Abstract
Investigations of the molecular events involved in activation of genomic target genes by peroxisome proliferator-activated receptors (PPARs) have been hampered by the inability to establish a clean on/off state of the receptor in living cells. Here we show that the combination of adenoviral delivery and chromatin immunoprecipitation (ChIP) is ideal for dissecting these mechanisms. Adenoviral delivery of PPARs leads to a rapid and synchronous expression of the PPAR subtypes, establishment of transcriptional active complexes at genomic loci, and immediate activation of even silent target genes. We demonstrate that PPARγ2 possesses considerable ligand-dependent as well as independent transactivation potential and that agonists increase the occupancy of PPARγ2/retinoid X receptor at PPAR response elements. Intriguingly, by direct comparison of the PPARs (α, γ, and β/δ), we show that the subtypes have very different abilities to gain access to target sites and that in general the genomic occupancy correlates with the ability to activate the corresponding target gene. In addition, the specificity and potency of activation by PPAR subtypes are highly dependent on the cell type. Thus, PPAR subtype-specific activation of genomic target genes involves an intricate interplay between the properties of the subtype- and cell-type-specific settings at the individual target loci.
The peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors belonging to the nuclear receptor superfamily. All members of the PPAR family, PPARα, -γ, and -β/δ, bind as heterodimers with the retinoid X receptor (RXR) to conserved PPAR response elements (PPREs), i.e., a direct repeat of the hexameric sequence (A/G)GGTCA spaced by one nucleotide (18). The different PPAR subtypes display highly diverse biological functions in vivo. Thus, PPARα (30) and PPARβ/δ (11, 67) activate primarily genes encoding enzymes involved in lipid oxidation (catabolic pathways), whereas PPARγ activation leads to induction of genes involved in lipogenesis (anabolic pathways) (44).
PPARs are activated by a variety of fatty acids and fatty acid derivatives such as prostaglandins and leukotrienes. In addition, members of the PPAR family are important drug targets for the treatment of insulin resistance, type II diabetes, dyslipidemia, hypertension, atherosclerosis, and some cancers (8, 20).
In spite of the pharmacological and biological importance, the mechanisms by which PPARs induce endogenous target genes in their natural chromatin context has not been thoroughly examined. Approaches such as stable overexpression of the PPAR subtypes or administration of subtype-selective agonists to activate endogenous PPARs in tissues and cell lines have been used to identify novel PPAR target genes (1, 19, 67, 70). However, while such experiments have contributed significantly to the identification of target genes, they are not useful for dissecting the molecular mechanisms by which PPAR subtypes activate endogenous target genes. This is due to the fact that PPARs, as a result of endogenous ligands and/or ligand-independent transactivation, activate target genes in the absence of exogenous ligands. This has been demonstrated in transient transfections (25) as well as in retroviral transductions (7). The relatively high basal expression of endogenous target genes precludes experiments analogous to the ones performed with the steroid receptors, which are kept inactive either by nuclear exclusion and/or by interaction with heat shock protein complexes (28, 38, 56, 57). Thus, in contrast to steroid receptors, the addition of exogenous agonists cannot be used to define clean on/off states of the PPARs with respect to the establishment of a transcriptionally active complex at the promoters.
The different PPAR subtypes show limited specificity in their binding to different PPREs in in vitro mobility shift assays. A preference for PPARγ/RXR heterodimers has been shown for weak PPREs, but for most PPREs no clear subtype specificity has been demonstrated by such assays (27). Similarly, the different PPAR subtypes differ only little in their ability to activate artificial promoter reporter constructs in transient transfections (17, 24, 29, 55). Intriguingly, the limited difference between PPAR subtypes in electrophoretic mobility shift assays and in transient transfections is in sharp contrast to results obtained by in vivo activation of endogenous PPARs using subtype-selective agonists or by experiments with knockout models. The differential biological actions of PPAR subtypes are undoubtedly due in part to the differential expression patterns of the different PPAR subtypes. Thus, PPARα is mainly expressed in tissues with high fatty acid β-oxidation rates, such as liver, heart, skeletal muscle, and brown fat, while PPARγ is abundantly expressed in brown and white adipose tissues and macrophages (6). However, adenoviral expression of PPARγ1 in mouse liver leads to induction of several genes which are not readily activated by PPARα, i.e., genes involved in lipid accumulation and adipogenesis (73). Importantly, results from the few comparative analyses have shown that the PPARs are also biochemically distinct and behave differently when ectopically expressed in the same cell. Thus, PPARγ is by far the most adipogenic of the PPAR subtypes in fibroblasts (7). In addition, we have recently shown that PPARγ and PPARα induce distinct subsets of genes in pancreatic β-cells. In these cells ectopic expression of PPARα results in increased fatty acid uptake and fatty acid oxidation and in promotion of glucose-stimulated insulin secretion, whereas ectopic expression of PPARγ results in increased fatty acid uptake and increased lipid accumulation and inhibition of glucose-stimulated insulin secretion (49). The molecular mechanisms underlying this subtype-specific activation of gene expression in a particular cell are not understood.
Here we show that acute ectopic expression of the PPARs by adenoviral vectors is an ideal approach to investigate PPAR subtype-specific activation of target genes in individual cell types. This system allows us to tightly control the timing and expression level of the PPARs and thereby go from a clean off to a clean on state of PPAR target loci. By combining adenoviral delivery and chromatin immunoprecipitation (ChIP), we investigate the rapid establishment of an active transcriptional complex on PPAR target genes immediately following the ectopic expression of PPARs. We show that the PPARs retain significant subtype specificity in this system and induce target genes in a highly subtype- and cell-type-specific manner. This PPAR subtype-specific activation was shown to be correlated with PPAR occupancy at the given promoters, indicating that PPAR binding to chromatin-embedded target sites is a major determinant of PPAR subtype-specific activation of endogenous target genes.
MATERIALS AND METHODS
Cell culture and differentiation.
Mouse NIH-3T3 (ATCC CRL-1658), NIH-CAR, and 3T3-L1 fibroblasts (ATCC CL-173) were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% bovine serum (Gibco). Mouse AML-12 hepatocytes (ATCC CRL-2254) were cultured in Ham's nutrient mixture F-12/DMEM (1:1) with 2.5 mM l-glutamine, 1.2 g/liter sodium bicarbonate, 15 mM HEPES, and 0.5 mM sodium pyruvate (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) [5 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml selenium (Sigma-Aldrich)] and 0.1 μM dexamethasone. Mouse MIN6 pancreatic β-cells (kindly provided by J. Miyazaki) were grown in DMEM supplemented with 15% heat-inactivated FBS (Gibco) and 50 μM β-mercaptoethanol (Sigma-Aldrich). 293-HEK cells (ATCC CRL-1573) were cultured in DMEM supplemented with 0.1 mM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), and 10% FBS (Gibco). Phoenix cells (ATCC SD3444) were grown in DMEM supplemented with 10% FBS (Gibco). All cell lines were kept in medium supplemented with streptomycin (100 μg/ml) and penicillin (62.5 μg/ml). The 3T3-L1 fibroblasts were differentiated to adipocytes by stimulation with 3-isobutyl-1-methylxanthine, dexamethasone, and insulin as described previously (25).
Mouse embryonic fibroblasts (MEFs) were isolated from embryonic day 13.5 C57Bl/6J-Bom embryos. The embryos were transferred to cold sterile phosphate-buffered saline (PBS); placenta, membranes, heads, and inner organs were removed, and the rest of the bodies were dissected into small pieces. After centrifugation (1,200 rpm for 5 min at room temperature), the supernatant was removed, and the pellet was resuspended in 25% collagenase (Sigma), 1× PBS, and 20% FBS. Following a 1-h digestion at 37°C, the cells were harvested by centrifugation and resuspended in MEF culture medium (AmnioMax C-100 basal medium [Gibco] supplemented with 7.5% AmnioMax C-100 supplement [Gibco], 7.5% FBS, 1% l-glutamine, 62.5 μg/ml penicillin, and 100 μg/ml streptomycin). Cells were counted and plated on Falcon plates at 37°C with 95% humidity and 7.5% CO2. Medium was changed every day.
Retroviral transduction of NIH 3T3 fibroblasts.
Phoenix cells were transfected using the calcium-phosphate technique with a retroviral LXSN-hCARΔ1 vector expressing the truncated coxsackie-adenovirus receptor (CARΔ1) (47) at 50% confluence. Two days after transfection, virus supernatant was harvested and centrifuged to remove living phoenix cells. NIH-3T3 cells at 50% confluence were transduced with a 1:1 dilution of virus supernatant and fresh growth medium in the presence of 6 μg/ml Polybrene (Sigma-Aldrich). The following day, the cells were split and subjected to neomycin (G418; 0.7 mg/ml; Bie and Berntsen) selection. Approximately 5 days later, the selected NIH 3T3 (CARΔ1) cells were pooled and replated for single colony isolation. Single clones were isolated by replating 80% confluent cells at a 103-fold dilution and culturing them for 3 to 5 days before trypsinization in sterile O-rings. The level of CARΔ1 expression in the individual clones was evaluated by fluorescence-activated cell sorter (FACS) analysis. A high CARΔ1-expressing clone, termed NIH-CAR, was chosen for further studies.
FACS analysis.
For CAR FACS analysis, 105 cells [NIH 3T3 and NIH 3T3(CARΔ1)] were trypsinized, washed in PBS, and incubated for 30 min with 100 μl of PBS containing 0.5% bovine serum albumin (BSA) and CAR antibody (05-644; Upstate) at 1:200 at 4°C. The cells were washed twice in PBS and incubated for 30 min with 150 μl of PBS including 0.5% BSA and fluorescein isothiocyanate-conjugated anti-mouse antibody (12-487; Upstate) at 1:150 at 4°C. The cells were washed with PBS, fixed in 100 μl of PBS containing 1% paraformaldehyde, and analyzed by FACS.
For green fluorescence protein (GFP) FACS analysis, 106 cells [NIH 3T3, NIH 3T3(CARΔ1), and NIH-CAR] were transduced with adenoviral GFP (47) (AdGFP; 55 PFU/cell). Cells were washed twice with PBS, trypsinized, fixed in 100 μl of PBS containing 1% paraformaldehyde, and analyzed by FACS.
Adenovirus generation and purification.
Recombinant adenoviruses containing full-length hemagglutinin (HA)-tagged mouse PPARα, PPARγ2, and PPARβ/δ were generated using the AdEasy cloning system (Stratagene). The three PPAR subtypes were initially N-terminally HA-tagged and cloned into pShuttle-CMV (Stratagene) (where CMV is cytomegalovirus) using the NotI/XhoI (PPARα) and KpnI/EcoRV (PPARγ2 and PPARβ/δ) restriction sites. The CMV-HA-PPAR cassettes were transferred to the AdEasy-1 vector by homologous recombination in Escherichia coli to generate AdHA-PPAR constructs. Plasmids were linearized and transfected into 293-HEK cells using the calcium-phosphate procedure. The adenoviruses were amplified and purified using CsCl gradients, and viral titers were estimated by a plaque assay-based approach as recommended by the AdEasy protocol (Stratagene).
Adenoviral transduction of cell lines.
For adenoviral transduction fresh medium containing AdHA-PPARs (55 PFU/cell) was given to NIH-CAR, MIN6, or AML-12 cells at 80% confluence. After 2 h of viral transduction the virus-containing medium was removed, and new medium containing the following PPAR-specific agonists was added for an additional 2 to 10 h: 1 μM rosiglitazone/BRL49653 (Novo Nordisk), 30 μM Wy14.643 (Calbiochem), 1 μM L-165041 (Merck), and/or PPARγ antagonist 1 μM GW9662 (Sigma-Aldrich).
RNA purification and cDNA synthesis.
Adenovirally transduced NIH-CAR, MIN6, and AML-12 cells were harvested in guanidium thiocyanate at the indicated time points (see figures), and RNA was isolated according to a modified Chomczynski-Sacchi protocol (12). RNA preparations were subjected to DNase I (Invitrogen) treatment, and cDNA was synthesized using random deoxynucleic acid hexamers and reverse transcriptase (First-Strand Kit; Invitrogen) as previously described (23). All experiments were performed in duplicates.
Protein analysis.
Adenovirally transduced NIH-CAR, MIN6, and AML-12 cells were harvested in a hypotonic sodium dodecyl sulfate (SDS) sample buffer as described previously (23) and separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were blotted onto polyvinylidene difluoride membranes (Millipore Corp.) and probed with specific antibodies. The following primary antibodies were used: anti-PPARγ (sc-7273; Santa Cruz), anti-TFIIB (sc-225; Santa Cruz) and anti-HA (12CA5 [see reference 40]). The secondary antibodies used were horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (P0447; DAKO) and swine anti-rabbit immunoglobulin G (P0339; DAKO). Enhanced chemiluminescence (ECL; Amersham Phamacia Biotech) was used for detection.
Transient transfection of AdPPAR-transduced NIH-CAR cells.
NIH-CAR cells were transfected at 50 to 60% confluence with a PPRE3-TK-Luc (where TK is thymidine kinase and Luc is luciferase) reporter construct (34) and a pCMV-β-galactosidase control (Promega) in six-well plates using the calcium-phosphate procedure (10). Following 6 h of incubation with DNA, the medium was changed to DMEM supplemented with 10% resin-charcoal-stripped calf serum containing AdHA-PPARs (55 PFU/cell) for 2 h together with the PPAR-specific ligands 1 μM rosiglitazone/BRL49653 (Novo Nordisk), 30 μM Wy14.643 (Calbiochem), and 1 μM L-165041 (Merck). The transduced cells were subsequently incubated additionally for 10 h in the presence of PPAR-specific ligands before the cells were harvested in a lysis solution (Tropix). Luciferase and β-galactosidase assays were performed as described previously (24). All experiments were performed in triplicate, and luciferase and β-galactosidase measurements were done in duplicate.
ChIP.
NIH-CAR (106 to 107) cells were cross-linked in 1% formaldehyde for 20 min. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M. Cross-linked cells were removed from the plate and washed once in cold PBS and once in cross-link buffer 1 (0.25% Triton, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl, pH 8.0) at 4°C. Finally, the cross-linked cells were washed in cold cross-link buffer 2 (0.2 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl, pH 8.0). Cell extract was diluted in 2 ml of chromatin dilution buffer (0.05% SDS, 1 mM EDTA, 0.5 mM EGTA, 50 mM HEPES, pH 8.0, and 1×complete protease inhibitor cocktail [Boehringer Mannheim]) and sonicated according to the manufacturer’s protocol using the Diagenode Bioruptor. The amount of DNA in each sample was determined by the absorbance at 260 nm and diluted to equal amounts. Protein A (Amersham Pharmacia Biotech), protein G (Amersham Pharmacia Biotech), or protein A/G (Santa Cruz) beads were prepared by three washes in immunoprecipitation (IP) dilution buffer (1% Triton, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris, pH 8.0) and subsequently incubated in IP dilution buffer containing 1 μg/μl BSA and 1× complete protease inhibitor mixture. Chromatin was diluted in IP dilution buffer (2% triton, 0.15% SDS, 300 mM NaCl, 4 mM EDTA, 2 mM EGTA, 40 mM Tris, pH 8.0, 2 μg/μl BSA, and 1× complete protease inhibitors) and incubated with prepared protein A, G, or A/G beads at 4°C for 1 h. Beads were removed and supernatant was incubated with antibody over night at 4°C on a rotating wheel, followed by 3 h incubation with protein A, G or A/G beads at 4°C, rotating. Beads were washed at 4°C once with IP wash buffer 1 (1% triton, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris, pH 8.0), twice with IP wash buffer 2 (1% triton, 0.1% SDS, 500 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM Tris, pH 8.0), once with IP wash buffer 3 (0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris, pH 8.0), and finally once with Tris-EDTA buffer, all at 4°C. DNA-protein complexes were eluted with 200 μl of elution buffer (1% SDS and 0.1 M NaHCO3) and decross-linked by adding 0.2 M NaCl and shaking overnight at 65°C. DNA was purified by a PCR purification kit (Amersham Pharmacia Biotech) and dissolved in 50 μl of water. Immunoprecipitated DNA and 1% input DNA were analyzed by real-time PCR. Antibodies used for IP were anti-PPARγγ (sc-7273; Santa Cruz), anti-RXRα (sc-774; Santa Cruz), anti-CREB binding protein (anti-CBP; sc-369; Santa Cruz), anti-thyroid hormone receptor-associated protein 220; (anti-TRAP220) (sc-8998; Santa Cruz), anti-TAT a box binding protein (anti-TBP) (SL30 [previously described in reference 52]), anti-RNA polymerase II (anti-Pol II) (8WG16; Covance Research Products), anti-H3-Ac (06-599; Upstate Technologies), anti-H4-Ac (06-866; Upstate Technologies), and anti-HA (12CA5 [previously described in reference 40]).
Real-time PCR.
Quantitative three-step real-time PCR was performed on the ABI-7700 PRISM real-time PCR instrument (Applied Biosystems) using 2× SYBR Green Master Mix and Sigma passive reference (Sigma-Aldrich) according to the instructions from the manufacturer. All measurements were performed in duplicate. Primers for real-time PCR (URL: http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html) were designed using Primer Express 2.0 (Applied Biosystems), and specificity and efficacy were validated before use.
RESULTS
Adenoviral delivery in NIH 3T3 fibroblasts.
To investigate the molecular events involved in PPAR-mediated induction of endogenous target genes and to compare PPAR subtype-specific activation of target genes, we sought to identify a system which could be used to define a true “off” as well as “on” state of PPAR target genes. For this we needed a cell line with low endogenous levels of all PPAR subtypes and a vector system that could be used for efficient delivery and acute ectopic expression of PPAR subtypes. The murine NIH 3T3 fibroblast cell line is known to have very low levels of endogenous PPARs, and this cell line can be efficiently transduced with retroviral vectors. However, these vectors give rise to a slow and nonsynchronous induction of ectopic protein expression and therefore are not well suited to investigate the sequence of events involved in activation of PPAR target genes (data not shown).
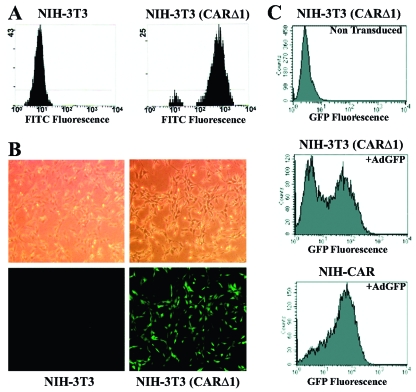
We therefore turned to the adenoviral vector system, which mediates fast and efficient ectopic expression of proteins. However, adenoviral infection of this fibroblastic cell line is known to be very inefficient due to the low expression of the CAR (47). To overcome this problem, we obtained a retroviral vector expressing a truncated version of the CAR protein, CARΔ1, which lacks the cytoplasmic tail of CAR. Orlicky and coworkers have previously described and used this elegant system for rendering fibroblasts susceptible to adenoviral infection (47). Using this retroviral vector for expression of CARΔ1, the amount of CARΔ1 receptors at the cell surface, as determined by FACS analysis, was increased approximately 100-fold compared to parental NIH 3T3 cells (Fig. 1A). The expression of the CARΔ1 protein greatly facilitated efficient adenoviral-mediated entry of transgenes into NIH 3T3 (CARΔ1) as exemplified by infection with the AdGFP vector (Fig. 1B). To ensure as fast and synchronous uptake of the transgene as possible, we isolated single clones from the pool of CARΔ1-transduced NIH 3T3 cells. These clones were screened for their expression of CARΔ1 by FACS analysis, and one clone with a high level of CARΔ1 expression, termed NIH-CAR, was selected for further use. As illustrated in Fig. 1C, this high-expressing clone (NIH-CAR) exhibited almost 100% efficiency in AdGFP uptake and expression after only 2 h of infection, whereas the mixed NIH 3T3 (CARΔ1) population had only 50% efficiency at this early time point.
FIG. 1.
Adenoviral delivery in NIH 3T3 fibroblasts. (A) FACS analysis of CARΔ1 expression on the cell surface of NIH 3T3 and NIH 3T3 retrovirally transduced with CARΔ1. Cells were incubated with a primary antibody against CAR and a secondary fluorescein isothiocyanate-conjugated antibody, fixed with formaldehyde, and subjected to FACS analysis. (B) Recombinant AdGFP (55 PFU/cell for 8 h) were used to transduce NIH 3T3 and NIH 3T3 (CARΔ1) cells. Top frames show phase-contrast microscopy. Lower frames show GFP expression visualized by fluorescence microscopy. (C) NIH-CAR cells show homogenous uptake of AdGFP 2 h posttransduction compared to the nonselected mixed population of NIH 3T3 (CARΔ1) cells. Cells were fixed, and GFP expression was determined by FACS analysis.
Adenoviral expression of HA-tagged PPAR subtypes in NIH-CAR cells.
Having established the NIH-CAR clone, we used adenoviral vectors for acute, effective, and synchronous expression of the PPARs in fibroblasts. Murine PPARα, PPARγ2, and PPARβ/δ were cloned into adenoviral vectors as previously described (49), except that an HA tag was added to the N terminus of all PPAR subtypes. This tag allowed us to directly compare the expression level as well as recruitment to target sites of all PPAR subtypes by Western blotting and ChIP, respectively.
Initially, the infection ratio of the AdHA-PPARγ2 was adjusted so that the expression of PPARγ after 8 h of infection was equivalent to the expression of PPARγ in differentiated 3T3-L1 adipocytes as determined by Western blotting and probing with a PPARγ antibody (Fig. 2A). Subsequently, using an antibody directed against the HA epitope, the infection ratios of AdHA-PPARα and AdHA-PPARβ/δ were adjusted to give protein expression equivalent to that of HA-PPARγ2. Using these infection ratios, elevated PPAR mRNA level could be detected 2 h after adenoviral transduction (Fig. 2B), and prominent PPAR protein expression was observed 4 h after adenoviral transduction (Fig. 2C). Protein and mRNA expression increased between 4 to 8 h and decreased slightly between 8 and 10 h (Fig. 2B and C).
FIG. 2.
Equal expression of transcriptionally active HA-PPAR subtypes in NIH-CAR cells. (A) The expression of AdHA-PPARγ2 in NIH-CAR is equal to PPARγ2 in differentiated 3T3-L1 adipocytes. Whole-cell lysates were prepared from differentiated 3T3-L1 adipocytes (day 8 and 10) and from NIH-CAR cells 8 h after transduction with AdHA-PPARγ2 (55 PFU/cell). Cell extracts were subjected to SDS-PAGE and immunoblotted using antibodies against PPARγ and TFIIB. (B) Elevated PPAR mRNA levels can be detected 2 h after adenoviral transduction. NIH-CAR cells were transduced with HA-PPARs (55 PFU/cell), and RNA was isolated at 0, 2, 4, 6, 8, and 10 h after adenoviral transduction. The PPARα, γ2, and β/δ mRNA levels were determined by real-time PCR and normalized to the corresponding TFIIB levels. (C) A distinct expression of HA-PPAR protein is present 4 h after adenoviral transduction. Whole-cell extracts were submitted to SDS-PAGE and Western blotting using antibodies against the HA epitope and TFIIB, respectively. (D) The transcriptional activity of all PPAR subtypes was verified by transient transfection of NIH-CAR cells with a multimerized PPRE luciferase reporter construct (3xPPRE-Tk-Luc) and subsequent transduction with adenoviral HA-PPARα, γ2, or β/δ (55 PFU/cell) for 12 h. Experiments were performed in the presence or absence of 30 μM WY14.643, 1 μM rosiglitazone/BRL49653, or 1 μM L-165041, respectively. Luciferase values were normalized to β-galactosidase activity and plotted relative to activity of the reporter in nontransduced NIH-CAR cells. All results are representative of a minimum of three independent experiments.
To confirm that the adenovirally expressed HA-PPARs were transcriptionally active in NIH-CAR cells, we transiently transfected the NIH-CAR cells with a multimerized PPRE luciferase reporter construct; cells were subsequently transduced with AdHA-PPARs (Fig. 2D). Although HA-PPARα was the most potent inducer of transcription, all HA-PPARs were transcriptionally active. Thus, the combination of the NIH-CAR cells and the AdHA-PPAR vectors provides a very useful tool for acute and synchronous ectopic expression of equal amounts of the HA-PPAR subtypes in a background with very low endogenous levels of PPAR. This makes it possible to directly investigate and compare PPAR subtype-specific activation of endogenous PPAR target genes.
Kinetics of AdHA-PPAR activation of target gene expression in NIH-CAR cells.
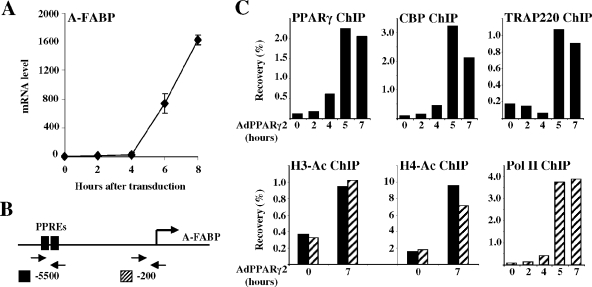
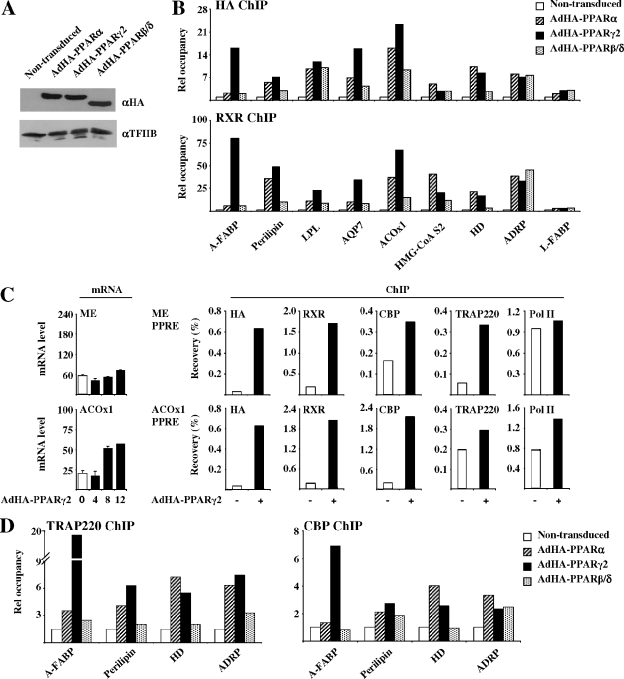
To investigate the kinetics of PPAR-mediated activation of endogenous target genes in our model system, we chose to focus on PPARγ2-mediated induction of the adipocyte fatty acid binding protein (A-FABP, also called aP2) gene, which is a well-characterized PPARγ target gene (63). Adenoviral expression of PPARγ2 in the presence of the PPARγ-specific agonist rosiglitazone/BRL49653 led to a significant induction of the A-FABP transcript from the endogenous gene after 4 h (Fig. 3A). The fast induction of the A-FABP gene by PPARγ2 is reflected in a significant recruitment of PPARγ2 to the A-FABP enhancer PPREs (Fig. 3B) within 4 h after transduction (Fig. 3C). The coactivators CREB-binding protein (CBP) and TRAP220 are recruited to the enhancer PPREs, and the RNA polymerase II is recruited to the A-FABP promoter within 4 to 5 h. Maximal occupancy is obtained at 5 h following transduction. The activation of the A-FABP gene by PPARγ2 and the subsequent recruitment of coactivators are associated with increased histone H3K9/14di- and H4 tetra-acetylation (Fig. 3C). These data show that the establishment of a transcriptionally active complex and induction of the classical PPARγ target gene, A-FABP, are fast-occurring processes immediately following the expression of PPARγ2 in fibroblasts. This indicates that PPARγ2 is sufficient to trigger activation of the A-FABP gene in NIH-CAR cells.
FIG. 3.
Activation of the endogenous A-FABP gene by PPARγ2. (A) PPARγ2 induces the A-FABP gene following 4 h of adenoviral transduction. NIH-CAR cells were transduced with 55 PFU/cell recombinant adenoviral PPARγ2 in the presence of 1 μM rosiglitazone/BRL49653, and RNA was purified from cells 0, 2, 4, 6, and 8 h after transduction. The A-FABP mRNA levels were determined by real-time PCR, normalized to the corresponding TFIIB levels and shown relative to the A-FABP/TFIIB level of nontransduced cells. (B) Schematic illustration of the A-FABP promoter. The relative positions of the A-FABP −5500 and −200 primers are indicated. (C) Activation of the A-FABP gene correlates with PPARγ2, CBP, TRAP220, and RNA Pol II recruitment to the A-FABP promoter and increased H3 and H4 acetylation. Chromatin was prepared 0, 2, 4, 5, and 7 h after transduction and subsequently subjected to IP using antibodies against PPARγ, CBP, TRAP220, RNA Pol II, acetylated K9 and K14 H3, and tetra-acetylated H4, respectively. Enriched DNA was analyzed using real-time PCR with primers positioned at the A-FABP PPREs (−5500) (black bars) and the proximal promoter (−200) (hatched bars). Results are shown as percent recovery relative to chromatin input. Results are representative of at least three independent experiments.
PPARγ2 displays ligand-dependent as well as ligand-independent transactivation potential.
Up to this point, we have expressed the PPARs in the presence of their selective agonists. A fundamental question in PPAR transactivation that has remained unanswered is the extent to which PPARs display ligand-independent transactivation of endogenous target genes. Thus, it is unknown whether, e.g., the high basal expression of PPAR target genes in adipocytes is due to endogenous PPARγ agonists, ligand-independent activity of PPARγ, or the activity of other transcription factors that activate the expression from these promoters once PPARγ has triggered the induction. In transient transfections all PPARs, and in particular PPARα, show significant activation of a PPRE reporter construct in the absence of exogenous ligand (Fig. 2D) (25). However, in most experiments it is unclear to what extent this is due to the presence of endogenous ligand or to ligand-independent transactivation. We therefore addressed the question of ligand-independent transactivation by the PPARs using adenoviral delivery of PPARγ2 in NIH-CAR cells in the presence or absence of the specific PPARγ agonist rosiglitazone or the PPARγ antagonist GW9662. The latter binds covalently to PPARγ, thereby permanently abolishing binding of agonists to PPARγ (37).
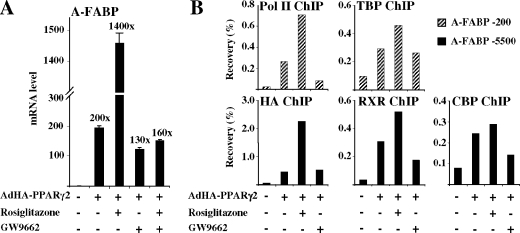
As seen in Fig. 4A, adenoviral expression of PPARγ2 for 8 h in the presence of the PPARγ-specific agonist rosiglitazone leads to a 1,400-fold induction of A-FABP gene expression relative to nontransduced cells. However, in the absence of exogenous agonist, PPARγ2 in itself leads to a significant (200-fold) induction of the A-FABP gene. The majority of this transactivation appears to be due to ligand-independent transactivation, as the PPARγ antagonist GW9662 decreased PPARγ2-induced A-FABP expression only slightly. In contrast, GW9662 totally abolishes rosiglitazone-dependent PPARγ2 transactivation. Induction of other PPARγ target genes like CD36, perilipin, and lipoprotein lipase (LPL) followed a pattern similar to A-FABP (see Fig. 7A; data not shown). These results clearly demonstrate that PPARγ2 possesses significant ligand-independent as well as ligand-dependent transactivation.
FIG. 4.
Induction of the endogenous A-FABP gene by PPARγ2 ligand-dependent and -independent transactivation. (A) PPARγ2 exhibits a pronounced ligand-independent as well as ligand-dependent transcriptional potential. RNA from NIH-CAR cells transduced for 8 h with recombinant AdHA-PPARγ2 (55 PFU/cell) in the presence of 1 μM rosiglitazone/BRL49653 and/or 1 μM GW9662 was purified. The A-FABP mRNA levels were determined by real-time PCR, normalized to the corresponding TFIIB levels and shown relative to the A-FABP/TFIIB level of nontransduced cells. (B) The ligand-dependent and -independent induction of the A-FABP gene correlates with PPARγ2/RXR, CBP, TBP, and RNA Pol II recruitment to the A-FABP promoter. Chromatin was prepared 8 h after transduction and subsequently subjected to IP with antibodies against HA tag, RXR, CBP, TBP, and RNA Pol II, respectively. DNA recovery was determined by real-time PCR with primers positioned at the A-FABP PPREs (−5500) and the proximal promoter (−200), respectively. Results are representative of at least three independent experiments.
FIG. 7.
The role of agonists in PPAR subtype-specific activation of endogenous target genes. (A) Activation of PPAR target genes in the presence and absence of agonists. NIH-CAR cells were transduced and treated as in described in the legend of Fig. 5A, except that the experiment was carried out in the presence as well as the absence of agonists. RNA was harvested 12 h after transduction, and expression of target genes was determined by real-time PCR, normalized to TFIIB and shown relative to the A-FABP/TFIIB level of nontransduced cells. Results are representative of at least four independent experiments. (B) Relative occupancy of HA-PPARs and RXR on PPAR target promoters in the presence and absence of agonists. NIH-CAR cells were transduced and treated as described in the legend of Fig. 6B except that the experiment was carried out in the presence as well as the absence of agonists. Relative occupancy at the indicated target genes is indicated. Results are representative of at least three independent experiments.
To investigate how agonists and antagonists affect the recruitment of PPARs, RXR, and cofactors to target PPREs and the recruitment of basal transcription factors at the promoter, we performed ChIP (Fig. 4B). The results show a significant recruitment of PPAR and RXR to the A-FABP enhancer in the presence of vehicle alone, as well as in the presence of antagonist. In keeping with the significant transactivation induced by PPARγ2 under these conditions, cofactors and basal transcription factors were recruited to the promoter. When the agonist is added, recruitment of PPARγ2 and RXR to the PPRE and recruitment of TBP and Pol II to the promoter are further increased, while the effect on CBP recruitment is more modest. Thus, there is a strong correlation between activation of the PPAR target gene and recruitment of PPARγ2/RXR and auxiliary transcriptional cofactors.
Subtype-specific activation of endogenous PPAR target genes.
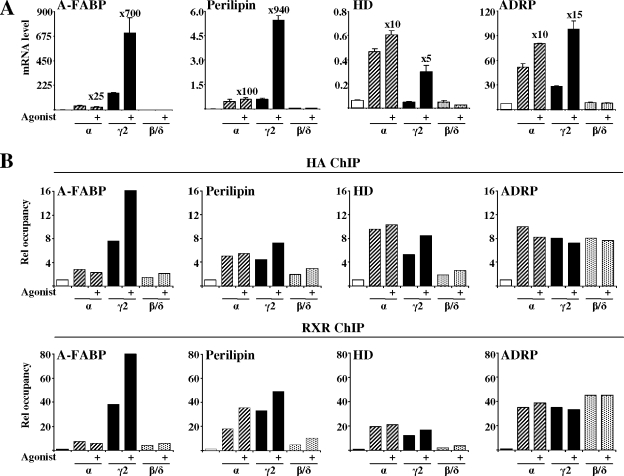
To compare the transactivation potential of PPAR subtypes in fibroblasts, NIH-CAR cells were transduced with AdHA-PPARα, -γ, or -β/δ virus in the presence of selective PPAR agonists. RNA was isolated for cDNA synthesis 4, 8, and 12 h posttransduction, and the mRNA expression levels of a large set of PPAR target genes chosen from the literature were evaluated by real-time PCR (Table 1 and Fig. 5A). The ability of the different subtypes to activate endogenous target genes was highly subtype dependent. Based on the previously reported induction by the PPAR subtypes, the selected target genes were divided into different subgroups (Table 1). The first group encompasses characteristic PPARγ target genes investigated: A-FABP, perilipin, and aquaporin-7 (AQP7). These genes were highly activated by ectopic PPARγ2, only modestly activated by PPARα, and not significantly activated by PPARδ in NIH-CAR cells. The genes reported to be activated by all PPAR subtypes followed (except for adipose differentiation-related protein [ADRP]) a very similar pattern as the PPARγ-type genes; i.e., they were most efficiently activated by PPARγ, less by PPARα, and not activated by PPARβ/δ. The reported PPARα/(δ/β) target genes, by contrast, could be divided into three subgroups. Six target genes, i.e., those encoding hydroxymethyl glutaryl coenzyme A synthetase 2 (HMG-CoA S2), the bifunctional enzyme enoyl-CoA hydrates/l-3hydroxyacyl-CoA dehydrogenase (HD), pyruvate dehydrogenase kinase 4, the long-chain acyl-CoA dehydrogenenase, and carnitine palmitoyltransferase II were most effectively induced by PPARα. Three of these (HMG-CoA S2, pyruvate dehydrogenase kinase 4, and HD) were induced more than fivefold. Surprisingly, the genes encoding medium chain acyl coenzyme A (acyl-CoA) dehydrogenase, peroxisomal biogenesis factor 11a (PEX11a), and acyl-CoA oxidase 1 (ACOx1) were most effectively induced by PPARγ2 rather than by PPARα or -δ. A large group of known PPARα/(β/δ) target genes, including, malic enzyme (ME), 3-ketoacyl-CoA thiolase B (thiolase B), cytochrome P450 A10, long chain acyl-CoA dehydrogenenase, and liver-FABP (L-FABP), were not induced by any of the PPARs in NIH-CAR cells. Notably, expression of endogenous PPAR genes was not affected by the acute ectopic expression of PPARs (data not shown). This clearly demonstrates that changes in gene expression cannot be ascribed to activation of endogenous PPARs. Furthermore, most target genes were significantly induced by PPARγ2 (and in some cases also PPARα) already at the 4-h time point, indicating that PPARγ2 is sufficient for activation of a large number of target genes in NIH-CAR cells.
TABLE 1.
Induction of PPAR target genes in NIH-CAR cells
| Gene | Level of induction by:
|
Reported PPAR activatorb
|
||||
|---|---|---|---|---|---|---|
| PPARαa | PPARγa | PPARδa | Cell typec | Subtype(s) | Reference | |
| A-FABP | ++++ | ++++++ | − | A | γ | 63 |
| Perilipin | +++ | +++++ | − | A | γ | 59 |
| AQP7 | − | +++ | − | A | γ | 32 |
| CD36 | − | ++++++ | − | A, M, H | α/γ/δ | 54, 66, 72 |
| LPL | + | +++ | − | A, M, H | α/γ/δ | 53, 55 |
| ADRP | ++ | ++ | − | A, T, H | α/γ/δ | 5, 62 |
| HMG-CoA S2 | ++++++ | +++ | + | H | α | 51 |
| PDK4 | +++ | ++ | − | H, S | α/δ | 26, 61, 72 |
| HD | ++ | + | − | H | α | 13 |
| CPT-II | + | + | − | H | α | 4 |
| MCAD | − | + | − | H | α | 22 |
| LCAD | + | - | − | H | α/δ | 3, 16, 67 |
| PEX11a | − | + | − | H | α | 59 |
| ACOx1 | − | − | − | H | α/δ | 64, 67 |
| ME | − | − | − | H | α | 9 |
| Thiolase B | − | − | − | H | α/δ | 36, 61 |
| L-FABP | − | − | − | H | α | 48 |
| CYP4A10 | − | − | − | H | α | 2 |
Target genes are considered induced (+) after 12 h by a PPAR subtype when activated more than threefold over background (nontransduced cells). In addition, the following indications were used: ++, >32-fold induction; +++, >33-fold induction; ++++, >34-fold induction; +++++, >35-fold induction; ++++++, >36-fold induction; −, noninduced target genes. For a full account of target gene expression, please refer to http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html.
Reported PPAR activator as described in the corresponding references.
The cell type in which the reported PPAR target is determined: A, adipocyte; M, macrophage; H, hepatocyte; T, trophoblast; S, skeletal muscle cells.
FIG. 5.
PPAR subtype-specific activation of endogenous genes in NIH 3T3 cells and MEFs. The PPARs activate endogenous target genes in a highly subtype-specific manner. NIH-CAR cells (A) and MEFs (B) were transduced with AdHA-PPARα, -γ2, or -β/δ (55 PFU/cell) in the presence of 30 μM WY14.643, 1 μM rosiglitazone/BRL49653, or 1 μM L-165041, respectively. RNA was purified 0, 4, 8, and 12 h after transduction, and expression of target genes was determined by real-time PCR, normalized to TFIIB expression and shown relative to the A-FABP/TFIIB level of nontransduced cells. A subset of genes representing different types of target genes is shown. The increase in induction (n-fold) of each gene at the 12-h time point is indicated. For the complete set of target genes, please refer to http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html. Results are representative of at least four independent experiments.
To investigate whether the gene activation profile of NIH-CAR cells reflected that in primary fibroblasts, we isolated MEFs, infected these with the adenoviral vectors, and investigated the induction of the same set of target genes (Fig. 5B and data not shown). Notably, although the difference between potency of PPARα and PPARγ2 is slightly less pronounced in MEFs compared to NIH-CAR cells, the PPAR subtypes display very similar induction profiles as observed in NIH-CAR cells. The only major difference is that the L-FABP gene can be activated by PPARα in MEFs, whereas it is insensitive to PPAR transactivation in NIH-CAR cells. Thus, it appears that during the establishment of the NIH 3T3 cell line from MEFs, the L-FABP gene has been more permanently inactivated.
In summary, despite the equal expression levels of the PPAR subtypes and the lower transactivation potential of PPARγ2 compared to PPARα as determined by transient transfection (Fig. 2D), PPARγ2 is the most efficient subtype in activating the majority of endogenous chromatin-embedded PPAR target genes in NIH-CAR fibroblasts and in primary MEFs. A small number of genes is more susceptible to PPARα-mediated transactivation, whereas PPARβ/δ is a poor activator of the genes investigated (Fig. 5A and B and Table 1), even though PPARβ/δ is transcriptionally competent in NIH-CAR cells, as shown in transient transfections (Fig. 2D).
PPAR/RXR binding to PPREs is a major determinant of subtype specificity.
The observed inability of the PPAR subtypes to activate certain target genes in the NIH-CAR cells could be due to a reduced potential of the PPARs to gain access to the target sites in the promoters/enhancers or due to the reduced ability of the bound PPAR/RXR heterodimer to induce formation of a transcriptionally active complex. Thus, to investigate how acute activation of PPAR target genes correlates with PPAR occupancy at the corresponding PPREs, we compared subtype-specific recruitment to target sites using ChIP. NIH-CAR cells were transduced with one of the AdHA-PPARs (α, γ or β/δ) for 8 h in the presence of specific agonists. Equal expression of the PPAR subtypes at the 8-h time point was verified by Western blotting (Fig. 6A). ChIP was performed with antibodies directed against RXR and the HA epitope of the HA-PPARs, respectively. Input as well as immunoprecipitated DNA was quantified by real-time PCR using primers (available at http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html) positioned at the PPREs of a number of different PPAR target genes (Table 2). To be able to compare the different target sites, recoveries were normalized to recoveries for nontransduced NIH-CAR cells (background). Although there is a slightly higher background when the HA antibody is used, there is a good correlation between the ChIP obtained with the HA and the RXR antibodies (Fig. 6B).
FIG. 6.
PPAR subtype-specific binding to endogenous PPREs in the presence of specific agonists. (A) HA-PPARs are expressed at equal levels. Whole-cell extracts from NIH-CAR cells transduced with 55 PFU/cell of AdHA-PPARα, -γ2, or -β/δ for 8 h show equal protein expression of the PPAR subtypes. Proteins were separated by SDS-PAGE and immunoblotted using antibodies against the HA tag and TFIIB, respectively. (B) The ability of the different PPAR subtypes to bind to target sites in the presence of their specific agonists (Fig. 5) was determined by ChIP. Chromatin was harvested 8 h following transduction, and ChIP was performed using antibodies against the HA epitope and RXR. Relative occupancy (recovery in the presence of PPAR expression over recovery in the nontransduced cells) was determined using primers positioned at the PPREs of the indicated genes (Table 2). (C) PPARγ2 fails to further activate the transcription of the ME gene and only slightly activates that of the ACOx1 gene although receptors and cofactors are significantly recruited to the corresponding PPREs. Expression of the ME and ACOx1 genes following adenoviral transduction was determined as described in the legend of Fig. 5A. Chromatin was harvested 8 h following transduction, and ChIP was performed using antibodies against the HA tag, RXR, CBP, TRAP220, and RNA Pol II. Recovery at the ME and ACOx1 PPREs is indicated. (D) The PPAR-induced recruitment of the cofactors TRAP220 and CBP to PPREs is correlated with binding of the respective subtype to the target sites. ChIP experiments were performed as described for panel B except that antibodies against TRAP220 and CBP were used. Relative occupancy is indicated. Results are representative of three independent experiments.
TABLE 2.
Sequence and position of PPREs in the mouse genome
| Gene and/ or PPRE | Sequencea | Positionb | Reference |
|---|---|---|---|
| A-FABP, ARE6 | CTCTCT GGGTGA A ATGTGC | ∼−5460 | 63 |
| A-FABP, ARE7 | CTTACT GGATCA G AGTTCA | ∼−5360 | 63 |
| ACOx1, PPRE A | GGAAAC AGGACA A TGGGAA | ∼−550 | 64 |
| ACOx1, PPRE B | AAAGCA AGGTAA A AGGTCA | ∼−260 | 35 |
| ME | CTTTCT GGGTCA A AGTTGA | ∼−250 | 9 |
| LPL | AGAAGA AGGGGA A AGGGCA | ∼−160 | 55 |
| L-FABP | AGATAT AGGCCA T AGGTCA | ∼−60 | 48 |
| AQP7 | TTCTCC AGGGGA G AGGTCA | ∼−80 | 31 |
| HD | AAATGT AGGTAA T AGTTCA | ∼−2800 | 13 |
| HMG-CoA S2 | AAAACT GGGCCA A AGGTCT | ∼−100 | 51 |
| ADRP | TTTTGT AGGTGA A AGGGCA | ∼−2000 | 60 |
| Perilipin | GGATGT GGGTGA A AGGTGA | ∼−1970 | 59 |
| Consensus | AAAACT AGGTCA A AGGTCA | 27 | |
| GG T G T |
Annotated PPRE (boldface) and 5′ flanking region.
Position of mouse PPRE relative to the transcriptional start site.
The ChIP data showed efficient recruitment of the PPARγ/RXR heterodimer to the PPREs of the A-FABP, perilipin, LPL, AQP7, ACOx1, HMG-CoA S2, HD, and ADRP genes, whereas there was only minor or no recruitment to the L-FABP gene (Fig. 6B). Interestingly, for most genes there was a good correlation between the ability of the subtypes to bind to target sites and their ability to activate transcription of the corresponding genes. The PPARα/RXR and especially the PPARβ/δ/RXR heterodimer are much less efficiently bound to PPREs of the PPARγ-specific genes investigated, like A-FABP, perilipin, and AQP7, than is the PPARγ2/RXR heterodimer. This indicates that the failure of PPARα and PPARβ/δ to potently activate transcription of these genes in NIH-CAR cells is due in part to poor binding to these target sites in chromatin. By contrast, PPARα binds most efficiently to the HMG-CoA S2 and HD PPREs and is also the best activator of these genes. In the case of the silent gene L-FABP, which cannot be activated by PPARs in NIH-CAR cells, we did not detect significant PPAR/RXR binding to the corresponding PPRE. Thus, the ability to gain access to the target sites appears to be a major limiting factor in determining whether a gene can be activated by a particular PPAR subtype.
Although the general observation is that binding and activation are correlated, there are clear exceptions. For example, we detect significant binding of PPARβ/δ to target genes where PPARδ is unable to activate transcription. The most notable example is ADRP, which shows equal binding of all PPAR subtypes, yet the gene can be activated by only PPARα and PPARγ2. In addition, the already actively transcribed ACOx1 and ME genes respond very modestly (twofold increase) (ACOx1) or not at all (ME) to ectopic PPAR expression (Fig. 6C and Table 1). Yet these genes display significant binding of the PPARγ2/RXR heterodimer to their proximal PPREs (Fig. 6B and C). To investigate whether the failure to further increase expression of the ME and ACO genes was due to the inability of PPARγ2 to increase cofactor occupancy, we determined CBP and TRAP220 recruitment to the PPREs. As seen in Fig. 6C, ectopic expression of PPARγ2 led to a significant increase in cofactor occupancy at both genes. However, in keeping with the high basal transcriptional activity and little PPAR responsiveness of the genes, Pol II occupancy at the proximal promoter was high in the nontransduced cells and only marginally increased by ectopic PPAR expression. This contrasts to the significant PPARγ2-dependent recruitment of Pol II at the A-FABP proximal promoter (Fig. 3C).
Taken together, PPARγ is the PPAR subtype that most efficiently gains access to a large number of PPREs in NIH-CAR cells. Furthermore, although exceptions exist, there is a good correlation between PPAR/RXR binding and target gene activation. Thus, the ability to gain access to the target PPREs appears to be a major, but clearly not the only, determinant in PPAR subtype specific activation of target gene expression.
It is likely that the differential ability of the PPAR subtypes to recruit cofactors to target sites plays an important role in determining PPAR subtype specificity at these sites. To make a first step toward elucidating the role of cofactors, we therefore compared the ability of the subtypes to recruit CBP and TRAP220 to different types of PPAR target genes in the presence of specific agonists. We tested PPARγ-selective target genes (A-FABP and perilipin), a PPARα-selective target gene (HD), and a gene equally activated by PPARγ2 and PPARα (ADRP) (Fig. 6D). Notably, recruitment of CBP and TRAP220 to the PPREs correlates to a large extent with binding of the different PPAR subtypes; i.e., subtypes that bind well also efficiently recruit CBP and TRAP220. Interestingly, however, PPARβ/δ is unable to efficiently recruit TRAP220 to the ADRP PPRE, notwithstanding the efficient binding of the PPARβ/δ/RXR heterodimer, and this may in part explain why PPARβ/δ is incapable of activating ADRP (Fig. 5A).
Target genes show different dependencies on agonists for transactivation.
The results presented in Fig. 4 for the A-FABP gene (as well as parallel analyses of the LPL, perilipin, and CD36 genes [data not shown]) clearly demonstrated that PPARγ2 displays significant agonist-independent as well as agonist-dependent binding to target sites and activation of target genes. To investigate to what degree exogenous agonists contribute to PPAR subtype specificity, we compared the activation of target genes by the different PPAR subtypes in the presence and absence of their respective agonists. Selected target genes are shown in Fig. 7A. Interestingly, the relative contribution of the agonists to the transactivation is highly dependent on the target gene and the PPAR subtype. In general, PPARα transactivation is less dependent on exogenous agonist than PPARγ. Thus, in the absence of exogenous agonists, PPARα and PPARγ activate perilipin equally well. In addition, the slight preference for PPARγ in the activation of the ADRP gene is reversed in the absence of agonists. However, for genes where the subtype specificity is very pronounced, like the A-FABP gene, subtype specificity is maintained also in the absence of exogenous agonists.
In keeping with the mRNA analysis, agonists had limited effect on binding of PPARα/RXR and PPARβ/δ/RXR to the corresponding PPREs under the conditions investigated, while the effect on binding of PPARγ2/RXR is highly dependent on the target gene and to a large extent reflects the effects on gene expression (Fig. 7B). Interestingly, however, we did not observe any effect of agonists on PPAR/RXR binding to the ADRP PPRE. Thus, overall these results are in keeping with our general finding that the ability to gain access to target PPREs is a major, but clearly not the only, determinant in PPAR subtype-specific activation of target gene expression.
PPAR subtype specificity is affected by cell type.
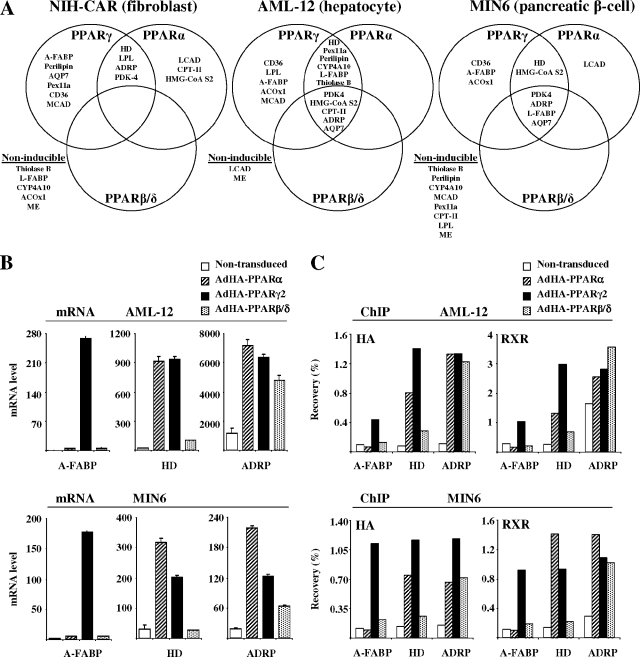
To investigate to what extent cell type affects PPAR subtype-specific transactivation of target genes, we compared the ability of HA-PPAR subtypes to induce target genes in NIH-CAR cells with the ability to induce these genes in two other murine cell culture models, the AML-12 hepatocyte cell line (69) and the MIN6 pancreatic β-cell line (41). Experiments were performed in the presence of specific agonists. In contrast to NIH 3T3 fibroblasts, the AML-12 and MIN6 cells naturally express high levels of the CAR receptor and are therefore efficiently infected with adenoviruses (data not shown). For each cell line the infection ratios of the AdHA-PPARs were adjusted so that the level of HA-PPAR protein after 8 h of infection was similar to the one obtained in NIH-CAR cells (data not shown).
The ability of the PPAR subtypes to induce target genes following 12 h of transduction are summarized in Fig. 8A (for details, see the data at http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html). It is evident that PPAR subtype specificity is highly cell-type dependent. In MIN6 and NIH-CAR, a few distinct target genes cannot be activated by any of the PPARs. These genes might be subject to permanent inactivation in the particular cell line (e.g., L-FABP in NIH-CAR and perilipin in MIN6), or they might be actively transcribed but insensitive to further activation by PPARs (e.g., ME in NIH-CAR and MIN6 cells and medium chain acyl-CoA dehydrogenase in MIN6 cells). However, among the target genes that are susceptible to PPAR transactivation, the cell type has great impact on subtype specificity. Thus, whereas PPARβ/δ is largely unable to directly induce endogenous target gene expression in NIH-CAR cells, it potently induces several target genes, such as ADRP and AQP7, in both AML-12 and MIN-6 cells. PPARα is also significantly more active in inducing target genes, such as L-FABP and AQP7, in AML-12 and MIN-6 cells compared to NIH-CAR cells. Notably, in all three cell lines PPARγ2 is the only subtype able to potently activate the A-FABP and CD36 genes, suggesting that, irrespective of the subtype, these genes are highly PPARγ specific. By contrast, AQP7 which is a highly PPARγ2-specific target in NIH-CAR cells, is activated by all PPAR subtypes in AML-12 and MIN6 cells. Taken together, our data show that the cell type plays a determining role in rendering a particular target gene susceptible to different PPAR subtypes.
FIG. 8.
Cell-type-specific modulation of PPAR subtype specificity. (A) Grouping of target genes according to their relative subtype specificity in NIH-CAR fibroblasts, AML-12 hepatocytes, and MIN6 pancreatic β cells. Cells were transduced and treated with agonists, and the expression of target genes was determined as described in the legend of Fig. 5A. For a full account of target gene expression, please refer to http://www.sdu.dk/Nat/bmb/faculty/pubs/MCB06_supp_data.html. The investigated target genes are considered to be induced by a particular PPAR subtype, when the gene is activated more than threefold over background (nontransduced cells), and when the induction is at least 15% of the induction obtained by the most potent PPAR subtype. (B) Expression of selected target genes in AML-12 and MIN6 cells following transduction with HA-PPARs. Cells were transduced and treated with agonists, and expression of target genes was determined as described in the legend of Fig. 5A. (C) Relative occupancy of HA-PPARs and RXR on PPAR target promoters in AML-12 and MIN 6 cells. Cells were transduced and treated as described in the legend of Fig. 6B. Relative occupancy at the indicated target genes is indicated. Results are representative of three independent experiments.
To investigate whether the ability of PPAR subtypes to transactivate target genes in AML-12 and MIN6 cells also correlates with binding to target sites, we performed ChIP on selected targets. For the NIH-CAR cells (Fig. 6B) there was a good correlation between the RXR and HA-PPAR ChIP. However, in the case of the ADRP gene, which is highly active in AML-12 cells, we observed a significant binding of RXR in the absence of ectopic PPAR expression. It is possible that this is due to RXR homodimer or heterodimer binding with other nuclear receptor partners at or close to the PPRE. Importantly, in keeping with the data from the NIH-CAR cells, there was a good correlation between the ability of a subtype to bind to target sites (Fig. 8C) and the ability to activate the corresponding gene in AML-12 and MIN6 cells (Fig. 8B). This correlation is seen for PPARγ-specific genes (exemplified by A-FABP), PPARγ/α target genes (exemplified by HD), and genes activated by all PPAR subtypes (exemplified by ADRP).
DISCUSSION
Detailed investigations of the molecular events involved in activation of endogenous chromatin-embedded target genes by nuclear receptors have so far been limited to steroid receptors, such as the progesterone, glucocorticoid, estrogen, and androgen receptors (28, 38, 45, 50, 56), and a few RXR-dependent nuclear receptors such as the thyroid hormone and vitamin D3 receptors (31, 58). In the absence of agonists these receptors are either transcriptionally inactive due to interaction with heat shock proteins or nuclear exclusion or they are kept in a repressive state due to interaction with transcriptional corepressors. Addition of agonists promotes release from heat shock protein complexes, import to the nucleus, and/or exchange of transcriptional corepressors with coactivators (43, 58, 71). Thus, exogenous agonists can be used to set clean on/off states of these receptors, and the sequence of events of transcription factor recruitment to target promoters can be closely followed using ChIP. However, since many of the nuclear receptors, including the PPARs, are considered to be metabolic sensors activated by lipid metabolites produced within the cells (14, 68), the agonist concentration is difficult to control. Furthermore, transcriptional activity may not be strictly agonist dependent. These built-in difficulties have hampered the investigations of the molecular events involved in the activation of genomic target genes by this large group of nuclear receptors.
Here, we demonstrate that the combination of adenoviral delivery and ChIP is ideal for studying the acute molecular effects of PPAR activation on PPAR target sites and promoters. We show that adenoviral transduction of the HA-PPARs in NIH-CAR fibroblasts results in increased PPAR mRNA levels after 2 h and potent PPAR protein expression within 4 h posttransduction. The HA-PPAR proteins reach maximal levels, similar to that of PPARγ protein in 3T3-L1 adipocytes, 4 to 8 h after transduction (Fig. 2). Binding of PPARγ, RXR, cofactors, and basal transcription factors to the enhancer of the well-established PPARγ target gene, A-FABP, is maximal at 5 h. At this point, the expression from the A-FABP gene is significantly activated. Recruitment of PPARγ to the locus also leads to a significant increase in histone H3 and H4 acetylation at the promoter as well as at the enhancer (Fig. 3). Thus, the adenoviral transgene delivery provides a unique system to tightly control the expression of PPARs and to study the acute and direct effects of PPAR binding to target sites of endogenous genes.
As PPARs are activated by fatty acids and derivatives thereof, the “basal” transcriptional activity (i.e., activity in the absence of exogenous agonists) of PPARs is often ascribed to endogenous ligands or ligands present in the serum. In addition, the ligand-independent activation function of the receptors may contribute to this activity; however, the relative contribution of endogenous ligands and ligand-independent function to “basal” transcriptional activity is unknown. Using the adenoviral delivery system, we show here that PPARγ2 displays significant ligand-dependent as well as -independent transcriptional activity. PPARγ2 leads to profound activation of the A-FABP gene and other target genes like LPL and perilipin in the absence of exogenous ligands. This activation is only slightly decreased by the addition of a specific PPARγ antagonist, GW9662 (Fig. 4A and results not shown). Thus, the considerable transcriptional potential of PPARγ2 in NIH-CAR fibroblasts in the absence of exogenous agonists is predominantly a consequence of the high ligand-independent activity of PPARγ2.
The ligand-independent transcriptional activity of PPARγ is likely to be mediated at least in part by ligand-independent activation function 1 (AF1) in the N-terminal part of the receptor. However, the AF2 of the ligand binding domain (LBD) may also contribute to the ligand-independent transactivation. Helix 12 has been reported to be in an “agonist position” in crystal structures of apo-PPARγLBD (46), and recent investigations indicated that this is due to the stabilization of helix 12 in the agonist position by specific charged amino acids in the LBD (42). In keeping with this, PPARγLBD has recently been shown to interact with CBP/p300 (33) as well as p160 CoA (42) in the absence of ligands. Presence of a synthetic ligand further induces stabilization of helix 12, thereby leading to an increased interaction with coactivators (42). Future investigations in our adenoviral system with truncated and mutated PPARs will help clarify whether the ligand-independent transactivation of endogenous genes is due primarily to the AF1 or the AF2 function and whether these effects are promoter specific. Recent data from the Lazar laboratory indicate that there is indeed promoter specificity in the dependency of ligand for cofactor recruitment (21).
Intriguingly, our data show that PPARγ occupancy of the A-FABP PPRE is considerably increased in the presence of the thiazolidinedione-type agonist, rosiglitazone. The increased occupancy is paralleled by increased recruitment of basal transcription factors and of coactivators such as CBP. This ligand-dependent increase in occupancy may in part be explained by a stabilization of the PPAR/RXR/DNA interaction, as suggested by electrophoretic mobility shift assays of PPARα/RXR (15, 17). In addition, ligand-induced stabilization of receptor-cofactor interactions may lead to a longer residence time of the receptor at the locus. Interestingly, however, the relative importance of agonist for PPARγ2/RXR binding to target sites is highly dependent on the target gene, and for some genes like ADRP, agonists did not appear to affect PPARγ2/RXR binding although it potentiated expression of the gene (Fig. 7A and B).
In general, the effect of the agonist Wy14.643 on PPARα binding to target sites and activation of target genes is minor (Fig. 7A and B). However, the lack of efficient synthetic PPARα antagonists for cell culture experiments precludes investigations similar to those shown in Fig. 4 for PPARγ to determine whether the activity in the absence of exogenous agonist is due to agonist-independent transactivation or to the presence of endogenous agonists.
It is clear from numerous in vivo and ex vivo experiments that the different PPAR subtypes serve different physiological functions. However, it is unclear to what extent these different actions are due to intrinsic biochemical properties or to differences in the cellular context in which the PPARs are expressed. Only a few studies have sought to mutually compare PPAR subtype-specific activation of endogenous genes in a particular cell type (7, 60). Although these comparisons have been very useful, they were performed with stable ectopic expression, which makes it difficult to distinguish between direct and indirect target genes. Furthermore, constitutive expression of the receptors hampers investigations of the molecular mechanism involved in target gene activation. The combination of adenoviral delivery and ChIP provides a unique possibility to investigate the molecular mechanisms underlying PPAR subtype specificity by comparing the acute actions of the different PPAR subtypes on endogenous chromatin-embedded target genes. We show that, when adenovirally expressed at equal levels, HA-tagged PPARs have highly different potentials to activate endogenous target genes in the NIH-CAR cells. While PPARα had more transcriptional activity than PPARγ in transient transfections with a reporter construct consisting of a mulitmerized ACOx1 PPRE, PPARγ was the most potent activator of the majority of the endogenous genes investigated. However, a small number of genes were most potently activated by PPARα. Surprisingly, however, in NIH-CAR cells as well as in MEFs, PPARβ/δ was either unable to activate or a poor activator of all genes investigated (Fig. 5A and B and Table 1). The reason for this is unclear, as adenovirally expressed PPARβ/δ was active in transient transfections of NIH-CAR cells and able to activate endogenous genes in AML-12 and MIN6 cells. Previous experiments with stable retroviral expression of PPARβ/δ in NIH 3T3 cells have shown that, although PPARβ/δ is by far the least potent PPAR subtype in these cells, constitutive PPARβ/δ expression induces low levels of several PPAR target genes (7). However, from these long-term experiments it is impossible to conclude whether the induction is due to direct but slow induction of the target genes or due to indirect activation of the target genes via induction of another transcription factor, e.g., PPARγ. The lack of target gene activation by adenovirally expressed PPARβ/δ suggests the existence of a global mechanism (e.g., posttranslational modification or lack of a particular cofactor) that renders PPARβ/δ less transcriptionally active on endogenous genes in the NIH 3T3 fibroblasts.
To address the question how the PPARs retain gene specificity, we used ChIP to investigate if PPAR subtype-specific transcriptional activation is a consequence of differential PPAR binding to promoters or to PPAR subtype-specific recruitment of coactivators. Interestingly, ChIP analysis demonstrated that, in general, the transcriptional potential of the PPAR subtypes is correlated with the level of PPAR/RXR heterodimer binding to the promoters of activated genes (Fig. 6B). There are clear exceptions where a subtype binds without further activating; however, for all target genes investigated, the subtype that binds best is also the subtype that activates most efficiently. This indicates that a major, but clearly not the only, rate-limiting step in establishing a transcriptionally active complex at the target genes investigated is the ability of the receptor subtypes to gain access to the chromatin-embedded binding sites.
Some target genes remained transcriptionally silent in NIH-CAR cells irrespective of the PPAR subtype. As exemplified by the L-FABP gene, we did not observe PPAR/RXR binding to these target sites, and hence these are likely to be embedded in inaccessible chromatin or to require synergy with other cell-type-specific transcription (co)factors to gain access. Other genes (e.g., ME and thiolase B) were transcriptionally active but insensitive to further activation by PPARs. Interestingly, however, PPARγ2/RXR displays significant binding and PPARγ2-dependent cofactor recruitment to the PPREs of the ME gene (Fig. 6C). This suggests that other (co)factors than PPAR/RXR, CBP, and TRAP220 are limiting the expression of this gene. Further detailed ChIP analyses of factor recruitment to this and other loci will be required to understand why receptor binding is unable to further enhance the transcription of such genes.
The differential binding of PPAR subtypes to different PPREs may in part be explained by the sequence composition of the PPREs. In vitro experiments performed in the absence of ligands have shown that PPREs with a relatively low match to the consensus have a preference for PPARγ heterodimers (27). Such “weak” PPREs include the two PPREs of the A-FABP enhancer. However, the LPL PPRE has been shown to bind both PPARα and -γ in vitro and is a recognized PPARα as well as PPARγ target gene (39, 55). In addition, the ACOx1 PPRE(s) match the consensus very well and is generally regarded as a PPARα-type PPRE (64, 65). For both the LPL and ACO PPREs, we find that they preferably bind PPARγ heterodimers in the context of chromatin in NIH-CAR cells, indicating that factors other than the affinity for the naked DNA sequence play a role. Furthermore, as discussed below, PPAR subtype specificity differs between the cell lines. Thus, the abilities of the PPAR subtypes to gain access to PPREs in vivo cannot be explained only by their differential affinity for a given PPRE in vitro. It is likely that an important determining factor is the synergy with other transcription factors and the joint ability to effectively recruit the right combination of chromatin remodeling and modifying complexes present in the cell.
In addition to the ability of the PPAR/RXR heterodimer to gain access to target sites, the ability of the PPAR/RXR heterodimer to bind cofactors prior to DNA-binding or to recruit cofactors to target genes once bound to DNA is likely to greatly influence the level of expression of the target genes. An exhaustive analysis of this aspect is beyond the scope of this article; however, we have investigated the differential ability of the PPAR subtypes to recruit TRAP220 and CBP to a small number of selected target genes in NIH-CAR cells (Fig. 6D). Importantly, for most genes the recruitment of these more general cofactors correlates with binding of the PPAR subtypes and therefore does not add to the subtype specificity. An exception may be the ADRP gene to which all PPAR subtypes bind, whereas only PPARα and PPARγ2 are able to significantly recruit TRAP220. Interestingly, this may in part explain why PPARβ/δ is unable to activate the ADRP gene. Further insight into the role of cofactors in PPAR subtype specificity for different genes awaits large-scale ChIP analysis of more specific cofactors.
The poor PPARα and PPARβ/δ transcriptional activity of endogenous genes in NIH 3T3 fibroblasts prompted us to investigate the ability of PPARα and PPARβ/δ to activate endogenous genes in other cell lines such as AML-12 hepatocytes and MIN6 pancreatic β cells. From these experiments it is evident that the cell type is indeed a very important parameter of PPAR subtype specificity toward several genes (Fig. 8A). PPARγ remains the subtype able to activate the largest number of genes in the cell types investigated. However, for the target genes investigated PPARα and PPARβ/δ are on average more transcriptionally active in AML-12 and MIN6 cells than in NIH-CAR cells. Irrespective of the cell line, the A-FABP and CD36 genes remain highly PPARγ specific, but other genes that are PPARγ specific in NIH-CAR cells can be induced by PPARα and/or PPARβ/δ in AML-12 and MIN6 cells. Similarly, HMG-CoA S2 and HD, which are PPARα selective in NIH-CAR cells, are induced by other PPAR subtypes in AML-12 and MIN6 cells. These results show that PPAR subtype specificity is a highly complex phenomenon likely to be dependent on the cell-type-specific setting of the individual target sites, i.e., a combination of other transcription factors, cofactors, and chromatin modifications in a cell. Importantly, however, as observed for the NIH-CAR cells, there was a good correlation between the ability of the PPAR subtypes to bind to target sites and their ability to activate the corresponding gene (Fig. 8B and C).
In conclusion, we demonstrate that the combination of adenoviral delivery and ChIP is an ideal method for investigating the basic molecular events involved in the activation of genomic target genes by PPARs in a broad range of cell lines. Using this approach, we show that PPARγ2 possesses considerable agonist-dependent as well as -independent transactivation potential and that agonist increases the occupancy of PPARγ2/RXR at most but not all PPREs. The ability of a particular PPAR subtype to gain access to a PPRE is highly dependent on the subtype as well as the cell type. However, in general (although there are clear exceptions) there is a good correlation between the extent of PPAR binding and the magnitude of the target gene induction. Future challenges will be to investigate why the PPAR subtypes differ in their ability to gain access to endogenous chromatin-embedded PPREs and how the cell type regulates this ability. In addition, large-scale ChIP analyses will be required to define the role of different cofactors in PPAR subtype-specific transactivation.
Acknowledgments
This work was supported by grants to the EU FP6 STREP project X-TRA-NET and grants from the Danish Diabetes Association and the Lundbeck Foundation.
We are grateful to D. J. Orlicky for the LXSN-hCARΔ1 and AdGFP constructs, R. M. Evans for the PPRE3-TK-LUC reporter, Jun-ichi Miyazaki for the MIN6 cell line, and Novo Nordisk for the rosiglitazone/BRL49653 ligand. We thank members of the Mandrup and Stunnenberg laboratory for fruitful discussions and comments on the manuscript, D. Neess for isolation of the MEFs, and K. Andersen for expert technical assistance.
REFERENCES
- 1.Albrektsen, T., K. S. Frederiksen, W. E. Holmes, E. Boel, K. Taylor, and J. Fleckner. 2002. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes 51:1042-1051. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, T. C., J. D. Tugwood, and S. Green. 1995. Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem. J. 306:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyama, T., J. M. Peters, N. Iritani, T. Nakajima, K. Furihata, T. Hashimoto, and F. J. Gonzalez. 1998. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor alpha (PPARalpha). J. Biol. Chem. 273:5678-5684. [DOI] [PubMed] [Google Scholar]
- 4.Barrero, M. J., N. Camarero, P. F. Marrero, and D. Haro. 2003. Control of human carnitine palmitoyltransferase II gene transcription by peroxisome proliferator-activated receptor through a partially conserved peroxisome proliferator-responsive element. Biochem. J. 369:721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bildirici, I., C. R. Roh, W. T. Schaiff, B. M. Lewkowski, D. M. Nelson, and Y. Sadovsky. 2003. The lipid droplet-associated protein adipophilin is expressed in human trophoblasts and is regulated by peroxisomal proliferator-activated receptor-gamma/retinoid X receptor. J. Clin. Endocrinol. Metab. 88:6056-6062. [DOI] [PubMed] [Google Scholar]
- 6.Braissant, O., F. Foufelle, C. Scotto, M. Dauca, and W. Wahli. 1996. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 137:354-366. [DOI] [PubMed] [Google Scholar]
- 7.Brun, R. P., P. Tontonoz, B. M. Forman, R. Ellis, J. Chen, R. M. Evans, and B. M. Spiegelman. 1996. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 10:974-984. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, I. W. 2005. The clinical significance of PPAR gamma agonism. Curr. Mol. Med. 5:349-363. [DOI] [PubMed] [Google Scholar]
- 9.Castelein, H., T. Gulick, P. E. Declercq, G. P. Mannaerts, D. D. Moore, and M. I. Baes. 1994. The peroxisome proliferator-activated receptor regulates malic enzyme gene expression. J. Biol. Chem. 269:26754-26758. [PubMed] [Google Scholar]
- 10.Chen, C. A., and H. Okayama. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques 6:632-638. [PubMed] [Google Scholar]
- 11.Chevillotte, E., J. Rieusset, M. Roques, M. Desage, and H. Vidal. 2001. The regulation of uncoupling protein-2 gene expression by omega-6 polyunsaturated fatty acids in human skeletal muscle cells involves multiple pathways, including the nuclear receptor peroxisome proliferator-activated receptor beta. J. Biol. Chem. 276:10853-10860. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 13.Chu, R., Y. Lin, M. S. Rao, and J. K. Reddy. 1995. Cooperative formation of higher order peroxisome proliferator-activated receptor and retinoid X receptor complexes on the peroxisome proliferator responsive element of the rat hydratase-dehydrogenase gene. J. Biol. Chem. 270:29636-29639. [DOI] [PubMed] [Google Scholar]
- 14.Desvergne, B., and W. Wahli. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20:649-688. [DOI] [PubMed] [Google Scholar]
- 15.Elholm, M., I. Dam, C. Jorgensen, A. M. Krogsdam, D. Holst, I. Kratchmarova, M. Gottlicher, J. A. Gustafsson, R. Berge, T. Flatmark, J. Knudsen, S. Mandrup, and K. Kristiansen. 2001. Acyl-CoA esters antagonize the effects of ligands on peroxisome proliferator-activated receptor alpha conformation, DNA binding, and interaction with Co-factors. J. Biol. Chem. 276:21410-21416. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, M., M. You, M. Matsumoto, and D. W. Crabb. 2003. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. J. Biol. Chem. 278:27997-28004. [DOI] [PubMed] [Google Scholar]
- 17.Forman, B. M., J. Chen, and R. M. Evans. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 94:4312-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gearing, K. L., M. Gottlicher, M. Teboul, E. Widmark, and J. A. Gustafsson. 1993. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc. Natl. Acad. Sci. USA 90:1440-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhold, D. L., F. Liu, G. Jiang, Z. Li, J. Xu, M. Lu, J. R. Sachs, A. Bagchi, A. Fridman, D. J. Holder, T. W. Doebber, J. Berger, A. Elbrecht, D. E. Moller, and B. B. Zhang. 2002. Gene expression profile of adipocyte differentiation and its regulation by peroxisome proliferator-activated receptor-gamma agonists. Endocrinology 143:2106-2118. [DOI] [PubMed] [Google Scholar]
- 20.Giannini, S., M. Serio, and A. Galli. 2004. Pleiotropic effects of thiazolidinediones: taking a look beyond antidiabetic activity. J. Endocrinol. Investig. 7:982-991. [DOI] [PubMed] [Google Scholar]
- 21.Guan, H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 19:453-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulick, T., S. Cresci, T. Caira, D. D. Moore, and D. P. Kelly. 1994. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl. Acad. Sci. USA 91:11012-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, J. B., R. K. Petersen, B. M. Larsen, J. Bartkova, J. Alsner, and K. Kristiansen. 1999. Activation of peroxisome proliferator-activated receptor gamma bypasses the function of the retinoblastoma protein in adipocyte differentiation. J. Biol. Chem. 274:2386-2393. [DOI] [PubMed] [Google Scholar]
- 24.Helledie, T., M. Antonius, R. V. Sorensen, A. V. Hertzel, D. A. Bernlohr, S. Kolvraa, K. Kristiansen, and S. Mandrup. 2000. Lipid-binding proteins modulate ligand-dependent trans-activation by peroxisome proliferator-activated receptors and localize to the nucleus as well as the cytoplasm. J. Lipid Res. 41:1740-1751. [PubMed] [Google Scholar]
- 25.Helledie, T., L. Grontved, S. S. Jensen, P. Kiilerich, L. Rietveld, T. Albrektsen, M. S. Boysen, J. Nohr, L. K. Larsen, J. Fleckner, H. G. Stunnenberg, K. Kristiansen, and S. Mandrup. 2002. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J. Biol. Chem. 277:26821-26830. [DOI] [PubMed] [Google Scholar]
- 26.Huang, B., P. Wu, M. M. Bowker-Kinley, and R. A. Harris. 2002. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-alpha ligands, glucocorticoids, and insulin. Diabetes 51:276-283. [DOI] [PubMed] [Google Scholar]
- 27.Juge-Aubry, C., A. Pernin, T. Favez, A. G. Burger, W. Wahli, C. A. Meier, and B. Desvergne. 1997. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J. Biol. Chem. 272:25252-25259. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Z., O. A. Janne, and J. J. Palvimo. 2004. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 18:2633-2648. [DOI] [PubMed] [Google Scholar]
- 29.Kassam, A., J. Hunter, R. A. Rachubinski, and J. P. Capone. 1998. Subtype- and response element-dependent differences in transactivation by peroxisome proliferator-activated receptors alpha and gamma. Mol. Cell Endocrinol. 141:153-162. [DOI] [PubMed] [Google Scholar]
- 30.Kersten, S., B. Desvergne, and W. Wahli. 2000. Roles of PPARs in health and disease. Nature 405:421-424. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S., N. K. Shevde, and J. W. Pike. 2005. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J. Bone Miner. Res. 20:305-317. [DOI] [PubMed] [Google Scholar]
- 32.Kishida, K., I. Shimomura, H. Nishizawa, N. Maeda, H. Kuriyama, H. Kondo, M. Matsuda, H. Nagaretani, N. Ouchi, K. Hotta, S. Kihara, T. Kadowaki, T. Funahashi, and Y. Matsuzawa. 2001. Enhancement of the aquaporin adipose gene expression by a peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 276:48572-48579. [DOI] [PubMed] [Google Scholar]
- 33.Klein, F. A., R. A. Atkinson, N. Potier, D. Moras, and J. Cavarelli. 2005. Biochemical and NMR mapping of the interface between CREB-binding protein and ligand binding domains of nuclear receptor: beyond the LXXLL motif. J. Biol. Chem. 280:5682-5692. [DOI] [PubMed] [Google Scholar]
- 34.Kliewer, S. A., K. Umesono, D. J. Mangelsdorf, and R. M. Evans. 1992. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature 355:446-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krey, G., A. Mahfoudi, and W. Wahli. 1995. Functional interactions of peroxisome proliferator-activated receptor, retinoid-X receptor, and Sp1 in the transcriptional regulation of the acyl-coenzyme-A oxidase promoter. Mol. Endocrinol. 9:219-231. [DOI] [PubMed] [Google Scholar]
- 36.Latruffe, N., V. Nicolas-Frances, V. K. Dasari, and T. Osumi. 1999. Studies on regulation of the peroxisomal beta-oxidation at the 3-ketothiolase step. Dissection of the rat liver thiolase B gene promoter. Adv. Exp. Med. Biol. 466:253-259. [DOI] [PubMed] [Google Scholar]
- 37.Leesnitzer, L. M., D. J. Parks, R. K. Bledsoe, J. E. Cobb, J. L. Collins, T. G. Consler, R. G. Davis, E. A. Hull-Ryde, J. M. Lenhard, L. Patel, K. D. Plunket, J. L. Shenk, J. B. Stimmel, C. Therapontos, T. M. Willson, and S. G. Blanchard. 2002. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry 41:6640-6650. [DOI] [PubMed] [Google Scholar]
- 38.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 39.Michaud, S. E., and G. Renier. 2001. Direct regulatory effect of fatty acids on macrophage lipoprotein lipase: potential role of PPARs. Diabetes 50:660-666. [DOI] [PubMed] [Google Scholar]
- 40.Mitsiou, D. J., and H. G. Stunnenberg. 2000. TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAαβ precursor and the TFIIAγ subunit. Mol. Cell 6:527-537. [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki, J., K. Araki, E. Yamato, H. Ikegami, T. Asano, Y. Shibasaki, Y. Oka, and K. Yamamura. 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126-132. [DOI] [PubMed] [Google Scholar]
- 42.Molnar, F., M. Matilainen, and C. Carlberg. 2005. Structural determinants of the agonist-independent association of human peroxisome proliferator-activated receptors with coactivators. J. Biol. Chem. 280:26543-26556. [DOI] [PubMed] [Google Scholar]
- 43.Morimoto, R. I. 2002. Dynamic remodeling of transcription complexes by molecular chaperones. Cell 110:281-284. [DOI] [PubMed] [Google Scholar]
- 44.Mueller, E., S. Drori, A. Aiyer, J. Yie, P. Sarraf, H. Chen, S. Hauser, E. D. Rosen, K. Ge, R. G. Roeder, and B. M. Spiegelman. 2002. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J. Biol. Chem. 277:41925-41930. [DOI] [PubMed] [Google Scholar]
- 45.Nagaich, A. K., D. A. Walker, R. Wolford, and G. L. Hager. 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell 14:163-174. [DOI] [PubMed] [Google Scholar]
- 46.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 47.Orlicky, D. J., J. DeGregori, and J. Schaack. 2001. Construction of stable coxsackievirus and adenovirus receptor-expressing 3T3-L1 cells. J. Lipid Res. 42:910-915. [PubMed] [Google Scholar]
- 48.Poirier, H., O. Braissant, I. Niot, W. Wahli, and P. Besnard. 1997. 9- cis-Retinoic acid enhances fatty acid-induced expression of the liver fatty acid-binding protein gene. FEBS Lett. 412:480-484. [DOI] [PubMed] [Google Scholar]
- 49.Ravnskjaer, K., M. Boergesen, B. Rubi, J. K. Larsen, T. Nielsen, J. Fridriksson, P. Maechler, and S. Mandrup. 2005. Peroxisome proliferator-activated receptor alpha (PPARalpha) potentiates, whereas PPARgamma attenuates, glucose-stimulated insulin secretion in pancreatic beta-cells. Endocrinology 146:3266-3276. [DOI] [PubMed] [Google Scholar]
- 50.Rayasam, G. V., C. Elbi, D. A. Walker, R. Wolford, T. M. Fletcher, D. P. Edwards, and G. L. Hager. 2005. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol. Cell. Biol. 25:2406-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez, J. C., G. Gil-Gomez, F. G. Hegardt, and D. Haro. 1994. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J. Biol. Chem. 269:18767-18772. [PubMed] [Google Scholar]
- 52.Ruppert, S. M., V. McCulloch, M. Meyer, C. Bautista, M. Falkowski, H. G. Stunnenberg, and N. Hernandez. 1996. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-react with TBP from other species. Hybridoma 15:55-68. [DOI] [PubMed] [Google Scholar]
- 53.Sartippour, M. R., and G. Renier. 2000. Differential regulation of macrophage peroxisome proliferator-activated receptor expression by glucose: role of peroxisome proliferator-activated receptors in lipoprotein lipase gene expression. Arterioscler. Thromb. Vasc. Biol. 20:104-110. [DOI] [PubMed] [Google Scholar]
- 54.Sato, O., C. Kuriki, Y. Fukui, and K. Motojima. 2002. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J. Biol. Chem. 277:15703-15711. [DOI] [PubMed] [Google Scholar]
- 55.Schoonjans, K., J. Peinado-Onsurbe, A. M. Lefebvre, R. A. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15:5336-5348. [PMC free article] [PubMed] [Google Scholar]
- 56.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 57.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 58.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu, M., A. Takeshita, T. Tsukamoto, F. J. Gonzalez, and T. Osumi. 2004. Tissue-selective, bidirectional regulation of PEX11α and perilipin genes through a common peroxisome proliferator response element. Mol. Cell. Biol. 24:1313-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tachibana, K., Y. Kobayashi, T. Tanaka, M. Tagami, A. Sugiyama, T. Katayama, C. Ueda, D. Yamasaki, K. Ishimoto, M. Sumitomo, Y. Uchiyama, T. Kohro, J. Sakai, T. Hamakubo, T. Kodama, and T. Doi. 2005. Gene expression profiling of potential peroxisome proliferator-activated receptor (PPAR) target genes in human hepatoblastoma cell lines inducibly expressing different PPAR isoforms. Nucl. Recept. 3:3. [Online.] http://www.nuclear-receptor.com/content/3/1/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka, T., J. Yamamoto, S. Iwasaki, H. Asaba, H. Hamura, Y. Ikeda, M. Watanabe, K. Magoori, R. X. Ioka, K. Tachibana, Y. Watanabe, Y. Uchiyama, K. Sumi, H. Iguchi, S. Ito, T. Doi, T. Hamakubo, M. Naito, J. Auwerx, M. Yanagisawa, T. Kodama, and J. Sakai. 2003. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA 100:15924-15929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Targett-Adams, P., M. J. McElwee, E. Ehrenborg, M. C. Gustafsson, C. N. Palmer, and J. McLauchlan. 2005. A PPAR response element regulates transcription of the gene for human adipose differentiation-related protein. Biochim. Biophys. Acta 1728:95-104. [DOI] [PubMed] [Google Scholar]
- 63.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224-1234. [DOI] [PubMed] [Google Scholar]
- 64.Tugwood, J. D., I. Issemann, R. G. Anderson, K. R. Bundell, W. L. McPheat, and S. Green. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varanasi, U., R. Chu, Q. Huang, R. Castellon, A. V. Yeldandi, and J. K. Reddy. 1996. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J. Biol. Chem. 271:2147-2155. [DOI] [PubMed] [Google Scholar]
- 66.Vosper, H., L. Patel, T. L. Graham, G. A. Khoudoli, A. Hill, C. H. Macphee, I. Pinto, S. A. Smith, K. E. Suckling, C. R. Wolf, and C. N. Palmer. 2001. The peroxisome proliferator-activated receptor delta promotes lipid accumulation in human macrophages. J. Biol. Chem. 276:44258-44265. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y. X., C. H. Lee, S. Tiep, R. T. Yu, J. Ham, H. Kang, and R. M. Evans. 2003. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 113:159-170. [DOI] [PubMed] [Google Scholar]
- 68.Willson, T. M., P. J. Brown, D. D. Sternbach, and B. R. Henke. 2000. The PPARs: from orphan receptors to drug discovery. J. Med. Chem. 43:527-550. [DOI] [PubMed] [Google Scholar]
- 69.Wu, J. C., G. Merlino, and N. Fausto. 1994. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. USA 91:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamazaki, K., J. Kuromitsu, and I. Tanaka. 2002. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator-activated receptor alpha agonists. Biochem. Biophys. Res. Commun. 290:1114-1122. [DOI] [PubMed] [Google Scholar]
- 71.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu, S., W. Q. Cao, P. Kashireddy, K. Meyer, Y. Jia, D. E. Hughes, Y. Tan, J. Feng, A. V. Yeldandi, M. S. Rao, R. H. Costa, F. J. Gonzalez, and J. K. Reddy. 2001. Human peroxisome proliferator-activated receptor α (PPARα) supports the induction of peroxisome proliferation in PPARα-deficient mouse liver. J. Biol. Chem. 276:42485-42491. [DOI] [PubMed] [Google Scholar]
- 73.Yu, S., K. Matsusue, P. Kashireddy, W. Q. Cao, V. Yeldandi, A. V. Yeldandi, M. S. Rao, F. J. Gonzalez, and J. K. Reddy. 2003. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J. Biol. Chem. 278:498-505. [DOI] [PubMed] [Google Scholar]