Abstract
The biogenesis of iron-sulfur (Fe/S) proteins in eukaryotes is a complex process involving more than 20 components. So far, functional investigations have mainly been performed in Saccharomyces cerevisiae. Here, we have analyzed the role of the human cysteine desulfurase Nfs1 (huNfs1), which serves as a sulfur donor in biogenesis. The protein is located predominantly in mitochondria, but small amounts are present in the cytosol/nucleus. huNfs1 was depleted efficiently in HeLa cells by a small interfering RNA (siRNA) approach, resulting in a drastic growth retardation and striking morphological changes of mitochondria. The activities of both mitochondrial and cytosolic Fe/S proteins were strongly impaired, demonstrating that huNfs1 performs an essential function in Fe/S protein biogenesis in human cells. Expression of murine Nfs1 (muNfs1) in huNfs1-depleted cells restored both growth and Fe/S protein activities to wild-type levels, indicating the specificity of the siRNA depletion approach. No complementation of the growth retardation was observed, when muNfs1 was synthesized without its mitochondrial presequence. This extramitochondrial muNfs1 did not support maintenance of Fe/S protein activities, neither in the cytosol nor in mitochondria. In conclusion, our study shows that the essential huNfs1 is required inside mitochondria for efficient maturation of cellular Fe/S proteins. The results have implications for the regulation of iron homeostasis by cytosolic iron regulatory protein 1.
Iron-sulfur (Fe/S) clusters are versatile cofactors of numerous proteins involved in electron transport, enzyme catalysis, and regulation of gene expression. In eukaryotes, Fe/S proteins are localized in the mitochondria, cytosol, and nucleus. The synthesis of Fe/S clusters and their insertion into apoproteins is a surprisingly complex process and involves several multicomponent systems (reviewed by references 6, 32, and 42). Most of our functional knowledge on this process in eukaryotes comes from studies in the yeast Saccharomyces cerevisiae. To date, some 20 yeast components have been identified that execute a direct function in Fe/S protein maturation. Mitochondria perform a central role in biogenesis in that they contain the so-called ISC assembly machinery (30). Most of the 14 known mitochondrial ISC proteins exhibit sequence similarities to bacterial proteins encoded by the isc operon which is crucial for Fe/S cluster assembly in bacteria (7, 22, 55). In yeast the mitochondrial ISC assembly machinery is not only required for the maturation of mitochondrial but also for cytosolic and nuclear Fe/S proteins (13, 25, 27, 29, 34). In addition, maturation of extramitochondrial Fe/S proteins needs the function of the mitochondrial ISC export machinery and of cytosolic factors comprising the cytosolic Fe/S protein assembly (CIA) machinery. Most components of the mitochondrial ISC and cytosolic CIA machineries are conserved in eukaryotes (31, 32). Therefore, one might expect the mechanisms of Fe/S protein maturation in higher eukaryotes to be similar to those defined in yeast. However, only few experimental data are available yet on Fe/S protein assembly in higher eukaryotes. A role of energized mitochondria was suggested by a study on the assembly of IRP1 (8), a cytosolic Fe/S protein with aconitase activity (19, 40, 42). A functional study on a presumed ISC assembly component in higher eukaryotes was performed with human HeLa cells by depleting the mitochondrial ISC protein frataxin with the RNAi technology (46). Low levels of frataxin caused defects in mitochondrial Fe/S proteins as well as in the maturation of IRP1. Most recently, a crucial function in cellular Fe/S protein biogenesis was shown for the scaffold protein huIsu1 in HeLa cells (49), while genetic ablation of the mitochondrial ABC transporter ABCB7 in mouse liver led to a specific defect in cytosolic Fe/S proteins (41). These findings provide a first hint that in human cells mitochondria and the ISC systems also play an important role in the biogenesis of cellular Fe/S proteins.
In human cells, low levels of some mitochondrial ISC assembly proteins have been detected in the cytosol and nucleus, namely huNfs1, huIsu1, huNfu1, and under special conditions, also frataxin (1, 26, 47, 48). The cytosolic forms of huNfs1 or huIsu1 proteins are generated by alternative usage of an internal start codon or by alternative splicing, respectively, from the genes that encode the mitochondrial forms of the proteins. The presence of these ISC proteins in the cytosol and nucleus suggests that they are involved in the generation of Fe/S proteins in these compartments. Recent RNAi depletion studies for huIsu1 showed an important role for the mitochondrial version of the protein, while no effect was observed on the steady-state levels of Fe/S proteins upon depletion of the cytosolic version (49). From regeneration studies after treatment of cells with H2O2 or an iron chelator, it appeared that cytosolic huIsu1 might have an auxiliary role in the repair of Fe/S proteins. In the present study, we chose the highly conserved huNfs1 protein to examine its presumed role in the biogenesis of Fe/S proteins in both the mitochondria and the cytosol of human cells.
Yeast Nfs1 and its bacterial homologs IscS, NifS, and SufS are central components of Fe/S cluster assembly (25, 28, 33, 34, 37-39, 45, 53). In both yeast mitochondria and in bacteria, these proteins function as cysteine desulfurases, thus serving as the sulfur donors for Fe/S cluster synthesis. The pyridoxal phosphate-dependent enzymes initially generate a covalently bound persulfide (23, 54) which then is transferred to the so-called scaffold proteins (Isu1 in eukaryotes and IscU, NifU, or SufU in bacteria) for de novo synthesis of the Fe/S clusters (7, 22, 32). In mitochondria, this step has been shown to involve further ISC assembly proteins like the yeast adrenodoxin homolog Yah1, the yeast frataxin homolog Yfh1, and the recently identified small protein Isd11 which forms a tight complex with Nfs1 (2, 35, 52). Small amounts of yeast Nfs1 are localized in the nucleus where the protein performs an essential function, presumably as a sulfur donor for thiouridine modification of tRNAs (34, 37, 38).
In this work, we analyzed the role of huNfs1 in the biogenesis of mitochondrial and cytosolic Fe/S proteins in a human cell culture model. Using a vector-based RNAi approach, we depleted endogenous huNfs1 in HeLa cells and analyzed the phenotypic effects on cell growth and activity of cellular Fe/S proteins. We also complemented huNfs1-depleted cells with a full-length and a presequence-lacking Nfs1 homolog of mice (muNfs1) to address the question of whether the mitochondrial and/or cytosolic/nuclear isoforms of Nfs1 were required for the maturation of Fe/S proteins in the respective compartments. Our findings suggest that the mitochondrial isoform of huNfs1 is essential for the maturation of Fe/S proteins both inside and outside mitochondria, whereas the cytosolic version of muNfs1 alone did not support maturation of IRP1.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: β-ME, β-mercaptoethanol; CS, citrate synthase; huNFS1-R1, huNFS1-R2, or huNFS1-R3, human NFS1-siRNA construct 1, 2, or 3; IRE, iron-responsive element; IRP, iron regulatory protein; ISC, iron-sulfur cluster; LDH, lactate dehydrogenase; MnSOD, manganese superoxide dismutase; huNfs1, human Nfs1; muNfs1, murine Nfs1; RNAi, RNA interference; SC, synthetic minimal medium; SDH, succinate dehydrogenase; siRNA, small interfering RNA.
siRNA vector design and muNFS1 expression constructs.
Nfs1 was depleted in human HeLa cells by RNAi using a pSuper vector-based approach (9). Three 19-nucleotide-long NFS1 gene-specific targeting sequences corresponding to positions 501 to 520 (GCTGAGGGCTTTCAGGTCAT; huNFS1-R1), 564 to 583 (CTAGAGGCTGCTATCCAGC; huNFS1-R2), and 1040 to 1059 (GCACCATTATCCCGGCTGT; huNFS1-R3) of the coding region were cloned into the vector pSuper as described for the human frataxin-RNAi construct (46). Expression was directed by the H1 promoter.
The cDNA of murine full-length NFS1 (muNFS1) (39) was subcloned into the pEGFP-N3-derived mammalian expression vector pMCS-HA (kindly provided by G. Suske, IMT, Marburg, Germany) using the EcoRI restriction site. The ΔN48-muNFS1 cDNA encoding muNfs1 lacking its N terminus including the mitochondrial presequence was amplified from the pMCS-HA/muNFS1 using as forward primer 5′-TTACGCGTATGGTTCACTCAGAGGCAGAGGCA-3′ and as reverse primer 5′-ATACGCGTCCGCGGCGCTTGCAATGCCT-3′. After incubation with the MluI restriction enzyme (NEB), the PCR product was religated and transformed.
Targeting of N-terminally truncated ΔN48-muNfs1 into mitochondria was achieved by attaching the presequence of the Neurospora crassa beta subunit of F1-ATPase. For synthesis of this fusion protein (termed F1β-ΔN48-muNfs1), the corresponding cDNA (StuI/XhoI restriction fragment of ΔN48-muNFS1) was amplified by PCR and inserted into the HpaI/XhoI site of pBluescript II KS+ containing the F1-β presequence cDNA. The F1β-ΔN48-muNFS1 construct was cloned into the pMCS-HA vector via HindIII/XhoI and HindIII/SalI restriction sites, respectively, and expressed in HeLa cells. For synthesis of muNfs1 proteins in yeast cells, ΔN48-muNFS1 (as a StuI/XhoI restriction fragment) was cloned into the EcoRV/XhoI restriction sites of pBluescript II KS+ (Stratagene). Both ΔN48-muNFS1 and F1β-ΔN48-muNFS1 constructs were cloned from the respective pBluescript II KS+ vectors into the yeast expression vector p426TDH3 via EcoRI/XhoI restriction sites to yield p426-ΔN48-muNFS1 and p426-F1β-ΔN48-muNFS1.
Cell culture, transfection, and cell fractionation.
Human cervix carcinoma cells (HeLa) were cultured in Dulbecco's modified Eagle medium supplemented with 7.5% fetal calf serum, 1 mM glutamine, and 50 μg/ml gentamicin. Cells were grown on an area of 25 cm2 (6 × 106 cells), harvested by trypsination, washed twice with transfection buffer (21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, and 6 mM dextrose), resuspended in 500 μl of transfection buffer, and supplemented with 10 μg of plasmid. Transfections were carried out by electroporation (250 V, 1500 μF, 25- to 30-ms duration) using an EASYJect+ device (46). Cells were immediately cultured in complete Dulbecco's modified Eagle medium and retransfected every third or fourth day in order to prolong the time of huNfs1 depletion and promote muNfs1 expression. In the experiments shown in Fig. 5 and 6, the efficiency of huNfs1 depletion was further optimized by transfection with 30 μg of plasmid and a second transfection after 3 days. This optimized procedure allowed a faster detection of the effects of huNfs1 depletion. Transfected cells were harvested by trypsination and washed twice with phosphate-buffered saline, and cell pellets were shock-frozen in liquid nitrogen and stored at −80°C.
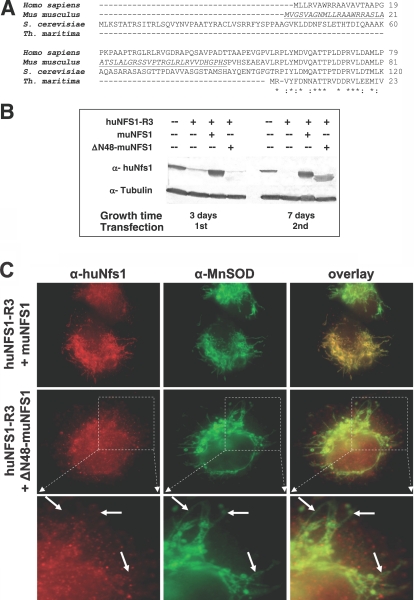
FIG. 5.
Expression and localization of Δ48-muNfs1 lacking a mitochondrial presequence. (A) Sequence alignment of the N-terminal regions of Nfs1-like proteins from the indicated species. The segment deleted in ΔN48-muNfs1 is underlined. Asterisks indicate conserved amino acid residues within the N-terminal part of Nfs1-like proteins including the NifS-like protein of the bacterium Thermotoga maritima (23) which lacks the typical eukaryotic mitochondrial targeting sequence. (B) HeLa cells were cotransfected as indicated with the RNAi vector huNFS1-R3, an expression vector encoding full-length muNfs1 (muNFS1) or N-terminally truncated muNfs1 (ΔN48-muNFS1), or the corresponding empty vectors. After 3 or 7 days, cells were analyzed by Western blotting of cell lysates for Nfs1 and tubulin (see Fig. 4A). (C) Immunofluorescence detection of muNfs1 and MnSOD 7 days after the first transfection. Arrows indicate the absence (left column) or presence (middle column) of mitochondrial staining. The lower panel shows cellular sections magnified from the panel in the middle. In contrast to the full-length muNfs1, ΔN48-muNfs1 does not colocalize with mitochondrial MnSOD. (D) HeLa cells cotransfected with the vectors as indicated were harvested 3 or 7 days after the first transfection, and their protein content was determined as a measure for cell growth (mean + standard deviation; n = 4). (E) Mitochondria-localized ΔN48-muNfs1 can restore growth of huNfs1-depleted HeLa cells. To verify the functionality of ΔN48-muNfs1, HeLa cells were cotransfected as indicated with the RNAi vector huNFS1-R3 and with an expression vector containing the coding information of mitochondria-targeted ΔN48-muNfs1 (F1β-ΔN48-muNFS1), or the corresponding empty vectors. Cells were harvested 3 or 7 days after the first transfection, and their protein content was determined as a measure of cell growth. One representative experiment is shown. (F) Mitochondria-localized ΔN48-muNfs1 can restore growth of huNfs1p-depleted yeast cells. To verify the enzymatic functionality of ΔN48-muNfs1, yeast Gal-NFS1 cells were transformed with expression vectors containing the coding information of ΔN48-muNfs1 either with (p426-F1β-ΔN48-muNFS1) or without (p426-ΔN48-muNFS1) an additional mitochondrial presequence, or the vector alone (p426TDH). To deplete endogenous yeast Nfs1p, cells were first cultured on SC supplemented with dextrose and further cultured on SC supplemented with glycerol at 30°C.
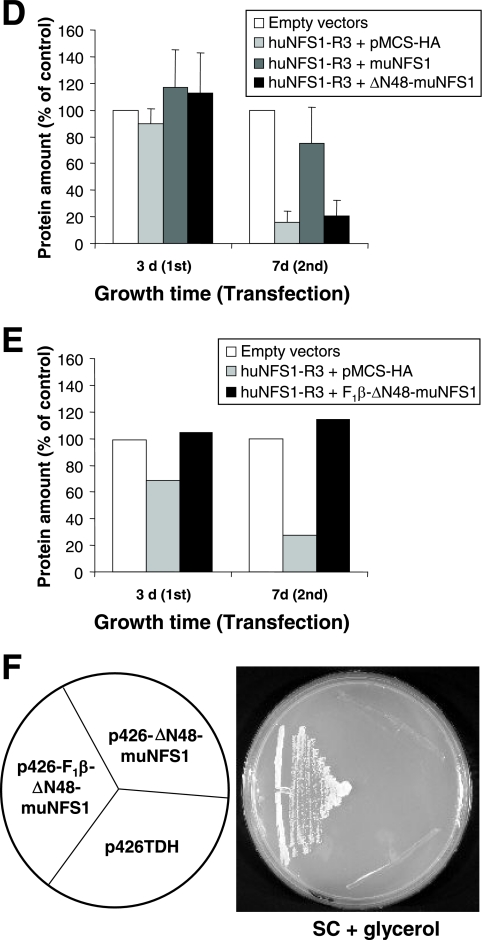
FIG.6.
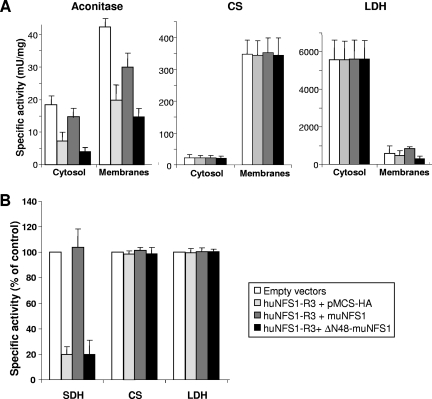
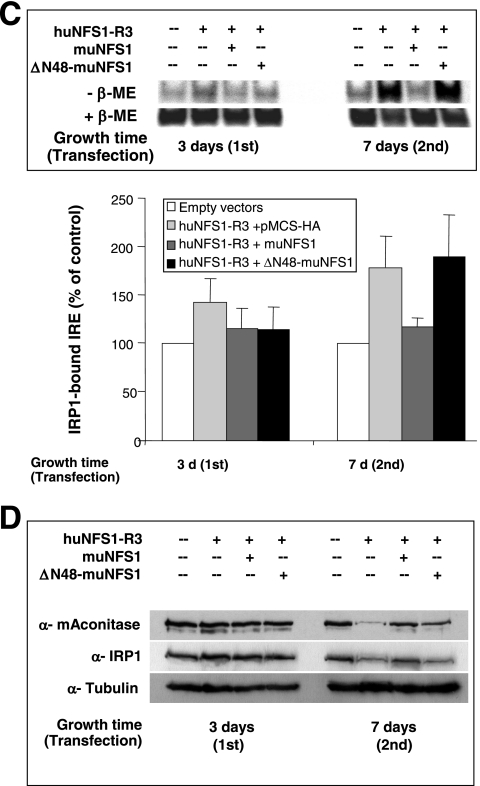
Extramitochondrial ΔN48-muNfs1 cannot reverse the defects in cytosolic and mitochondrial Fe/S proteins. (A) HeLa cells were transfected as described in the legend for Fig. 5 and harvested 7 days after the first transfection. Soluble cytosolic and membrane fractions were prepared and used to measure the specific activities (mean + standard deviation; n = 5) of aconitase and two non-Fe/S cluster-containing enzymes (CS and LDH). Aconitase activity is composed of mitochondrial aconitase and cytosolic IRP1. (B) Whole-cell lysates were used to determine the specific activities of the mitochondrial Fe/S cluster-containing enzyme SDH and of CS and LDH. Data were normalized to the control vector-transfected cells (mean + standard deviation; n = 3). (C) Transfected cells were harvested 3 or 7 days after the first transfection and analyzed for total binding activity of IRP1 to 32P-labeled IRE of human ferritin mRNA. After native gel electrophoresis, binding of IREs to IRP1 was visualized by autoradiography in the absence (−β-ME) or presence (+β-ME) of β-mercaptoethanol (upper panel) and quantified using a phosphorimager. The amounts of IRP1-bound IRE in the various transfected cells were normalized to the maximal binding capacity of β-ME-treated samples and expressed relative to the amount of IRP1-bound IRE in control vector-transfected cells (mean + standard deviation; n = 3) (lower panel). (D) Cell lysates from a representative experiment were analyzed by immunostaining for mitochondrial aconitase (mAconitase), cytosolic IRP1, and tubulin (see also Fig. 4A).
For fractionation of soluble and membrane-associated proteins, freshly harvested cells were incubated for 8 min at 4°C in 5 mM Tris/HCl, pH 7.4, 250 mM sucrose, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 3 mM phenylmethylsulfonyl fluoride, and 0.007% digitonin (11). Lysates were centrifuged at 1,800 × g and 4°C, and the mitochondrion-containing pellet was washed once at 15,000 × g with the above buffer lacking digitonin and used as the “membrane” fraction. The supernatant of the lysate corresponding to soluble cytosolic proteins was subjected to another centrifugation step at 15,000 × g and 4°C to remove residual membranes and was designated the “cytosol.” Fractions were shock-frozen in liquid nitrogen and stored at −80°C.
Yeast strains and cell growth.
The following strains of S. cerevisiae were used: the promoter exchange mutant Gal-NFS1 (35) and the corresponding wild-type strain W303-1A (MATa ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112). Cells were grown in rich (yeast extract-peptone) medium or SC containing galactose, dextrose, or glycerol as carbon sources. Yeast cells were transformed with plasmid DNA by the lithium acetate method.
Immunostaining and electron microscopy.
Transfected HeLa cells were grown on coverslips, washed with phosphate-buffered saline and fixed with 4% paraformaldehyde. After permeabilization with Triton X-100 and blocking with bovine serum albumin and goat serum (DAKO, Germany), huNfs1 and muNfs1 proteins were labeled with a polyclonal rabbit antibody raised against a C-terminal peptide of huNfs1 and were detected by indirect immunofluorescence using a tetramethyl rhodamine isothiocyanate-labeled goat anti-rabbit secondary antibody (Dianova, Germany). MnSOD was detected using a monoclonal anti-human MnSOD antibody (Alexis Biochemicals) and an Alexis Fluor-488 coupled goat anti-mouse secondary antibody (Molecular Probes). For reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequent electroblotting, 50 to 150 μg of HeLa cell protein was applied per lane. huNfs1 was immunostained by rabbit anti-Nfs1 antiserum, and α-tubulin was detected using a mouse anti-α-tubulin monoclonal antibody (DM1α; Sigma). Visualization of antigens was performed by peroxidase-conjugated secondary antibodies in combination with 3′,3′,5′,5′-tetramethylbenzidine substrate solution (Seramun Diagnostica GmbH, Dolgenbrodt, Germany). Transmission electron microscopy was performed as described previously (12).
Enzyme assays.
Aconitase activity was determined by a coupled aconitase/isocitrate dehydrogenase assay (11). SDH activity was assessed by the 2,6-dicholorophenol-indophenol assay in combination with decyl ubiquinone (46).
IRE binding of IRP1.
The RNA-binding capacity of IRP1 was determined by RNA electrophoretic mobility shift assay, essentially as described previously (36). In brief, [α-32P]CTP-labeled IREs of human ferritin mRNA were incubated with HeLa cell lysates, and unbound RNA was removed by digestion with RNase T1. α-IRP2 antiserum (Alpha Diagnostic, Texas) was added to the samples for 45 min at room temperature to achieve the “supershift” separation of IRP2-bound IREs from IRP1-bound IREs by native polyacrylamide gel electrophoresis (15). IRP1 binding to ferritin IRE was determined by autoradiography and quantified using a phosphorimager. Control samples were pretreated with 2% β-mercaptoethanol in order to disassemble the Fe/S cluster of IRP1 to achieve maximum IRE binding.
RESULTS
Depletion of huNfs1 by RNAi leads to impaired cell growth.
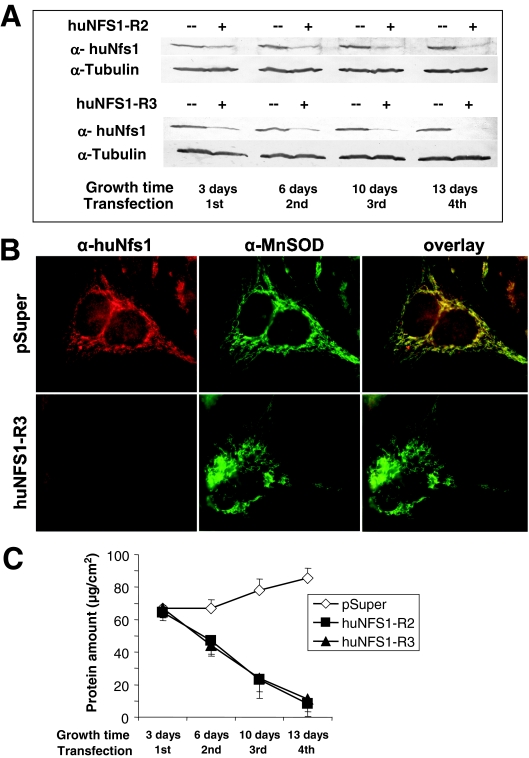
In order to deplete huNfs1 in cultured HeLa cells, we cloned three different NFS1 mRNA-specific sequences into the siRNA vector pSuper (3, 9). The resulting plasmids termed huNFS1-R1, huNFS1-R2, and huNFS1-R3 or the pSuper vector was transfected into HeLa cells, and cells were allowed to grow. The desired decrease of the cellular amount of huNfs1 was verified by both immunoblot analysis and immunofluorescence microscopy using a polyclonal anti-huNfs1 antiserum that was raised against a C-terminal peptide of huNfs1. Western blotting of cell lysates obtained 3 days after transfection revealed a decreased level of huNfs1 for cells carrying the siRNA constructs huNFS1-R2 and huNFS1-R3 (Fig. 1A). A further decrease of the huNfs1 levels was achieved by retransfection of the cells with the same vectors and continued cell growth (46). After the forth transfection round huNfs1 almost completely disappeared, while the amount of tubulin as a control did not change significantly. The efficiency of the depletion of huNfs1 was also followed by fluorescence microscopy. huNfs1 showed a perfect colocalization with the mitochondrial matrix protein MnSOD (Fig. 1B, upper panels). Furthermore, the faint staining in the nuclear area may indicate the dual subcellular localization of huNfs1 (26). In cells transfected with huNFS1-R2 (not shown) or huNFS1-R3 (Fig. 1B, lower panels), both the mitochondrial and the nuclear staining disappeared almost completely, while the labeling of MnSOD identified the elongated mitochondrial network even after 12 days of growth under depletion conditions. In lysates of cells transfected with plasmid huNFS1-R1 we detected only a slight decrease of the huNfs1 protein levels, even after repeated transfections (not shown). Due to the weak effect, the huNFS1-R1 plasmid was not used in further experiments.
FIG. 1.
Depletion of huNfs1 by RNAi leads to impaired cell growth. HeLa cells were transfected by electroporation with RNAi vectors pSuper, huNFS1-R2, or huNFS1-R3. After the time points indicated cells were harvested, and a fraction was retransfected. (A) Depletion of huNfs1 was analyzed by Western blotting of cell lysates using antibodies against huNfs1 and tubulin as a control. (B) Immunofluorescence detection of huNfs1 and mitochondrial matrix protein MnSOD in HeLa cells 12 days after first transfection. (C) At each time point the protein content of the cells was determined as a measure of cell growth (mean ± standard deviation; n = 4).
The strong depletion of huNfs1 in cells transfected with plasmids huNFS1-R2 and huNFS1-R3 was accompanied by a strongly impaired cell growth (Fig. 1C). huNfs1-deficient cells did not become confluent as fast as the control vector-transfected cells, and the total protein content (used as a measure of cell growth) was diminished. Significant growth retardation was evident after the second transfection. Thirteen days after the first transfection the total cell protein amount had dropped to 12% of the control cells, indicating that huNfs1 is essential for normal growth of HeLa cells. Our data show that huNfs1 plays an essential role in the human cell.
Alteration of the mitochondrial inner membrane structure after huNfs1 depletion.
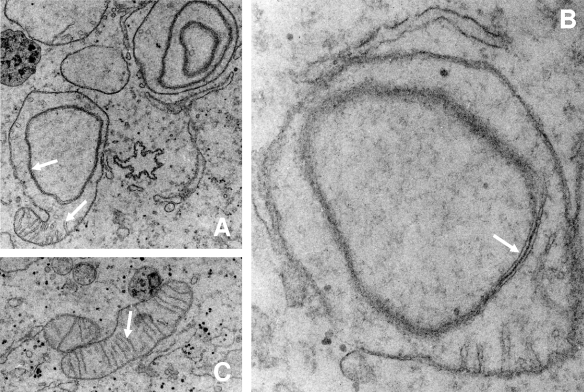
We next investigated the ultrastructural consequences of the huNfs1 depletion by electron microscopy. Significant alterations were observed for the morphology of mitochondria but not for other compartments of the cell. Different types of mitochondrial structures were detected in huNfs1-depleted cells (Fig. 2). A set of mitochondria appeared as elongated structures with normal cristae membranes similar to those seen in control vector-transfected HeLa cells. Thus, these organelles are similar to wild-type structures (Fig. 2C). Another set showed up as spherical mitochondria with a conspicuously altered inner membrane structure. They almost entirely lacked cristae membranes and exhibited onion-shaped inner membrane structures that were not detectable in control cells (Fig. 2). The characteristic onion-shaped structure depicted in Fig. 2 appears to be morphologically similar to altered mitochondria in the fuzzy onion mutant of Drosophila spermatids (16). In some cases, both wild-type and mutant morphologies were observed in a single organelle (Fig. 2A). Mitochondria with altered structures were evident after the second transfection and increased in number, correlating well with the time course of huNfs1 depletion (data not shown). A loss of cristae membranes has been described for human cell cultures that are devoid of mitochondrial DNA (rho0 cells) and thus are respiration deficient (14, 20). However, DAPI (4′,6′-diamidino-2-phenylindole) staining revealed the presence of mitochondrial DNA in huNfs1-depleted HeLa cells (not shown), indicating that these cells did not lose their mitochondrial DNA.
FIG. 2.
Alteration of mitochondrial inner membrane structure after huNfs1 depletion. HeLa cells were repeatedly transfected with huNFS1-R3 (A, B) or pSuper (C) as described in the legend for Fig. 1 and analyzed by electron microscopy (total growth time, 10 days). (A) Some regions of huNFS1-depleted mitochondria still contained cristae (lower arrow), while other regions had an altered mitochondrial inner membrane structure (upper arrow). (B) huNFS1-depleted mitochondria with altered mitochondrial inner membrane structure (arrow) in higher magnification than the one in panel A. (C) Mitochondrion of a pSuper-treated control cell with intact cristae (arrow). Magnifications: panels A and C, ×7,000; panel B, ×20,000.
Defects in mitochondrial Fe/S protein activities upon huNfs1 depletion.
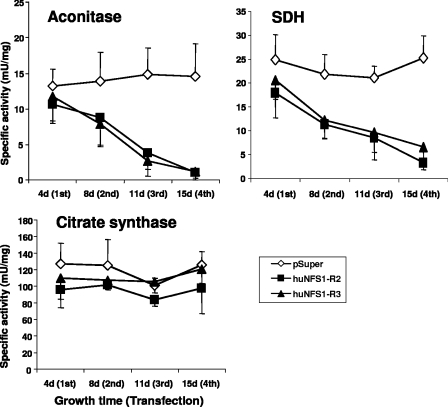
To assess the importance of huNfs1 in human HeLa cells for the function of Fe/S proteins, we measured the activities of the Fe/S cluster-containing enzymes aconitase and SDH in cell lysates. Upon depletion of huNfs1 by RNAi, the activities of aconitase and SDH decreased strongly, and after four rounds of transfection, the enzyme activities had dropped to 14% and 20%, respectively, of the values determined in control cells (Fig. 3). In contrast, the enzyme activities measured in control vector-transfected cells remained constant around 15 and 25 mU/mg, respectively. The non-Fe/S cluster-containing mitochondrial matrix enzyme citrate synthase was not affected in its activity by the huNfs1 depletion, indicating the specificity of the effect on Fe/S proteins. In conclusion, a deficiency in functional huNfs1 results in specific defects in mitochondrial Fe/S protein activities in human cells.
FIG. 3.
Defects in mitochondrial Fe/S proteins upon huNfs1 depletion. HeLa cells were repeatedly transfected with RNAi vectors pSuper, huNFS1-R2, or huNFS1-R3 and harvested at the time points indicated. Specific activities of mitochondrial Fe/S proteins aconitase and SDH and of the non-Fe/S cluster-containing enzyme citrate synthase were measured in whole cell lysates (mean ± standard deviation; n = 3).
Expression of muNfs1 in huNfs1-depleted HeLa cells reversed cell growth impairment and defects in mitochondrial Fe/S proteins.
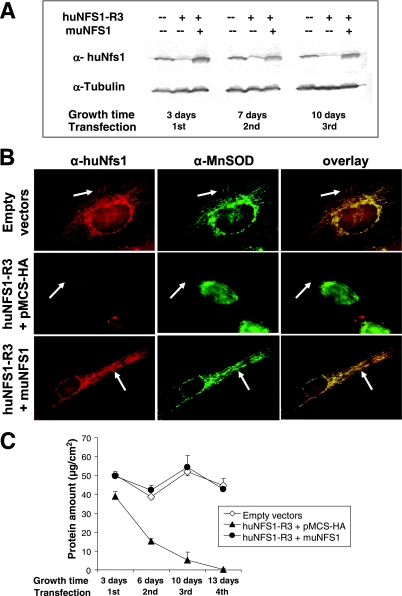
The murine and human Nfs1 protein sequences are highly similar (26, 39), yet the respective DNA sequences differ in the region corresponding to the siRNA sequence selected for huNfs1 depletion by vector huNFS-R3. This allowed us to test whether muNfs1 can functionally replace huNfs1. To this end, we cloned the full-length muNFS1 cDNA into the mammalian expression vector pMCS-HA and cotransfected HeLa cells repeatedly with the resulting plasmid (designated muNFS1) and with huNFS1-R3. After cell growth, the synthesis of muNfs1 and the decrease of huNfs1 were analyzed by Western blotting of cell lysates (Fig. 4A). The C-terminal sequence of muNfs1 is identical to that of huNfs1, allowing detection of muNfs1 by the α-huNfs1 antiserum. The expressed muNfs1 had a molecular mass of 45 kDa, similar to that of huNfs1. The murine protein was already detectable from its overproduction 3 days after the first transfection and remained constant in its concentration at the later time points of the experiment (Fig. 4A). To investigate the subcellular localization of muNfs1 by immunofluorescence microscopy, transfected cells were coimmunostained with α-huNfs1 antiserum and α-human MnSOD antibody (Fig. 4B). Since 10 days after the first transfection round the huNfs1 was nearly completely depleted (Fig. 4A), the antibody almost exclusively detected muNfs1. The latter protein colocalized perfectly with MnSOD, which labeled the mitochondrial network. In addition, muNfs1-containing cells exhibited a faint staining of the nuclear area reminiscent of the localization of huNfs1 in control vector-transfected cells. We conclude that muNfs1 exhibits the same subcellular distribution as huNfs1.
FIG. 4.
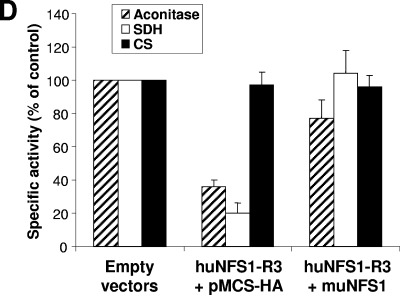
Expression of muNfs1 in huNfs1-depleted HeLa cells restores cell growth and mitochondrial Fe/S protein activities. (A) HeLa cells were cotransfected with an expression vector containing full-length murine NFS1 cDNA (muNFS1) and the RNAi vector huNFS1-R3, or the corresponding empty vectors. After the time points indicated cells were harvested, and a fraction was retransfected. Expression of muNfs1 and depletion of huNfs1 were analyzed by Western blotting of cell lysates. Blots were probed for Nfs1 and tubulin. The antiserum against huNfs1 also recognizes muNfs1. (B) Immunofluorescence detection of Nfs1 and mitochondrial matrix protein MnSOD 10 days after the first transfection. Arrows indicate areas of mitochondrial staining. (C) Cellular protein content was determined at each harvest (mean ± standard deviation; n = 4). (D) The specific activities of mitochondrial Fe/S proteins aconitase and SDH and of the non-Fe/S cluster-containing enzyme CS were determined in lysates of cells harvested 7 days after the first transfection (mean + standard deviation; n = 6).
A crucial aspect of our investigation was whether synthesis of muNfs1 in huNfs1-depleted HeLa cells was able to complement their growth defect. After repeated transfections and cell cultivation, the protein content was determined as a measure of cell growth. While huNfs1-depleted cells showed hardly any residual growth after the fourth round of transfection, the huNfs1-depleted cells synthesizing muNfs1 grew as fast as control cells, indicating the functional replacement of the depleted human protein by muNfs1 (Fig. 4C). We further measured the enzyme activities of SDH, aconitase, and citrate synthase to determine to what extent the Fe/S protein activities were normalized by functional muNfs1. Synthesis of muNfs1 in huNfs1-depleted cells could fully restore SDH activity and about 80% of the aconitase activities of control cells (Fig. 4D). As expected, the non-Fe/S cluster-containing enzyme citrate synthase was unchanged under all conditions. Together, the functional replacement of huNfs1 by its murine relative demonstrates that the proteins perform an orthologous function in Fe/S protein biogenesis. Importantly, these findings document the specificity of our RNAi approach.
Only mitochondrion-localized muNfs1 can restore growth of huNfs1-deficient cells.
The possibility of depleting huNfs1 in human cell cultures by the RNAi technology enabled us to investigate in which compartment the Nfs1 protein is required for functional participation in mammalian Fe/S protein biogenesis. For this purpose, we created an expression plasmid (designated ΔN48-muNFS1, based on vector pMCS-HA) that encodes a mutant version of muNfs1 lacking part of the nonconserved N terminus (underlined in the Nfs1 multisequence alignment shown in Fig. 5A), which includes the putative mitochondrial targeting sequence. Synthesis of ΔN48-muNfs1 in huNfs1-depleted HeLa cells was analyzed by Western blotting of cell lysates. The higher mobility of truncated ΔN48-muNfs1 compared to full-length muNfs1 indicated that presequence cleavage to the mature form of muNfs1 occurs in front of the chosen truncation site (Fig. 5B). Efficient synthesis of ΔN48-muNfs1 comparable to the levels of full-length muNfs1 was achieved after two rounds of transfection of the HeLa cells. Tubulin stained as a control for equal protein loading remained unchanged. To analyze the subcellular localization of ΔN48-muNfs1, transfected HeLa cells were coimmunostained with both the Nfs1-specific antiserum and the anti-MnSOD antibody and then analyzed by fluorescence microscopy. Colocalization of ΔN48-muNfs1 with the MnSOD-labeled elongated mitochondrial network was not detectable (Fig. 5C). Instead, the N-terminally truncated ΔN48-muNfs1 was observed mainly in the nuclear area and in the cytosol of huNfs1-depleted cells, while full-length muNfs1 was predominantly located in mitochondria.
We then examined whether the nuclear/cytosolic ΔN48-muNfs1 could restore growth of the huNfs1-deficient cells. HeLa cells were cotransfected with both the ΔN48-muNFS1 plasmid and with huNFS1-R3 in order to deplete the endogenous huNfs1. For comparison, cells were cotransfected in parallel either with the empty vectors, with the huNFS1-R3 plasmid and pMCS-HA expression vector, or with huNFS1-R3 and pMCS-HA encoding full-length muNfs1 (Fig. 5D). In huNfs1-depleted cells, the total protein content as a measure for cell growth decreased to 15% of control cells 7 days after the first transfection. Cells producing the truncated ΔN48-muNfs1 protein did not show any improvement of growth compared to huNfs1-depleted cells. However, the cells producing full-length muNfs1 reached almost wild-type levels of growth. The failure to restore growth of huNfs1-depleted cells by synthesis of ΔN48-muNfs1 shows that huNfs1 performs an essential function inside mitochondria. This function apparently cannot be replaced by an extramitochondrial version of muNfs1.
The functionality of the truncated ΔN48-muNfs1 protein was verified both in HeLa cells and in yeast. The coding sequence of ΔN48-muNfs1 was cloned into a mammalian expression vector with a mitochondrial targeting sequence (residues 1 to 40 of the F1β-subunit from N. crassa), and the vector (designated F1β-ΔN48-muNFS1, based on vector pMCS-HA) was cotransfected with huNFS1-R3 into HeLa cells in order to deplete the endogenous huNfs1 (Fig. 5E). For comparison, cells were cotransfected in parallel either with the empty vectors or with the huNFS1-R3 plasmid and pMCS-HA expression vector. The mitochondrion-targeted F1β-ΔN48-muNfs1 restored growth of huNfs1-depleted cells to wild-type levels, indicating that ΔN48-muNfs1 is an active protein.
The function of the truncated ΔN48-muNfs1 protein was also analyzed in yeast. We used the conditional strain Gal-NFS1 that carries the yeast NFS1 gene under the control of the regulatable GAL1-10 promoter that is turned off in the absence of galactose (34). Gal-NFS1 cells growing on nonfermentable carbon sources such as glycerol produce strongly diminished amounts of Nfs1 and thus show a marked growth defect. The coding sequences of ΔN48-muNFS1 and F1β-ΔN48-muNFS1 were cloned into yeast expression vectors to yield plasmids p426-ΔN48-muNFS1 and p426-F1β-ΔN48-muNFS1. Yeast Gal-NFS1 cells were transformed with either of these plasmids or with the vector alone (p426TDH3) and cultured on synthetic minimal medium supplemented with glycerol. Cells harboring the mitochondrion-targeted version of ΔN48-muNfs1 showed wild-type growth under these conditions (Fig. 5F), indicating that ΔN48-muNfs1 could replace yeast Nfs1 as a cysteine desulfurase and therefore is an active protein. The findings further demonstrate that mammalian muNfs1 is orthologous to yeast Nfs1. The presequence-lacking ΔN48-muNfs1, however, did not support growth, confirming earlier results that in yeast the Nfs1 protein is required inside mitochondria to be functional in supporting cell growth and Fe/S protein biogenesis (25, 34, 37). Collectively, our results show that in human cells a mitochondrion-localized version of Nfs1 is essential for efficient cell growth.
ΔN48-muNfs1 cannot reverse defects in cytosolic or mitochondrial Fe/S proteins.
We next asked whether huNfs1 and in particular the extramitochondrial version of this protein is involved in cytosolic Fe/S protein biogenesis in human cells. As a cytosolic Fe/S marker protein we used IRP1, which possesses a dual function. As an Fe/S protein it functions as an aconitase. Dissociation of the Fe/S cluster enables binding to specific mRNA stem-loop structures called IREs, thereby regulating the expression of corresponding genes on a posttranscriptional level. To distinguish cytosolic aconitase activity of IRP1 from that of mitochondrial aconitase, the HeLa cell plasma membrane was opened with digitonin, and soluble proteins (containing IRP1) were separated from membrane fractions (containing mitochondria) by centrifugation. To evaluate the efficiency of this fractionation procedure, we measured the specific enzyme activities of cytosolic LDH and mitochondrial CS in both fractions. Less than 10% of cytosolic LDH activity was present in the membrane fraction, and even less mitochondrial CS activity was measured in the cytosol fraction, indicating a highly efficient separation procedure (Fig. 6A). The specific aconitase activity of the membrane fraction was twofold higher than that of the soluble fraction.
Upon depletion of huNfs1 by the RNAi vector huNFS1-R3, the specific aconitase activity of the cytosol decreased by 60% compared to control vector-transfected cells, demonstrating that huNfs1 was also required for IRP1 maturation in the cytosol (Fig. 6A). These data suggest that huNfs1, the only cysteine desulfurase encoded by human cells, is also responsible for maturation of cytosolic Fe/S proteins. We next tested whether the extramitochondrial version of huNfs1 is involved in cytosolic Fe/S protein biogenesis in human cells. No restoration of cytosolic aconitase function was observed upon synthesis of presequence-lacking ΔN48-muNfs1 in huNfs1-depleted cells, while in cells producing mitochondrion-targeted muNfs1 the activity was recovered up to 85% of that of control cells. Similar results were obtained for mitochondrial aconitase activity which was diminished by 60% (Fig. 6A) and for the activity of mitochondrial SDH which was decreased by 80% in total cell lysates (Fig. 6B). In contrast, no differences in the enzyme activities of the non-Fe/S cluster-containing enzymes CS and LDH could be observed in the various transfected cells (Fig. 6A and B). We noted that upon functional inactivation of huNfs1 both IRP1 and aconitase protein levels were decreased after longer depletion times (Fig. 6D). This result indicates that the apoforms of these proteins are prone to degradation, especially after longer times of huNfs1 depletion. Similar findings were made in yeast upon depletion of ISC and CIA proteins (4). Taken together, these results unequivocally suggest that assembly of the Fe/S cluster on IRP1 in the cytosol requires the activity of a mitochondrion-localized Nfs1.
We finally employed the IRE-binding capacity of IRP1 as an alternative assay for the presence of an Fe/S cluster on IRP1 in the cytosol. The RNA-binding activity of IRP1 increases under conditions of impaired Fe/S cluster assembly and can be analyzed by an RNA electrophoretic mobility shift assay using radiolabeled RNA containing an IRE. In order to distinguish the IRE-binding capacity of IRP1 from that of the non-Fe/S cluster-containing IRP2, we performed a supershift assay using an anti-IRP2 antibody (15). Therefore, the amount of IRP1 bound to an [α-32P]CTP-labeled IRE of ferritin mRNA reflected the amounts of Fe/S cluster-free IRP1 in the various transfected cells. IRE binding of IRP1 was strongly increased upon depletion of huNfs1 (Fig. 6C, upper panel). Mitochondrion-targeted muNfs1, but not the truncated version ΔN48-muNfs1, yielded an IRE-binding capacity of IRP1 similar to that found in control vector-transfected cells. Quantitation of the data by phosphorimager analysis revealed a 1.8-fold increase of IRE binding in cells lacking a mitochondrion-localized version of Nfs1 after 7 days of huNfs1 depletion (Fig. 6C, lower panel). These data fully support the findings made above for the cytosolic aconitase activities and suggest that the extramitochondrial version of Nfs1 does not suffice to assemble cytosolic Fe/S proteins. We conclude that the mitochondrion-localized form of Nfs1 is indispensable for efficient maturation of cytosolic IRP1 to the Fe/S cluster-containing aconitase.
DISCUSSION
In the present study, we depleted the NifS-like cysteine desulfurase huNfs1 in human HeLa cells by employing the RNAi technique in order to investigate the function of the protein, in particular its potential involvement in the maintenance of mitochondrial and cytosolic Fe/S proteins. NifS-like proteins are highly conserved in evolution and represent central components of the ISC assembly machinery in prokaryotes and in yeast (22, 24, 32). A strong decrease of the cellular huNfs1 content was achieved by repeated transfections with two specific siRNA vectors. This was accompanied by a dramatic lowering of the growth rates, indicating that huNfs1 performs a crucial function in human cells. Upon depletion of huNfs1, the enzyme activities of the mitochondrial Fe/S proteins aconitase and SDH decreased up to 10-fold, while non-Fe/S control enzymes remained unchanged. Both the growth and mitochondrial Fe/S enzyme defects could be complemented by synthesizing the corresponding muNfs1 protein from mice in these cells. These findings indicate the specificity of our RNAi approach and demonstrate the essential character of huNfs1 for mitochondrial Fe/S protein biogenesis in human cells.
Depletion of huNfs1 not only decreased the activities of the mitochondrial Fe/S proteins but also affected the aconitase function and IRE-binding capabilities of the cytosolic Fe/S protein IRP1. After 7 days of huNfs1 depletion, a 2.5-fold loss of the aconitase function of IRP1 was observed. This was accompanied by a nearly twofold increased RNA-binding activity of this protein, clearly indicating a decrease in bound Fe/S cluster. Apparently, huNfs1 is also required for the maturation of extramitochondrial proteins, suggesting a potential function in the biogenesis of all cellular Fe/S proteins. Our immunofluorescence analyses are compatible with a subcellular localization of both the huNfs1 and the heterologously expressed muNfs1 predominantly in mitochondria (39) with minor amounts in the nuclei of human HeLa cells (26). Several other members of the human mitochondrial ISC assembly machinery, namely, huIsu1, huNfu1, and frataxin, were detected in small amounts in the human cytosol and nucleus (1, 47, 48). The different isoforms of huNfs1, huIsu1, and huNfu1 were proposed to be generated by alternative translation initiation or alternative splicing of a common (pre-)mRNA. The human ISC assembly proteins are synthesized either with or without the mitochondrial presequence, and hence are targeted to either mitochondria or the cytosol/nucleus, respectively. Despite the small amounts of the ISC assembly proteins found outside mitochondria, these findings led to the suggestion that in human cells the mitochondrial ISC machinery may be duplicated in the cytosol/nucleus and function there in the maturation of extramitochondrial Fe/S proteins (42).
Our cell culture model allowed us to test whether the mitochondrial or cytosolic/nuclear isoforms of huNfs1 are required for the maturation of cytosolic IRP1. For this purpose, a truncated form of the orthologous muNfs1 (termed Δ 48-muNfs1) lacking its N-terminal mitochondrial targeting sequence was synthesized in huNfs1-depleted cells. This murine protein covered the entire portion of the cytosolic/nuclear version of huNfs1 (26) and was shown to be fully active, since a mitochondrion-targeted version complemented the growth defects of Nfs1 depletion in both human and yeast cells (25, 34, 37). Extramitochondrial ΔN48-muNfs1 was unable to restore the functional defect of IRP1 as a cytosolic aconitase. Likewise, the defect in Fe/S cluster assembly on IRP1 was evident from the increased binding of IRP1 to the IRE stem-loop structure of ferritin mRNA. Clearly, an extramitochondrial copy of Nfs1 does not suffice to assemble cytosolic Fe/S proteins. Rather, the mitochondrial version of Nfs1 is necessary for efficient Fe/S protein biogenesis in the cytosol. Similar findings were made recently for RNAi depletion of huIsu1 (49). While our findings clearly demonstrate the requirement of the mitochondrial isoform of Nfs1 in cytosolic Fe/S protein assembly, they do not exclude an additional function of its cytosolic/nuclear version. In fact, the cytosolic form of huIsu1 was shown to be required but not essential for regeneration/repair of IRP1 after Fe/S cluster damage by H2O2 or iron chelator treatment (49). It will be interesting to learn more about this process.
The findings made here for human cells are strikingly similar to the situation reported for yeast (25, 34, 37, 38). Nfs1 and Isu1/Isu2 were shown to be required inside yeast mitochondria for efficient Fe/S protein biogenesis in the mitochondria, cytosol, and nucleus (6, 13, 32). The extramitochondrial location of Nfs1 in yeast may be explained by its essential function in thiouridine modification of tRNAs (34, 38), but direct evidence for this idea is still lacking. This presumed mechanistic involvement of Nfs1 in tRNA nucleotide modification may be conserved in mammalian cells. At present, however, little is known about the mechanisms and additional factors involved in thio-modifications of tRNA in eukaryotes, in particular in mammalian cells (50).
Over the past few years several cytosolic/nuclear components have been identified in yeast with a function in Fe/S protein maturation. Known members of the so-called CIA machinery are the P-loop NTPase Cfd1, the related protein Nbp35, the iron-only hydrogenase-like Nar1, and the WD40 repeat protein Cia1 (4, 5, 18, 43). Depletion of these CIA components in yeast results in the loss of Fe/S clusters on cytosolic and nuclear Fe/S proteins, while mitochondrial Fe/S proteins are not affected in their assembly. Thus, the CIA proteins are specifically required for de novo assembly of cytosolic/nuclear Fe/S proteins. Conspicuously, all four CIA proteins are well conserved in virtually all eukaryotes including mammals such as mice and humans, suggesting that they might perform a similar function in other eukaryotes (31, 32). The potent RNAi technology will help in testing the functional roles of human CIA relatives.
Our studies demonstrate the importance of mitochondria and, in particular, of the mitochondrial version of huNfs1 for assembly of the Fe/S cluster on IRP1. Hence, mitochondria appear to be crucial for the mode of posttranscriptional regulation of iron homeostasis by IRP1. This idea recently received strong support from a targeted deletion of the gene encoding the mitochondrial ABC transporter ABCB7 (a homolog of yeast Atm1) in mouse liver. The defect in ABCB7 results in functional impairment of cytosolic but not mitochondrial Fe/S proteins (41). In particular, IRP1 was poorly converted to cytosolic aconitase, explaining the disturbance of cellular and systemic iron metabolism in the ABCB7 knockout liver. The obvious role of mitochondria in posttranscriptional regulation of iron homeostasis by IRP1 has some parallelism to the transcriptional iron regulation by the Aft1 protein in yeast. Activation of this transcription factor is triggered by defects in the mitochondrial ISC assembly machinery and in the mitochondrial ABC transporter Atm1 (10, 44). Hence, Aft1 may sense a product of the mitochondrial ISC assembly machinery that is exported by Atm1. Since Atm1 is also involved in Fe/S protein maturation in the cytosol (25), the molecule sensed by Aft1 and the compound needed for Fe/S cluster assembly on cytosolic Fe/S proteins such as IRP1 may be similar or even identical. We would like to emphasize though that the current knowledge does not allow us to decide which pool and which form of iron are sensed by IRP1. Clearly, mitochondrial iron necessary for Fe/S protein assembly inside the organelles is crucial, but since the source of iron for Fe/S protein assembly in the cytosol is unknown, it is well possible that the cytosolic iron pool also plays a regulatory role. IRP2 is closely related in sequence to IRP1, but its posttranscriptional iron-regulatory function is mediated by heme-dependent and/or oxygenase-dependent protein degradation (17, 21, 51). Since heme synthesis in mammals involves the mitochondrial Fe/S protein ferrochelatase, it is conceivable that impairment of huNfs1 has an impact on IRP2 stability and thus function. These considerations again point to the importance of human mitochondria in cellular Fe/S protein biogenesis and iron homeostasis.
Acknowledgments
We thank R. Eisenstein and L. Szweda for antibodies against IRP1 and aconitase and W. Suske for vector pMCS-HA.
Our work was supported by grants from Sonderforschungsbereiche 593 and TR1, Deutsche Forschungsgemeinschaft (Gottfried-Wilhelm Leibniz program), the European Commission (MitEURO), German-Israeli foundation GIF, and Fonds der chemischen Industrie.
REFERENCES
- 1.Acquaviva, F., I. De Biase, L. Nezi, G. Ruggiero, F. Tatangelo, C. Pisano, A. Monticelli, C. Garbi, A. M. Acquaviva, and S. Cocozza. 2005. Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J. Cell Sci. 118:3917-3924. [DOI] [PubMed] [Google Scholar]
- 2.Adam, A. C., C. Bornhövd, H. Prokisch, W. Neupert, and K. Hell. 2006. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25:174-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarzguioui, M., J. J. Rossi, and D. Kim. 2005. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 579:5974-5981. [DOI] [PubMed] [Google Scholar]
- 4.Balk, J., A. J. Pierik, D. Aguilar Netz, U. Mühlenhoff, and R. Lill. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balk, J., D. J. Aguilar Netz, K. Tepper, A. J. Pierik, and R. Lill. 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25:10833-10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balk, J., and R. Lill. 2004. The cell's cookbook for iron-sulfur clusters: recipes for fool's gold? Chembiochem 5:1044-1049. [DOI] [PubMed] [Google Scholar]
- 7.Barras, F., L. Loiseau, and B. Py. 2005. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 50:41-101. [DOI] [PubMed] [Google Scholar]
- 8.Bouton, C., M. J. Chauveau, S. Lazereg, and J. C. Drapier. 2002. Recycling of RNA binding iron regulatory protein 1 into an aconitase after nitric oxide removal depends on mitochondrial ATP. J. Biol. Chem. 277:31220-31227. [DOI] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 10.Chen, O. S., R. J. Crisp, M. Valachovic, M. Bard, D. R. Winge, and J. Kaplan. 2004. Transcription of the yeast iron regulon responds not directly to iron but rather to iron-sulfur cluster biosynthesis. J. Biol. Chem. 279:29513-29518. [DOI] [PubMed] [Google Scholar]
- 11.Drapier, J. C., and J. B. Hibbs, Jr. 1996. Aconitases: a class of metalloproteins highly sensitive to nitric oxide synthesis. Methods Enzymol. 269:26-36. [DOI] [PubMed] [Google Scholar]
- 12.Elsasser, H. P., U. Lehr, B. Agricola, and H. F. Kern. 1993. Structural analysis of a new highly metastatic cell line PaTu 8902 from a primary human pancreatic adenocarcinoma. Virchows Arch. B 64:201-207. [DOI] [PubMed] [Google Scholar]
- 13.Gerber, J., K. Neumann, C. Prohl, U. Mühlenhoff, and R. Lill. 2004. The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol. Cell. Biol. 24:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilkerson, R. W., D. H. Margineantu, R. A. Capaldi, and J. M. Selker. 2000. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 474:1-4. [DOI] [PubMed] [Google Scholar]
- 15.Guo, B., F. M. Brown, J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Characterization and expression of iron regulatory protein 2 (IRP2). Presence of multiple IRP2 transcripts regulated by intracellular iron levels. J. Biol. Chem. 270:16529-16535. [DOI] [PubMed] [Google Scholar]
- 16.Hales, K. G., and M. T. Fuller. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90:121-129. [DOI] [PubMed] [Google Scholar]
- 17.Hanson, E. S., M. L. Rawlins, and E. A. Leibold. 2003. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 278:40337-40342. [DOI] [PubMed] [Google Scholar]
- 18.Hausmann, A., D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Mühlenhoff, and R. Lill. 2005. The eukaryotic P-loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 102:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentze, M. W., M. U. Muckenthaler, and N. C. Andrews. 2004. Balancing acts: molecular control of mammalian iron metabolism. Cell 117:285-297. [DOI] [PubMed] [Google Scholar]
- 20.Holmuhamedov, E., A. Jahangir, M. Bienengraeber, L. D. Lewis, and A. Terzic. 2003. Deletion of mtDNA disrupts mitochondrial function and structure, but not biogenesis. Mitochondrion 3:13-19. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa, H., M. Kato, H. Hori, K. Ishimori, T. Kirisako, F. Tokunaga, and K. Iwai. 2005. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell 19:171-181. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser, J. T., T. Clausen, G. P. Bourenkow, H. D. Bartunik, S. Steinbacher, and R. Huber. 2000. Crystal structure of a NifS-like protein from. Thermotoga maritima: implications for iron-sulfur cluster assembly. J. Mol. Biol. 297:451-464. [DOI] [PubMed] [Google Scholar]
- 24.Kessler, D. Enzymatic activation of sulfur for incorporation in biomolecules. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 25.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are required for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Land, T., and T. A. Rouault. 1998. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell 2:807-815. [DOI] [PubMed] [Google Scholar]
- 27.Lange, H., G. Kispal, A. Kaut, and R. Lill. 2000. A mitochondrial ferredoxin is essential for biogenesis of intra- and extra-mitochondrial Fe/S proteins. Proc. Natl. Acad. Sci. USA 97:1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, J., M. Kogan, S. A. Knight, D. Pain, and A. Dancis. 1999. Yeast mitochondrial protein Nfs1p coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem. 274:33025-33034. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., S. Saxena, D. Pain, and A. Dancis. 2001. Adrenodoxin reductase homolog (Arh1p) of yeast mitochondria required for iron homeostasis. J. Biol. Chem. 276:1503-1509. [DOI] [PubMed] [Google Scholar]
- 30.Lill, R., and G. Kispal. 2000. Maturation of cellular Fe/S proteins: the essential function of mitochondria. Trends Biochem. Sci. 25:352-356. [DOI] [PubMed] [Google Scholar]
- 31.Lill, R., R. Dutkiewicz, H. P. Elsässer, A. Hausmann, D. J. A. Netz, A. J. Pierik, O. Stehling, E. Urzica, and U. Mühlenhoff. Mechanisms of iron-sulfur protein maturation in mitochondria, cytosol and nucleus of eukaryotes. Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 32.Lill, R., and U. Mühlenhoff. 2005. Iron-sulfur protein biogenesis in eukaryotes. Trends Biochem. Sci. 30:133-141. [DOI] [PubMed] [Google Scholar]
- 33.Loiseau, L., S. Ollagnier-de-Choudens, L. Nachin, M. Fontecave, and F. Barras. 2003. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 278:38352-38359. [DOI] [PubMed] [Google Scholar]
- 34.Mühlenhoff, U., J. Balk, N. Richhardt, J. T. Kaiser, K. Sipos, G. Kispal, and R. Lill. 2004. Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 279:36906-36915. [DOI] [PubMed] [Google Scholar]
- 35.Mühlenhoff, U., J. Gerber, N. Richhardt, and R. Lill. 2003. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22:4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullner, E. W., B. Neupert, and L. C. Kuhn. 1989. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell 58:373-382. [DOI] [PubMed] [Google Scholar]
- 37.Nakai, Y., M. Nakai, H. Hayashi, and H. Kagamiyama. 2001. Nuclear localization of yeast Nfs1p is required for cell survival. J. Biol. Chem. 276:8314-8320. [DOI] [PubMed] [Google Scholar]
- 38.Nakai, Y., N. Umeda, T. Suzuki, M. Nakai, H. Hayashi, K. Watanabe, and H. Kagamiyama. 2004. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 279:12363-12368. [DOI] [PubMed] [Google Scholar]
- 39.Nakai, Y., Y. Yoshihara, H. Hayashi, and H. Kagamiyama. 1998. cDNA cloning and characterization of mouse nifS-like protein, m-Nfs1: mitochondrial localization of eukaryotic NifS-like proteins. FEBS Lett. 433:143-148. [DOI] [PubMed] [Google Scholar]
- 40.Pantopoulos, K. 2004. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 1012:1-13. [DOI] [PubMed] [Google Scholar]
- 41.Pondarre, C., B. B. Antiochos, D. R. Campagna, S. L. Clarke, E. L. Greer, K. M. Deck, A. McDonald, A. P. Han, A. Medlock, J. L. Kutok, S. A. Anderson, R. S. Eisenstein, and M. D. Fleming. 2006. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet., 15:953-964. [DOI] [PubMed] [Google Scholar]
- 42.Rouault, T. A., and W. H. Tong. 2005. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 6:345-351. [DOI] [PubMed] [Google Scholar]
- 43.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22:4826-4835, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutherford, J. C., L. Ojeda, J. Balk, U. Mühlenhoff, R. Lill, and D. R. Winge. 2005. Activation of the iron-regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135-10140. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stehling, O., H. P. Elsässer, B. Brückel, U. Mühlenhoff, and R. Lill. 2004. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 13:3007-3015. [DOI] [PubMed] [Google Scholar]
- 47.Tong, W. H., G. N. Jameson, B. H. Huynh, and T. A. Rouault. 2003. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA 100:9762-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong, W. H., and T. Rouault. 2000. Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19:5692-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong, W. H., and T. A. Rouault. 2006. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3:199-210. [DOI] [PubMed] [Google Scholar]
- 50.Umeda, N., T. Suzuki, M. Yukawa, Y. Ohya, H. Shindo, and K. Watanabe. 2005. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 280:1613-1624. [DOI] [PubMed] [Google Scholar]
- 51.Wang, J., G. Chen, M. Muckenthaler, B. Galy, M. W. Hentze, and K. Pantopoulos. 2004. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol. Cell. Biol. 24:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiedemann, N., E. Urzica, B. Guiard, H. Müller, C. Lohaus, H. E. Meyer, M. T. Ryan, C. Meisinger, U. Mühlenhoff, R. Lill, and N. Pfanner. 2006. Essential role of Isd11 in iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25:184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, L., R. H. White, V. L. Cash, and D. R. Dean. 1994. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33:4714-4720. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]