Abstract
We have previously demonstrated that nitrosylcobalamin (NO-Cbl), an analogue of vitamin B12 that delivers nitric oxide (NO), had potent antiproliferative activity against several human cancer cell lines. NO-Cbl induced apoptosis via a death receptor/caspase-8 pathway. In this study, we demonstrate that a functional Apo2L/TRAIL receptor was necessary for the induction of cell death by NO-Cbl. Furthermore, the Apo2L/TRAIL death receptor DR4 (TRAIL R1) was S nitrosylated following NO-Cbl treatment. Human melanoma (A375), renal carcinoma (ACHN), and ovarian carcinoma (NIH-OVCAR-3) cells were treated with NO-Cbl and subjected to the biotin switch assay; S-nitrosylated DR4 was detected in all three cell lines. NO-Cbl treatment did not cause S nitrosylation of DR5. The seven cysteine residues located in the cytoplasmic domain of DR4 were individually point mutated to alanines. NIH-OVCAR-3 cells expressing the DR4 C336A mutation lacked S nitrosylation following NO-Cbl treatment. Overexpression of wild-type DR4 sensitized cells to growth inhibition by NO-Cbl. Cells expressing the DR4 C336A mutant were more resistant to NO-Cbl and Apo2L/TRAIL than were the other six C-A mutations or wild-type cells. The C336A mutant also displayed blunted caspase-8 enzymatic activity following NO-Cbl treatment compared to the other mutants. Thus, DR4 residue C336 becomes S nitrosylated and promotes apoptosis following NO-Cbl treatment.
Mammalian cells have two major apoptotic pathways, the extrinsic and intrinsic pathways. The extrinsic pathway is triggered by members of the death receptor superfamily (Fas/CD95, TNFR1, and TRAIL receptors DR4 and DR5). Binding of ligands to their cognate death receptors induces receptor clustering and the formation of a death-inducing signaling complex. The intrinsic apoptotic pathway is controlled by mitochondrial events.
We have previously demonstrated the antitumor activity of nitrosylcobalamin (NO-Cbl), a prodrug based upon vitamin B12 that delivers nitric oxide (NO). NO-Cbl induces the expression of tumor necrosis factor-related apoptosis-inducing ligand (Apo2L/TRAIL) and its receptors (DR4 and DR5) in ovarian carcinoma cells (4). NO-Cbl induces tumor cell apoptosis through the extrinsic apoptotic pathway rather than the mitochondrion-dependent intrinsic pathway.
Nitric oxide is a pleiotropic short-lived free radical that regulates blood vessel and airway tone, inflammation, and apoptosis. NO covalently modifies heme groups (as in guanylyl cyclase) and also nitrosylates protein sulfhydryl groups (S nitrosylation), an important posttranslational modification that affects signal transduction (27). Nitrosylation of cellular proteins regulates the normal physiologic ventilatory response to hypoxia (21), ion channel activity and neurotransmission (6), smooth muscle relaxation (19), and blood pressure regulation (8).
In neurons, the plasma membrane N-methyl-d-aspartate receptor becomes S nitrosylated by neuronal nitric oxide synthase (NOS1), which modulates receptor activity (15). Recently, S nitrosylation of the estrogen receptor by synthetic NO donors such as (DETA)-NONOate {(z)-1-[2-(2-aminoethyl)-N-(ammonioethyl) amino] diazen-1-ium-1,2-diolate} has been demonstrated, resulting in decreased transcriptional activity of estradiol-stimulated genes; it has been postulated that endogenous endothelial NOS (NOS3) S nitrosylates the estrogen receptor as part of normal signal transduction following ligation of the estrogen receptor by estradiol (10). In endothelial cells, NOS3 appears to S nitrosylate itself, which inhibits NOS3 activity; this enzymatic repression is reversed by vascular endothelial growth factor (7). Thus, in many receptor- and membrane-associated signaling pathways, S nitrosylation plays an important physiologic role.
S-nitrosylated proteins regulate apoptosis in a complex fashion (5). GAPDH (glyceraldehyde-3-phosphate dehydrogenase), when S nitrosylated, translocates to the nucleus and induces apoptosis (24); conversely, caspase cleavage and activation are inhibited by S nitrosylation (13). The Apo2L/TRAIL receptor DR4 (TRAIL R1, TNFR superfamily 10A) is a transmembrane protein of 57 kDa that induces caspase-8 activation following receptor ligation (25). We hypothesized that treatment with NO-Cbl might result in S nitrosylation of DR4. We utilized the biotin switch assay (16) to demonstrate that DR4 became S nitrosylated following NO-Cbl treatment. Point mutagenesis was used to identify cysteine residues of DR4 that modulated receptor function. The specific aim of this study was to identify cysteine residues of the DR4 Apo2L/TRAIL receptor that might be important for apoptosis induction following NO-Cbl treatment.
MATERIALS AND METHODS
Reagents.
Recombinant human Apo2L/TRAIL (a generous gift from Avi Ashkenazi, Genentech Inc., San Francisco, CA) consisted of >9% trimeric protein with Zn2+, which is necessary for optimal biological activity (20). Antibody to FLAG (rabbit anti-FLAG) (Immunology Consultants Laboratory, Inc., Newberg, OR), anti-DR4 polyclonal antibody (EMDBiosciences, San Diego, CA), anti-DR5 monoclonal antibody (Imgenex, Sorrento Valley, CA), the nitric oxide donors DETA-NONOate (NOC-18) (a gift from Joseph Hrabie, NCI, Frederick, MD), and S-nitroso-l-acetyl-dl-penicillamine (SNAP) (Calbiochem, San Diego, CA) were utilized in this study.
Synthesis of nitrosylcobalamin.
Nitrosylcobalamin was synthesized as previously described (3, 4). Hydroxocobalamin (vitamin B12) acetate (Hebei Huarong Pharmaceutical Co., Hebei Province, People's Republic of China) was dissolved in dichloromethane (Burdick and Jackson, Muskegon, MI) and exposed to chemically pure NO gas (Praxair, Wickliff, OH) at 75 lb/in2. The reaction proceeded in a closed system within a high-pressure gas cylinder (Praxair, Cleveland, OH). The system was nitrogen purged daily and evacuated prior to NO exposure. The NO gas was scrubbed prior to entering the system using a stainless steel cylinder (Abbott valve and fitting; Swagelok, Solon, OH) containing NaOH pellets. The solid NO-Cbl product was collected following rotary evaporation of the solvent and stored at −80°C prior to use.
Cell culture.
Human melanoma tumor cell line A375 and human renal carcinoma tumor cell line ACHN were grown in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) supplemented with heat-inactivated 5% fetal bovine serum (Mediatech) and 1% antibiotic-antimycotic (Invitrogen, San Diego, CA). Human ovarian carcinoma tumor cell line NIH-OVCAR-3 was grown in RPMI 1640 medium (Mediatech) supplemented with heat-inactivated 5% fetal bovine serum (Mediatech) and 1% antibiotic-antimycotic (Invitrogen). Cells were maintained in a 5% CO2 atmosphere at 37°C in a humidified tissue culture incubator. Cells were confirmed to be mycoplasma free by PCR.
SRB cell growth assay.
Cells were plated in 96-well plates in 0.2-ml aliquots containing 3,000 cells. After 6 h, to allow for cell adherence, drug stock solutions were diluted in medium and added to the wells. The cells were cultured for 4 days in the continuous presence of the agents. Growth was monitored by the sulforhodamine B (SRB; Sigma Chemical, St. Louis, MO) colorimetric assay (26) as follows. After 4 days, the medium was removed, and the cells were fixed with 10% trichloroacetic acid and stained with SRB. Bound dye was eluted from the cells with 10 mM Tris-HCl (pH 10.5), and absorbance was measured at 570 nm using a Lab System Multiscan RC 96-well plate reader (Lab System Multiscan RC; Thermo Lab System, Franklin, MA). To quantify the growth of the cells, the experimental absorbance values (Aexp) were compared with initial absorbance readings representing the starting cell numbers (Aini). To determine the starting cell number, an additional 96-well plate was seeded with cells and fixed at the beginning of the experiment. After 4 days of growth, the untreated control cells and drug-treated cells were fixed and stained with SRB. The absorbances derived from the initial plate and from the untreated cells at the end of the growth period (Afin) were defined as 0% and 100% growth, respectively. Growth of cells was quantified as follows: % control growth = 100% × (Aexp − Aini)/(Afin − Aini). The percent control growth is expressed as a percentage of untreated controls. A decrease in cell number (death) is represented as a negative value on the y axis. Each treatment group contained eight replicates.
TUNEL assay.
NIH-OVCAR-3 cells transfected with vector and Flag-DR4 were cultured overnight and exposed to various treatments (isotype control antibody [Ab], anti-Flag monoclonal Ab [mAb], and Apo2L/TRAIL). Apoptotic cells were detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end-labeling (TUNEL) staining using a commercially available kit (APO-BRDU kit; BD PharMingen, San Diego, CA). Cells were processed according to the manufacturer's recommended protocol. The percentage of fluorescein isothiocyanate-positive cells was analyzed by fluorescence-activated cell scanning (Facsvantage; Becton Dickinson, San Diego, CA).
Construction of Flag-DR4 mutants.
Mutants of Flag-DR4 were generated by PCR-based site-directed mutagenesis using Flag-DR4 as a template in the pcDNA3 vector (provided by E. S. Alnemri, Thomas Jefferson University, Philadelphia, PA). Flag-DR4 lacks the 108 N-terminal residues of wild-type DR4; these residues have been replaced with the Flag epitope (DYKDDDK) that is preceded by the Fas signal peptide (22). The cytoplasmic cysteine residues of Flag-DR4 (C261, C262, C263, C268, C274, C279, and C336) were replaced with alanine to generate point mutants, named C1 through C7 for brevity. Thus, the wild-type sequences (in boldface type) VAVLIVCCCIGSG (DR4-C1), AVLIVCCCIGSGC (DR4-C2), VLIVCCCIGSGRG (DR4-C3), CCIGSGCGGDPKC (DR4-C4), CGGDPKCMDRVCF (DR4-C5), KCMDRVCFWRLGL (DR4-C6), and ADLTGQCLLGPAE (DR4-C7) were changed to VAVLIVACCIGSG (DR4-C1), AVLIVCACIGSGC (DR4-C2), VLIVCCAIGSGRG (DR4-C3), CCIGSGAGGDPKC (DR4-C4), CGGDPKAMDRVCF (DR4-C5), KCMDRVAFWRLGL (DR4-C6), and ADLTGQALLGPAE (DR4-C7), respectively. Mutants were amplified by PCR using the following primers: 5′-GTGGCTGTGCTGATTGTCGCTTGTTGCATCGGCTCAGGT-3′ and 5′-ACCTGAGCCGATGCAACAAGCGACAATCAGCACAGCCAC-3′ for DR4-C1, 5′-GCTGTGCTGATTGTCTGTGCTTGCATCGGCTCAGGTTGT-3′ and 5′-ACAACCTGAGCCGATGCAAGCACAGACAATCAGCACAGC-3′ for DR4-C2, 5′-GTGCTGATTGTCTGTTGTGCCATCGGCTCAGGTTGTGGA-3′ and 5′-TCCACAACCTGAGCCGATGGCACAACAGACAATCAGCAC-3′ for DR4-C3, 5′-TGTTGCATCGGCTCAGGTGCTGGAGGGGACCCCAAGTGC-3′ and 5′-GCACTTGGGGTCCCCTCCAGCACCTGAGCCGATGCAACA-3′ for DR4-C4, 5′-TGTGGAGGGGACCCCAAGGCCATGGACAGGGTGTGTTTC-3′ and 5′-GAAACACACCCTGTCCATGGCCTTGGGGTCCCCTCCACA-3′ for DR4-C5, 5′-AAGTGCATGGACAGGGTGGCTTTCTGGCGCTTGGGTCTC-3′ and 5′-GAGACCCAAGCGCCAGAAAGCCACCCTGTCCATGCACTT-3′ for DR4-C6, and 5′-TCCCCAGGGGAGGCACAGGCTCTGCTGGGACCGGCAGAA-3′ and 5′-TTCTGCCGGTCCCAGCAGAGCCTGTGCCTCCCCTGGGGA-3′ for DR4-C7. The PCR products were digested with BamHI and XhoI, ligated into the pcDNA3 vector, and then transformed into Escherichia coli DH5α. All mutations were confirmed by sequencing.
Transfection.
Cells were transfected with mutants using Cell Line Nucleofector Kit T (program T-27; AMAXA, Koeln, Germany) according to the manufacturer's protocol. Transfection efficiency was routinely 85 to 90%, as determined by transfection of enhanced green fluorescent protein reporter plasmid and quantitation by flow cytometry.
Biotin switch assay.
The biotin switch assay was performed as described previously by Jaffrey and Snyder (16), using low-light conditions and opaque tubes. Briefly, cells were washed in phosphate-buffered saline (PBS), homogenized in HEN buffer (250 mM HEPES-NaOH, pH 7.7, 1 mM EDTA, 0.1 mM neocuproine). Free thiols were blocked by methylation with methyl methanethiosulfonate (Sigma). Unreacted methyl methanethiosulfonate was removed by protein precipitation in 10 volumes of acetone (−20°C). Cysteine residues that had been S nitrosylated by NO-Cbl were converted to free thiols with sodium ascorbate (1 mM final concentration), which does not alter the methylated thiols. The free thiols were then biotinylated with biotin-hexyl pyridyldithiopropionamide (HPDP) at 25°C for 1 h. Thus, the S-nitrosylated cysteines were switched for biotin. In some reaction mixtures, biotin-HPDP was omitted as a negative control. Proteins were precipitated by chilled acetone, and the pellet was resuspended in HENS buffer (250 mM HEPES, pH 7.7, 1 mM EDTA, 0.1 mM neocuproine, 1% sodium dodecyl sulfate [SDS]). Biotinylated proteins were precipitated with streptavidin-agarose (Sigma) and eluted from the beads with a solution containing 20 mM HEPES-NaOH, pH 7.7, 100 mM NaCl, 1 mM EDTA, and 100 mM 2-mercaptoethanol.
Immunoblot analysis.
After biotinylation, proteins were separated by SDS-polyacrylamide gel electrophoresis using a 10% gel, transferred onto a nitrocellulose membrane (Millipore, Billerica, MA), and blocked in Blocker Blotto (Pierce, Rockford, IL) at room temperature for 2 h. Blots were incubated with primary antibody diluted in Blocker Blotto overnight at 4°C, washed two times for 10 min each with Tris-buffered saline-Tween, and then incubated with goat anti-rabbit secondary antibody diluted in Blocker Blotto at room temperature for 1 h. Bands were detected with SuperSignal West Pico chemiluminescent substrate (Pierce) according to the manufacturer's protocol.
RT-PCR.
Total RNA was isolated from NIH-OVCAR-3 cells using the SV total RNA isolation system (Promega, Madison, WI) according to the manufacturer's protocol. The primers used for Flag-DR4 reverse transcription-PCR (RT-PCR) were 5′-GACTACAAGGACGACGATGA-3′ and 5′-GTGACACCTGTCAAATCTGC-3′. The primers used for endogenous DR4 were 5′-ATGGCGCCACCACCAGCTAG-3′ and 5′-GCTGTGTTCCTGGTCGTGGT-3′. PCR for Flag-DR4 started with 1 cycle of 94°C for 2 min, 41°C for 45 s, and 72°C for 2 min followed by 94°C for 30 s, 41°C for 45 s, and 72°C for 2 min for 36 cycles. PCR for endogenous DR4 started with 1 cycle of 94°C for 2 min, 45°C for 45 s, and 72°C for 2 min followed by 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min for 36 cycles.
Caspase-8 activity.
The TruPoint caspase-8 assay kit (Perkin-Elmer, Turku, Finland) was used according to the manufacturer's protocol. The caspase-8 substrate was a hexapeptide with a fluorescent europium (Eu) chelate coupled to one end and a quencher of europium fluorescence coupled to the other end. Fluorescence is quenched when the labels are in close proximity in the intact hexapeptide. After substrate cleavage and separation of the labels, lanthanide fluorescence is measured by time-resolved fluorometry. The Eu signal was detected fluorometrically with a Wallac 1420 multilabel counter (Perkin-Elmer, Gaithersburg, MD) using a 340-nm excitation filter and a 615-nm emission filter. NIH-OVCAR-3 cells were transfected with vector or DR4 mutants; 48 h after transfection, cells were treated with NO-Cbl (50 μM) for 2 h. Cells were treated with 100 ng/ml Apo2L/TRAIL for 3 h and 6 h as positive controls for caspase-8 activation. Cells were washed twice with cold PBS and lysed with cell lysis buffer (BD Biosciences Clontech, Palo Alto, CA); 5 μg of total cell lysate was subjected to this assay. Background (fluorescence units generated by substrate and reaction buffer) averaged 905 units and was subtracted from all measurements. Each data point represents the mean ± standard error (SE) of triplicate measurements.
RESULTS
Effect of Apo2L/TRAIL blockade on NO-Cbl-induced death.
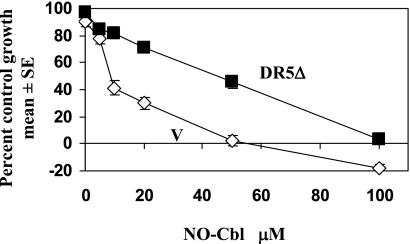
To examine whether Apo2L/TRAIL-mediated signaling was required for the induction of death by NO-Cbl, NIH-OVCAR-3 cells were transfected with a dominant negative DR5 mutant (DR5Δ) in which the intracellular death domain had been deleted (11, 22). Expression of the DR5Δ mutant interferes with signaling mediated by endogenous DR4 and DR5 death receptors by acting as a decoy receptor, binding ligand and thereby reducing the availability for ligand binding to DR4 and DR5. As shown in Fig. 1, vector-expressing cells (V) had a 50% inhibitory dose (ID50) of 10 μM, whereas cells expressing the DR5Δ mutant had an ID50 of 50 μM. Stably transfected NIH-OVCAR-3 cells expressing the DR5Δ construct were resistant to the antiproliferative effects of NO-Cbl. Thus, the pathway utilizing Apo2L/TRAIL and its receptors appears to be critical for the induction of cell death by NO-Cbl.
FIG. 1.
Effect of Apo2L/TRAIL blockade on NO-Cbl antiproliferative activity. NIH-OVCAR-3 cells were transiently transfected with vector alone (V) or DR5Δ, a dominant negative death receptor that lacks the cytoplasmic domain and interferes with signaling through DR4 and DR5. Cells were grown in the presence of NO-Cbl for 4 days, and the cell number was determined at the end of the assay (each data point represents n = 8).
S nitrosylation of Apo2L/TRAIL receptor DR4.
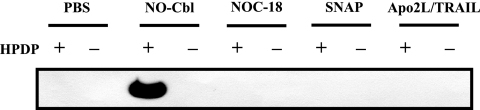
To determine whether DR4 was S nitrosylated following treatment with nitric oxide donors, NIH-OVCAR-3 cells were treated with PBS, NO-Cbl (50 μM), DETA-NONOate (NOC-18) (50 μM), or SNAP (50 μM) overnight. Apo2L/TRAIL (100 ng/ml) was also included to determine whether S nitrosylation occurred during Apo2L/TRAIL signal transduction. Cell lysates were subjected to the biotin switch assay followed by immunoprecipitation with streptavidin-agarose. Protein eluted from the agarose was subjected to SDS-polyacrylamide gel electrophoresis and then immunoblotted with anti-DR4. NO-Cbl treatment resulted in the detection of S-nitrosylated DR4 protein (Fig. 2, lane 3). When HPDP-biotin was eliminated from the assay, S-nitrosylated cysteine could not be switched for biotin, and no signal was detected (Fig. 2, lane 4). No immunoblot signal was detected when cells were treated with NOC-18, SNAP, or Apo2L/TRAIL. Thus, the other two NO donors did not induce S nitrosylation of DR4, nor did Apo2L/TRAIL, the natural ligand of DR4.
FIG. 2.
S nitrosylation of Apo2L/TRAIL receptor R1 (DR4) in NIH-OVCAR-3 cells. Cells were treated with PBS, NO-Cbl (50 μM), DETA-NONOate (NOC-18) (50 μM), SNAP (50 μM), or Apo2L/TRAIL (100 ng/ml) overnight. The biotin switch assay was performed, followed by immunoblotting for DR4. The band indicates the presence of S-nitrosylated DR4. Data are representative of three independent experiments.
To examine DR4 nitrosylation in other cell lines of different histologies, human melanoma (A375) and renal carcinoma (ACHN) cells were treated with NO-Cbl (50 μM) overnight. Cell lysates were subjected to the biotin switch assay followed by immunoprecipitation with streptavidin-agarose as in Fig. 2. A strong signal was detected when A375 and ACHN cells were treated with NO-Cbl and HPDP-biotin was included in the reaction mixture (Fig. 3, lanes 2 and 6), indicating that at least one cysteine residue of DR4 was S nitrosylated in all three cell lines.
FIG. 3.
S nitrosylation of Apo2L/TRAIL receptor R1 (DR4) in A375 and ACHN cells. Cells were treated with NO-Cbl (50 μM) overnight. Cells lysates were subjected to biotin switch assay followed by immunoprecipitation with streptavidin-agarose and then immunoblot analysis with an anti-DR4 monoclonal antibody. Bands were detected only in NO-Cbl-treated cells and when biotin-HPDP was included in the assay.
Overexpressed Flag-DR4 is functional.
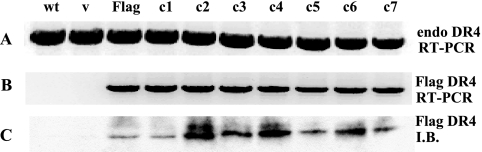
Flag-tagged DR4 was utilized for the expression of DR4 mutants. Thus, episomally expressed mutants could be distinguished from endogenously expressed DR4. Wild-type endogenous DR4 was expressed at similar levels in all cells expressing Cys-Ala point mutants as well as in vector-transfected and wild-type cells (Fig. 4A). Flag-specific mRNA (Fig. 4B) and Flag-tagged DR4 protein were detected only in cells transfected with Flag-DR4 (Fig. 4C, lane 3, Flag) or in cells transfected with point mutants (Fig. 4C, lanes c1 to c7).
FIG. 4.
Expression of DR4 mutants in NIH-OVCAR-3 cells. Cells were transiently transfected with vector (v), Flag-DR4, or Flag-DR4 mutants (c1 to c7). (A) After 48 h, RNA was isolated from cells, and RT-PCR was performed. Endogenous DR4 mRNA was detected at comparable levels in untransfected, vector-transfected, and mutant-transfected cells. (B) Expression of Flag-specific mRNA was detected only in cells transfected with wild-type (wt) Flag-DR4 and Flag-DR4 mutants. (C) Immunoblot (I.B.) with an anti-Flag monoclonal antibody detected fusion proteins only in lysates of Flag-DR4-transfected cells.
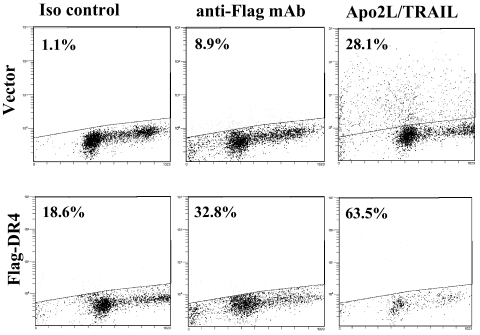
To determine if the Flag-DR4 Apo2L/TRAIL receptor could transduce a death signal in response to ligand, NIH-OVCAR-3 cells were transiently transfected with Flag-DR4 or the pcDNA3 vector. Cells were treated with anti-Flag mAb (1.67 mg/ml) or Apo2L/TRAIL (100 ng/ml) overnight and assayed for TUNEL staining by flow cytometry. Treatment of Flag-DR4-transfected cells with Apo2L/TRAIL resulted in 63.5% TUNEL-positive cells compared to 28.1% in cells expressing vector alone (Fig. 5). Cells expressing the Flag-DR4 fusion protein also displayed enhanced TUNEL staining following receptor ligation with anti-Flag mAb (32.8% and 8.9% for Flag-DR4 and vector, respectively). Thus, the Flag-DR4 receptor could transduce a death signal in response to natural ligand or following cross-linking with an anti-Flag antibody.
FIG. 5.
TUNEL assay. NIH-OVCAR-3 cells were transiently transfected with vector or Flag-DR4, and 48 h later, cells were treated with isotype control Ab (1.67 mg/ml) (Iso control), anti-Flag mAb (1.67 mg/ml), or Apo2L/TRAIL (100 ng/ml) overnight. Apoptotic cells were detected by flow cytometry using the APO-BRDU kit (BD PharMingen). Events above the line represent fluorescein isothiocyanate-stained apoptotic cells, and percentages of positive cells are indicated for each treatment group. Treatment of Flag-DR4-transfected cells with Apo2L/TRAIL resulted in the highest levels of apoptosis compared to cells expressing vector alone. Cells expressing the Flag-DR4 fusion protein also displayed enhanced TUNEL staining following receptor ligation with anti-Flag antibody. The experiment was performed three times, with similar results.
S nitrosylation of DR4.
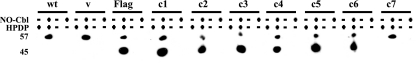
To identify which residues of DR4 were sensitive to S nitrosylation, cells were transfected with point mutants, treated with NO-Cbl overnight, and subject to the biotin switch assay. As in Fig. 2 and 3, S nitrosylation was not detected in untreated cells (Fig. 6, lane 1). Treatment with NO-Cbl induced S nitrosylation of endogenous DR4 (57-kDa band) in all transfectants and wild-type cells. Cells transfected with Flag-DR4 or with point mutants C1 to C6 displayed an additional band (45 kDa) consistent with the predicted molecular mass of Flag-DR4. However, the C7 mutant lacked the 45-kDa band, suggesting that the C336A mutation destroys a potential site for S nitrosylation.
FIG. 6.
S-nitrosylated DR4 protein expression in wild-type and transfected NIH-OVCAR-3 cells. NIH-OVCAR-3 cells were transiently transfected with vector (v), Flag-DR4, or Flag-DR4 point mutants. Two days after transfection, cells were treated with NO-Cbl (50 μM) overnight. Cell lysates were subjected to the biotin switch assay followed by immunoprecipitation with streptavidin-agarose and then immunoblot analysis with anti-DR4 polyclonal antibody. Immunoblot analysis demonstrated that wild-type (wt) and transfected cells all contained nitrosylated endogenous DR4 following NO-Cbl treatment. Cells transfected with DR4-C7 lacked nitrosylated Flag-DR4 following NO-Cbl treatment.
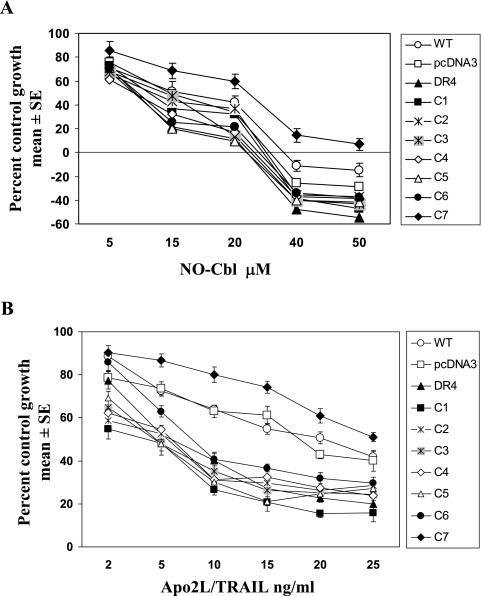
Effect of DR4 mutants on cell growth and sensitivity to NO-Cbl and Apo2L/TRAIL.
The biologic effect of NO-Cbl on cells transfected with DR4 mutants was determined by antiproliferative assays. NIH-OVCAR-3 cells transiently transfected with DR4 mutants were treated with NO-Cbl for 4 days. Cells that overexpressed Flag-DR4 and mutants C1 through C6 displayed enhanced sensitivity to NO-Cbl (Fig. 7A). The ID50 was reduced from 10 μM (vector) to 6 μM (Flag-DR4). Vector-transfected and untransfected cells displayed similar sensitivities to NO-Cbl. However, the C7 mutant was more resistant to NO-Cbl than were the other mutants; the ID50 was 25 μM. Thus, overexpression of Flag-DR4 enhanced sensitivity to growth inhibition by NO-Cbl, whereas the C7 mutation (C336A) conferred relative resistance to NO-Cbl. The sensitivities of individual mutants to growth inhibition by Apo2L/TRAIL displayed a similar pattern. The ID50 of wild-type and empty-vector-transfected cells was approximately 20 ng/ml Apo2L/TRAIL; for Flag-DR4 and mutants C1 to C6, it ranged from 5 to 10 ng/ml, whereas the C7 mutant displayed relative resistance, with an ID50 of 25 ng/ml (Fig. 7B).
FIG. 7.
Effect of the DR4 mutation on the antiproliferative activity of NO-Cbl. (A) NIH-OVCAR-3 cells were transiently transfected with vector, Flag-DR4, or Flag-DR4 mutants and then treated with NO-Cbl for 4 days. Growth was measured by the colorimetric sulforhodamine B assay. Data (n = 8 for each data point) are expressed as the mean percentages of control ± SE. The experiment was performed three times, with similar results. (B) Cells were transfected as described above and then plated and exposed to Apo2L/TRAIL for 4 days. Wild-type (WT) and vector-transfected cells displayed similar sensitivities to Apo2L/TRAIL. Mutants C1 to C6 exerted effects similar to that of Flag-DR4, whereas mutant C7 displayed relative resistance to Apo2L/TRAIL.
Effect of NO-Cbl on caspase-8 activity.
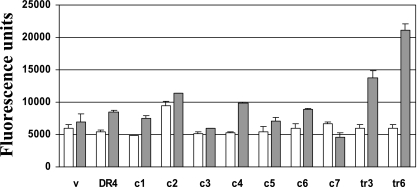
Since the ligation of Apo2L/TRAIL receptors results in caspase-8 activation, signaling through the mutant DR4 constructs was investigated using a time-resolved fluorometric energy transfer assay utilizing a hexapeptide substrate. After 2 h of treatment with NO-Cbl, elevated caspase-8 activity was detectable in vector and in cells transfected with constructs encoding wild-type DR4 or mutants C1 to C6 (Fig. 8). However, cells expressing the C7 mutant (C336A) displayed a blunted response to NO-Cbl; fluorescence in C7-expressing cells decreased 20 to 30% following NO-Cbl treatment. Data represent means ± SE of triplicate measurements.
FIG. 8.
Time-resolved fluorometric energy transfer caspase-8 assay. Vector-, Flag-DR4-, and mutant-transfected NIH-OVCAR-3 cells (C1 to C7) were subjected to NO-Cbl treatment (50 μM, 2 h). As positive controls for caspase-8 enzymatic activity, vector-transfected cells were exposed to Apo2L/TRAIL (100 ng/ml, 3 to 6 h) (lanes tr3 and tr6). Cell lysates were analyzed for their ability to cleave a europium-labeled hexapeptide and generate a fluorescence signal at 615 nm.
Effect of NO-Cbl on DR5.
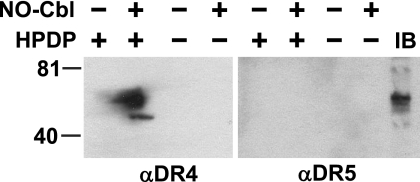
There are two agonistic Apo2L/TRAIL receptors, DR4 and DR5. Because NO-Cbl treatment resulted in S nitrosylation of DR4, the biotin switch assay was used to analyze DR5. Cell extracts that were immunoprecipitated with streptavidin-agarose and probed with anti-DR5 antibody demonstrated no detectable signal (Fig. 9, lane 6). A parallel blot probed with anti-DR4 confirmed the presence of S-nitrosylated DR4 (Fig. 9, lane 2). Simple immunoblot analysis (without lysate subjected to a biotin switch) showed that DR5 was present in the lysates.
FIG. 9.
Biotin switch assay of DR4 and DR5. NIH-OVCAR-3 cells were subjected to the biotin switch assay as described in the legend of Fig. 3. Lysates made from NO-Cbl-treated cells displayed a signal when biotin-HPDP was included in the assay and when probed with anti-DR4 (lane 2) but not when probed with anti-DR5 (lane 6). Immunoblot (IB) analysis detected DR5 in cell lysates that were not subjected to a biotin switch (lane 9).
DISCUSSION
In this study, we present evidence that NO-Cbl, a prodrug that releases NO following receptor-mediated endocytosis, nitrosylates the Apo2L/TRAIL R1 receptor DR4. NO-Cbl induced S nitrosylation, whereas two other NO donors (SNAP and NOC-18) did not. Even though NO diffuses freely, the absorption characteristics and kinetics of NO release of different donors, as well as the microenvironment and concentration of reducing agents, determine which protein targets become nitrosylated. Neighboring amino acids that affect cysteine reactivity and the presence of a hydrophobic environment promote the formation of S-nitrosylating species via the reaction between NO and O2 (14). Stamler et al. previously proposed that (K/R/H)C(D/E) is a consensus motif for the S nitrosylation of proteins by S transnitrosation with the acidic amino acid following the cysteine residue being the most important (28). This linear motif is lacking in DR4; however, it may exist in the three-dimensional structure of the receptor. Local hydrophobicity also promotes S nitrosylation (13). Indeed, 8 out of the 10 flanking residues surrounding C336 are nonpolar (PGEAQCLLGPA). S nitrosylation of DR4 was restricted to residue C336, indicating that there was some selectivity to the reaction. We propose that S nitrosylation of DR4, through conformational changes, may prime the receptor or render it more sensitive to activation by ligand. We believe that this paradox can be explained on the basis of a unique mode of NO delivery by NO-Cbl, via the receptor-mediated endosomal pathway. NO may be liberated from NO-Cbl while in close proximity to the plasma membrane.
Other death receptor pathways appear to be primed by nitric oxide. Induction of NOS2 (inducible NOS) can sensitize tumor cells to apoptosis. Gamma interferon induced the induction of NOS2 in ovarian carcinoma cell lines and sensitized the cells to the apoptotic effects of FasL (9). Blocking of NOS2 activity by NG-monomethyl-l-arginine reduced the sensitization. Cell stressors activate NOS, leading to the S nitrosylation of GAPDH, causing binding to SIAH 1 and nuclear translocation (12). However, protein S nitrosylation does not uniformly promote apoptosis. S nitrosylation of caspase-3, caspase-9, and c-Jun N-terminal kinase blocks their activity and inhibits apoptosis (13), whereas S nitrosylation of matrix metalloproteinase 9, IκB kinase beta (IKKβ), and nuclear factor κB (NF-κB) promotes cell death (13).
Comparisons have been drawn between S nitrosylation and phosphorylation as signaling mechanisms (23, 27, 28). Although they both involve posttranslational protein modifications, phosphorylation is a catalytic process, whereas S nitrosylation is noncatalytic. Nitrosothiols are exceptionally labile as a result of their reactivity with intracellular reducing agents such as ascorbic acid and glutathione as well as reduced metal ions, especially Cu(I), and have tissue half-lives ranging from seconds to minutes (17, 18).
Nitric oxide can cause both apoptosis and necrosis. SNAP and diethylenetriamine-NO adduct (NOC-18) induce cell death through mitochondrial damage characterized by a decrease in mitochondrial membrane potential, cytochrome c leakage, and caspase-9 activation, components of the intrinsic pathway (1, 2, 29-32). In a previous study, we showed that NO-Cbl induced cell death through the extrinsic apoptotic pathway; NO-Cbl increased the expression of Apo2L/TRAIL, DR4 and DR5 mRNAs, and caspase-8 enzymatic activity (4).
We have shown that the Cys-336 residue is a target for S nitrosylation by NO-Cbl. The C336A mutant was not S nitrosylated by NO-Cbl and was more resistant to growth inhibition by NO-Cbl. The identification of Apo2L/TRAIL receptor DR4 as a target for S nitrosylation may provide insights into the mechanism of death induction by NO-Cbl.
Acknowledgments
This work was supported by Public Health Service grant CA095020 to D.J.L. from the National Cancer Institute and gifts from the Mareb Foundation, the Reuter Foundation, and the Charles R. Jelm Foundation.
REFERENCES
- 1.Bal-Price, A., V. Borutaite, and G. C. Brown. 1999. Mitochondria mediate nitric oxide-induced cell death. Ann. N. Y. Acad. Sci. 893:376-378. [DOI] [PubMed] [Google Scholar]
- 2.Bal-Price, A., and G. C. Brown. 2000. Nitric-oxide-induced necrosis and apoptosis in PC12 cells mediated by mitochondria. J. Neurochem. 75:1455-1464. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, J. A. 1998. Synthesis, characterization and nitric oxide release profile of nitrosylcobalamin: a potential chemotherapeutic agent. Anticancer Drugs 9:239-244. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, J. A., B. H. Morrison, R. W. Grane, B. S. Jacobs, S. Dabney, A. Gamero, K. A. Carnevale, D. J. Smith, J. Drazba, B. Seetharam, and D. J. Lindner. 2002. Effects of interferon beta on transcobalamin II-receptor expression and antitumor activity of nitrosylcobalamin. J. Natl. Cancer Inst. 94:1010-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benhar, M., and J. S. Stamler. 2005. A central role for S-nitrosylation in apoptosis. Nat. Cell Biol. 7:645-646. [DOI] [PubMed] [Google Scholar]
- 6.Choi, Y. B., L. Tenneti, D. A. Le, J. Ortiz, G. Bai, H. S. Chen, and S. A. Lipton. 2000. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat. Neurosci. 3:15-21. [DOI] [PubMed] [Google Scholar]
- 7.Erwin, P. A., A. J. Lin, D. E. Golan, and T. Michel. 2005. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 280:19888-19894. [DOI] [PubMed] [Google Scholar]
- 8.Funai, E. F., A. Davidson, S. P. Seligman, and T. H. Finlay. 1997. S-nitrosohemoglobin in the fetal circulation may represent a cycle for blood pressure regulation. Biochem. Biophys. Res. Commun. 239:875-877. [DOI] [PubMed] [Google Scholar]
- 9.Garban, H. J., and B. Bonavida. 1999. Nitric oxide sensitizes ovarian tumor cells to Fas-induced apoptosis. Gynecol. Oncol. 73:257-264. [DOI] [PubMed] [Google Scholar]
- 10.Garban, H. J., D. C. Marquez-Garban, R. J. Pietras, and L. J. Ignarro. 2005. Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proc. Natl. Acad. Sci. USA 102:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, B., and A. Almasan. 2000. Apo2 ligand/TNF-related apoptosis-inducing ligand and death receptor 5 mediate the apoptotic signaling induced by ionizing radiation in leukemic cells. Cancer Res. 60:5754-5760. [PubMed] [Google Scholar]
- 12.Hara, M. R., N. Agrawal, S. F. Kim, M. B. Cascio, M. Fujimuro, Y. Ozeki, M. Takahashi, J. H. Cheah, S. K. Tankou, L. D. Hester, C. D. Ferris, S. D. Hayward, S. H. Snyder, and A. Sawa. 2005. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7:665-674. [DOI] [PubMed] [Google Scholar]
- 13.Hess, D. T., A. Matsumoto, S. O. Kim, H. E. Marshall, and J. S. Stamler. 2005. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6:150-166. [DOI] [PubMed] [Google Scholar]
- 14.Hogg, N. 2002. The biochemistry and physiology of S-nitrosothiols. Annu. Rev. Pharmacol. Toxicol. 42:585-600. [DOI] [PubMed] [Google Scholar]
- 15.Jaffrey, S. R., H. Erdjument-Bromage, C. D. Ferris, P. Tempst, and S. H. Snyder. 2001. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3:193-197. [DOI] [PubMed] [Google Scholar]
- 16.Jaffrey, S. R., and S. H. Snyder. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001:PL1. [DOI] [PubMed] [Google Scholar]
- 17.Kashiba-Iwatsuki, M., K. Kitoh, E. Kasahara, H. Yu, M. Nisikawa, M. Matsuo, and M. Inoue. 1997. Ascorbic acid and reducing agents regulate the fates and functions of S-nitrosothiols. J. Biochem. (Tokyo) 122:1208-1214. [DOI] [PubMed] [Google Scholar]
- 18.Kashiba-Iwatsuki, M., M. Miyamoto, and M. Inoue. 1997. Effect of nitric oxide on the ligand-binding activity of albumin. Arch. Biochem. Biophys. 345:237-242. [DOI] [PubMed] [Google Scholar]
- 19.Kuenzli, K. A., I. L. Buxton, and M. E. Bradley. 1998. Nitric oxide regulation of monkey myometrial contractility. Br. J. Pharmacol. 124:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, D., Z. Shahrokh, S. Marsters, K. Achilles, D. Shih, B. Mounho, K. Hillan, K. Totpal, L. DeForge, P. Schow, J. Hooley, S. Sherwood, R. Pai, S. Leung, L. Khan, B. Gliniak, J. Bussiere, C. A. Smith, S. S. Strom, S. Kelley, J. A. Fox, D. Thomas, and A. Ashkenazi. 2001. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat. Med. 7:383-385. [DOI] [PubMed] [Google Scholar]
- 21.Lipton, A. J., M. A. Johnson, T. Macdonald, M. W. Lieberman, D. Gozal, and B. Gaston. 2001. S-nitrosothiols signal the ventilatory response to hypoxia. Nature 413:171-174. [DOI] [PubMed] [Google Scholar]
- 22.MacFarlane, M., M. Ahmad, S. M. Srinivasula, T. Fernandes-Alnemri, G. M. Cohen, and E. S. Alnemri. 1997. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J. Biol. Chem. 272:25417-25420. [DOI] [PubMed] [Google Scholar]
- 23.Mannick, J. B., and C. M. Schonhoff. 2002. Nitrosylation: the next phosphorylation? Arch. Biochem. Biophys. 408:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Messmer, U. K., and B. Brune. 1996. Modification of macrophage glyceraldehyde-3-phosphate dehydrogenase in response to nitric oxide. Eur. J. Pharmacol. 302:171-182. [DOI] [PubMed] [Google Scholar]
- 25.Pan, G., J. Ni, Y. F. Wei, G. Yu, R. Gentz, and V. M. Dixit. 1997. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277:815-818. [DOI] [PubMed] [Google Scholar]
- 26.Skehan, P., R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney, and M. R. Boyd. 1990. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82:1107-1112. [DOI] [PubMed] [Google Scholar]
- 27.Stamler, J. S., S. Lamas, and F. C. Fang. 2001. Nitrosylation. The prototypic redox-based signaling mechanism. Cell 106:675-683. [DOI] [PubMed] [Google Scholar]
- 28.Stamler, J. S., E. J. Toone, S. A. Lipton, and N. J. Sucher. 1997. (S)NO signals: translocation, regulation, and a consensus motif. Neuron 18:691-696. [DOI] [PubMed] [Google Scholar]
- 29.Tamatani, M., S. Ogawa, G. Nunez, and M. Tohyama. 1998. Growth factors prevent changes in Bcl-2 and Bax expression and neuronal apoptosis induced by nitric oxide. Cell Death Differ. 5:911-919. [DOI] [PubMed] [Google Scholar]
- 30.Umansky, V., A. Ushmorov, F. Ratter, K. Chlichlia, M. Bucur, A. Lichtenauer, and M. Rocha. 2000. Nitric oxide-mediated apoptosis in human breast cancer cells requires changes in mitochondrial functions and is independent of CD95 (APO-1/Fas). Int. J. Oncol. 16:109-117. [DOI] [PubMed] [Google Scholar]
- 31.Ushmorov, A., F. Ratter, V. Lehmann, W. Droge, V. Schirrmacher, and V. Umansky. 1999. Nitric-oxide-induced apoptosis in human leukemic lines requires mitochondrial lipid degradation and cytochrome C release. Blood 93:2342-2352. [PubMed] [Google Scholar]
- 32.Yabuki, M., K. Tsutsui, A. A. Horton, T. Yoshioka, and K. Utsumi. 2000. Caspase activation and cytochrome c release during HL-60 cell apoptosis induced by a nitric oxide donor. Free Radic. Res. 32:507-514. [DOI] [PubMed] [Google Scholar]