detected in purified A complexes (i.e., FLJ10839, RBM5/LUCA15, E1B-APS, SMC1/2, and FLJ21007), some of which have no known function in splicing, were not detected in our B complexes, suggesting that they are lost during the transition from the A to B complex. Thus, they may associate solely with early splicesomal complexes, and, if they indeed contribute to the splicing reaction, they likely act prior to B complex formation.
A comparison of the proteomes of the A and B complexes further revealed that a large number of proteins are recruited to the pre-mRNA during B complex formation (Table 1). As expected for the B complex, nearly all U5-, U4/U6-, and tri-snRNP-specific proteins were detected. Exceptions were Lsm5 and the 27K protein, from which tryptic peptides are difficult to identify by MS (H. Urlaub, personal communication). Strikingly, over 50 non-snRNP proteins not previously detected in the A complex were also found, suggesting that they are first recruited at the time of B complex formation. These include, among others, all components of the hPrp19/CDC5 complex, a group of 10 hPrp19/CDC5-related proteins, subunits of the newly identified RES (retention and splicing) complex, and human homologues of the S. cerevisiae proteins Snu23 and Prp38, which were detected in yeast (but not human) tri-snRNP complexes (18, 36) (see below for detailed discussion). Thus, these proteins are recruited prior to spliceosome activation and thus potentially play a role in the transition from the B complex to the catalytically activated B* complex. In contrast, several proteins known to act during the second step of splicing (i.e., hPrp16, hPrp18, and hPrp22), as well as most components of the exon junction complex (EJC) that is deposited 20 nucleotides upstream of exon-exon junctions during pre-mRNA splicing (reviewed in reference 37), were not found. Thus, this group of ∼15 proteins appears to be recruited either after catalytic activation or after the first step of splicing.
EM of purified native B complexes.
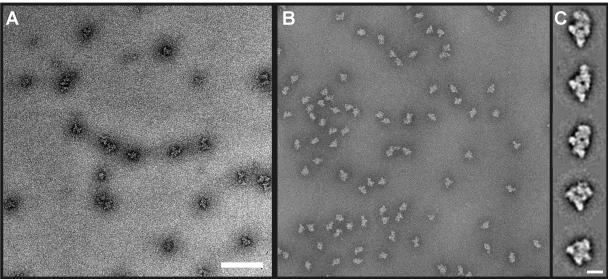
To determine the structure of native B complexes, MS2 affinity-purified particles were negatively stained with uranyl formate and analyzed by EM. A typical EM raw image of the B complex shows monodisperse particles of similar sizes (Fig. 5B). The particles appear rhombic with a maximum dimension of 420 Å (Fig. 5B). In comparison to images obtained with the heparin-treated BΔU1 complex (7) (Fig. 5A), several significant improvements can already be detected at the level of the raw electron microscopic image. Smaller particles that may represent dissociation products of the B complex cannot be detected, ensuring that subsequent image analysis is indeed performed with an intact spliceosome, rather than one or more breakdown products. Additionally, the concentration of particles has more than doubled, and the staining appears very homogeneous, which facilitates computerized image analysis. A total of 7,500 molecular images were subjected to image processing. Characteristic two-dimensional averages of the spliceosomal B complex were obtained by averaging ∼20 individual images after “reference-free” image alignment, MSA, and classification (Fig. 5C). As expected from the improved quality of the raw images, the eigenimages obtained by MSA clearly indicate a structurally more homogeneous preparation in comparison to previous images of the BΔU1 complex (data not shown). The most frequent class averages (the first three from the top in Fig. 5C) show a central density surrounded by four peripheral density elements. The central density and the three peripheral density elements in the lower part of the image are arranged in a triangular shape of similar size in these class averages (Fig. 5C). The shape and position of the fourth density in the upper part of the image is different. The size and shape of the lower part of the image are reminiscent of the projection of the triangular body of the BΔU1 complex, which was isolated under stringent conditions (7). The density in the upper part of the image corresponds to the head density of the BΔU1 complex. In spite of being structurally better defined than the BΔU1 complex, the head region of the MS2 affinity-purified B complexes displays conformational heterogeneity. However, due to improved structural definition of the entire complex, the head density of the native complex clearly reveals more fine structural details in the class averages compared to the BΔU1 complex and appears to be larger (Fig. 5C). This is also consistent with the fact that it contains significantly more proteins than the latter complex.
FIG. 5.
EM of the native spliceosomal complex B. (A) CCD image of negatively stained complex BΔU1. Particles were purified as previously described (7), adsorbed onto a carbon film, and sandwiched beneath another carbon film in 2% (wt/vol) uranyl formate. The images were taken at a magnification of ×27,500 under low-dose conditions. Bar, 1,500 Å. (B) CCD image of negatively stained complex B. Particles were adsorbed onto a carbon film and sandwiched beneath another carbon film in 2% (wt/vol) uranyl formate. The images were taken at a magnification of ×122,000 under low-dose conditions. Bar, 1,500 Å. (C) A gallery of individual class averages of the native B complex obtained by averaging ∼20 individual raw images. Bar, 200 Å.
DISCUSSION
Here, we have purified native B complexes under low-stringency conditions, using two independent affinity selection methods. Complexes isolated by both methods contained nearly stoichiometric amounts of unspliced pre-mRNA, U1, U2, U4, U5, and U6 snRNA, consistent with their designation as precatalytic spliceosomal B complexes. MS2 affinity-purified B complexes were shown to be functional, catalyzing splicing in nuclear extract depleted of one or more of the spliceosomal snRNPs. Subsequent characterization of the protein composition of both B complex preparations indicated that a large number of non-snRNP proteins are recruited to the spliceosome prior to its catalytic activation. Proteins detected in both B complex preparations included (i) nearly all proteins found in the human A complex (including U1 and U2 snRNP proteins), (ii) essentially all U4/U6.U5 tri-snRNP proteins, and (iii) a large number of non-snRNP proteins that were not previously detected in the A complex.
MS2 and tobramycin affinity-purified B complexes are highly pure.
A number of observations attest to the high purity of the B complexes that we have isolated. Our purified B complexes were not contaminated with post-second step spliceosomal/mRNP complexes as (i) no spliced mRNA was detected and (ii) almost no known components of the EJC complex, which associates with spliced mRNPs, were found. Only faint amounts of splicing intermediates (less than 2% of total pre-mRNA) were detected in our purified complexes, indicating that greater than 98% are precatalytic. In addition, the presence of stoichiometric amounts of both the U4 and U1 snRNAs further suggests that the vast majority of these complexes have not undergone catalytic activation. Finally, purified MS2 affinity-selected complexes appeared structurally homogeneous under the electron microscope, consistent with the idea that they consist predominantly of spliceosomes stalled at a single assembly stage.
A large number of B complex proteins are lost upon treatment with heparin.
Compared to B complexes previously isolated by immunoaffinity purification followed by glycerol gradient centrifugation under stringent conditions (i.e., in the presence of heparin; designated BΔU1) (27), the purified B complexes analyzed here contain considerably more proteins. This indicates that a large number of proteins that associate with the B complex are not stably bound; that is, their association does not withstand heparin treatment. These include, among others, most SR and hnRNP proteins and nearly all components of the Prp19/CDC5 complex and related proteins (discussed in more detail below). In contrast, those proteins that are present in both complexes—such as all 17S U2 core proteins, nearly all tri-snRNP proteins (excluding U5-100K, Lsm2/Lsm6, and the tri-snRNP-specific 65K protein), and ∼25 non-snRNP proteins—represent more stably integrated B complex components. As less stringent conditions were used in this study, proteins that were detected in BΔU1 but not in the MS2 or tobramycin affinity-selected B complexes (a total of 8 proteins) likely represent either nonspliceosomal contaminants (e.g., Ncor1, Npat, or TBLR1 which have not been detected in any other MS study of spliceosome-associated proteins) or are potentially components of B*/C spliceosomal complexes that may be present in low amounts in the BΔU1 preparation. Thus, a comparison of the proteomes of B complexes isolated at 150 mM either in the presence or absence of heparin provides useful information about the strength of association of B complex components identified in this study.
The vast majority of A complex proteins are also present in the B complex.
As both complexes were isolated under very similar conditions, a meaningful comparison of the proteome of the B complexes isolated here with that of the previously described A complex can be made (19). Like the spliceosomal A complex, the human B complex contains the U1 snRNP specific proteins, U1-70K, U1-A, and U1-C, and all core 17S U2 snRNP proteins (SF3a, SF3b, U2-A′, and U2-B"). Several U2-associated proteins that copurified with human 17S U2 snRNPs (e.g., hPrp43, SPF45, and SPF30) (49) were present in both A and B complexes. In contrast, the U2- and A complex-associated, DEAD-box protein hPrp5 was notably absent in all B complex preparations. Thus, it appears to be only transiently associated with the pre-mRNA prior to B complex formation. Prp5 plays an important role in bridging the U1 and U2 snRNPs within the A complex (51), and its absence in the B complex is consistent with the idea that its activity or presence is only required for early spliceosome assembly events.
Interestingly, the early splicing factors U2AF35 and U2AF65 were also present in the B complex, indicating that they remain associated at least until this stage of spliceosome assembly; previous studies with purified spliceosomes had suggested that both U2AF subunits are largely lost already during the transition from the E to A complex (4). A very similar, but not identical, set of SR and hnRNP proteins was also found in both the A and B complexes. Likewise, most of the other non-snRNP, A complex proteins were also present in our B complexes, with the exception of FLJ10839, RBM5/LUCA15, E1B-APS, SMC1/2, and FLJ21007. The apparent loss of the latter proteins is a first indication that they associate solely with early splicesomal complexes and function prior to B complex formation.
A large number of proteins, including the hPrp19/CDC5 complex, are recruited during B complex formation.
A comparison of the proteomes of the A and B complexes isolated under low-stringency conditions indicates that a large number of proteins are recruited to the pre-mRNA during B complex formation. As expected, essentially all proteins of the U4/U6.U5 tri-snRNP, which is recruited during B complex formation, were detected in the MS2 and tobramycin affinity-selected B complexes. Exceptions include only those proteins that are typically difficult to detect by MS (i.e., Lsm5 and the 27K tri-snRNP-specific protein). Unexpectedly, U5-52K, which was previously reported to be lost upon formation of the U4/U6.U5 tri-snRNP (25), was also found. Interestingly, human homologues of the yeast tri-snRNP proteins Snu23 and Prp38, as well as the human homologue of Spp382p (designated TFIP11), which is a genetic suppressor of yeast Prp38 (Shatakshi Pandit and Brian Rymond, personal communication), were also identified. None of these proteins is detected by MS in purified human U4/U6.U5 tri-snRNPs (data not shown). Like most of the human tri-snRNP proteins, hSnu23, hPrp38, and TFIP11 are recruited during B complex formation, and hSnu23 and TFIP11 are also stably integrated at this stage, as evidenced by their presence in heparin-treated BΔU1 complexes (27). Thus, one or more of these proteins may contribute to tri-snRNP integration during B complex formation and/or may be involved in subsequent spliceosome activation or catalysis. Indeed, yeast Prp38p is an essential splicing factor that is required for conformational changes leading to catalytic activation of the spliceosome, i.e., U4/U6 snRNA intermolecular helix unwinding and U4 snRNA release (5, 50). Consistent with these results, the association of the human Prp38 protein with the spliceosome is destabilized concomitantly with the U4 snRNA and U4/U6-associated proteins, namely, during spliceosome activation (26). Genetic depletion of yeast Snu23p leads to a block in pre-mRNA splicing in vivo, but its precise function remains unknown (18). Finally, human TFIP11, which remains stably associated with the spliceosome at least until step I (i.e., it is found in purified B* and C complexes) (21, 26), was also recently shown to function in pre-mRNA splicing, although its precise role also remains to be determined (47).
Unexpectedly, all recently identified subunits of the human 14S Prp19/CDC5 complex (i.e., hPrp19, CDC5L, SPF27, PRL1, CCAP1/hsp73, CCAP2/AD002, and β-like catenin 1) were found in both B complex isolates, as along with Npw38BP and Npw38, which also appear to copurify with the Prp19/CDC5 complex (27). Likewise, 10 additional proteins, which we have designated Prp19/CDC5-related proteins, based on evidence that they (or their yeast homologue) physically or genetically associate with Prp19/CDC5, were also found (Table 1). Most of these proteins are components of the 35S U5 snRNP, a form of U5 that is generated by RNP remodeling events within the spliceosome during its catalytic activation (26). Prp19/CDC5 and associated proteins were absent or highly underrepresented in the BΔU1 complex, indicating that they are recruited during B complex formation but stably integrated at a later stage. Indeed, previous studies confirmed that they are stable components of the B* complex (26). Thus, this large group of proteins interacts with the pre-mRNA prior to catalytic activation, but during subsequent RNP remodeling events accompanying catalytic activation, they become stably associated. Although the heptameric Prp19/CDC5 complex is recruited prior to activation, functional studies in humans indicate that components of this complex are not required for tri-snRNP addition (1, 27) but, rather, act at a subsequent step. However, several of the hPrp19/CDC5-related proteins could potentially act at this stage.
Indeed, that components of the yeast Prp19 complex function prior to activation (and thus also associate earlier) has been suggested by recent experiments. In yeast, Prp19p is associated with at least eight proteins, forming the NTC, which is involved in the catalytic activation of the spliceosome (reference 8 and references therein). Three NTC components, Prp19, Cef1/CDC5, and Snt309 (human SPF27), are also found in the human Prp19/CDC5 complex, whereas the remaining NTC components are among those human proteins that we have designated Prp19/CDC5-related (Table 1). Previous immunoprecipitation studies suggested that Prp19p first interacts with the yeast spliceosome concomitant with or just after catalytic activation (39). However, these studies did not rule out that Prp19 might interact at an earlier stage; its presence in early complexes may have escaped detection due to the limitations of the experimental methods employed. More recent data revealed that Clf1 (the yeast homologue of the human protein hSyf3), a component of the NTC, functions during the transition from complex A to B and thus associates with the spliceosome prior to its activation (10, 46). Thus, the recruitment of at least one Prp19/CDC5-related protein prior to the activation step appears to be conserved between yeast and man.
Components of the RES complex are found in the B complex.
Recently, a trimeric complex termed RES (for retention and splicing) containing three proteins, namely, Bud13p, Pml1p and Snu17p, was identified in yeast (16). This complex was shown to be required for efficient splicing and the retention of pre-mRNA in the yeast nucleus. Furthermore, pull-down experiments indicated that it associates with the yeast spliceosome prior to step I (16). Human homologues of the RES complex subunits, namely MGC13125, Snip1, and CGI-79, were also identified, but their function is currently unknown. Significantly, MGC13125, Snip1, and CGI-79 were detected in purified B complexes, providing a first indication that, like components of the yeast RES complex, they are involved in splicing and might act prior to the first catalytic step of splicing.
Most second-step factors and components of the EJC complex are recruited after B complex formation.
Although B complex formation entails the recruitment of a large number of spliceosomal proteins, key proteins that function at later stages of the splicing reaction appear to be recruited at subsequent steps of spliceosome assembly/function. Indeed, most second-step factors (i.e., hPrp16, hPrp18, and hPrp22) (20, 32, 53) were not detected in our purified B complexes. Furthermore, although hSlu7 was detected once in each preparation, only one hSlu7 peptide was identified in each instance, suggesting that it is present in very low amounts. In contrast, the second-step factor hPrp17, as well as the first-step factor hPrp2, were found in the MS2 but not in tobramycin affinity-selected complexes, suggesting that these factors are recruited prior to step I of splicing. Indeed, the yeast homologues of these proteins were also shown to associate with spliceosomes prior to catalytic step I (3, 24). Taken together, our results indicate that functionally important protein recruitment events take place after B complex formation.
During pre-mRNA splicing, a complex of proteins (designated the EJC) is deposited ∼20 nucleotides upstream of exon-exon junctions (reviewed in reference 37). This complex consists of a tetrameric core containing eIF4AIII, Magoh, Y14, and MLN51 and at least 12 additional proteins that include, among others, Pinin and SRm160 (38). At present the assembly pathway of the EJC during splicing is poorly understood. Most EJC-associated proteins were not found in our affinity-purified B complexes, consistent with the idea that they interact with the pre-mRNA at later stages of splicing. Exceptions included Y14, UAP56, Pinin, Acinus, SRm160, and eIF4AIII. However, these proteins were typically observed in only one out of four B complex preparations, and only a relatively low number of peptides was identified in most cases. Thus, these proteins appear to be highly underrepresented in our purified complexes. SRm160 and Pinin have been shown additionally to function in pre-mRNA splicing per se (17, 45). Thus, these proteins might associate with the spliceosome at a much earlier stage, independent of other components of the EJC complex. Previous studies indicated that the heparin-resistant (i.e., stable) association of most EJC factors occurs first at the time of catalytic activation or C complex formation (21, 26). The future isolation of the B* and C spliceosomal complexes in the absence of heparin should reveal the precise stage at which individual EJC and step II factors associate with the pre-mRNA during splicing.
Analysis of the factor requirements for catalytic activation and step I of splicing using MS2 affinity-selected B complexes.
MS2 affinity-purified B complexes catalyzed splicing in nuclear extract depleted of one or more of the spliceosomal snRNPs, indicating that they are not dead-end assembly intermediates. However, they were not capable of catalyzing splicing in the absence of nuclear extract when incubated under splicing conditions. This suggests that one or more factors required for catalytic activation and/or the catalytic steps of splicing is missing. Indeed, as described above, these complexes did not contain essential step II factors, such as hPrp16, hPrp18, or hPrp22. Thus, it is not unexpected that splicing products could not be formed. However, all currently known factors required for catalytic activation, as well as the first step of splicing (e.g., hPrp2), were detected in these complexes. As catalytic activation was not assayed per se, it is not clear whether our purified B complexes alone can undergo the conformational rearrangements that accompany this functionally decisive step. Nonetheless, splicing intermediates were not detected, suggesting that one or more factors required for subsequent spliceosome activation and/or catalysis are lacking. Potential candidates include those proteins previously identified by MS in purified B* and/or C complexes that were not found in our MS2 affinity-purified B complexes. Excluding step II factors and EJC components, this includes only a handful of proteins (21, 26).
Alternatively, MS2 affinity-purified B complexes could potentially contain a full protein complement required for at least the first step of splicing, but the incorrect posttranslational modification status of one or more spliceosomal proteins may block catalysis. Posttranslational modifications play an important role in splicing. Spliceosome assembly is dependent on the phosphorylation of spliceosomal proteins such as members of the SR protein family, and SF3b155 is hyperphosphorylated just prior to or during the first catalytic step of splicing (44). In contrast, the catalytic steps of splicing require protein dephosphorylation that is mediated by members of the PP1 and PP2 phosphatase families (28, 30). Significantly, only PP1 was detected in our MS2 affinity-selected B complexes. However, whether it alone is sufficient to catalyze the dephosphorylation events required for catalysis is currently not known.
To precisely determine which functionally important components are lacking in our purified B complexes, we are currently performing reconstitution experiments with fractionated HeLa nuclear extract. These studies may elucidate novel factors required for the catalytic activation of the B complex as well as for the first step of splicing. Thus, the availability of native B complexes for future studies represents an important step toward functionally dissecting the spliceosome's catalytic activation step in vitro.
EM of MS2 affinity-purified B complexes.
Negatively stained images of MS2 affinity-purified B complexes could be obtained with much improved image quality compared to previous spliceosome preparations. Visual inspection of EM raw images already revealed greatly enhanced and uniform image contrast and a significantly higher amount of particles per specimen area, which greatly facilitates computerized image analysis of these complexes. The most significant difference to previous EM work on spliceosomal complexes is the high level of structural preservation and homogeneity, which is evidenced by the fact that practically no dissociation products could be detected. This is also reflected by the high signal in the eigenimages, which can be used as a means to evaluate the level of structural features that can be detected reproducibly at the same place within the images of a given data set. In contrast to what one may have assumed, the mild conditions used for the purification of MS2 affinity-purified B complexes allows for the isolation of spliceosomal complexes that are more structurally homogeneous than those isolated under more stringent purification conditions. In summary, images of MS2 affinity-purified B complexes are at an unprecedented quality level, and thus they should be well suited for three-dimensional structure analyses, based on criteria such as their structural integrity and homogeneity.
However, in the initial image processing analysis we could still detect potential movements of the two major structural domains, the triangular body and the head domain, relative to each other. The level of conformational heterogeneity present in the data set appears to be slightly reduced compared to the BΔU1 complex. However, there is still a significant amount of conformational heterogeneity remaining which makes high-resolution structure determination difficult and which will require new computational tools to deal with. To what extent this conformational heterogeneity reflects dynamic movements in the spliceosome that are required for its function is presently not clear. There are many indications based on other macromolecular complexes such as ribosomes (12) or the anaphase promoting complex (14) that such flexibilities indeed play important roles in terms of function. Indeed, the observed conformational heterogeneity may represent different states where the spliceosome is waiting for an additional component to bind in order to finally lock it into the next structurally stable state that can perform the subsequent step of the reaction cycle. Additional studies, however, are required to clarify the significance of this structural flexibility.
Acknowledgments
We are grateful to Thomas Conrad, Peter Kemkes, Hossein Kohansal, and Irene Öchsner for excellent technical assistance. We also thank Monika Raabe and Uwe Plessmann for their excellent help in MS analysis, Christian Merz for helpful discussions regarding MS2 affinity selections, and Andreas Kuhn, Monika Golas, and Olex Dybkov for their comments on the manuscript. We are grateful to Robin Reed for kindly providing pAdML-M3 plasmid.
This work was supported by grants from the BMBF BioFuture (0311899) to H.S. and from the BMBF (031U215B), Fonds der Chemischen Industrie, and the Ernst-Jung Stiftung to R.L.
REFERENCES
- 1.Ajuh, P., B. Kuster, K. Panov, J. C. Zomerdijk, M. Mann, and A. I. Lamond. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19:6569-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azubel, M., S. G. Wolf, J. Sperling, and R. Sperling. 2004. Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Mol. Cell 15:833-839. [DOI] [PubMed] [Google Scholar]
- 3.Ben Yehuda, S., I. Dix, C. S. Russell, S. Levy, J. D. Beggs, and M. Kupiec. 1998. Identification and functional analysis of hPRP17, the human homologue of the PRP17/CDC40 yeast gene involved in splicing and cell cycle control. RNA 4:1304-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, M., S. Michaud, J. Kingston, and R. Reed. 1992. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 6:1986-2000. [DOI] [PubMed] [Google Scholar]
- 5.Blanton, S., A. Srinivasan, and B. C. Rymond. 1992. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol. Cell. Biol. 12:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blencowe, B. J., B. S. Sproat, U. Ryder, S. Barabino, and A. I. Lamond. 1989. Antisense probing of the human U4/U6. Cell 59:531-539. [DOI] [PubMed] [Google Scholar]
- 7.Boehringer, D., E. M. Makarov, B. Sander, O. V. Makarova, B. Kastner, R. Lührmann, and H. Stark. 2004. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat. Struct. Mol. Biol. 11:463-468. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. P., D. I. Kao, W. Y. Tsai, and S. C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. H., W. C. Yu, T. Y. Tsao, L. Y. Wang, H. R. Chen, J. Y. Lin, W. Y. Tsai, and S. C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, S., M. R. McLean, and B. C. Rymond. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5:1042-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, R., Z. Zhou, and R. Reed. 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell 5:779-787. [DOI] [PubMed] [Google Scholar]
- 12.Diaconu, M., U. Kothe, F. Schlunzen, N. Fischer, J. M. Harms, A. G. Tonevitsky, H. Stark, M. V. Rodnina, and M. C. Wahl. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121:991-1004. [DOI] [PubMed] [Google Scholar]
- 13.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dube, P., F. Herzog, C. Gieffers, B. Sander, D. Riedel, S. A. Muller, A. Engel, J. M. Peters, and H. Stark. 2005. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol. Cell 20:867-879. [DOI] [PubMed] [Google Scholar]
- 15.Dube, P., P. Tavares, R. Lurz, and M. van Heel. 1993. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dziembowski, A., A. P. Ventura, B. Rutz, F. Caspary, C. Faux, F. Halgand, O. Laprevote, and B. Séraphin. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23:4847-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldridge, A. G., Y. Li, P. A. Sharp, and B. J. Blencowe. 1999. The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl. Acad. Sci. USA 96:6125-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottschalk, A., G. Neubauer, J. Banroques, M. Mann, R. Lührmann, and P. Fabrizio. 1999. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18:4535-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horowitz, D. S., and A. R. Krainer. 1997. A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 11:139-151. [DOI] [PubMed] [Google Scholar]
- 21.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurica, M. S., D. Sousa, M. J. Moore, and N. Grigorieff. 2004. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat. Struct. Mol. Biol. 11:265-269. [DOI] [PubMed] [Google Scholar]
- 23.Kastner, B., and R. Lührmann. 1989. Electron microscopy of U1 small nuclear ribonucleoprotein particles: shape of the particle and position of the 5′ RNA terminus. EMBO J. 8:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, S. H., and R. J. Lin. 1993. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. USA 90:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laggerbauer, B., S. Liu, E. Makarov, H. P. Vornlocher, O. Makarova, D. Ingelfinger, T. Achsel, and R. Lührmann. 2005. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA 11:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 27.Makarova, O. V., E. M. Makarov, H. Urlaub, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2004. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermoud, J. E., P. Cohen, and A. I. Lamond. 1992. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 20:5263-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, M. J., and P. A. Sharp. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365:364-368. [DOI] [PubMed] [Google Scholar]
- 30.Murray, M. V., R. Kobayashi, and A. R. Krainer. 1999. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 13:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsen, T. W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing, p. 279-307. In R. W. Simons and A. M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Ono, Y., M. Ohno, and Y. Shimura. 1994. Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol. Cell. Biol. 14:7611-7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ségault, V., C. L. Will, B. S. Sproat, and R. Lührmann. 1995. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 14:4010-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 36.Stevens, S. W., and J. Abelson. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. USA 96:7226-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tange, T. O., A. Nott, and M. J. Moore. 2004. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16:279-284. [DOI] [PubMed] [Google Scholar]
- 38.Tange, T. O., T. Shibuya, M. S. Jurica, and M. J. Moore. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11:1869-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. USA 90:10821-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tarn, W. Y., K. R. Lee, and S. C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13:1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Heel, M. 1989. Classification of very large electron microscopical image data sets. Optik 82:114-126. [Google Scholar]
- 42.van Heel, M., and J. Frank. 1981. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6:187-194. [DOI] [PubMed] [Google Scholar]
- 43.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 44.Wang, C., K. Chua, W. Seghezzi, E. Lees, O. Gozani, and R. Reed. 1998. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 12:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, P., P. J. Lou, S. Leu, and P. Ouyang. 2002. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun. 294:448-455. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Q., K. Hobbs, B. Lynn, and B. C. Rymond. 2003. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 278:7875-7883. [DOI] [PubMed] [Google Scholar]
- 47.Wen, X., Y. P. Lei, Y. L. Zhou, C. T. Okamoto, M. L. Snead, and M. L. Paine. 2005. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11). Cell Mol. Life Sci. 62:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Will, C. L., and R. Lührmann. 2006. Spliceosome structure and function, p. 369-400. In R. F. Gesteland, T. R. Cech, and A. J. F. Atkins (ed.), The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Will, C. L., H. Urlaub, T. Achsel, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 21:4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie, J., K. Beickman, E. Otte, and B. C. Rymond. 1998. Progression through the spliceosome cycle requires Prp38p function for U4/U6 snRNA dissociation. EMBO J. 17:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, Y. Z., C. M. Newnham, S. Kameoka, T. Huang, M. M. Konarska, and C. C. Query. 2004. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 23:376-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, Z., and R. Reed. 1998. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 17:2095-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zillmann, M., M. L. Zapp, and S. M. Berget. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]