Abstract
The nicotinic acetylcholine receptor (nAChR) β4/α3/α5 gene cluster encodes several heteromeric transmitter receptor subtypes that are essential for cholinergic synaptic transmission in adrenal gland, autonomic ganglia, pineal gland, and several nuclei in the central nervous system. However, the transcriptional mechanisms coordinating expression of these subunit genes in different cell populations are unknown. Here, we used transgenic methods to investigate long-range transcriptional control of the cluster. A 132-kb P1-derived artificial chromosome (PAC) encoding the rat cluster recapitulated the neurally- and endocrine-restricted expression patterns of the endogenous β4/α3/α5 genes. Mutation of ETS factor binding sites in an enhancer, β43′, embedded in the β4 3′-untranslated exon resulted in greatly diminished β4, α3, and α5 expression in adrenal gland and to a lesser extent in the superior cervical ganglion (SCG) but not in other tissues. Phylogenetic sequence analyses revealed several conserved noncoding regions (CNRs) upstream of β4 and α5. Deletion of one of them (CNR4) located 20 kb upstream of β4 resulted in a dramatic decrease in β4 and α3 expression in the pineal gland and SCG. CNR4 was sufficient to direct LacZ transgene expression to SCG neurons, which express the endogenous β4α3α5 subunits, and pineal cells, which express the endogenous β4α3 combination. Finally, CNR4 was able to direct transgene expression to major sites of expression of the endogenous cluster in the brain. Together, our findings support a model in which cell type-specific shared long-range regulatory elements are required for coordinate expression of clustered nAChR genes.
Recent evidence from whole-genome studies has led to the conclusion that gene clustering is common in the eukaryotic genome (8, 26). Two well-studied clusters are the β-globin and Hox loci, in which clustering is thought to facilitate interactions between gene-proximal elements in or near individual promoters and shared long-range regulatory elements. These interactions are necessary for correct cell type- and stage-specific switching of β-globin gene expression during erythropoiesis and coordinate expression of Hox genes in order for them to fulfill their roles in limb development and in patterning the main body axis (34, 52). Many gene clusters are expressed either exclusively or predominantly in the nervous system, but the mechanisms involved are not understood (19, 51, 67). The prevalence of eukaryotic gene clusters suggests that the identification of the transcriptional mechanisms governing their expression will be important for understanding the differentiation of specific cell types and perhaps the molecular basis of disease (29).
A phylogenetically conserved cluster of nicotinic acetylcholine receptor (nAChR) subunit genes, β4/α3/α5, encodes heteromeric neurotransmitter-gated cation channels that are critically important for fast cholinergic synaptic transmission (7, 11, 13). Several observations indicate that the clustered genes are coordinately regulated. First, all three subunit genes are coexpressed in adrenal medulla and neurons of the sympathetic and parasympathetic nervous systems to enable assembly of β4α3 and β4α3α5 receptor subtypes (16, 37, 57). Second, the cluster is also expressed in numerous neuronal cell types of the brain and retina, where it likely encodes heteromeric nAChRs that are also involved in synaptic transmission and presynaptic neuromodulation (22). Major sites of β4/α/3α5 coexpression in the brain include neurons of the medial habenula, the interpeduncular nucleus, and the inferior colliculus (21, 40, 50, 55, 58, 69). The β4α3 heteromer is also the major nAChR subtype in the pineal gland, which is responsible for melatonin production and secretion in the brain (24, 44). Third, β4/α3/α5 transcripts are coordinately upregulated during synaptogenesis in autonomic ganglia (12, 32). All three clustered genes are coordinately upregulated by presynaptic innervation, while β4/α3 but not α5 transcript levels are upregulated by target tissue interactions (31, 47). The effect of presynaptic input on β4/α3/α5 transcript levels can be mimicked by treatment of developing autonomic neurons with neuregulin isoforms (62). Fourth, axotomy of adult autonomic neurons results in a coordinate decrease in levels for β4/α3/α5 transcripts (32, 68). Together, these expression characteristics suggest that cell type-specific transcriptional programs that can be influenced through presynaptic and target-derived extrinsic cues may coordinately regulate the cluster. However, the mechanisms involved are poorly understood.
Several laboratories, including ours, have used cell culture transfection methods to characterize cis regulatory elements in the cluster. The β4, α3, and α5 promoters are all able to direct expression of reporters in neural cell lines, and each can be transactivated by the ubiquitously expressed family of Sp transcription factors (5, 6, 9, 10, 54, 61) and the neurally restricted factors SCIP/Tst-1/Oct-6 and Sox10 (20, 35, 63). These data raise the possibility that the interaction of ubiquitous and cell type-restricted transcription factors with cis elements in or near the three cluster promoters contributes to coordinate expression of the cluster. However, these short-range interactions are not likely to be sufficient for proper transcriptional control of the cluster, as transgenic studies of the isolated α3 and β4 promoters indicate that they are not able to direct expression to neuronal cell types that express the cluster (65). In a search for other regulatory elements within the cluster, we identified an enhancer (β43′) located in the 3′ untranslated region of the rat β4 gene (38, 39). Reporter analyses in cell lines and in primary neural cell cultures have demonstrated that β43′ is preferentially active in neurons and that this activity depends on ETS factor interactions (18, 39). Its location between the β4 and α3 promoters led us to hypothesize that it may be a shared long-range regulatory element (14).
Here, we describe the use of a P1-derived artificial chromosome (PAC) transgenic approach to investigate the transcriptional mechanisms that function to coordinate expression of the nAChR cluster. Mutagenesis of the β43′ ETS sites suggests that it is important for coordinating expression of all three clustered genes in the adrenal gland. We then searched for additional regulatory information outside the cluster and found several conserved noncoding regions (CNRs) extending far upstream of both the β4 and the α5 genes. Deletion of one such element, CNR4, from a PAC resulted in a dramatic decrease in the expression levels of the β4 and α3 transgenes in the pineal gland and sympathetic neurons. Moreover, CNR4 was able to direct transgene reporter expression to sympathetic neurons and the pineal gland as well as several neuronal cell types in the brain that strongly express the clustered genes. Our findings suggest that gene- and cell type-specific long-range control coordinately regulate the nAChR gene cluster. We hypothesize that the several other conserved noncoding regions upstream of β4 and α5 may contain additional long-range regulatory information needed for proper spatiotemporal control of the cluster.
MATERIALS AND METHODS
Isolation of a 132-kb PAC.
A PAC, RP31-224F1, was identified by screening Roswell Park Cancer Institute rat PAC genomic library 31 (60) with a 0.5-kb probe corresponding to the intragenic region between the rat β4 and α3 genes. The identity of the PAC was further confirmed by restriction enzyme digestion and by end sequencing with primers recognizing the SP6 and T7 promoters.
Targeting vectors and LacZ reporter construct.
A chloramphenicol resistance cassette (CaM) was amplified from pPCR-Script Cam SK(+) vector (Strategene, CA) with the following primers: forward, 5′-GGAATTCGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCTGTGACGGAAGATCACTTCG-3′ (EcoRI); reverse, 5′-GGCGCGCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCTGCCATTCATCCGCTTATT-3′ (AscI). Underlined sequences represent the restriction enzyme digestion sites described in the parentheses and used in the cloning strategy. Italic sequences represent FRT sites. The PCR product was cloned in pPCR-Script Amp SK(+) vector (Stratagene, CA) to generate the plasmid named FRTCaM. All oligonucleotides used in this study were ordered from Integrated DNA Technologies, Inc. (Iowa). Primers longer than 40 bp were purified by polyacrylamide gel electrophoresis. All PCRs for the purpose of cloning were done with high-fidelity Pfu Taq polymerase (Stratagene, CA).
β43′ ETS site targeting vector.
To mutate the β43′ enhancer, the left homologous arm (0.5 kb) was amplified from the wild-type (WT) PAC with primers (forward, 5′-TCCCCGCGGGAACTCCATGA CCAGAACCAA-3′ [SacII]; reverse, 5′-GGCGCGCCGCCCTGAATCCCAGGCTGGTCTC-3′ [AscI]) and then cloned into FRTCaM. The right homologous arm that includes the enhancer (1.5 kb) was amplified with primers (forward, 5′-GGAATTCCGACTGCCACATCAGTGCCTA-3′ [EcoRI]; reverse, 5′-GGGGTACCCCGGGGAGAGAGTCCAGGAGAT-3′ [KpnI]) and cloned into the vector BGZA (66). Three ETS binding sites in β43′ (see Fig. 3) were then mutated by three rounds of mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, CA) using the following oligonucleotides duplexed with their complementary strands: left ETS site, 5′-CTCTGCTCCAATGCCACGCGTTTGTATAAGCCTTCCC-3′; spacer ETS site, 5′-GTATAAGCCTTCCCATGGCCATCTGCTCCAGTGTCATTTC-3′; right ETS site, 5′-GGCCATCTGCTCCAGTGTCATATGGTTGTATAAGCCTTCCCTAG-3′. The underlined sequences indicate the mutated sequences. The resulting triple-ETS-mutated right homologous arm was then cloned into FRTCaM. The final targeting construct was released from the vector by double digestion with KpnI and SacII.
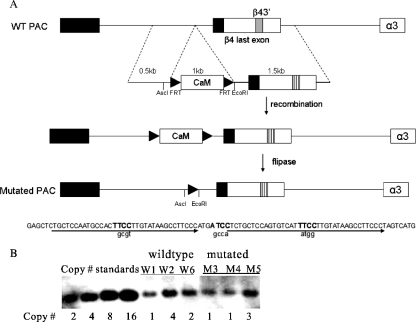
FIG. 3.
Mutagenesis of PAC β43′ by bacterial recombination. (A) Targeting strategy to change WT nucleotides in the three β43′ ETS core sites. Black boxes represent protein-coding exons, and white boxes represent 3′ untranslated exonic sequences. The shaded box represents the wild-type enhancer, and the hatched box represents the mutated enhancer, in which the three ETS binding sites were replaced with sequences shown in lowercase letters below. After induction of λ prophage expression and recombination between the homologous arms, the endogenous β43′ was replaced by the mutated β43′ and the chloramphenicol resistance cassette was inserted into the last intron of the β4 gene. Then, flipase was induced, which recombined between the two FRT sites (black triangles) of the same orientation to excise the CaM cassette, leaving an FRT scar in the β4 intron about 100 bp from the splice junction. (B) Semiquantitative Southern blotting to determine the copy numbers of the wild-type (W) and mutated (M) transgenic lines.
CNR4 targeting vector.
To delete 487 bp of the CNR4 core region, homologous arms were amplified with primers (left arm, forward, 5′-CCATCGATCCCCCTGGTCCTATTTCATT-3′ [ClaI]; left arm, reverse, 5′-GGAATTCTACAAAACCCAGGGAAGCAG-3′ [EcoRI]; right arm, forward, 5′-GGCGCGCCCACTGCCCTGTGGTGTACTG-3′ [AscI]; right arm, reverse, 5′-TCCCCGCGGCCCCTGCTCTAGGCTCTTTT-3′ [SacII]) and cloned into FRTCaM. The targeting construct was released by double digestion with XhoI and SacII.
CNR4Z reporter.
To make the CNR4Z LacZ reporter construct, a 700-bp fragment spanning the entire CNR4 region was amplified from mouse genomic DNA with primers (forward, 5′-GACTAGTCCCACTAGGCACTGATCACA-3′ [SpeI]; reverse, 5′-GACTAGTCTATCGAGCTGACACCCACA-3′ [SpeI]) and cloned into a LacZ reporter plasmid (BGZA) (23, 66) immediately upstream of the β-globin minimal promoter. CNR4Z with CNR4 in its natural orientation was confirmed by sequencing and double digested with HindIII and NotI to release the 4.4-kb construct for pronuclear injection.
Bacterial recombination.
PAC modification was done using the bacteriophage λ red recombination system as previously described (30). Briefly, the wild-type PAC was introduced into Escherichia coli strain EL250 (a gift from Neal Copeland), which has a λ prophage under the control of a temperature-sensitive repressor and an arabinose-inducible flipase gene. A single kanamycin-resistant colony was picked and grown overnight, diluted 1:20 into 10 ml fresh LB medium the next day, and grown to an optical density at 600 nm of 0.4 to 0.6. Cultures were then incubated with constant shaking at 42°C for 15 min to induce the λ-prophage expression, followed by 5 min on wet ice. Recombination-competent cells were centrifuged at 4,000 rpm for 5 min and washed with sterile ice-cold water three times. Cells were then resuspended into 50 μl sterile cold water and mixed with 100 to 200 ng linearized targeting vector. The mixture was electroporated at 1.8 kV, 25 μF, and 200 Ω using a Bio-Rad gene pulser and incubated at 37°C for 1 h in 1 ml of fresh LB. Recombinant clones were selected by plating of the cells on LB plates with 70 μg/ml of kanamycin and 25 μg/ml of chloramphenicol. To excise the chloramphenicol resistance cassette, recombinant cells were grown overnight, diluted 1:50 into 10 ml fresh LB, and grown until the optical density at 600 nm was 0.4 to 0.6. One hundred microliters of 10% l-(+)-arabinose (Sigma) was added to the culture to a final concentration of 0.1%. Cultures were incubated for another hour at 37°C and then diluted 1:1,000 and plated on LB plates with 70 μg/ml of kanamycin. Clones with desired mutations were confirmed by restriction enzyme digests (see Fig. S1 and S3 in the supplemental material), PCR, and sequencing.
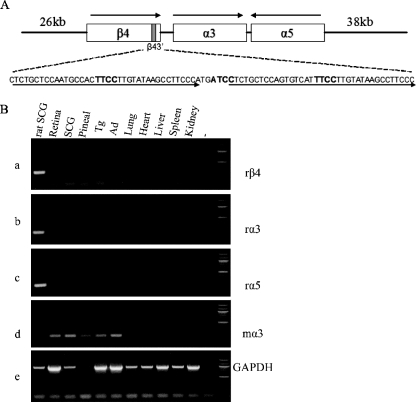
FIG. 1.
PAC transgenic strategy. (A) Schematic (not to scale) of a 132-kb PAC encoding the rat nAChR gene cluster. The PAC extends 26 kb upstream of the β4 gene and 38 kb upstream of α5. Arrows above the schematic indicate the transcriptional orientation of each gene. The shaded box in β4 indicates the position of β43′, with its sequences shown below. Arrows below the sequences indicate the two 37-bp repeats of the enhancer, and ETS factor binding site core sequences are highlighted in bold letters. (B) RT-PCR analysis of rat versus mouse β4, α3, and α5 amplification primers. Total RNA extracted from the indicated rat (far left lane) or mouse tissues was used to determine primer specificity. The results show that the rat primers, rβ4, rα3, and rα5, were able to amplify the appropriate product from rat SCG RNA but not when combined with any of the various mouse RNA samples (panels a, b, and c). Conversely, the mouse-specific α3 primer, mα3, was able to amplify product when combined with mouse tissue RNA but not when combined with rat SCG RNA (panel d). Pineal, pineal gland; Tg, trigeminal ganglia; Ad, adrenal gland; −, no RT control; rightmost lane, markers. The specific primers used in each experiment are indicated on the right. r, rat-specific primer; m, mouse-specific primer; GAPDH, RT-positive control.
Transgenic mice.
The rat PAC clone with the pPAC4 vector backbone attached was purified as previously described (64) for injection. All constructs were injected into hybrid C57B6/SJL F2 zygotes by the Case Transgenic and Targeting Facility. PAC founders were screened with SP6, T7, rα3, rβ4, and rα5 primers. The CNR4Z founders were screened with the primers that recognize the β-galactosidase (β-Gal) gene: forward, 5′-AAAAGTCAGGGCAGAGCCATC-3′; reverse, 5′-TGTGCTGCAAGGCGATTAAG-3′. All founders were subsequently confirmed by Southern blot analyses.
Semiquantitative Southern blotting.
Ten micrograms of genomic DNA was digested overnight with EcoRI. Copy number standards were prepared by mixing various amounts of pPAC4 vector with 10 μg wild-type mouse genomic DNA. The probe was prepared by labeling a 1.7-kb AgeI fragment of pPAC4 with [α-32P]dCTP using a Rediprime II kit (Amersham, United Kingdom). Hybridization was carried out with Rapid-Hyp buffer (Amersham, United Kingdom) according to the vendor's protocol. The membrane was exposed to the X-ray film overnight at −80°C, and the band intensities were analyzed using the Versadoc 3000 imaging system (Bio-Rad, CA).
RT-PCR and quantitative RT-PCR.
Tissues were dissected from gender- and age-matched 5- to 7-week-old wild-type and mutated PAC transgenic animals. Total RNA was extracted in TRIzol according to the vendor's protocol (Invitrogen, CA). Genomic DNA contamination was eliminated by DNase I digestion using DNA-free (Ambion, TX). Total RNA (0.5 to 1 μg) was used to synthesize first-strand cDNA using an Advantage RT-for-PCR kit (BD Bioscience). For conventional reverse transcription (RT)-PCR, 5 μl of diluted cDNA was used in each PCR (35 cycles total), with species-specific primers at the annealing temperatures described in Table 1 or with GAPDH primers provided by the kit. For real-time RT-PCR, 40 cycles were performed in a 25-μl PCR mixture in the presence of SYBR green (Molecular Probes, OR), fluorescein calibration dye (Bio-Rad, CA), platinum Taq (Invitrogen), and gene-specific primers. All real-time experiments were done in triplicate using the iCycler system (Bio-Rad, CA). Relative quantification of transcript levels was done based on a mathematical model previously described (45). Briefly, the expression level (e) of transgene, normalized to endogenous mouse α3 transcript levels, was calculated by the following equation: 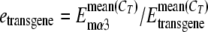 . E stands for efficiency that was determined in a separate experiment (see Table 1). CT is the threshold cycle value deduced using the Optical system software, version 3.1 (Bio-Rad). Relative expression levels of mutant transgenic lines (R) compared to those of wild-type transgenic lines was calculated using the following equation: R = mean [etransgene(mutant)/copy number]/mean [etransgene(WT)/copy number]. To confirm the validity of the method, all of the data were also analyzed using the relative expression software tool (REST) (46), with similar results obtained.
. E stands for efficiency that was determined in a separate experiment (see Table 1). CT is the threshold cycle value deduced using the Optical system software, version 3.1 (Bio-Rad). Relative expression levels of mutant transgenic lines (R) compared to those of wild-type transgenic lines was calculated using the following equation: R = mean [etransgene(mutant)/copy number]/mean [etransgene(WT)/copy number]. To confirm the validity of the method, all of the data were also analyzed using the relative expression software tool (REST) (46), with similar results obtained.
TABLE 1.
RT-PCR primers
| Gene | Upstream primers | Downstream primers | Temp (°C) | Product size (bp) | Efficiency |
|---|---|---|---|---|---|
| Rat β4 | 5′-TTCTGCTGATCCCTCTAGTGC-3′ | 5′-TGAGGTTTGGACGGTAGCAGT-3′ | 61 | 271 | 1.98 |
| Rat α3 | 5′-AAGAAATGTGGGAGACAAATG-3′ | 5′-GCGCTGCTGAATACACTACG-3′ | 58 | 241 | 2.03 |
| Rat α5 | 5′-ATCCTTGGAGTGCTGGAAAACT-3′ | 5′-AATGGGCCTACCTCCTATAAATA-3′ | 61 | 188 | 1.96 |
| Mouse α3 | 5′-AAGAAATGTGGGAGAAAAACA-3′ | 5′-GGGTGCTGCTGGAATACTT-3′ | 58 | 191 | 1.99 |
Luciferase assays.
Luciferase reporter CNR4-luc was prepared by amplification of CNR4 sequences (700 bp) from mouse genomic DNA with the following primers: forward, 5′-CCGCTCGAGCCCACTAGGCACTGATCACA-3′ (XhoI); reverse, 5′-GAAGATCTCTATCGAGCTGACACCCACA-3′ (BglII). The resulting product was ligated upstream of the SV40 promoter in pGL3 vector (Promega, WI). The PC12 cell line was grown at 37°C in a humidified 5% CO2 incubator with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 5% heat-inactivated horse serum, 100 U/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate (Invitrogen). Transfections were carried out using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Luciferase assays were done according to the vendor's protocol (ProMega Corp., WI). Briefly, 24 to 48 h after transfection, cells were lysed in 200 μl of lysis buffer per well of six-well plates. Lysate (1 μl) was analyzed in 100 μl of luciferin substrate using a Lumat LB9501 luminometer (EG&G Berthold, NH).
Histology.
Animals were perfused with 4% paraformaldehyde in phosphate-buffered saline (PBS), pH 7.4 (Electron Microscopy Science, PA), and dissected tissues postfixed in the same solution for 15 min for X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) histology and 4 h for immunohistochemistry. Tissues were placed in 20% sucrose (wt/vol), 1× PBS overnight at 4°C and sectioned on a freezing microtome or on a cryostat at 20 μm.
X-Gal staining.
X-Gal (Alexis Biochemicals, San Diego, CA) was dissolved into DMFA (N,N-dimethylformamide) (Sigma) and stored as a 20-mg/ml stock at −20°C. Whole-mount tissues or tissue sections were incubated with 1 ml of staining solution composed of 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, and 1 mg/ml X-Gal in 1× PBS at 37°C overnight. Tissue sections were then counterstained with 1% neutral red (Sigma).
Immunohistochemistry.
Tissue sections were double stained with a sheep polyclonal anti-tyrosine hydroxylase (TH) antibody (Pel-Freeze, Rogers, Arkansas) at 1:100 dilution and a rabbit polyclonal anti-β-Gal (Biogenesis, Sandown, NH) at 1:2,500 overnight and then stained with Cy2-labeled donkey anti-sheep (1:100) secondary antisera and Texas Red-labeled donkey anti-rabbit (1:400) secondary antisera the next day.
Sequence comparison.
Phylogenic sequence comparisons were performed using ECR browser (43), with sequences 76611222 to 76790069 from human chromosome 15 as the base sequence.
RESULTS
PAC transgenic lines encoding the rat β4α3α5 gene cluster.
To begin an investigation of the mechanisms involved in coordinating expression of the nAChR cluster, we isolated a 132-kb PAC clone from a rat genomic library, which includes the cluster as well as 26 kb upstream of the β4 gene and 38 kb upstream of the α5 gene. Also present in this PAC is the coding sequence of a ubiquitously expressed peroxisome subunit gene, PMSA4, upstream of α5 (Fig. 1A; see also Fig. 5A). PAC DNA was purified and injected into fertilized oocytes to generate transgenic mice as described previously (65).
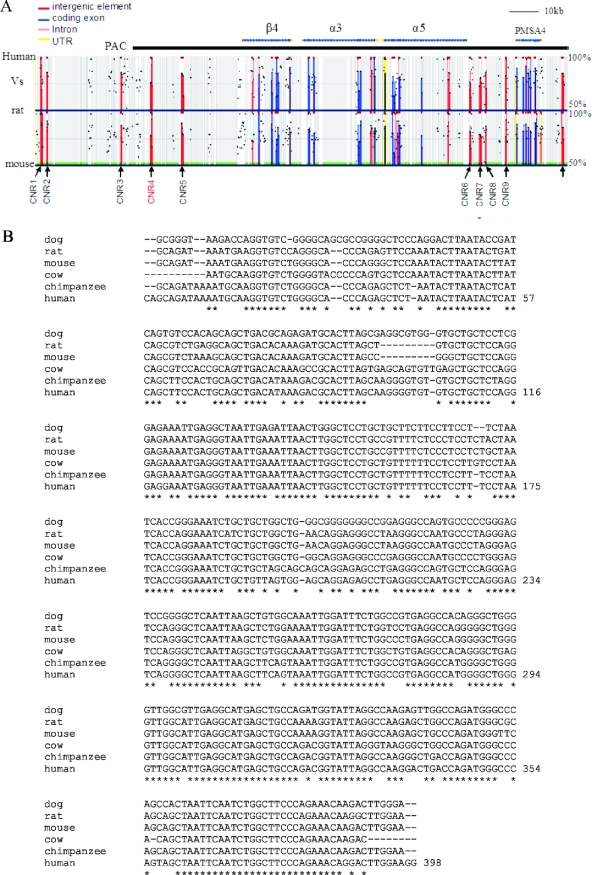
FIG. 5.
Identification of conserved noncoding regions by phylogenetic sequence comparisons. (A) Human, mouse, and rat sequence comparisons within the nAChR gene cluster. A 178-kb region of human chromosome 15 was compared to mouse and rat genomic sequences using ECR browser. The black bar on the top indicates the subregion present in the PAC insert. The criteria for identifying evolutionarily conserved regions were set as 250-bp minimum length and 75% minimum sequence identity. The cutoff line for the graph is 50%. Shaded backgrounds indicate the repetitive sequences. Vertical lines indicate regions conserved between the species: blue, coding sequences; red, intergenic sequences; pink, introns; and yellow, untranslated (UTR) exons. Black arrows point to the CNRs upstream of β4 and α5. CNR4 is highlighted in red letters. (B) Alignment of the 400-bp core sequence of vertebrate CNR4s.  , bases that are present in all analyzed species.
, bases that are present in all analyzed species.
The amplification efficiency of Taq polymerase is decreased when nucleotide mismatches occur at the 3′ termini of primers and templates (25). Thus, to discriminate between the evolutionarily related endogenous mouse and transgene-transcribed rat sequences, we designed PCR primers whose 3′ termini match sequences for one but not the other species (Table 1). Indeed, when these primers were tested by conventional RT-PCR using mouse or rat RNAs under the conditions described in Table 1, amplification products were detected only when the rat-specific primers were combined with rat transcripts (Fig. 1B, panels a, b, and c) or when mouse α3 primers were combined with mouse transcripts (Fig. 1B, panel d). GAPDH primers that do not discriminate between the two species were able to generate comparable levels of product with all samples (Fig. 1B, panel e). The efficiencies of all primer pairs described in Table 1 were determined as previously described (45), by performing real-time RT-PCR on triplicate 1:10 serial dilutions of neural cDNAs from various tissues.
Founders were identified with rat-specific primer pairs (Table 1) and then bred to establish three wild-type PAC transgenic lines, W1, W2, and W6. The integrity of the PAC DNA in each line was further tested with genomic DNA and PCR primers that recognize β4, α3, or α5 gene sequences or sequences at each end of the PAC (see Fig. S1 in the supplemental material). These studies indicated that all three wild-type lines had an intact PAC genomic insert. To estimate transgene copy numbers, semiquantitative Southern blotting was carried out using a probe that corresponds to the pPAC4 backbone sequences. These analyses showed that lines W1, W2, and W6 have one, four, and two copies of the 132-kb PAC transgene, respectively (see Fig. 3B).
Expression patterns of β4, α3, and α5 PAC transgenes.
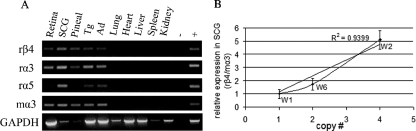
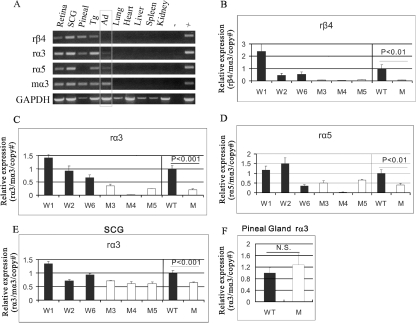
To determine whether the β4, α3, and α5 PAC transgenes were expressed in a pattern that recapitulated the patterns of their endogenous mouse counterparts, total RNA was extracted from various tissues and RT-PCR was carried out using rat-specific β4, α3, and α5 primers. Lines W1, W2, and W6 showed restricted expression of all three clustered genes in a pattern that correlated well with the endogenous mouse gene expression patterns (Fig. 2A). For example, comparable levels of expression were detected for all three transgenes in the superior cervical autonomic ganglion (SCG), adrenal gland, retina, and trigeminal ganglion, which is consistent with their endogenous mouse expression patterns (40, 69). The endogenous β4 and α3 genes are also strongly expressed in the pineal gland; however, endogenous α5 expression is not detectable there (24). Similarly, β4 and α3 but not α5 PAC transgene expression was detected in pineal gland. Furthermore, as expected from the endogenous expression patterns, neither β4, α3, nor α5 transgene expression was detected in lung, liver, heart, spleen, or kidney.
FIG. 2.
Wild-type PAC transgene expression. (A) RT-PCR analysis of β4/α3/α5 transgene expression in the indicated neural, endocrine, and nonneural tissues. Primers used are indicated on the left, with GAPDH as the positive control. Pineal, pineal gland; Tg, trigeminal ganglia; Ad, adrenal gland; −, no RT control; +, rat SCG cDNA for rβ4, rα3, and rα5 and mouse SCG cDNA for mα3; r, rat-specific primer; m, mouse-specific primer. Wild-type PAC line W2 was used in this experiment, and similar results were obtained with lines W1 and W6. (B) Copy number-dependent expression of the β4 transgene in SCG. Relative expression levels of rat β4 in SCG normalized to the level of endogenous mouse α3 were measured in four animals from each of the indicated transgenic lines by real-time RT-PCR. The means were calculated and plotted against the copy numbers, which were determined by Southern blotting. The expression level of line W1, which has one copy, was set at relative level 1 for comparison to the expression levels of lines W2 and W6. Error bars indicate the standard error of the mean.
We next used real time RT-PCR to quantify β4, α3, and α5 transgene expression levels. Rat β4, α3, and α5 transcript levels in SCG were normalized to the level for endogenous mouse α3 in four animals from each of the three PAC lines. A linear correlation (R2 values equal to 0.94, 0.58, and 0.84 for β4, α3, and α5, respectively) was obtained when the mean transcript levels were plotted against copy number for each line (Fig. 2B). The reproducible expression patterns between independent transgenic lines and copy number-dependent transgene expression levels suggest that the 132-kb PAC includes crucial transcriptional elements that are required for proper expression of all three nAChR genes in the cluster.
Functional analysis of β43′ in vivo.
Previous cell culture studies showed that the mutation of three ETS sites in the β43′ enhancer nearly eliminated its activity (18, 39). To determine whether or not β43′ is important for regulating transcription of the nAChR gene cluster in vivo, we used bacterial recombination to mutate the three critical enhancer ETS core motifs in the 132-kb PAC (Fig. 3A). The identity and integrity of this mutated PAC were confirmed by endonuclease restriction digests of the cloned DNA and sequence analysis of the PAC ends, junctions of the targeted sequences, and enhancer region (see Fig. S1 in the supplemental material). Pronuclear injection of this mutated PAC led to the identification of three β43′-mutated PAC founder lines, M3, M4, and M5. The integrity of PAC DNA in each of these lines was examined by PCR analysis of sequences within each of the clustered genes and sequences at each end of the PAC (see Fig. S1 in the supplemental material). No gross deletions or rearrangements were identified in any of the three lines. Southern analysis indicated that lines M3, M4, and M5 had one, one, and three copies, respectively (Fig. 3B).
β43′ activity is required for β4, α3, and α5 transgene expression in adrenal gland.
Conventional RT-PCR analyses of lines M3, M4, and M5 indicated that elimination of β43′ activity did not dramatically alter the β4/α3/α5 transgene expression patterns (Fig. 4A) relative to those of the wild-type PAC lines, W1, W2, and W6. These data confirm that the remaining FRT site in the last β4 intron, which is located about 100 bp away from a splice junction, does not have a major impact on PAC gene expression. However, there was a noticeable and reproducible reduction in β4/α3/α5 transgene expression in the M3, M4, and M5 lines in the adrenal gland (Fig. 4A).
FIG. 4.
β43′ is required for expression of the β4, α3, and α5 transgenes in adrenal gland. (A) RT-PCR analysis of M5 transgene expression in various tissues indicated above. Similar results were obtained from lines M3 and M4. Primers used in each experiment are indicated on the left, with GAPDH as the positive control. The gray box highlights the reduction of transgene expression in the adrenal gland (Ad). Pineal, pineal gland; Tg, trigeminal ganglia; −, no RT control; +, rat SCG cDNA for rβ4, rα3, and rα5 and mouse SCG cDNA for mα3; r, rat-specific primer; m, mouse-specific primer. (B to D) Real-time RT-PCR to quantitate the reduction in β4 (B), α3 (C), and α5 (D) transgene expression levels in the adrenal glands of each of the three mutant PAC lines. On the right-side portions of panels B to D are the mean levels of expression of the WT (W) lines versus those of the mutant (M) lines, where the wild-type level was set to 1. (E and F) Real-time RT-PCR analysis to quantitate WT and β43′-mutated α3 transgene expression levels in the SCG (E) and in the pineal gland (F). The data in panel F are the mean levels of expression of the three WT lines versus those of the three mutant lines, where the wild-type level was set to 1. Total RNA was extracted from adrenal glands, SCG, or pineal glands obtained from four animals from each line. Relative transgene expression levels, normalized to endogenous mouse α3 expression level and copy number, were determined for each wild-type and mutant line. Error bars indicate the standard errors of the means. Student's t test was used for statistical analyses. N.S., not significantly different.
To quantify the reduced expression in adrenal gland, we used quantitative real-time RT-PCR. As shown in Fig. 4B, the levels for β4 were dramatically reduced in all three mutated lines to about 5% of the wild-type PAC transgene level. The levels for the α3 transgene were also reduced in all three mutant lines to about 20% of the wild-type level (Fig. 4C), while the mean levels of α5 transgene expression were reduced to about 40% of the wild-type level (Fig. 4D). Similar experiments with the SCG indicated a reduction of about 36% in α3 transgene expression in the β43′-mutated PAC lines (Fig. 4E), while no reproducible differences between wild-type and β43′-mutated PAC lines could be detected in expression of the β4 and α5 transgenes in this ganglion (data not shown). We also could not detect reproducible quantitative differences in β4 or α3 transgene expression between wild-type and β43′-mutated PAC lines in pineal gland (Fig. 4F and data not shown). Taking these data together, we conclude that β43′ and its interactions with the ETS factors are required for the coordinate transcriptional regulation of all three clustered nAChR subunit genes in adrenal gland. β43′ also appears to be required for optimal α3 transgene expression in the SCG but not its partners in the cluster.
Identification of conserved noncoding regions.
The finding that mutation of β43′ did not eliminate nAChR gene expression in all tissues suggests that other cis elements are present in the PAC and these control other aspects of nAChR cluster expression. Recently, comparative genomic analysis, or phylogenetic footprinting, has become a powerful tool in identifying regulatory DNA elements (15, 28, 36, 42, 59). To search for other regulatory elements, we compared human, mouse, and rat genomic sequences around and within the nAChR gene cluster. This analysis revealed several interesting features of the nAChR cluster genomic landscape (Fig. 5 and Table 2). Beginning about 20 kb upstream of rat β4 and distributed over about 30 kb are four conserved noncoding regions (CNR1 to CNR4), 200 to 700 bp in length, whose human, mouse, and rat sequences are at least 70% similar to one another and whose relative spacing is phylogenetically conserved. CNR3 and CNR4 are present as single copies in the genome, while there are three and eight additional copies of CNR1 and CNR2, respectively, at different locations in the human genome. The distribution of these CNRs relative to the β4 gene is very similar in the mouse genome. However, in the human genome, CNR4 is positioned about 30 kb upstream of β4 and a fifth CNR (CNR5) is present 20 kb upstream of β4. Six additional copies of CNR5 are present in the human genome. Rat and mouse CNR5-related sequences are present only on other chromosomal locations. One copy of mouse CNR5 is located on chromosome 11, 22 kb downstream of Hoxb9, at the very end of the Hoxb gene cluster. Another copy of mouse CNR5 is located on chromosome 2, between olfactory receptor 50 and olfactory receptor 3 in the middle of an extended olfactory receptor gene cluster. No CNR5 is similarly positioned in the human genome. Interestingly, just upstream of CNR1, the most distant CNR from the β4 gene, the synteny between human and mouse is interrupted, suggesting that sequences in or near CNR1 may be a regulatory region boundary for the β4 side of the cluster. On the α5 side of the cluster are four additional CNRs (CNR6 to CNR9), each >200 bp in length, positioned 1 to 13 kb upstream of α5 and whose sequence conservation is at least 70% in human, mouse, and rat (Fig. 5 and Table 2). Each of these is present as single copies in the mammalian genome. Roughly 18 kb upstream of α5 and about 3 kb upstream of the most distal CNR is the ubiquitously expressed PMSA gene, suggesting a second regulatory region boundary on the α5 side.
TABLE 2.
Human and mouse CNRsa
| Name | Length (bp) | Identity (%) | Copy no. | Distance to β4 (kb) | Distance to BP (kb) | Distance to α5 (kb) | Distance to PMSA (kb) |
|---|---|---|---|---|---|---|---|
| CNR1 | 414 | 75 | 4 | 67.1 (53.7) | 1.8 (1.6) | ||
| CNR2 | 247 | 77 | 9 | 65.3 (50.6) | 3.8 (5.0) | ||
| CNR3 | 438 | 78 | 1 | 40.2 (27.3) | 28.6 (28.0) | ||
| CNR4 | 599 | 80 | 1 | 30.0 (18.3) | 38.6 (36.9) | ||
| CNR5 | 655 | 80 | 7 | 19.5 | 49.1 | ||
| CNR6 | 388 | 74 | 1 | 1.3 (0.5) | 15.5 (23.1) | ||
| CNR7 | 544 | 76 | 1 | 4.8 (4.7) | 12.1 (19.0) | ||
| CNR8 | 298 | 76 | 1 | 6.5 (7.8) | 10.1 (15.6) | ||
| CNR9 | 251 | 74 | 1 | 13.2 (20.0) | 3.3 (3.4) |
The numbers in parentheses represent the relative locations of mouse CNRs. Percent identity was determined from comparisons between mouse, rat, and human sequences. BP, breakpoint.
The clustering of evolutionarily conserved noncoding sequences on either side of β4/α3/α5 led us to hypothesize that these elements may function as long-range transcriptional regulatory elements for the vertebrate nAChR gene cluster. To begin to test this idea, we decided to focus on CNR4 for several reasons. First, its level of conservation (80%) is among the highest of the nine CNRs (Fig. 5B and Table 2). Second, this CNR is single copy in both the mouse and the human genomes. Third, it is present in the 132-kb PAC (Fig. 5A), which allowed us to investigate its function by eliminating it from the PAC using bacterial recombination.
CNR4 is required for β4/α3/α5 expression in pineal gland and SCG.
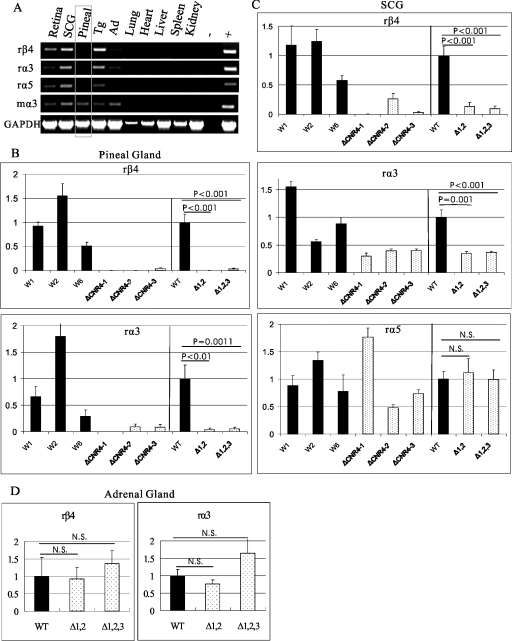
We constructed a targeting vector designed to remove 487 bp of the conserved core region of CNR4, leaving an FRT scar (see Fig. S2 in the supplemental material). This CNR4-mutated PAC clone was purified and then used to generate transgenic mice. Three lines, ΔCNR4-1, ΔCNR4-2, and ΔCNR4-3, were initially identified by PCR. Southern blotting indicated that each of these lines has one copy of the transgene (see Fig. S3 in the supplemental material). Genomic PCR with primers recognizing sequences in each of the clustered genes and sequences at each end of the PAC revealed that lines ΔCNR4-1 and ΔCNR4-2 have a complete copy of the transgene while line ΔCNR4-3 has a copy that is partially truncated upstream of α5 (see Fig. S3 in the supplemental material). More-detailed PCR analysis revealed that the breakpoint in ΔCNR4-3 is between CNR8 and CNR9, resulting in a 20-kb to 25-kb deletion that removes the PMSA gene (data not shown).
Conventional RT-PCR revealed that elimination of CNR4 had no noticeable effect on the expression pattern of the clustered transgenes in ΔCNR4-1, ΔCNR4-2, and ΔCNR4-3 compared to the wild-type PAC expression pattern in most tissues (compare Fig. 2 and 6). However, there was an apparent absence of β4/α3 transgene expression in the pineal gland (Fig. 6A). We then used real-time RT-PCR to quantitate the reduction of β4/α3 in pineal gland. As shown in Fig. 6B, the expression levels of both β4 and α3 transgenes in the CNR4-mutated PAC lines were dramatically decreased to 0.2% and 4%, respectively, in pineal gland. We also used real-time RT-PCR to determine whether there might be a quantitative effect on expression of the β4/α3/α5 transgenes in SCG. Indeed, the levels for the β4 and α3 transgenes but not α5 were reduced to 13% and 35%, respectively, in the SCG. Similar experiments were carried out to compare the transgene expression levels in the adrenal glands of wild-type and CNR4-mutated PAC transgenic animals. However, no detectable differences were found (Fig. 6D and data not shown). It is possible that the decreased expression levels of β4 and α3 in pineal gland and SCG of ΔCNR4-3 were impacted by the unintended deletion upstream of α5. However, this may not be the case, as the pattern of β4/α3/α5 transgene expression among the tissues examined in this line was not different from that in the other CNR4 mutant lines and the levels of α5 transgene expression were comparable among the CNR4 mutant lines in the SCG. Thus, our analysis of the three CNR4-mutated PAC lines suggests that CNR4 functions to coordinate the expression of β4 and α3, but not α5, in the pineal gland and SCG.
FIG. 6.
CNR4 is required for β4 and α3 transgene expression in the pineal gland and SCG. (A) RT-PCR analysis of ΔCNR4-2 transgene expression in various indicated tissues. Similar results were obtained for ΔCNR4-1 and ΔCNR4-3 transgenic lines. Pineal, pineal gland; Tg, trigeminal ganglia; Ag, adrenal gland; −, no RT control; +, rat SCG cDNA for rβ4, rα3, and rα5 and mouse SCG cDNA for mα3; r, rat-specific primer; m, mouse-specific primer; GAPDH, RT-positive control. (B) Real-time RT-PCR to quantitate β4 (top) and α3 (bottom) wild-type and CNR4-mutated transgene expression levels in the individual wild-type and CNR4-mutated PAC lines in pineal gland. (C) Real-time RT-PCR to quantitate β4 (top), α3 (middle), and α5 (bottom) transgene expression levels in the individual wild-type and CNR4-mutated PAC lines in SCG. (D) Real-time RT-PCR to quantitate transgene expression levels of β4 (left) and α3 (right) in adrenal gland. The data presented in panel D are the mean levels of expression of the three WT lines versus those of the three mutant (Δ) lines, where the wild-type level was set to 1. Total RNA was extracted from either pineal gland or SCG from four animals of each line. Relative transgene expression levels, normalized to endogenous mouse α3 expression level and copy number, were determined for each wild-type and mutant line. On the right-side portions of panels B and C are the mean levels of expression of the wild-type lines versus the mean levels of expression of the mutant lines, where the wild-type levels were set to 1. WT, means ± standard errors of the means for W1, W2, and W6; Δ1,2,3, means ± standard errors of the means for ΔCNR4-1, ΔCNR4-2, and ΔCNR4-3; Δ1,2, means ± standard errors of the means for ΔCNR4-1 and ΔCNR4-2. Error bars indicate the standard errors of the means. Student's t test was used for statistical analyses. N.S., not significantly different.
CNR4 is sufficient to direct transgene reporter expression in SCG neurons and pineal gland.
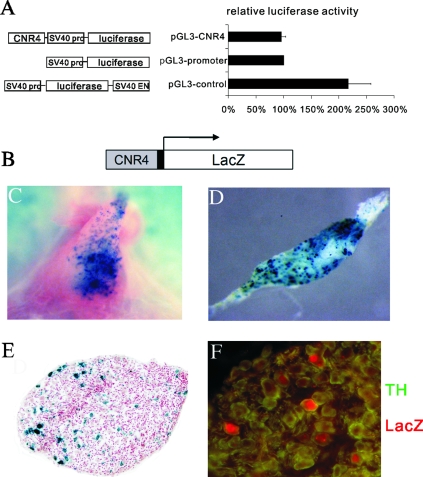
To provide additional experimental support for a role for CNR4 in transcriptional control of the cluster, we next investigated whether CNR4 was sufficient to direct reporter expression in cell types that express the endogenous β4/α3/α5 genes. The pheochromocytoma line PC12 expresses all three clustered genes, and therefore, we transfected into these cells a reporter in which CNR4 was placed upstream of a minimal promoter-luciferase cassette. However, CNR4 failed to either activate or repress luciferase expression in this cell type (Fig. 7A).
FIG. 7.
CNR4 is sufficient to direct LacZ reporter expression in pineal gland and SCG. (A) Transfection analysis of CNR4 activity in the adrenal gland-derived PC12 cell line. Luciferase reporters used in the transfection assays are indicated on the left. Data are presented as luciferase activity relative to that from the control pGL3-promoter transfection experiment. Error bars indicate the standard errors of the means from six independent experiments. (B) Schematic of the CNR4Z transgene reporter construct. The solid black box represents a 40-bp β-globin minimal promoter. (C) Whole-mount X-Gal staining of pineal gland. A single founder was analyzed for CNR4Z expression in the pineal gland. (D) Whole-mount SCG X-Gal staining. (E) X-Gal staining of SCG section followed by counterstaining with neutral red. (F) Coimmunostaining of TH (green) and β-Gal (red) in SCG. TH immunostaining was cytoplasmic, while β-Gal was concentrated in the nucleus. Eight out of a total of 17 independent lines showed CNR4Z expression in SCG.
We then investigated whether CNR4 could direct transgene expression in either the pineal gland or SCG neurons in vivo by generating transgenic mice carrying a construct, CNR4Z, in which a 700-bp fragment spanning the mouse CNR4 was placed immediately upstream of the β-globin minimal promoter and the bacterial LacZ gene encoding the β-galactosidase (Fig. 7B). In corroboration of our CNR4 PAC loss-of-function findings, expression of CNR4Z was detected in both the pineal gland and SCG (Fig. 7C, D, and E). Double immunostaining with an anti-TH antibody and anti-β-galactosidase antibody showed that CNR4Z expression in the SCG was restricted to TH+ neurons that express the endogenous β4/α3/α5 gene (Fig. 7F). Together, the PAC loss-of-function and CNR4Z studies show that CNR4 is not only necessary but also sufficient for transgene expression in pineal gland and SCG.
CNR4 is sufficient to direct transgene expression to brain neuronal cell types that express the nAChR gene cluster.
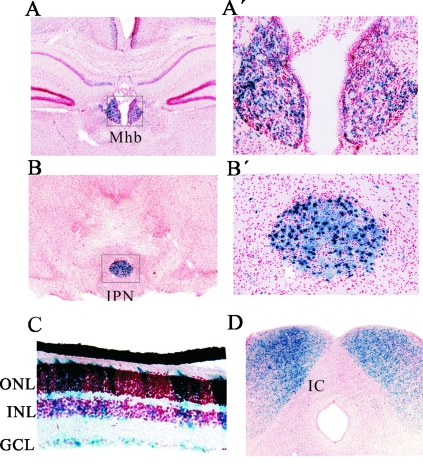
Having demonstrated that CNR4 was sufficient to direct transgene reporter expression in the pineal gland and SCG, we next wanted to investigate whether it might also be sufficient for reporter expression in β4/α3/α5 expressing neuronal cell types of the brain. Two well-documented and major sites of β4/α3/α5 expression in the brain are the medial habenula and interpeduncular nucleus (50, 55, 58), but to date, nAChR regulatory elements capable of directing transcription in these nuclei have not been reported. Interestingly, CNR4Z expression was detected in both of these nuclei (Fig. 8). Moreover, CNR4Z expression was also detected in the inferior colliculus, ganglion cell layer, and inner and outer nuclear layers of the retina, which are also sites of β4/α3/α5 expression (Fig. 8C and D). Similar β-globin LacZ reporters without CNR4 sequences fail to direct lacZ expression to these cell types (49). These findings strongly suggest that CNR4 is also involved in coordinating transcription of the cluster in various nAChR cluster-expressing neuronal cell types of the central nervous system (CNS).
FIG. 8.
CNR4 is sufficient to direct LacZ expression to neurons in the retina, medial habenula, interpeduncular nucleus, and inferior colliculus. (A to D) X-Gal staining of CNR4Z tissue sections. (A and A′) Medial habenula (Mhb). (B and B′) interpeduncular nucleus (IPN). Panels A′ and B′ are high-power images of the black-box regions in panels A and B. (C) Retina, ganglion cell layer (GCL), inner nuclear layer (INL), and outer nuclear layer (ONL). (D) Inferior colliculus (IC). Counterstain is neutral red. Four out of a total of 15 independent lines showed similar CNR4Z expression in these nuclei.
DISCUSSION
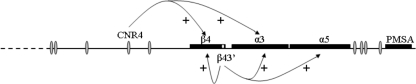
Although cell culture studies have provided detailed characterization of promoters and an ETS-dependent enhancer in the nAChR β4/α3/α5 subunit gene cluster, these studies have not provided insight into the mechanisms through which the cluster is coordinately regulated in the nervous system. To begin to investigate these mechanisms, we used a combination of PAC transgenesis and bioinformatics approaches to determine whether long-range control elements operate to coordinate expression of the β4/α3/α5 subunit genes. Our functional genomics analyses of cis elements in vivo and the identification of conserved noncoding regions flanking both sides of the cluster support a model in which expression of the β4/α3/α5 genes is regulated by shared gene- and cell type-specific long-range cis regulatory elements (Fig. 9).
FIG. 9.
Model for organization of the nAChR gene cluster regulatory terrain. The model proposes a chromatin domain bounded by CNRs (ovals) that operate together with β43′ to coordinate expression of β4/α3/α5 in various neural and endocrine cell types. The model predicts that control of the cluster is achieved through long-range interactions between individual gene-proximal promoters and the distant enhancers (β43′ and the individual CNRs) in a gene- and cell type-specific manner. The curved arrows indicate the cell type- and gene-specific interactions that are supported by the data presented in Results. We speculate that some of the CNRs may function as boundary elements to block inappropriate cross talk between regulatory elements for nAChR, PMSA, and other nearby genes. Dashed line, synteny break between mouse and human genomes; +, positive effects on transcription.
PAC transgenic approach.
Our investigation necessitated the simultaneous readouts of all three clustered genes in different neural and endocrine tissues. Thus, we chose to analyze bulk transcript levels in tissues as readouts of PAC transgene expression rather than introduce histological markers into each of the clustered genes. The use of an unmodified PAC also eliminated the possibility of adventitious effects of introduced heterologous sequences on cis element interactions within the cluster. However, the expression patterns of the clustered genes in the nervous system are complex, especially in the brain, and therefore, a limitation of our experimental plan is that it would be difficult to analyze PAC transgene expression in specific brain nuclei that express the endogenous cluster by measurement of bulk transcript levels. By contrast, expression of the clustered genes in different neural cell types of the peripheral nervous system and pineal gland provided a convenient approach to test our hypothesis. The nAChR cluster is coordinately expressed in chromaffin cells of the adrenal medulla, neurons of the SCG, and endocrine pinealocytes. Thus, we focused our analyses on these more accessible tissues in which the cluster is expressed in a single cell type within each of these tissues. With this approach, we could determine the impact of regulatory sequences on each gene within the cluster in tissues whose cholinergic synapses depend on the function of a β4α3α5 heteromeric receptor subtype.
The 132-kb PAC described in this study was used to generate several wild-type and mutated transgenic lines. Each of the wild-type lines expressed the three clustered rat transgenes in the adrenal gland, autonomic ganglia, and pineal gland as well as other tissues that express the endogenous mouse cluster. Furthermore, none of the PAC transgenes were expressed in several nonneural tissues that do not express the endogenous genes. Thus, a critical finding was that the 132-kb PAC includes sufficient regulatory information to accurately and reproducibly recapitulate the expression pattern of the cluster. However, we did notice variation in transgene transcript levels among the wild-type PAC lines in the tissues examined. These more subtle quantitative differences likely arise from position effects at the site of PAC integration, which is often observed with small transgenes as well as large BAC transgenes (1, 27). Nevertheless, the reproducible restricted expression patterns of the β4/α3/α5 PAC transgenes suggested that this approach could be used to investigate the impact of individual cis regulatory elements on each subunit gene in the cluster in different cell types in vivo.
Function of β43′ in vivo.
Our analysis in vivo showed that mutagenesis of the three ETS binding site core motifs in the β43′ enhancer led to significant decreases in the levels of β4, α3, and α5 PAC transgene expression in the adrenal gland. These findings are consistent with the previously defined properties of β43′ in cell culture studies: (i) its preferential activity in adrenal medullary-derived PC12 cells compared to that in other neural and nonneural lines (38), (ii) its ETS binding site dependence in PC12 cells (18, 39), and (iii) its differential activities in different neuronal cell types (18).
In addition to these confirmatory findings, we also present evidence in favor of β43′ operating as a shared regulatory element that helps to coordinate the expression of all three clustered nAChR genes in the adrenal gland. Because β43′ is part of the 3′ untranslated exon sequences of the β4 gene, it is possible that the decreased levels for mutated transgene-derived β4 transcripts seen in adrenal gland resulted from decreased posttranscriptional regulation rather than decreased β4 transcription. Although we cannot rule out a posttranscriptional effect on β4 expression, we note that in cell culture studies, β43′ is able to enhance minimal promoter activity of reporters when placed outside the reporter transcription unit, which provides support for the idea that it is a bona fide transcriptional enhancer. However, if β43′ ETS binding sites also influence posttranscriptional regulation of β4, then it may function at both the transcriptional and the posttranscriptional levels to coordinate expression of the cluster in the adrenal gland.
It is somewhat surprising that loss of β43′ function had only a modest effect on α3 transgene expression but no noticeable effect on β4/α5 transgene expression in the SCG, as previous transient-transfection-based analyses of β43′ indicated that it was able to preferentially drive reporter expression in dissociated SCG neurons compared to nonneuronal cells of the ganglion (39). This may be explained by the observation that the enhancer activity of β43′ on a minimal promoter is relatively weak in reporter assays performed in dissociated SCG neurons (unpublished data). Alternatively, β43′ activity in the SCG may be redundant to other cis elements, such as CNR4. It is possible that the interline variation in expression among the wild-type and mutated PAC lines precluded detection of a similar modest reduction in β4 and α5 transgene expression levels in the absence of β43′ activity in SCG.
We also reported previously that β43′ was preferentially active in retinal ganglion and amacrine cell types in dissociated retinal culture transfection assays (18). However, because β43′ is active in more than one retinal cell type, we did not attempt to quantitate PAC transgene expression in this tissue. Perhaps embryonic stem cell knock-in approaches or additional PAC transgenic lines that include histological markers could be used in future studies of β43′ function in tissues, such as retina and brain, which have complex patterns of β4/α3/α5 expression. The findings presented here provide a foundation with which to pursue these more in-depth and complex analyses of β43′ function.
Conserved noncoding regions.
Bioinformatics was used to identify conserved noncoding sequences flanking both sides of the cluster (Table 2). The results of this analysis were informative and immediately raised novel and testable hypotheses concerning the location and organization of additional cis regulatory information for the nAChR cluster. Based on the distribution, organization, and synteny of CNRs and other genes near the cluster, we hypothesize that the chromosomal region bounded by CNR1 and CNR9 is a phylogenetically conserved 150-kb chromatin expression domain that coordinates expression of the nAChR gene cluster in neural and endocrine cell types of diverse tissues (Fig. 9).
To begin to investigate this hypothesis, we selected CNR4 for functional characterization, as it is present as a single copy in the genome and it has the highest sequence conservation of all CNRs on the β4 side. Our findings suggest that CNR4 is a shared long-range control element that is involved in coordinating nAChR cluster expression in the peripheral and central nervous systems. First, mutagenesis of CNR4 showed that it is essential for both β4 and α3 transgene expression in the pineal gland, as levels of expression of these transgenes were nearly undetectable in the CNR4-mutated PAC lines. Second, the levels of β4 and α3 but not α5 transgene expression in CNR4-mutated PAC lines were also significantly reduced in the SCG. Third, CNR4 was able to direct lacZ transgene expression to SCG, pineal gland, and several CNS neuronal cell types in which prominent expression of all three clustered genes is well documented (55, 56, 69). To date, this is the only nAChR-associated sequence that has been shown to direct reporter expression in vivo to any of these major sites of nAChR cluster expression.
What might account for the effects of CNR4 on β4/α3 expression but not α5 expression in SCG? Perhaps CNR4 function is position dependent with respect to genes in the cluster. Additional support of this idea is the observation that β4 transgene expression was affected to a greater extent than that of α3 (Fig. 6). An alternative explanation is that CNR4 function may be determined by its relative orientation with respect to individual promoters in the cluster. Transcription of α5 on the opposite strand relative to β4 and α3 may prevent its interaction with CNR4. A third possibility is that CNR4 function is determined through promoter-specific interactions. It seems that none of these possibilities are necessarily mutually exclusive, and therefore, each may be important for the magnitude of the CNR4 effect on individual genes in the cluster. It will be interesting to determine whether the mechanism through which CNR4 controls transcription of the cluster explains why expression of α5 is not always coordinate with that of the β4 and α3 genes, for example, in the pineal gland.
In some ways, the characteristics we have determined so far for CNR4 are reminiscent of the core region of individual hypersensitive sites (HSs) that comprise the β-globin locus control region (LCR). Five evolutionarily conserved DNase I HS core sequences, each 200 to 400 bp long, spread out over a 14-kb region upstream of the ɛ-globin gene make up the LCR (33, 34). The LCR is required for physiological levels of erythroid cell-specific transcription of all genes in the β-globin locus, and each HS appears to function in an additive manner to achieve this level of transcription (3, 4). Although the LCR has an enhancing effect on transcription, it does not function like a classical orientation-independent and distance-independent enhancer, as LCR function is dependent on gene position and orientation (2, 53). Deletions of individual HSs differentially impact the expression of members of the β-globin gene cluster in a developmental-stage-specific manner (41). Our findings showing that CNR4 differentially impacts the expression of the clustered transgenes in different tissues suggests that it may be a component of an LCR-like regulatory domain. The CNRs may also possess another property of some β-globin HSs, that of insulating boundary elements that prevent inappropriate cross talk between β-globin gene regulatory elements and those of other nearby genes (17, 48).
Coordinate control of β4α3α5 heteromer expression.
The coordinate control of subunit genes in the nAChR cluster is essential for development of cholinergic synapses at many sites in nervous system. Our findings support a model in which β4α3α5 heteromers are made through the operation of at least two shared cis regulatory elements within and upstream of the β4 gene (Fig. 9). However, CNR4 and β43′ are not likely to be the only cis elements involved in the expression of the cluster, as CNR4Z expression was not detected in several brain stem nuclei that express the cluster, and a similar β43′-containing transgene construct failed to direct significant levels of reporter expression in the brain (unpublished data). We speculate that the other eight CNRs upstream of β4 and α5 may be the missing cis elements within a 150-kb nAChR chromatin domain.
In conclusion, our findings identify a route with which to further investigate the intrinsic transcriptional program that governs the expression of the nAChR subunit gene cluster. It will be of particular interest to determine the role of the various CNRs in the brain and to determine whether they are involved in the response of the cluster to presynaptic and target tissue-derived extrinsic cues. If human genetic variation exists within the highly conserved CNRs, they may impact nicotinic responses in the nervous system by affecting the level of β4α3α5 expression. Our findings suggest that the nAChR gene cluster may provide a relatively compact genetic model to investigate the transcriptional mechanisms that coordinate expression of clustered genes in nervous system.
.
Supplementary Material
Acknowledgments
We thank Neal Copeland for the EL250 bacterial strain. We thank Richard Zigmond for important comments on the final draft.
This work was supported by Public Health Service grant R01NS29123 from the NINDS.
Footnotes
Supplemental material for this article may be found at http:://mcb.asm.org/.
REFERENCES
- 1.Antoch, M. P., E. J. Song, A. M. Chang, M. H. Vitaterna, Y. Zhao, L. D. Wilsbacher, A. M. Sangoram, D. P. King, L. H. Pinto, and J. S. Takahashi. 1997. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89:655-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauchwitz, R., and F. Costantini. 2000. Developmentally distinct effects on human epsilon-, gamma- and delta-globin levels caused by the absence or altered position of the human beta-globin gene in YAC transgenic mice. Hum. Mol. Genet. 9:561-574. [DOI] [PubMed] [Google Scholar]
- 3.Bender, M. A., M. Bulger, J. Close, and M. Groudine. 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5:387-393. [DOI] [PubMed] [Google Scholar]
- 4.Bender, M. A., J. N. Roach, J. Halow, J. Close, R. Alami, E. E. Bouhassira, M. Groudine, and S. N. Fiering. 2001. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNaseI hypersensitive sites. Blood 98:2022-2027. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, C., E. A. Casanova, and P. D. Gardner. 1996. Transcriptional regulation of neuronal nicotinic acetylcholine receptor genes. Functional interactions between Sp1 and the rat beta4 subunit gene promoter. J. Biol. Chem. 271:32842-32848. [DOI] [PubMed] [Google Scholar]
- 6.Bigger, C. B., I. N. Melnikova, and P. D. Gardner. 1997. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor subunit gene. J. Biol. Chem. 272:25976-25982. [DOI] [PubMed] [Google Scholar]
- 7.Boulter, J., A. O'Shea-Greenfield, R. M. Duvoisin, J. G. Connolly, E. Wada, A. Jensen, P. D. Gardner, M. Ballivet, E. S. Deneris, D. McKinnon, S. Heinemann, and J. Patrick. 1990. α3, α5, and β4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J. Biol. Chem. 265:4472-4482. [PubMed] [Google Scholar]
- 8.Boutanaev, A. M., A. I. Kalmykova, Y. Y. Shevelyov, and D. I. Nurminsky. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420:666-669. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, R. T. 1996. Transcriptional regulation and cell specificity determinants of the rat nicotinic acetylcholine receptor α3 gene. Neurosci. Lett. 208:73-76. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Caro, A., C. Carrasco-Serrano, L. M. Valor, S. Viniegra, J. J. Ballesta, and M. Criado. 1999. Multiple functional Sp1 domains in the minimal promoter region of the neuronal nicotinic receptor alpha5 subunit gene. J. Biol. Chem. 274:4693-4701. [DOI] [PubMed] [Google Scholar]
- 11.Cordero-Erausquin, M., L. M. Marubio, R. Klink, and J. P. Changeux. 2000. Nicotinic receptor function: new perspectives from knockout mice. Trends Pharmacol. Sci. 21:211-217. [DOI] [PubMed] [Google Scholar]
- 12.Corriveau, R. A., and D. K. Berg. 1993. Coexpression of multiple acetylcholine receptor genes in neurons: quantification of transcripts during development. J. Neurosci. 13:2662-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couturier, S., L. Erkman, S. Valera, D. Rungger, S. Bertrand, J. Boulter, M. Ballivet, and D. Bertrand. 1990. α5, α3, and non-α3: three clustered avian genes encoding neuronal nicotinic acetylcholine receptor-related subunits. J. Biol. Chem. 265:17560-17567. [PubMed] [Google Scholar]
- 14.Deneris, E. S., N. Francis, J. McDonough, D. Fyodorov, T. Miller, and X. Yang. 2000. Transcriptional control of the neuronal nicotinic acetylcholine receptor gene cluster by the beta43′ enhancer, Sp1, SCIP and ETS transcription factors. Eur. J. Pharmacol. 393:69-74. [DOI] [PubMed] [Google Scholar]
- 15.de Souza, F. S., A. M. Santangelo, V. Bumaschny, M. E. Avale, J. L. Smart, M. J. Low, and M. Rubinstein. 2005. Identification of neuronal enhancers of the proopiomelanocortin gene by transgenic mouse analysis and phylogenetic footprinting. Mol. Cell. Biol. 25:3076-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Angelantonio, S., C. Matteoni, E. Fabbretti, and A. Nistri. 2003. Molecular biology and electrophysiology of neuronal nicotinic receptors of rat chromaffin cells. Eur. J. Neurosci. 17:2313-2322. [DOI] [PubMed] [Google Scholar]
- 17.Farrell, C. M., A. G. West, and G. Felsenfeld. 2002. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 22:3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francis, N., and E. S. Deneris. 2002. Retinal neuron activity of ETS domain-binding sites in a nicotinic acetylcholine receptor gene cluster enhancer. J. Biol. Chem. 277:6511-6519. [DOI] [PubMed] [Google Scholar]
- 19.Frank, M., and R. Kemler. 2002. Protocadherins. Curr. Opin. Cell Biol. 14:557-562. [DOI] [PubMed] [Google Scholar]
- 20.Fyodorov, D., and E. Deneris. 1996. The POU domain of SCIP/Tst-1/Oct-6 is sufficient for activation of an acetylcholine receptor promoter. Mol. Cell. Biol. 16:5004-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gahring, L. C., K. Persiyanov, and S. W. Rogers. 2004. Neuronal and astrocyte expression of nicotinic receptor subunit beta4 in the adult mouse brain. J. Comp. Neurol. 468:322-333. [DOI] [PubMed] [Google Scholar]
- 22.Gotti, C., and F. Clementi. 2004. Neuronal nicotinic receptors: from structure to pathology. Prog. Neurobiol. 74:363-396. [DOI] [PubMed] [Google Scholar]
- 23.Helms, A. W., A. L. Abney, N. Ben-Arie, H. Y. Zoghbi, and J. E. Johnson. 2000. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development 127:1185-1196. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez, S. C., S. Vicini, Y. Xiao, M. I. Davila-Garcia, R. P. Yasuda, B. B. Wolfe, and K. J. Kellar. 2004. The nicotinic receptor in the rat pineal gland is an alpha3beta4 subtype. Mol. Pharmacol. 66:978-987. [DOI] [PubMed] [Google Scholar]
- 25.Huang, M. M., N. Arnheim, and M. F. Goodman. 1992. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 20:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurst, L. D., C. Pal, and M. J. Lercher. 2004. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5:299-310. [DOI] [PubMed] [Google Scholar]
- 27.Kaufman, R. M., C. T. Pham, and T. J. Ley. 1999. Transgenic analysis of a 100-kb human beta-globin cluster-containing DNA fragment propagated as a bacterial artificial chromosome. Blood 94:3178-3184. [PubMed] [Google Scholar]
- 28.Khandekar, M., N. Suzuki, J. Lewton, M. Yamamoto, and J. D. Engel. 2004. Multiple, distant Gata2 enhancers specify temporally and tissue-specific patterning in the developing urogenital system. Mol. Cell. Biol. 24:10263-10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinjan, D. A., and V. van Heyningen. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76:8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 31.Levey, M. S., C. L. Brumwell, S. E. Dryer, and M. H. Jacob. 1995. Innervation and target tissue interactions differentially regulate acetylcholine receptor subunit mRNA levels in developing neurons in situ. Neuron 14:153-162. [DOI] [PubMed] [Google Scholar]
- 32.Levey, M. S., and M. H. Jacob. 1996. Changes in the regulatory effects of cell-cell interactions on neuronal AChR subunit transcript levels after synapse formation. J. Neurosci. 16:6878-6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levings, P. P., and J. Bungert. 2002. The human beta-globin locus control region. Eur. J. Biochem. 269:1589-1599. [DOI] [PubMed] [Google Scholar]
- 34.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Q., I. N. Melnikova, M. Hu, and P. D. Gardner. 1999. Cell type-specific activation of neuronal nicotinic acetylcholine receptor subunit genes by Sox10. J. Neurosci. 19:9747-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loots, G. G., R. M. Locksley, C. M. Blankespoor, Z. E. Wang, W. Miller, E. M. Rubin, and K. A. Frazer. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136-140. [DOI] [PubMed] [Google Scholar]
- 37.Mandelzys, A., B. Pié, E. S. Deneris, and E. Cooper. 1994. The developmental increase in ACh current densities on rat sympathetic neurons correlates with changes in nicotinic ACh receptor a-subunit gene expression and occurs independently of innervation. J. Neurosci. 14:2357-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonough, J., and E. Deneris. 1997. β43′: an enhancer displaying neural-restricted activity is located in the 3′-untranslated exon of the rat nicotinic acetylcholine receptor β4 gene. J. Neurosci. 17:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonough, J., N. Francis, T. Miller, and E. S. Deneris. 2000. Regulation of transcription in the neuronal nicotinic receptor subunit gene cluster by a neuron-selective enhancer and ETS domain factors. J. Biol. Chem. 275:28962-28970. [DOI] [PubMed] [Google Scholar]
- 40.Moretti, M., S. Vailati, M. Zoli, G. Lippi, L. Riganti, R. Longhi, A. Viegi, F. Clementi, and C. Gotti. 2004. Nicotinic acetylcholine receptor subtypes expression during rat retina development and their regulation by visual experience. Mol. Pharmacol. 66:85-96. [DOI] [PubMed] [Google Scholar]
- 41.Navas, P. A., K. R. Peterson, Q. Li, E. Skarpidi, A. Rohde, S. E. Shaw, C. H. Clegg, H. Asano, and G. Stamatoyannopoulos. 1998. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 18:4188-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nobrega, M. A., I. Ovcharenko, V. Afzal, and E. M. Rubin. 2003. Scanning human gene deserts for long-range enhancers. Science 302:413. [DOI] [PubMed] [Google Scholar]
- 43.Ovcharenko, I., M. A. Nobrega, G. G. Loots, and L. Stubbs. 2004. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32:W280-W286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry, D. C., Y. Xiao, H. N. Nguyen, J. L. Musachio, M. I. Davila-Garcia, and K. J. Kellar. 2002. Measuring nicotinic receptors with characteristics of alpha4beta2, alpha3beta2 and alpha3beta4 subtypes in rat tissues by autoradiography. J. Neurochem. 82:468-481. [DOI] [PubMed] [Google Scholar]
- 45.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenberg, M. M., R. C. Blitzblau, D. P. Olsen, and M. H. Jacob. 2002. Regulatory mechanisms that govern nicotinic synapse formation in neurons. J. Neurobiol. 53:542-555. [DOI] [PubMed] [Google Scholar]
- 48.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken b-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, M. M., K. C. Krueger, and E. S. Deneris. 2005. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. J. Neurosci. 25:2628-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheffield, E. B., M. W. Quick, and R. A. Lester. 2000. Nicotinic acetylcholine receptor subunit mRNA expression and channel function in medial habenula neurons. Neuropharmacology 39:2591-2603. [DOI] [PubMed] [Google Scholar]
- 51.Simon, J., H. Wakimoto, N. Fujita, M. Lalande, and E. A. Barnard. 2004. Analysis of the set of GABA(A) receptor genes in the human genome. J. Biol. Chem. 279:41422-41435. [DOI] [PubMed] [Google Scholar]
- 52.Spitz, F., C. Herkenne, M. A. Morris, and D. Duboule. 2005. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 37:889-893. [DOI] [PubMed] [Google Scholar]
- 53.Tanimoto, K., Q. Liu, J. Bungert, and J. D. Engel. 1999. Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature 398:344-348. [DOI] [PubMed] [Google Scholar]
- 54.Valor, L. M., A. Campos-Caro, C. Carrasco-Serrano, J. A. Ortiz, J. J. Ballesta, and M. Criado. 2002. Transcription factors NF-Y and Sp1 are important determinants of the promoter activity of the bovine and human neuronal nicotinic receptor beta 4 subunit genes. J. Biol. Chem. 277:8866-8876. [DOI] [PubMed] [Google Scholar]
- 55.Wada, E., D. McKinnon, S. Heinemann, J. Patrick, and L. W. Swanson. 1990. The distribution of RNA encoded by a new member of the neuronal nicotinic acetylcholine receptor gene family (a5) in the rat central nervous system. Brain Res. 526:45-53. [DOI] [PubMed] [Google Scholar]
- 56.Wada, E., K. Wada, J. Boulter, E. Deneris, S. Heinemann, J. Patrick, and L. W. Swanson. 1989. Distribution of alpha 2, alpha 3, alpha 4, and beta 2 neuronal nicotinic receptor subunit mRNAs in the central nervous system: a hybridization histochemical study in the rat. J. Comp. Neurol. 284:314-335. [DOI] [PubMed] [Google Scholar]
- 57.Wang, N., A. Orr-Urtreger, and A. D. Korczyn. 2002. The role of neuronal nicotinic acetylcholine receptor subunits in autonomic ganglia: lessons from knockout mice. Prog. Neurobiol. 68:341-360. [DOI] [PubMed] [Google Scholar]
- 58.Winzer-Serhan, U. H., and F. M. Leslie. 1997. Codistribution of nicotinic acetylcholine receptor subunit α3 and β4 mRNAs during rat brain development. J. Comp. Neurol. 386:540-554. [DOI] [PubMed] [Google Scholar]
- 59.Woolfe, A., M. Goodson, D. K. Goode, P. Snell, G. K. McEwen, T. Vavouri, S. F. Smith, P. North, H. Callaway, K. Kelly, K. Walter, I. Abnizova, W. Gilks, Y. J. Edwards, J. E. Cooke, and G. Elgar. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3:e7. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 60.Woon, P. Y., K. Osoegawa, P. J. Kaisaki, B. Zhao, J. J. Catanese, D. Gauguier, R. Cox, E. R. Levy, G. M. Lathrop, A. P. Monaco, and P. J. de Jong. 1998. Construction and characterization of a 10-fold genome equivalent rat P1-derived artificial chromosome library. Genomics 50:306-316. [DOI] [PubMed] [Google Scholar]
- 61.Yang, X., D. Fyodorov, and E. S. Deneris. 1995. Transcriptional analysis of acetylcholine receptor alpha 3 gene promoter motifs that bind Sp1 and AP2. J. Biol. Chem. 270:8514-8520. [DOI] [PubMed] [Google Scholar]
- 62.Yang, X., Y. Kuo, P. Devay, C. Yu, and L. Role. 1998. A cystein-rich isoform of neuregulin controls the level of expression of neuronal nicotinic receptor channels during synaptogenesis. Neuron 20:255-270. [DOI] [PubMed] [Google Scholar]
- 63.Yang, X., J. McDonough, D. Fyodorov, M. Morris, F. Wang, and E. S. Deneris. 1994. Characterization of an acetylcholine receptor alpha 3 gene promoter and its activation by the POU domain factor SCIP/Tst-1. J. Biol. Chem. 269:10252-10264. [PubMed] [Google Scholar]
- 64.Yang, X., P. Model, and N. Heintz. 1997. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat. Biotech. 15:859-865. [DOI] [PubMed] [Google Scholar]
- 65.Yang, X., F. Yang, D. Fyodorov, F. Wang, J. McDonough, K. Herrup, and E. Deneris. 1997. Elements between the protein-coding regions of the adjacent β4 and α3 acetylcholine receptor genes directs neuron-specific expression in the central nervous system. J. Neurobiol. 32:311-324. [PubMed] [Google Scholar]
- 66.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, X., and S. Firestein. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5:124-133. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, Y., E. Deneris, and R. E. Zigmond. 1998. Differential regulation of levels of nicotinic receptor subunit transcripts in adult sympathetic neurons after axotomy. J. Neurobiol. 34:164-178. [DOI] [PubMed] [Google Scholar]
- 69.Zoli, M., N. L. Novére, J. A. Hill, and J.-P. Changeux. 1995. Developmental regulation of nicotinic ACh receptor subunit mRNAs in the rat central and peripheral nervous systems. J. Neurosci. 15:1912-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.