Abstract
The ubiquitin-proteasome pathway (UPP) is involved in regulation of multiple cellular processes. Hypoxia-inducible factor 1α (HIF-1α) is a prototypic target of the UPP and, as such, is stabilized under conditions of proteasomal inhibition. Using carbonic anhydrase IX (CAIX) and vascular endothelial growth factor (VEGF) expression as paradigmatic markers of HIF-1 activity, we found that proteasomal inhibitors (PI) abrogated hypoxia-induced CAIX expression in all cell lines tested and VEGF expression in two out of three. Mapping of the inhibitory effect identified the C-terminal activation domain (CAD) of HIF-1α as the primary target of PI. PI specifically inhibited the HIF-1α CAD despite activating the HIF-1α coactivator p300 and another p300 cysteine/histidine-rich domain 1-dependent transcription factor, STAT-2. Coimmunoprecipitation and glutathione S-transferase pull downs indicated that PI does not disrupt interactions between HIF-1α and p300. Mutational analysis failed to confirm involvement of sites of known or putative posttranslational modifications in regulation of HIF-1α CAD function by PI. Our data provide evidence for the counterintuitive hypothesis that inhibition of HIF-1 function could be responsible for at least some of the antitumor effects of proteasomal inhibition. Further studies of the mechanism of the PI-induced attenuation of HIF-1α will provide important, potentially novel insight into regulation of HIF-1 activity and possibly identify new targets for HIF-directed therapy.
Cells experiencing lower-than-physiological O2 levels undergo a variety of biological responses in order to adapt to these unfavorable conditions. At a molecular level, hypoxic cells respond by increased expression of a number of gene products that will facilitate survival under these conditions. The master switch, orchestrating the cellular response to low O2 levels, is generally considered to be the transcription factor hypoxia-inducible factor 1 (HIF-1) (16, 58). HIF-1 is a heterodimer that consists of the regulated 100- to 120-kDa HIF-1α and the constitutively expressed 91- to 94-kDa HIF-1β subunits (62). Functionally, HIF-1α can be divided into several distinct domains. The basic helix-loop-helix domain at the N terminus, together with the N-terminal half of the PAS domain, is necessary for heterodimerization and DNA binding (57). The unique O2-dependent degradation domain directs rapid degradation of HIF-1α under normoxic conditions. HIF-1α also possesses two transcriptional activation domains (ADs), the N-terminal AD (NAD) and the C-terminal AD (CAD) (26, 52).
Tight regulation of HIF-1α by O2 is suggested by the half-life of HIF-1α under normoxia (<5 min) and its almost instantaneous stabilization and accumulation under hypoxic conditions (21). Under normoxic conditions, P402 and P564 within the O2-dependent degradation domain are hydroxylated by a family of prolyl-4-hydroxylases that require O2, Fe(II), and 2-oxoglutarate for activity (8, 11). Hydroxylated prolines enable specific recognition of HIF-1α by the von Hippel-Lindau protein (VHL) (23, 25), which, in a complex with elongin B, elongin C, and Cul2 (22), functions as an E3 ubiquitin ligase for HIF-1α (49). Polyubiquitylated HIF-1α is then targeted for degradation by the ubiquitin-proteasome pathway (UPP). In the absence of O2, prolyl hydroxylases cannot modify HIF-1α, and consequently the protein rapidly accumulates under hypoxic conditions (47). Stabilized HIF-1α is translocated to the nucleus where, upon dimerization with HIF-1β and interaction with cofactors, the HIF-1 complex binds to hypoxia-responsive elements (HRE) within the promoters of target genes and activates transcription (57, 58).
The NAD and the CAD are distinctly transcriptionally regulated. The CAD appears to be essential for transcriptional activation of HIF target genes, whereas the NAD was found to be dispensable (20). The primary function of the CAD is to recruit widely employed transcriptional coactivators p300/CBP (4). The cysteine/histidine-rich domain 1 (CH1) of p300/CBP serves as a scaffold for folding of the leucine-rich CAD through extensive hydrophobic and polar interactions (10, 12). Though the CAD is stable, its transcriptional activity is hypoxia inducible. Factor inhibiting HIF-1 (FIH-1) interacts with the C-terminal part of HIF-1α and represses its activity (46). The underlying mechanism appears to be regulation of p300/CBP accessibility to CAD through hydroxylation of N803 by FIH-1 (39). In parallel to prolyl hydroxylases, the asparaginyl hydroxylase FIH-1 was also found to require Fe(II) and O2 for optimal activity (38). Thus, FIH-1-mediated hydroxylation of N803 interferes with p300/CBP binding in normoxia, whereas hypoxic inhibition of FIH-1 enhances interaction between the nonhydroxylated CAD and p300/CBP and thereby HIF-1 transcriptional activity.
The UPP is the major proteolytic system in the cytosol and nucleus of all eukaryotic cells. Proteins are targeted for recognition and subsequent degradation by the proteasome via tagging with multiple ubiquitin molecules (18). The 26S proteasome is a cylindrical complex consisting of a 20S core catalytic component with a 19S regulatory component attached to one or both ends (1). The 19S component recognizes and binds polyubiquitylated proteins and then cleaves the ubiquitin chain off the protein substrate. In the 20S component, two outer (α) rings surround two internal (β) rings that carry out the proteolysis of unfolded proteins. Each β ring comprises three active enzymatic sites with trypsin-like, chymotrypsin-like, and post-glutamyl peptide hydrolase-like (caspase-like) activities (1, 34).
Studies involving proteasomal inhibitors (PI) have revealed that the UPP, by breaking down a large variety of cellular proteins with regulatory functions, is essential for many cellular mechanisms (34). A number of proteasome substrates have modulatory effects on pathways that are dysregulated in neoplastic progression. Examples of these molecules are wild-type p53 (17, 35) and IκB (33). It was reasoned that inhibition of the proteasome-mediated degradation of these molecules might arrest or retard cell growth and could be potentially clinically relevant if it was specific to or preferentially targeted neoplastic cells (3). Many types of actively proliferating malignant cells were indeed more sensitive to proteasome blockade than noncancerous cells, although the downstream mechanism responsible for this increased susceptibility has not been conclusively determined (1).
Due to the involvement of the UPP in regulation of multiple cellular processes, inhibition of the proteasome in the cellular context elicits a pleiotropic response, and some aspects of this response may not be fully understood. For instance, several preclinical studies reported that bortezomib (PS-341, Velcade) inhibits tumor angiogenesis, which is required for cancer progression and metastasis (40, 54). These observations were explained in terms of the selective effect of proteasome inhibition on proliferating endothelial cells (2). However, there may be an alternative mechanism which has not been considered. The role of HIF-1 in angiogenesis via up-regulation of vascular endothelial growth factor (VEGF) is well documented (13, 16), and it was also noted that HIF-1 in the presence of PI did not activate transcription (27, 55). This provides the groundwork for the counterintuitive hypothesis that PI, despite increasing the overall HIF-1α levels, inhibit HIF-1 function and that this inhibition could contribute to the antitumor effects of proteasomal inhibition.
In this study, we have analyzed the effect of PI on expression of hypoxia-inducible genes and HIF-1α transcriptional activity. We have also considered some mechanisms that could be responsible for the lack of HIF-1 transcriptional activity in the presence of PI.
MATERIALS AND METHODS
Sequences are written in the 5′-to-3′ direction, and numbers in brackets indicate each position relative to the carbonic anhydrase 9 (CA9) transcription start or the position in the appropriate database entry. Kits, enzymes, antibodies (Abs), and reagents were used according to the manufacturers' recommendations.
Plasmid constructions.
The [−46;+14] CA9 fragment was cloned in the pGL2 basic vector (Promega). pLuc-MCS, containing a minimal TATA box, and its derivatives pAP-1-Luc, pCRE-Luc, and pNF-κB-Luc were obtained from Stratagene. The heterologous HRE constructs were prepared by cloning the CA9 [−46;+14] sequence or HRE from the Glut-1 (only the top strand is shown) (CCACAGGCGTGCTGGCTGACACGCATCAG), VEGF (GTGCATACGTGGGCTCCAACAGGTCCTC), and murine phosphoglycerate kinase-1 (TTTGTCACGTCCTGCACGACGCG) genes into the pLuc-MCS vector and verified by sequencing. Gal4-HIF-1α 529-826, Gal4-HIF-1α 529-778, Gal4-HIF-1α 740-826, and Gal4-HIF-1α 787-826 (expressing the Gal4 DNA-binding domain [DBD] fused to the indicated HIF-1α fragments [amino acids]) and pFR-Luc (a reporter vector in which the firefly luciferase gene is under the control of a minimal E1B promoter and upstream four copies of a Gal4 binding site) constructs (56) were kind gifts from Nianli Sang (Thomas Jefferson University). The Gal4-STAT-2 and Gal4-HIF-1α/VP16 constructs express the fusion of the Gal4 DBD with the STAT-2 670-851 fragment (5) and the fusion of the HIF-1α 529-778 fragment with the herpes simplex virus VP16 AD, respectively. The Gal4-p300 construct was kindly provided by Antonio Giordano (Temple University). The HIF CAD-pGEX-2T construct contains the fragment coding for HIF-1α 787-826 in pGEX-2T (Amersham Biosciences). The p300 CH1-pGEX-2T and p300 CH1-VP16 AD constructs were prepared by cloning the CH1 domain of p300 in frame into pGEX-2T and pcDNA3 containing the VP16 AD, respectively. The hemagglutinin-tagged p300 in the pCMV5 expression plasmid (p300-pCMV5) was purchased from Upstate. The mutant Gal4-HIF CAD 787-826 constructs with individual T796D, S790D, S797D, S809D, Y798F, Y798D, N803A, and C800V mutations were prepared with PCR and verified by sequencing. The allmut construct contains all of the above-mentioned mutations with the exception of Y798F. The pRL-CMV vector was obtained from Promega.
Cell lines and culture.
Human breast carcinoma MCF-7, osteosarcoma Saos-2, and glioma M006 cell lines were grown in Dulbecco's modified Eagle's medium (BioWhittaker) supplemented with 10% fetal calf serum (Life Technologies), 1 × 102 U/ml penicillin (Sigma), 1 × 102 μg/ml streptomycin (ICN), and 125 ng/ml amphotericin B (Sigma). The following PI were used: carbobenzoxy-l-leucyl-l-leucyl-l-norvalinal (LLNV), lactacystin (both from Sigma), and bortezomib (Millennium Pharmaceuticals). The effect of PI on carbonic anhydrase IX (CAIX), VEGF, and HIF-1α expression was tested with cells that had been seeded at 10,000/cm2 and grown for 3 days. The cells were plated at 40,000/cm2, pretreated with PI (control for LLNV, dimethyl sulfoxide) for 30 min, and exposed to normoxia or a 0.5% O2 environment in a ProOx in vitro chamber (BioSpherix), controlled by ProOx model 110 (BioSpherix), for 24 h in the presence of PI.
Western blot analysis.
Western blot analysis of HIF-1α, CAIX, and β-actin expression was performed as described previously (32). Anti-p300 and anti-Gal4 DBD Abs were from Upstate and Clontech, respectively. Antiubiquitin Ab was a gift from Peter Kaiser (University of California, Irvine).
VEGF assay.
VEGF levels in cell culture media were assayed with a VEGF sandwich enzyme-linked immunosorbent assay kit (Chemicon) and expressed as pg protein/mg of total protein.
MTT assay.
Saos-2 cells (20,000/100 μl) were plated into 96-well microtiter plates, incubated overnight, pretreated with PI for 30 min, and exposed to normoxia or 0.5% O2 for 24 h in the presence of PI. Each control and PI concentration was run in triplicate. Cells were rinsed with phosphate-buffered saline (PBS) and incubated for 4 h at 37°C in 100 μl medium containing 1 mg/ml 3-(4,5-dimethylthiazolyl-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma). Upon lysis with 100 μl of 2% (wt/vol) sodium dodecyl sulfate, 50% (vol/vol) N,N-dimethylformamide, and 0.4% (vol/vol) glacial acetic acid (1 h at room temperature), the color development was read at 595 nm with a SpectraMax 340 microplate reader (Molecular Devices). The data were expressed as the percentages (±standard deviations [SD]) of the untreated control.
Reverse transcriptase PCR (RT-PCR).
MCF-7 and Saos-2 cells (6 × 105 plated at 40,000/cm2), pretreated with PI for 30 min, were exposed to normoxia or 0.5% O2 for 16 h in the presence of PI. Total RNA was isolated with an RNeasy mini kit (QIAGEN), and cDNA was synthesized with a ProtoScript first-strand cDNA synthesis kit (New England Biolabs). cDNA fragments were amplified with the following primer pairs: for HIF-1α (GenBank accession no. U22431), GCCGAGGAAGAACTATGAAC (residues 558 to 577), sense, and ATATTTGATGGGTGAGGAATGG (residues 726 to 704), antisense; for CA9 (GenBank accession no. NM_001216), CTGTCACTGCTGCTTCTGAT (residues 121 to 140), sense, and TCCTCTCCAGGTAGATCCTC (residues 321 to 301), antisense; for VEGF (GenBank accession no. M32977), GCCTTGCTGCTCTACCTC (residues 93 to 110), sense, and GGCACACAGGATGGCTTG (residues 292 to 275), antisense; for CDKN1A (GenBank accession no. NM_00389), CCAGTGGACAGCGAGCAG (residues 164 to 181), sense, and CCACATGGTCTTCCTCTGC (residues 428 to 410), antisense; for p300 (GenBank accession no. NM_001429), ATGGGTTCTGGAGCACAT (residues 1369 to 1386), sense, and CTTATCACCAGCATTTTTGAG (residues 1671 to 1651), antisense; and for β-actin (GenBank accession no. NM_001101), ACAACGGCTCCGGCATGTGCAA (residues 105 to 126), sense, and CGGTTGGCCTTGGGGTTCAG (residues 420 to 402), antisense. PCRs were performed with a GeneAmp PCR system 9700 (PE Applied Biosystems) for 32 cycles (with the exception of β-actin [25 cycles]) at 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s. Products were analyzed on a 1.5% agarose gel.
Transient transfection assay.
Cells were cotransfected with a firefly luciferase reporter construct driven by the tested promoter and the pRL-CMV expressing Renilla luciferase (internal control for transfection efficiency) as described previously (32). The Gal4 fusion constructs were cotransfected with pFR-Luc, pRL-CMV, and p300 CH1-VP16 AD or p300-pCMV5 expression vector (where appropriate) in the same way. After exposure to the transfection mixture for 16 h, the cells were trypsinized, plated at 40,000/cm2, and allowed to adhere for 5 h. The cells were then pretreated with PI for 30 min and exposed to normoxia or 0.5% O2 for 24 h in the presence of PI. Reporter assays were performed as described previously (32). Promoter activities were expressed as the average ratios of firefly to Renilla luciferase activities (±SD) from at least three independent experiments. Western blotting with anti-Gal4 DBD Ab was used to probe levels of Gal4-HIF-1α fusion proteins in MCF-7 and Saos-2 cells, transiently transfected with Lipofectamine 2000 (Invitrogen) and GenePorter 2 (Gene Therapy Systems) transfection reagents, respectively.
Coimmunoprecipitations of HIF-1α and p300.
MCF-7 cells (1 × 106) were treated as described in “Reverse transcriptase PCR” above. Cells were rinsed with ice-cold 1× PBS and 1 mM EDTA, and nuclear extracts (NE) were isolated with NE-PER nuclear and cytoplasmic extraction reagents (Pierce). NE (700 μg) were diluted to 1 ml in 1× cell lysis buffer (Cell Signaling Technology) supplemented with Complete EDTA-free protease inhibitor cocktail (Roche), 25 mM NaF, and 1 mM phenylmethylsulfonyl fluoride. Following preclearing with normal rabbit immunoglobulin G (Santa Cruz Biotechnology) and protein A Sepharose (Amersham Biosciences) for 1 h at 4°C, the samples were immunoprecipitated with 15 μl agarose-conjugated p300 (N-15) Ab (Santa Cruz Biotechnology) overnight at 4°C. Immunoprecipitates were washed five times with 1× cell lysis buffer (500 μl) and analyzed by Western blotting with HIF-1α Ab. As input, 1/20 of original NE was used.
GST pull down.
pGEX-2T constructs were transformed into Escherichia coli BL21, and glutathione S-transferase (GST) proteins were purified with glutathione-Sepharose 4B (Amersham Biosciences). MCF-7 cells (1 × 106) were treated as described in “Reverse transcriptase PCR” above, rinsed with ice-cold 1× PBS, and lysed in 1× cell lysis buffer (supplemented with the protease inhibitor cocktail and phenylmethylsulfonyl fluoride) for 15 min on ice. Cell lysates (500 μg) were agitated with GST only (negative control) or GST-p300 CH1 immobilized on glutathione-Sepharose 4B for 3 h at room temperature. Recovered complexes were washed three times with ice-cold 1× cell lysis buffer (500 μl) with inhibitors and analyzed by Western blotting with HIF-1α Ab. For GST-HIF CAD pull down, NE (100 μg) from MCF-7 cells were used and analyzed by Western blotting with p300 Ab.
RESULTS
Effect of PI treatment on CAIX, VEGF, and HIF-1α expression.
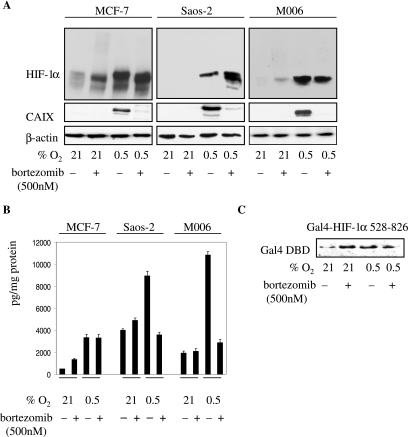
Initially, we tested the effect of PI treatment on expression of the hypoxia marker CAIX. Normoxic MCF-7, Saos-2, and M006 cells did not express detectable levels of CAIX, and addition of the PI bortezomib to normoxic cells did not activate CAIX expression (Fig. 1A). In all cell lines, hypoxia induced CAIX expression that was almost completely abrogated by bortezomib treatment (Fig. 1A). These data suggest that not only is PI treatment insufficient to activate CAIX expression under normoxia but, more importantly, it strongly inhibits hypoxia-induced CAIX expression as well. Bortezomib treatment had more-diverse effects on VEGF expression. In Saos-2 and M006 cells, it did not significantly influence normoxic VEGF levels, but it inhibited hypoxia-induced VEGF expression (Fig. 1B). In contrast, bortezomib moderately increased VEGF secretion by normoxic MCF-7 cells but had no effect on hypoxia-induced VEGF (Fig. 1B). Hypoxic induction of STRA13 and glyceraldehyde-3-phosphate dehydrogenase was also inhibited by bortezomib treatment in Saos-2 cells (data not shown).
FIG. 1.
(A) Effect of bortezomib treatment on HIF-1α and CAIX expression. Cells were seeded at 40,000/cm2, allowed to attach overnight, pretreated with bortezomib for 30 min, and exposed to 21% (normoxia) or 0.5% O2 for 24 h in the presence of bortezomib. Total protein lysates (40 μg) were tested for HIF-1α, CAIX, and β-actin by Western blotting. (B) Effect of bortezomib treatment on VEGF expression. VEGF, secreted by cells treated as described for panel A, was assayed with a VEGF sandwich enzyme-linked immunosorbent assay kit. VEGF levels are expressed as pg protein/mg of total protein, and each of the bars represents the mean value (±SD) from at least three individual experiments. (C) Effect of bortezomib treatment on the mobility of the Gal4-HIF-1α 529-826 fusion protein in MCF-7 cells. Cells were transfected with the Gal4-HIF-1α 529-826 construct, and 24 h later, they were treated as described for panel A and analyzed by Western blotting with anti-Gal4 DBD Ab.
HIF-1α also responded differently to PI treatment. A residual HIF-1α level was detected only in normoxic MCF-7 cells, and PI treatment further increased this level (Fig. 1A). PI stabilized HIF-1α in normoxic M006 cells but not in Saos-2 cells (Fig. 1A). PI also had various effects on mobility of HIF-1α: compared to hypoxia-stabilized HIF-1α, complete and partial conversions to a faster-migrating species were observed with MCF-7 and Saos-2 cells, respectively (Fig. 1A). On the other hand, the mobility of HIF-1α was not affected by PI in M006 cells (Fig. 1A). Thus, the appearance of the faster-migrating species is not a prerequisite for the loss of HIF-1α function. The transcriptionally important NAD and CAD reside in the C-terminal part of HIF-1α, and we asked whether PI affect the mobility of this region. To this end, we found that PI did not affect the mobility of the Gal4-HIF-1α 529-826 fusion protein in MCF-7 cells (Fig. 1C). We therefore conclude that the apparently different molecular weight of the faster-migrating HIF-1α species (presumably due to a differential posttranslational modification[s] outside the region from residues 529 to 826), induced by PI in some cell lines, appears to be an epiphenomenon and lacks significance with respect to the observed PI-mediated inhibitory effect.
Polyubiquitylation of HIF-1α was confirmed by immunoprecipitation and Western blotting with ubiquitin Ab. This Ab detected higher-molecular-weight species in cells treated with PI but not in the control normoxic and hypoxic cells (data not shown). It should also be noted that the vast majority of HIF-1α is observed as a distinct band and that only a small fraction is detected as the slower-migrating polyubiquitylated HIF-1α in the PI-treated cells (Fig. 1A). Even when cells were lysed in 8 M urea (to inhibit deubiquitinases), only a small increase in polyubiquitylated HIF-1α was observed (data not shown).
Effect of PI treatment on transcription of hypoxia-regulated genes.
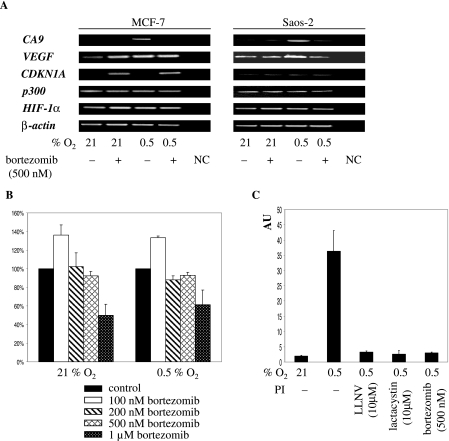
Above, we described that PI inhibited CAIX expression in all tested cell lines whereas regulation of VEGF was cell type specific. Therefore, we studied regulation of CA9 and VEGF by RT-PCR in MCF-7 and Saos-2 cells. Data for CA9 confirmed the tight control of transcription by hypoxia and the strong inhibitory effect of bortezomib in both cell lines (Fig. 2A). VEGF responded differently to PI treatment. The basal level of transcription of VEGF in normoxic cells was moderately up-regulated in MCF-7 cells but not in Saos-2 cells exposed to PI (Fig. 2A). In contrast, hypoxia-induced transcription was inhibited in Saos-2 cells but not in MCF-7 cells (Fig. 2A). Together, RT-PCR data correspond well with the data shown in Fig. 1A and confirm the opposite regulation of CA9 and VEGF in MCF-7 cells. In comparison with CA9, the inconsistent effect of PI on VEGF expression could be explained in terms of previous findings that show that VEGF is transcriptionally activated by multiple transcription factors (9, 48, 51). RT-PCR analysis of some other genes of interest further underscored the specificity of PI treatment on transcription. PI did not influence levels of p300, HIF-1α, and β-actin transcripts in either cell line (Fig. 2A). Transcription of CDKN1A in MCF-7 cells was activated only in the presence of PI, whereas low constitutive transcription was observed with Saos-2 cells (Fig. 2A). This is consistent with the stabilization and activation of wild-type p53 in MCF-7 cells and the lack of expression of p53 in Saos-2 cells (data not shown). In summary, these data provide evidence that PI exerts a selective inhibitory effect on hypoxia-induced transcription.
FIG. 2.
(A) RT-PCR analysis of transcription of selected genes in bortezomib-treated cells. Cells were seeded and treated as described in the legend for Fig. 1A and harvested 16 h later, and total RNA was isolated, reverse transcribed, and amplified with gene-specific primers. NC, negative control. (B) MTT assay of Saos-2 cells treated with various concentrations of bortezomib. The cells were seeded in triplicate and treated as described in the legend for Fig. 1A, and the MTT assay was performed 24 h later. The results are expressed as the percentages (±SD) of the untreated control, set as 100%. (C) Effects of various PI on activity of the [−46;+14] CA9 fragment in Saos-2 cells. Cells were cotransfected with the [−46;+14] CA9 reporter construct and pRL-CMV for 16 h, trypsinized, replated, and treated as described in the legend for Fig. 1A. Activity of the CA9 fragment is expressed as the ratio of firefly/Renilla activities in arbitrary units (AU). Each of the bars represents the mean value (±SD) from at least three individual experiments.
Toxicity and comparison of various PI.
Toxicity of 24-h bortezomib treatment on Saos-2 cells was tested by MTT assay. Under these conditions, only the highest concentration (1 μM) was toxic (Fig. 2B). We also compared the effects of various types of PI on activity of the [−46;+14] CA9 fragment. LLNV (a peptide aldehyde), lactacystin (a nonpeptide Streptomyces metabolite), and bortezomib (a peptide boronate) all suppressed CA9 promoter activity to comparable levels in transiently transfected hypoxic Saos-2 cells (Fig. 2C). This proves that, regardless of the chemical nature of the pharmacophore, inhibition of the proteasome also inhibits HIF-1-dependent CA9 promoter activity.
Effect of PI treatment on hypoxia and other response elements.
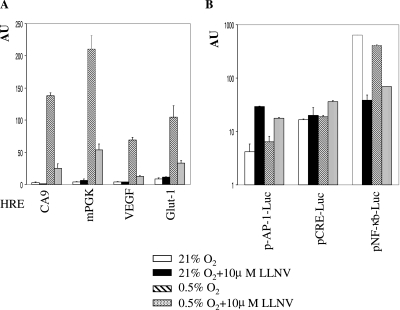
Next, we investigated whether transcription driven by heterologous promoter constructs with an isolated HRE upstream of a minimal TATA box is affected by PI. This approach allows direct comparison of individual HRE, as each is positioned in the same orientation and at the same distance with respect to the transcription start site and the contribution of other transcription factors is eliminated. In normoxia, all tested constructs had background activity that was not further modulated in the presence of PI (Fig. 3A). Each construct displayed hypoxia-dependent inducibility of various magnitudes, and in each case, PI markedly repressed hypoxia-induced activity. The residual reporter activity was proportional to the magnitude of hypoxic induction (Fig. 3A). We also wished to determine what effect PI would have on other transcription factors. To this end, we tested reporter constructs driven by the same minimal promoter as the HRE constructs and multimerized response elements for AP-1, CREB, and NF-κB transcription factors. In agreement with previous reports, PI strongly inhibited activity of the NF-κB construct (19). Activity of the AP-1 construct, on the other hand, was clearly up-regulated in the presence of the inhibitor (Fig. 3B), presumably due to the stabilization of c-jun in a functional form (18). No effect on activity of the CREB construct was observed. In summary, down-regulation of HRE-driven transcription in the presence of PI is not restricted to a particular type of HRE, and proteasomal inhibition will, therefore, interfere with hypoxic induction of multiple genes and not only CA9. Furthermore, proteasomal inhibition modulates activities of various transcription factors over a wide range, refuting the concern about general transcription factor inhibition. In keeping with this, activation of some transcriptional factors may compensate for inhibition of HIF-1 in the presence of PI and thus be responsible for the apparent lack of inhibitory effect on hypoxia-induced activity (e.g., VEGF in MCF-7 cells).
FIG. 3.
Effect of LLNV treatment on activities of various HIF-1 inducible (A) and noninducible (B) response elements in Saos-2 cells. Cotransfection (indicated construct and pRL-CMV) and expression of promoter activities are described in the legend for Fig. 2C. mPGK, murine phosphoglycerate kinase.
Effect of PI treatment on HIF-1α and other p300-dependent transcriptional activities.
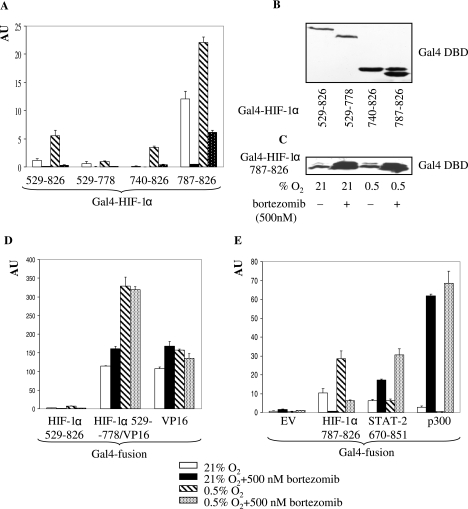
Having demonstrated that PI treatment affects HRE-driven transcriptional activity, we were next interested in the corresponding mechanism. First, we mapped the effect of PI by using HIF-1α deletion mutants fused to the Gal4 DBD. Data obtained with transiently transfected Saos-2 cells show that activities of all constructs under normoxic and hypoxic conditions were inhibited by PI (Fig. 4A). Among these constructs, the HIF-1α 787-826 (the constitutive CAD) construct produced the highest activity, whereas the HIF-1α 529-778 construct (lacking the CAD region) produced the lowest. It is noteworthy that under conditions that did not significantly stabilize HIF-1α in Saos-2 cells (normoxia plus PI), the activities of all constructs were still considerably inhibited (Fig. 4A). Expression of Gal4 fusion proteins was probed by Western blotting with anti-Gal4 DBD Ab, and results are shown in Fig. 4B. Our finding that bortezomib stimulated levels of the fusion proteins in Saos-2 cells (Fig. 4C) rules out the possibility that decreased activity of these constructs is due to inhibition of their expression. The Gal4-HIF-1α construct, with the VP16 AD in place of the CAD, retained hypoxic inducibility but, just like the control Gal4-VP16 AD, was not inhibited by PI (Fig. 4D). Although NAD is also inhibited by PI, our data are consistent with the notion that PI inhibits HIF-1α function primarily via interfering with activity of the relatively small constitutive HIF-1α CAD.
FIG. 4.
(A) Effect of bortezomib treatment on activities of the Gal4-HIF-1α deletion mutants in Saos-2 cells. Cotransfection (indicated Gal4-HIF-1α deletion mutant, pFR-Luc, and pRL-CMV) and expression of fusion construct activities are described in the legend for Fig. 2C. (B) Levels of Gal4-HIF-1α fusion proteins and (C) effect of bortezomib treatment on levels of Gal4-HIF-1α 787-826 protein in transiently transfected Saos-2 cells were tested by Western blotting with anti-Gal4 DBD Ab. (D) Effect of bortezomib treatment on activities of the Gal4-VP16 fusion construct and (E) Gal4-STAT-2 and Gal4-p300 constructs in Saos-2 cells. Cotransfection (indicated Gal4 fusion construct, pFR-Luc, and pRL-CMV) and expression of fusion construct activities are described in the legend for Fig. 2C. EV, empty vector.
The recruitment of transcriptional coactivator p300/CBP to HIF-1α CAD plays an essential role in activation of HIF-1α (4). Numerous reports have convincingly established that regulation of p300/CBP function and/or p300/CBP-HIF-1α interaction modulates HIF-1 activity (36, 39, 56). This prompted us to investigate the role of p300/CBP in the PI-mediated inhibition of HIF-1. Two alternative modes were considered: (i) PI inhibits the ability of p300 to activate transcription, or (ii) PI disrupts HIF-1α-p300/CBP interactions. We used the Gal4-p300 fusion construct to test the effect of PI on p300 transcriptional activation. Quite unexpectedly, a massive induction of p300 activity was observed in the presence of PI in normoxia and hypoxia (Fig. 4E). As multiple transcription factors share the p300/CBP CH1 domain (12), evaluation of their activities would provide information about the generality of the PI effect on p300/CBP CH1-dependent transcription. In this group, STAT-2 (5) is frequently compared with HIF-1α (6, 37). Activities of the HIF-1α CAD and STAT-2 670-851 fusion constructs were oppositely regulated under all experimental conditions used. Activity of STAT-2, as expected, did not change in hypoxia but was considerably stimulated in the presence of PI (Fig. 4E). The opposite regulation of the HIF-1α CAD and STAT-2 suggests a possible mechanism that competition of STAT-2 for p300/CBP binding is responsible for inhibition of HIF-1. However, decreased STAT-2 levels following PI treatment, as observed by us (data not shown) and others (61), do not support this mechanism.
Thus, of the three constructs tested, PI treatment inhibited only the HIF-1α CAD, whereas it had a potent stimulatory effect on STAT-2 and p300. The opposite regulation of these constructs proves that PI relatively specifically inhibits the HIF-1α CAD; moreover, it does so despite the concomitant activation of p300, the transcriptional coactivator of HIF-1α. Therefore, PI either selectively interferes with the HIF-1α CAD-p300 interaction or targets a p300-unrelated function of HIF-1α, either directly on HIF-1α CAD or indirectly through an interacting partner.
Effect of PI treatment on HIF-1α-p300 interaction.
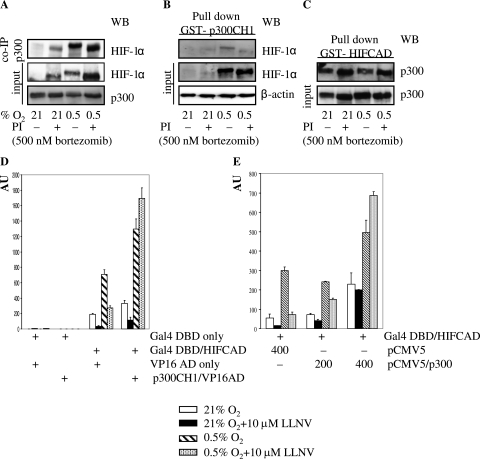
The observation of simultaneous inhibition of the HIF-1α CAD and stimulation of p300 by PI could be reconciled by invoking the disruption of the HIF-1α CAD-p300 interaction. To probe the effect of PI on the interaction between the HIF-1α CAD and p300, we used coimmunoprecipitation and GST pull-down assays. Coimmunoprecipitation of HIF-1α was performed with immobilized p300 Ab by using NE from MCF-7 cells. p300 Ab brought down HIF-1α from all three samples where HIF-1α was stabilized (Fig. 5A). This was further supported by pull-down experiments with bacterially expressed GST fusion proteins. GST-p300 CH1 pulled down endogenous hypoxia-stabilized HIF-1α from control and PI-treated MCF-7 cells (Fig. 5B) and Saos-2 cells (data not shown). In the reverse experiment, GST-HIF-1α CAD pulled down constitutively expressed p300 from control and PI-treated MCF-7 cells equally effectively (Fig. 5C). GST only did not bring down HIF-1α or p300 (data not shown). Thus, by using two different approaches, we were able to show that interaction between p300 and HIF-1α is not compromised in the presence of PI.
FIG. 5.
(A) Effect of bortezomib treatment on coimmunoprecipitation (co-IP) of endogenous HIF-1α with p300 in MCF-7 cells. NE were prepared from cells treated as described in the legend for Fig. 1A, immunoprecipitated with p300 Ab, and tested for HIF-1α by Western blotting (WB). Equal loading was verified by Ponceau S staining. (B and C) Effect of bortezomib treatment on pull down of HIF-1α with GST-p300 CH1 (B) and p300 with GST-HIF CAD (C) in MCF-7 cells. Total protein lysates (B) or NE (C), prepared from cells treated as described in the legend for Fig. 1A, were pulled down with bacterially expressed GST fusion proteins and glutathione-Sepharose 4B and tested for HIF-1α/p300 by Western blotting. (C) Equal loading was verified by Ponceau S staining. (D) Mammalian two-hybrid assay of HIF-1α CAD and p300 CH1 in Saos-2 cells. Gal4-HIF-1α CAD and p300 CH1-VP16 AD (or controls) were cotransfected with pFR-Luc and pRL-CMV as described in the legend for Fig. 2C. The interaction between fusion constructs is expressed as the ratio of firefly/Renilla activity in arbitrary units (AU). Each of the bars represents the mean value (±SD) from at least three individual experiments. (E) Effect of overexpression of p300 on LLNV-inhibited HIF-1α CAD activity in Saos-2 cells. Cotransfection (Gal4-HIF-1α CAD, pFR-Luc, indicated amounts of p300-pCMV5, and pRL-CMV) and expression of HIF-1α CAD activity are described in the legend for Fig. 2C.
To gain further insight into the role of the interaction between the HIF-1α CAD and p300 CH1, we employed the mammalian two-hybrid assay. Gal4-HIF-1α CAD was cotransfected with the p300 CH1-VP16 AD fusion construct or VP16 AD alone into Saos-2 cells. In the presence of VP16 AD alone, PI still inhibited HIF-1α CAD activity under normoxic and hypoxic conditions (Fig. 5D). The p300 CH1-VP16 AD fusion, on the other hand, not only stimulated HIF-1α CAD activity under hypoxia but counteracted the inhibitory effect of PI under hypoxia as well (Fig. 5D). This indicated that overexpression of p300 CH1 fused to the potent AD of VP16 is sufficient to rescue HIF-1α CAD function. However, the AD of p300 was equally effective and overexpression of the full-length p300 also rescued hypoxia-induced HIF-1α CAD activity (Fig. 5E). Thus, although PI does not seem to compromise the HIF-1α CAD-p300 interaction, overexpression of p300 CH1 in the context of native p300 or a chimeric protein prevents the inhibitory effect of PI and restores HIF-1α CAD activity.
Mutational analysis of the HIF-1α CAD.
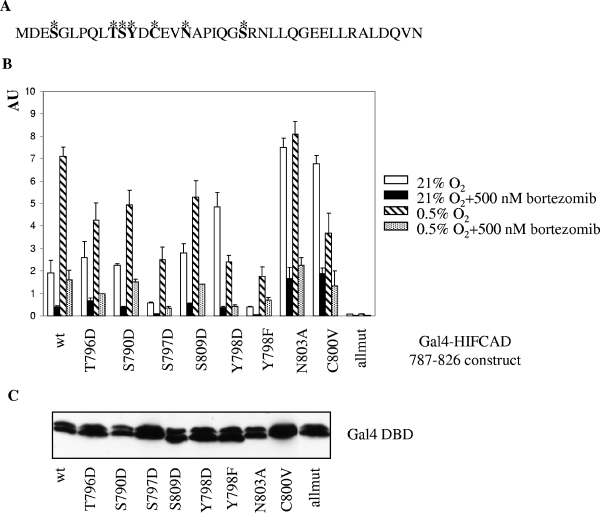
A number of posttranslational modifications of the HIF-1α CAD, encompassing 40 amino acids (Fig. 6A), have been described, and their role in modulation of transcriptional activity is being elucidated gradually. Hydroxylation of N803 (39), phosphorylation of T796 (14), and modifications of C800 (15) modulate HIF-1α CAD activity by regulating its access to p300/CBP coactivators. It is formally possible that PI interfere with an activating posttranscriptional modification(s) or, alternatively, induce/retain an inhibitory modification(s). For example, activation of FIH-1 in the presence of PI could keep N803 hydroxylated even in hypoxia, provide a facile mechanism explaining attenuation of HIF-1 activity, and still be compatible with the observed selective loss of HIF-1α function but not p300 and STAT-2 activities. To elucidate the role of known and putative posttranslational modifications in the regulation of HIF-1α CAD activity by PI, we performed mutational analysis of T796, S790, S797, S809, Y798, N803, and C800. Regulatable residues were individually replaced with nonregulatable ones, which either mimicked the constitutive modification (T→D, S→D, and Y→D) or prevented it (Y→F, N→A, and C→V). The activity of each of these mutants was tested in the context of Gal4 fusion constructs, and Fig. 6B shows a typical result obtained by use of transiently transfected Saos-2 cells. In normoxia, all mutants generated at least as much activity as the wild-type construct, with the exception of the S797D and Y798F mutations, which were detrimental to HIF-1α CAD activity (Fig. 6B). This suggests that either S797 is not phosphorylated or D is not a good mimic in this context and suggests the functional importance of the polar OH group on Y798. Activities of all constructs were stimulated under hypoxia, with the notable exceptions of Y798D, N803A, and C800V mutants (Fig. 6B). The lack of hypoxic activation of the N803A mutant due to high normoxic activity was expected and is the consequence of relieving the negative regulation exerted by N hydroxylation (39). The other two mutants generated high normoxic activity that significantly decreased in hypoxia (Fig. 6B). Apparently, this could reflect the complex regulation of HIF-1α CAD activity that may, in these particular cases, reflect simultaneous effects of mutated residues on FIH-1 and interaction with coactivators. Most importantly, however, PI invariably down-regulated activity of all constructs in normoxia and hypoxia (Fig. 6B). Basal activity of the construct harboring all mutations was severely impaired, but it was further inhibited by PI (Fig. 6B). Comparable expression levels of the mutant constructs were documented by Western blotting with Gal4 DBD Ab (Fig. 6C). Therefore, the mutation(s) (individual or cumulative) of residues that are known to be or could potentially be posttranslationally modified is not sufficient to prevent down-regulation of HIF-1α CAD activity by PI. We conclude that the negative effect of PI is not the result of interference with or induction of posttranslational modifications of HIF-1α CAD at the tested positions.
FIG. 6.
(A) Sequence of HIF-1α CAD (amino acids 787 to 826). Residues mutated in this study are indicated in bold and marked with asterisks. (B) Effect of bortezomib treatment on activities of the mutant Gal4-HIF-1α CAD fusion constructs in Saos-2 cells. Cotransfection (indicated Gal4-HIF-1α CAD mutant, pFR-Luc, and pRL-CMV) and expression of mutant construct activities are described in the legend for Fig. 2C. Allmut contains all mutations except the Y798F mutation. (C) The levels of mutant Gal4-HIF-1α CAD proteins in transiently transfected Saos-2 cells were tested by Western blotting with anti-Gal4 DBD Ab. wt, wild type.
DISCUSSION
The UPP regulates degradative turnover of a large number of proteins. In the absence of any systematic analysis to date, at least some of the proteins accumulated upon inhibition of the UPP appear to remain functionally active. Intuitively, functional activity of stabilized proteins, such as cell cycle regulators, proapoptotic factors, IκB, the inhibitor of the NF-κB pathway, and p53, would be required for the antineoplastic activity of the clinically approved PI bortezomib. It has been mentioned, although not systematically studied, that HIF-1α, another target of the UPP, accumulated in the presence of PI in a functionally inactive form. The first report that PI stabilized HIF-1α protein under normoxic conditions predated the finding that the VHL E3 ligase complex targeted HIF-1α for proteasomal degradation in normoxia (55). The authors ascribed the phenomenon of accumulation of inactive HIF-1 to a nonspecific toxic effect of PI. Of the two hypotheses formulated later, the first presumed that stabilization of the HIF-1α protein on its own is not sufficient to generate a functional form because additional regulatory steps are required to induce transcriptional activity of HIF-1α (27). This theory correctly anticipated the regulation of HIF-1α transcriptional activity by hydroxylation of N803 (39), but it fails to provide an explanation for inactive HIF-1 in hypoxia. In the second theory, polyubiquitylation of HIF-1α interfered with the ability of HIF-1 to mediate hypoxic signal transduction and impeded nuclear transport (27).
The cancer-related transmembrane CAIX (24, 63) catalyzes the conversion of carbon dioxide into bicarbonate and protons that consequently acidify the extracellular milieu (24). Correlation of CAIX expression with lowered O2 in vivo (43) and in vitro (63) and extensive colocalization of CAIX and the chemical hypoxia marker pimonidazole (50), together with high protein stability (53), an extremely tight transcriptional control (29, 63), and expression under mild hypoxic conditions (29), set CAIX apart from other hypoxia-regulated gene products (e.g., VEGF and erythropoietin) and make it one of the best intrinsic markers of cellular hypoxia (43, 50). Although regulation of CA9 promoter activity is complex (28, 31), we have found that juxtaposed HRE and SP1/SP3 sites (30, 31) are critical for transcriptional activation of CA9. We have shown previously that CAIX expression/transcription is an excellent tool for monitoring HIF-1 activity in vitro (29, 30, 32). In the course of the study of CA9 regulation, we noticed that PI specifically prevented hypoxic induction of CA9 transcription, despite the presence of substantial levels of HIF-1α. This counterintuitive observation drew our attention to regulation of HIF-1 activity by proteasomal inhibition and led us to evaluate the previously proposed theories about the loss of HIF-1 activity.
Our data do not support the conclusion that a nonspecific toxic effect is responsible for PI-mediated inactivation of HIF-1 (55). Treatment with PI under the described conditions was not significantly toxic to the cell lines used, and RT-PCR analysis did not show a nonspecific inhibitory effect on transcription in general. We have observed that only a small fraction of HIF-1α in PI-treated cells is polyubiquitylated. The majority of HIF-1α was detected as a distinct, nonubiquitylated band, even when the cells were lysed by 8 M urea to inactivate deubiquitinases. The relatively low levels of polyubiquitylated HIF-1α can be explained as follows. Initially, inhibition of the proteasome leads to accumulation of polyubiquitylated proteins from which ubiquitin is not recycled. Due to the limited pool, cells “run out” of ubiquitin and subsequently stabilized proteins will be mostly nonpolyubiquitylated. The fact that the bulk of HIF-1α is nonubiquitylated is at odds with the hypothesis that polyubiquitylation of HIF-1α could be primarily responsible for disrupting HIF-1 transcriptional activity (27). Furthermore, ubiquitin, a small, 8-kDa protein, is attached to a free amino group on the substrate, generally through an isopeptide bond to the amino group of a lysine side chain (18). The lack of lysines in the HIF-1α CAD (Fig. 6A), the minimal fragment inhibited by PI, argues further against the involvement of polyubiquitylation in regulation of HIF-1α activity. According to the “activation by destruction” theory, sustained transcription mediated by certain transcription factors requires proteasomal activity to remove “spent” activators and to reset the promoter (41). By the same token as outlined above, this theory cannot satisfactorily explain inhibition of the HIF-1α CAD by PI.
The original proposition about inactivation of HIF-1 by impaired translocation of HIF-1α to the nucleus in PI-treated cells (27) was not upheld in other studies (44, 55) or in the present studies, as large amounts of HIF-1α were detected in NE prepared from cells treated with PI. We have also confirmed efficient nuclear translocation of HIF-1α by immunostaining (data not shown). Moreover, HIF-1 in NE from normoxic and hypoxic cells treated with PI binds to HRE, as demonstrated in electromobility shift assays (55), suggesting that the interaction between HIF-1α and HIF-1β and the DNA binding affinity of HIF-1 per se is not compromised in the presence of PI.
We found that PI potently inhibited hypoxia-induced CAIX expression in all tested cell lines, whereas VEGF expression was inhibited in two out of three. This inhibition appears to be relatively specific, as a number of other genes were not affected by PI. The absence of VEGF inhibition in MCF-7 cells could be explained by the existence of other transcription factors controlling prominently transcription of VEGF, e.g., SP1 (51) and AP-1 (9). PI presumably differentially regulates activities of these factors and HIF-1, and activation of other factors outweighs decreased HIF-1 activity. In support of this, we found diverse effects of PI on activity of transcription factors, but heterologous constructs with isolated HRE from various hypoxia-inducible genes, including VEGF, were invariably inhibited. Therefore, inhibition of HIF-1-dependent transcription in the presence of PI appears to be a general phenomenon but contributions from other transcription factors could obscure inhibition of some hypoxia-inducible genes in certain cell types. Two recent studies reported on PI-mediated inhibition of CAIX and VEGF. Despite increased labeling of HIF-1α, there was a decreased level of CAIX in bortezomib-treated patients with metastatic colorectal cancer (45). In another study, bortezomib targeted angiogenesis in multiple myeloma and inhibited VEGF expression (54). Although neither study elaborated on the corresponding mechanism, they independently corroborate our findings about the inhibitory effect of PI on expression of hypoxia-inducible genes.
Cell-dependent inhibition of VEGF expression may have potentially important implications for modulation of angiogenic activity by PI in vivo. It suggests that PI-mediated decrease of HIF activity may not necessarily manifest in decreased proangiogenic activity but that, instead, the final outcome with a particular cell type depends on regulation of other transcription factors. More-comprehensive studies will be required for establishing the role of PI in cancer angiogenesis.
The mapping of the inhibitory effect of PI on HIF-1 activity with deletion and replacement mutants identified the HIF-1α CAD as the primary target. Numerous reports have convincingly established that regulation of p300/CBP-HIF-1α CAD interaction modulates HIF-1 activity (36, 39, 56). p300 and CBP are paralogous, multidomain proteins that serve as transcriptional coactivators by simultaneously binding the transactivation domains of a vast array of transcription factors and other proteins that comprise the general transcriptional apparatus (60). A single p300/CBP domain is shared with multiple transcription factors, e.g., the CH1 domain binds to HIF-1, STAT-2, p53, p73, and Ets-1 (12), to name just a few. The obvious possibility that PI down-regulates HIF-1 activity through targeting its transcriptional coactivator, either by silencing the ability of p300/CBP to activate transcription or by disrupting interactions between the HIF-1α CAD and p300/CBP, is not supported by our data. PI considerably stimulated transcriptional activity of p300 itself and did not decrease HIF-1α CAD-p300 interaction in coimmunoprecipitation and GST pull-down assays. The stimulation of CBP activity by PI MG132, reported earlier (42), provides further weight to the conclusion that coactivator activity is not compromised in the presence of PI.
Given the opposite regulation of the HIF-1α CAD (inhibition) and STAT-2 (activation), PI differs from other HIF-1 inhibitors. CITED2 (CBP/p300 interacting transactivator with ED-rich tail 2, previously p35srj/Mrg1) binds p300 CH1 with high affinity and down-regulates both hypoxia- and STAT-2-activated transcription through competitive inhibition of p300 CH1-HIF-1α/STAT-2 interactions (6). Chetomin, a small-molecule inhibitor of HIF activity, also functions as a general disrupter of p300-CH1 interactions and significantly attenuated both HIF-1α CAD and STAT-2 (37). Apparently, PI inhibits HIF-1 activity distinctly from CITED2 and chetomin via a specific mechanism that does not target binding to the p300 CH1 domain in general.
Because of the large number of partners that interact with p300, the amount of available p300 could become limiting, e.g., p53 decreases HIF-1α-dependent transcription by sequestering p300 (7). Data from the mammalian two-hybrid assay and cotransfection of p300 also confirm that overexpression of p300 CH1 in the context of full-length p300, or in combination with a heterologous AD, can relieve the inhibitory effect of PI and rescue HIF-1 activity. The relieving effect of p300 overexpression could be explained in two ways. An excess of p300, strongly activated in the presence of PI, is forced onto the HIF-1α CAD, and this eventually overrides inhibition of HIF-1α CAD transcriptional activity. Alternatively, the excess of p300 could restore the HIF-1α CAD function by sequestering a PI-induced protein (putative repressor) away from the HIF-1α CAD.
There is evidence that inhibition of HIF-1α CAD activity can be executed either by preventing a posttranslational modification that is required for optimal function or by inducing a posttranslational modification with a negative effect. Phosphorylation of T796 (14) and hydroxylation of N803 (39) are examples of positively and negatively acting modifications, respectively. Phospho-T796 was required for optimal interaction with p300/CBP and transcriptional activation (14). As some PI attack sulfhydryl groups in addition to hydroxy groups (34), C800, as an integral part of the leucine-rich hydrophobic interface (15), could also play a role in inhibition of HIF-1α CAD. Individual replacements of phosphorylatable residues with phosphomimetic residues, N803 with nonhydroxylatable A, and C800 with V all failed to alleviate PI-imposed inhibition. Introduction of all mutations into one construct severely compromised HIF-1α CAD activity that was still inhibited by PI in a way similar to that for the wild type/individual mutants. Therefore, modulation of posttranslational modifications at the tested residues is not responsible for the inhibitory effect of PI on HIF-1α CAD.
As far as the inhibition of HIF-2α is concerned, neither MCF-7 nor Saos-2 cells expressed this isoform (data not shown). It appears that in non-renal carcinoma cells expression of hypoxia-inducible genes critically depends on HIF-1α but not HIF-2α (59). However, the high homology between the two CADs (14) suggests that HIF-2α could be affected in a way similar to the way HIF-1α is affected.
Two apparently unrelated but coregulated molecular switches control HIF-1α activity. The first is the regulation of HIF-1α stability by prolyl hydroxylases/VHL/UPP, and the second is the modulation of transcriptional activity of the HIF-1α CAD. The effect of PI on HIF-1α demonstrates that these two switches can be uncoupled. The accumulation of HIF-1α is the expected outcome of inhibition of the UPP (first switch on), but the loss of HIF-1α CAD activity (second switch off) in hypoxia is not. In this study, we have discounted a number of mechanisms that could be involved in PI-mediated repression of HIF-1 activity. We have shown that toxicity, nonspecific effect on transcription in general, polyubiquitylation, or disruption of interaction with p300 is not responsible for the loss of HIF-1α CAD activity. We have also provided evidence that the loss of HIF-1α CAD activity is not due to maintaining the negative regulation (hydroxylation of N803) or interfering with some positively acting modification(s) of the HIF-1α CAD. In our opinion, our data are compatible with an intriguing mechanism in which PI stabilizes a novel HIF-1α-specific repressor. This putative repressor may be present under normoxic conditions, assisting negative regulation of HIF-1 activity. Activation of HIF-1 via degradation of a putative repressor of HIF-1α would represent an alternative to the “activation by destruction” theory proposed by Lipford et al. (41).
Due to the involvement of the UPP in regulation of multiple cellular processes, inhibition of proteasome function elicits a pleiotropic response in the cellular context, and some aspects of this response are not fully understood. By using the hypoxia markers CAIX and VEGF as a paradigm, we show that PI abrogates hypoxia-induced expression of endogenous genes. Therefore, inhibition of HIF-1 function by PI is physiologically relevant and could be responsible for at least some of the antitumor effects of proteasomal inhibition. Further investigations of the corresponding mechanism will provide both a novel insight into regulation of HIF-1 activity and a possible specific therapeutic target. These studies are in progress.
Acknowledgments
This study was supported by grants from the California Cancer Research Program (00-00789V-20240) and The Avon Foundation.
We thank J. Pastorek and S. Pastoreková for the CAIX Ab. We also thank N. Sang for the Gal4-HIF-1α and pFR-Luc constructs and A. Giordano for the Gal4-p300 construct. P. Kaiser provided the ubiquitin Ab.
REFERENCES
- 1.Adams, J. 2003. The proteasome: structure, function, and role in the cell. Cancer Treat. Rev. 29(Suppl. 1):3-9. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. 2004. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5:417-421. [DOI] [PubMed] [Google Scholar]
- 3.Adams, J. 2004. The proteasome: a suitable antineoplastic target. Nat. Rev. Cancer 4:349-360. [DOI] [PubMed] [Google Scholar]
- 4.Arany, Z., L. E. Huang, R. Eckner, S. Bhattacharya, C. Jiang, M. A. Goldberg, H. F. Bunn, and D. M. Livingston. 1996. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. USA 93:12969-12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signaling induced by interferon-α. Nature 383:344-347. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, S., C. L. Michels, M. K. Leung, Z. P. Arany, A. L. Kung, and D. M. Livingston. 1999. Functional role of p35srj, a novel p300/CBP binding protein during transactivation by HIF-1. Genes Dev. 13:64-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blagosklonny, M. V., W. G. An, L. Y. Romanova, J. Trepel, T. Fojo, and L. Neckers. 1998. p53 inhibits hypoxia-inducible factor-stimulated transcription. J. Biol. Chem. 273:11995-11998. [DOI] [PubMed] [Google Scholar]
- 8.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 9.Damert, A., E. Ikeda, and W. Risau. 1997. Activator-protein-1 binding potentiates the hypoxia-inducible factor-1-mediated hypoxia-induced transcriptional activation of vascular-endothelial growth factor expression in C6 glioma cells. Biochem. J. 327:419-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dames, S. A., M. Martinez-Yamout, R. N. De Guzman, H. J. Dyson, and P. Wright. 2002. Structural basis for Hif-1 alpha/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. USA 99:5271-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epstein, A. C., J. M. Gleadle, L. A. McNeil, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 12.Freedman, S. J., Z. Y. Sun, F. Poy, A. L. Kung, D. M. Livingston, G. Wagner, and M. J. Eck. 2002. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc. Natl. Acad. Sci. USA 99:5367-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giaccia, A., B. G. Siim, and R. S. Johnson. 2003. HIF-1 as a target for drug development. Nat. Rev. Drug Discov. 2:803-811. [DOI] [PubMed] [Google Scholar]
- 14.Gradin, K., C. Takasaki, Y. Fujii-Kuriyama, and K. Sogawa. 2002. The transcriptional activation function of the HIF-like factor requires phosphorylation at a conserved threonine. J. Biol. Chem. 277:23508-23514. [DOI] [PubMed] [Google Scholar]
- 15.Gu, J., J. Milligan, and L. E. Huang. 2001. Molecular mechanism of hypoxia-inducible factor 1α-p300 interaction. J. Biol. Chem. 276:3550-3554. [DOI] [PubMed] [Google Scholar]
- 16.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-46. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 19.Hideshima, T., D. Chauhan, P. Richardson, C. Mitsiades, N. Mitsiades, T. Hayashi, N. Munschi, L. Dang, A. Castro, and V. Palombela. 2002. NF-κB as a therapeutic target in multiple myeloma. J. Biol. Chem. 277:16639-16647. [DOI] [PubMed] [Google Scholar]
- 20.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, L. E., and H. F. Bunn. 2003. Hypoxia-inducible factor and its biomedical relevance. J. Biol. Chem. 278:19575-19578. [DOI] [PubMed] [Google Scholar]
- 22.Ivan, M., and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11:27-34. [DOI] [PubMed] [Google Scholar]
- 23.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, S., S. Y. Liao, A. Ivanova, A. Danilkovitch-Miagkova, N. Tarasova, G. Weirich, M. J. Merril, M. A. Proescholdt, E. H. Oldfield, J. Lee, J. Zavada, A. Waheed, W. Sly, M. I. Lerman, and E. J. Stanbridge. 2001. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am. J. Pathol. 158:905-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 27.Kallio, P. J., W. J. Wilson, S. O'Brien, Y. Makino, and L. Poellinger. 1999. Regulation of the hypoxia-inducible transcription factor 1 alpha by the ubiquitin-proteasome pathway. J. Biol. Chem. 274:6519-6525. [DOI] [PubMed] [Google Scholar]
- 28.Kaluz, S., M. Kaluzová, R. Opavský, S. Pastoreková, A. Gibadulinová, F. Dequiedt, R. Kettmann, and J. Pastorek. 1999. Transcriptional regulation of the MN/CA 9 gene coding for the tumor-associated carbonic anhydrase IX. Identification and characterization of a proximal silencer element. J. Biol. Chem. 274:32588-32595. [DOI] [PubMed] [Google Scholar]
- 29.Kaluz, S., M. Kaluzová, A. Chrastina, P. L. Olive, S. Pastoreková, J. Pastorek, M. I. Lerman, and E. J. Stanbridge. 2002. Lowered oxygen tension induces expression of the hypoxia marker MN/carbonic anhydrase IX in the absence of hypoxia-inducible factor 1α stabilization: a role for phosphatidylinositol 3′-kinase. Cancer Res. 62:4469-4477. [PubMed] [Google Scholar]
- 30.Kaluz, S., M. Kaluzová, and E. J. Stanbridge. 2003. Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on SP1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res. 63:917-922. [PubMed] [Google Scholar]
- 31.Kaluzová, M., S. Pastoreková, E. Švastová, J. Pastorek, E. J. Stanbridge, and S. Kaluz. 2001. Characterization of the MN/CA 9 promoter proximal region—a role for SP and AP1 factors. Biochem. J. 359:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaluzová, M., S. Kaluz, M. I. Lerman, and E. J. Stanbridge. 2004. DNA damage is a prerequisite for p53-mediated proteasomal degradation of HIF-1α in hypoxic cells and downregulation of the hypoxia marker carbonic anhydrase IX. Mol. Cell. Biol. 24:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karin, M., Y. Cao, F. R. Greten, and Z. W. Li. 2002. NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2:301-310. [DOI] [PubMed] [Google Scholar]
- 34.Kisselev, A. F., and A. L. Goldberg. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739-758. [DOI] [PubMed] [Google Scholar]
- 35.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 36.Kung, A. L., S. Wang, J. M. Klco, W. G. Kaelin, and D. M. Livingston. 2000. Suppression of tumor growth through disruption of hypoxia-inducible signaling. Nat. Med. 6:1335-1340. [DOI] [PubMed] [Google Scholar]
- 37.Kung, A. L., S. D. Zabludoff, D. S. France, S. J. Freedman, E. A. Tanner, A. Vieira, S. Cornell-Kennon, J. Lee, B. Wang, J. Wang, K. Memmert, H. U. Naegeli, F. Petersen, M. J. Eck, K. W. Bair, A. W. Wood, and D. M. Livingston. 2004. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible pathway. Cancer Cell 6:33-43. [DOI] [PubMed] [Google Scholar]
- 38.Lando, D., D. J. Peet, J. J. Gorman, D. A. Whelan, M. L. Whitelaw, and R. K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 40.LeBlanc, R., L. P. Catley, T. Hideshima, S. Lentzsch, C. S. Mitsiades, N. Mitsiades, D. Neuburg, O. Goloubeva, C. S. Pien, J. Adams, D. Gupta, P. G. Richardson, N. C. Munshi, and K. C. Anderson. 2002. Proteasome inhibitor PS-341 inhibits human myeloma cell growth in vivo and prolongs survival in a murine model. Cancer Res. 62:4996-5000. [PubMed] [Google Scholar]
- 41.Lipford, J. R., G. T. Smith, Y. Chi, and R. J. Deshaies. 2005. A putative stimulatory role for activator turnover in gene expression. Science 438:113-116. [DOI] [PubMed] [Google Scholar]
- 42.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 43.Loncaster, J. A., A. L. Harris, S. E. Davidson, J. P. Logue, R. D. Hunter, C. C. Wykoff, J. Pastorek, P. J. Ratcliffe, I. J. Stratford, and C. M. L. West. 2001. Carbonic anhydrase (CAIX) expression, a potential new intrinsic marker of hypoxia: correlations with tumor oxygen measurements and prognosis in locally advanced carcinoma of the cervix. Cancer Res. 61:6394-6399. [PubMed] [Google Scholar]
- 44.Mabjeesh, N. J., D. E. Post, M. T. Willard, B. Kaur, E. G. Van Meir, J. W. Simmons, and H. Zhong. 2002. Geldanamycin induces degradation of hypoxia-inducible factor 1α protein via the proteasome pathway in prostate cancer cells. Cancer Res. 62:2478-2482. [PubMed] [Google Scholar]
- 45.Mackay, H., D. Hedley, P. Major, C. Townsley, M. Mackenzie, M. Vincent, P. Degendorfer, M. S. Tsao, T. Nicklee, D. Birle, J. Wright, L. Siu, M. Moore, and A. Oza. 2005. A phase II trial with pharmacodynamic endpoints of the proteasome inhibitor bortezomib in patients with metastatic colorectal cancer. Clin. Cancer Res. 11:5526-5533. [DOI] [PubMed] [Google Scholar]
- 46.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maxwell, P. H., M. Wiesener, G.-W. Chang, S. Clifford, E. Vaux, M. Cockman, C. C. Wykoff, C. Pugh, E. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature (London) 399:271-275. [DOI] [PubMed] [Google Scholar]
- 48.Mizukami, Y., J. Li, X. Zhang, M. A. Zimmer, O. Iliopoulos, and D. C. Chung. 2004. Hypoxia-inducible factor-1-independent regulation of vascular endothelial growth factor by hypoxia in colon cancer. Cancer Res. 64:1765-1772. [DOI] [PubMed] [Google Scholar]
- 49.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin, Jr. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 50.Olive, P. L., C. Aquino-Parsons, S. H. MacPhail, S.-Y. Liao, J. A. Raleigh, M. I. Lerman, and E. J. Stanbridge. 2001. Carbonic anhydrase 9 as an endogenous marker for hypoxic cells in cervical cancer. Cancer Res. 61:8924-8929. [PubMed] [Google Scholar]
- 51.Pal, S., K. P. Claffey, H. T. Cohen, and D. Mukhopadhyay. 1998. Activation of Sp-1 mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase C ξ. J. Biol. Chem. 273:26277-26280. [DOI] [PubMed] [Google Scholar]
- 52.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the alpha subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 53.Rafajová, M., M. Zat'ovičová, R. Kettman, J. Pastorek, and S. Pastoreková. 2004. Induction by hypoxia combined with low glucose or low bicarbonate and high posttranslational stability upon reoxygenation contribute to carbonic anhydrase IX expression in cancer cells. Int. J. Oncol. 24:995-1004. [PubMed] [Google Scholar]
- 54.Rocarro, A. M., T. Hideshima, N. Raje, S. Kumar, K. Ishitsuka, H. Yasui, N. Shiraishi, D. Ribatti, B. Nico, A. Vacca, F. Dammacco, P. G. Richardson, and K. C. Anderson. 2006. Bortezomib mediates antiangiogenesis in multiple myeloma via direct and indirect effects on endothelial cells. Cancer Res. 66:184-191. [DOI] [PubMed] [Google Scholar]
- 55.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 56.Sang, N., D. P. Stiehl, J. Bohensky, I. Leschinsky, V. Srinivas, and J. Caro. 2003. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J. Biol. Chem. 278:14013-14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 58.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 59.Sowter, H. M., R. R. Raval, J. Moore, P. J. Ratcliffe, and A. Harris. 2003. Predominant role of hypoxia-inducible transcription factor (HIF)-1α versus HIF-2α in regulation of the transcriptional response to hypoxia. Cancer Res. 63:6130-6134. [PubMed] [Google Scholar]
- 60.Vo, N., and R. H. Goodman. 2001. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276:13505-13508. [DOI] [PubMed] [Google Scholar]
- 61.Wang, D., R. Moriggl, D. Stravopodis, N. Carpino, J.-C. Marine, S. Teglund, J. Feng, and J. N. Ihle. 2000. A small amphipathic α-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 19:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wykoff, C. C., N. J. Beasley, P. H. Watson, K. J. Turner, J. Pastorek, A. Sibtain, G. D. Wilson, H. Turley, K. L. Talks, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe, and A. L. Harris. 2000. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 60:7075-7083. [PubMed] [Google Scholar]