Abstract
The monocyte chemoattractant protein 1 gene (MCP-1) is regulated by TNF through an NF-κB-dependent distal enhancer and an Sp1-dependent promoter-proximal regulatory region. In the silent state, only the distal regulatory region is accessible to transcription factors. Upon activation by tumor necrosis factor, NF-κB binds to the distal regulatory region and recruits CBP and p300. CBP and p300 recruitment led to specific histone modifications that ultimately enabled the binding of Sp1 to the proximal regulatory region. During this process, a direct interaction between the distal and proximal regulatory regions occurred. Sp1, NF-κB, CBP, and p300 were required for this interaction. CBP/p300-mediated histone modifications enhanced the binding of the coactivator CARM1 to the distal regulatory region. CARM1, which is necessary for MCP-1 expression, was not required for distal-proximal region interactions, suggesting that it plays a later downstream activation event. The results describe a model in which the separation of the distal enhancer from the promoter-proximal region allows for two independent chromatin states to exist, preventing inappropriate gene activation at the promoter while at the same time allowing rapid induction through the distal regulatory region.
By definition, enhancers function to regulate genes from a distance in a position- and orientation-independent manner (6, 7, 54). Yet despite this simple definition, enhancers are extremely complex and have the capacity to assemble a multitude of regulatory factors, coactivators, and chromatin remodeling proteins. How such factors function as a unit to ultimately recruit an RNA polymerase complex to the promoter of a gene so that it can initiate transcription from a distance has yet to be fully understood. Here, we use a model tumor necrosis factor (TNF)-inducible enhancer system to explore the mechanism of coactivator action when recruited to a distal enhancer.
TNF-α is a potent primary mediator of inflammatory responses and can induce apoptosis in a number of tumor cell types (9, 11, 13). TNF-mediated cell survival and gene induction responses occur through the activation and nuclear translocation of the transcription factor NF-κB (3, 5, 19, 27, 39, 41). NF-κB is composed of homo- or heterodimeric complexes of RelA (p65), c-Rel, RelB, p50, and p52. The most common form is the heterodimeric RelA/p50 complex. The RelA subunit contains the canonical Rel homology domain that characterizes the family of proteins as well as a transcriptional activation domain (TAD). The transcriptional activity of RelA has been reported to be controlled in part through posttranslational modifications of each of the above domains (reviewed in reference 29). Phosphorylation of S276 and S311 is associated with the ability of RelA to interact with the transcriptional coactivators CREB-binding protein (CBP) and p300 (65). Whereas both sites can be modified in response to TNF, mitogen- and stress-activated kinase 1 and protein kinase Cζ modify S276 and 311, respectively (29). The TAD modifications are believed to be associated with the recruitment of general transcription factors (43).
In several genes, such as IκBα (17) and human immunodeficiency virus long terminal repeat (45), the NF-κB regulatory regions are located close to the promoter and contain Sp1 sites. RelA and Sp1 have been shown to interact, and such interactions are important for gene activation. The monocyte chemoattractant protein 1 gene (MCP-1) is distinct from this arrangement. The MCP-1 gene encodes a CC chemokine (also known as CCL2) responsible for the recruitment of monocytes, T lymphocytes, natural killer cells, and basophils to areas of inflammation and infection (4, 52, 53). Regulation of MCP-1 by TNF involves two regulatory regions—distal and proximal—separated by 2.2 kb of DNA (50). The two functional κB sites located on the distal regulatory region and GC box on the proximal regulatory region are critical in the regulation of MCP-1 induction by TNF (12, 47, 49, 50). The two MCP-1 κB sites are unoccupied in the absence of TNF stimulation. Moreover, in the uninduced state, the proximal region GC box, which binds Sp1, and two additional sites are unoccupied, despite the fact that Sp1 and the other factors are in the nucleus and able to bind DNA (47, 49, 50). These data suggested that the chromatin configuration of the proximal region of the MCP-1 gene is inaccessible to these factors until NF-κB is recruited to the distal regulatory region.
TNF-mediated induction of MCP-1 is accompanied by increases in histone acetylation at both the distal and proximal regions as well as within the intervening sequences separating the two regulatory regions (12). Histone acetylation was dependent on RelA but not NF-κB p50 (12). Infection of cells with adenovirus expressing wild-type E1a or transient expression of E1a resulted in inhibition of TNF-induced expression of MCP-1 (12), suggesting a role for CBP/p300, as E1a is known to squelch the activity of CBP/p300 (2, 23, 26, 46, 61). As cited above, phosphorylated NF-κB can recruit the histone acetyltransferases (HAT) CBP and p300. While the HAT activity of these factors could easily alter the local chromatin configuration of the distal regulatory region where NF-κB binds, it is not clear how such factors might alter the chromatin configuration within the proximal regulatory region located 2.2 kb away.
The coactivator-associated arginine methyltransferase 1, CARM1, was found to regulate several TNF-induced genes, including MCP-1 (18). CARM1 was initially found to modify histones and enhance gene expression (15). More recently, CARM1 has been shown to methylate CBP and p300 in multiple positions and modulate their activity (16, 35, 60). Intriguingly, the histone modifications catalyzed by CBP/p300 (histone H3 K14 and K18) create a substrate target for the binding of CARM1 and subsequent arginine methylation of histone H3 at R17 (1, 8, 20). The latter modification has been linked to gene activation and transcription. Thus, while it is easy to envision how the multiple coactivators may modulate nucleosome architecture at a promoter site, the question of how a distal regulatory region controls a proximal promoter at a distance of 2.2 kb is considerably more complex and becomes critical to understanding how NF-κB and distant enhancers function.
To investigate the mechanism by which the NF-κB distal MCP-1 regulatory enhancer region functions, we have taken advantage of DNA and protein-protein cross-linking technologies that permit the analysis of the local chromatin architecture (chromatin conformation capture [3C] assay) (22, 24, 34, 37, 55, 57, 58) and factor assembly and posttranslational modifications of nucleosomes associated with a regulatory region (chromatin immunoprecipitation [ChIP]) (33). We found that, in response to TNF, a specific interaction forms between the proximal and distal regulatory regions that is dependent on RelA and Sp1. Complementation of p65-deficient cells with a RelA S276A mutation did not restore the interaction, suggesting a specific role for CBP and p300 in the ability of the two regulatory regions to interact. Indeed, the loss of either CBP or p300 by small interfering RNA (siRNA) knockdown resulted in the loss of both the interaction and MCP-1 expression, suggesting that both of these factors are required for the formation of the interaction and that the interaction is critical to gene expression. Importantly, CBP/p300 knockdown results in the loss of specific modifications at the distal and proximal region and in the loss of Sp1 binding to the proximal region, suggesting that CBP/p300 recruitment controls the accessibility of the proximal regulatory region. CARM1 was found by ChIP to be associated with the distal regulatory region in response to TNF, as were modifications associated with its activity. In the absence of CARM1, distal and proximal region interactions occur but MCP-1 induction did not. This study therefore provides evidence for an ordered series of events that suggest that the MCP-1 distal NF-κB enhancer functions by recruiting HATs that open the local chromatin structure until it can form a stable interaction with proximal promoter element factors and then recruit the general transcription machinery.
MATERIALS AND METHODS
Cells and cell culture.
NIH 3T3 fibroblasts cells were purchased from the American Type Culture Collection (Manassas, VA). NFκB p65−/− (RelA) cells were kindly provided by D. Baltimore (California Institute of Technology, Pasadena, CA). Cells were grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% bovine calf serum (HyClone, Inc., Logan, UT), penicillin (50 U/ml), streptomycin (50 μg/ml), and l-glutamine (1 mM) (Life Technologies, Grand Island, NY). CARM1 knockout (3−/−) cells and wild-type (13+/+) murine embryonic fibroblasts (MEF) were a generous gift from M. T. Bedford (University of Texas, Smithville, TX) and grown as described elsewhere (18). Human recombinant TNF was purchased from Genzyme (Cambridge, MA) and used at 500 U/ml for 2 h unless other times are specified in the experiment details (12).
Reporter assay.
Plasmids expressing different mutants of p65 were initially obtained from Sankar Ghosh (Yale University, New Haven, CT) and recloned into the pcDNA3.1 expression vector. Wild-type p65 and mutant constructs were transiently cotransfected with an MCP-1 promoter-dependent chloramphenicol acetyltransferase (CAT) reporter construct (pJECAT) in p65−/− cells by electroporation methods described previously (48). One microgram of an alkaline phosphatase expression vector (pSV2AlkPhos) was cotransfected as a transfection efficiency control (51, 63). Cells were harvested 72 h posttransfection, and CAT activity was determined as previously described (28). Alkaline phosphatase activity was assayed using a kit from Bio-Rad (Richmond, CA). At least three transfections and CAT assays were performed for each experiment shown. The data were averaged, normalized to the alkaline phosphatase activity, and plotted with standard errors of the means.
Chromosome conformation capture assay.
A modified version of the 3C assay, adapted for mammalian cells (55, 57), was used. Cells (4 × 106) were resuspended in 50 ml DMEM supplemented with 10% bovine calf serum. The samples were cross-linked at room temperature for 10 min in 2% formaldehyde. The reaction was quenched by the addition of 0.125 M glycine. Cells were lysed in cold lysis buffer (0.34 M sucrose, 10 mM Tris, 10 mM NaCl, 1% NP-40) containing protease inhibitors, and the nuclei were collected. The nuclei were washed once with 1× restriction buffer (buffer 4; New England Biolabs, Beverly, MA), resuspended in restriction buffer containing 0.3% sodium dodecyl sulfate (SDS), and incubated for 1 h at 37°C while being shaken. To sequester the SDS, 2% Triton X-100 was added and the nuclei were incubated for 1 h at 37°C. The cross-linked DNA was digested overnight with 500 to 800 units NcoI restriction enzyme (New England Biolabs, Inc.). The restriction enzyme was inactivated by the addition of SDS to 1.6% and incubation at 65°C for 20 min. The reaction mixture was diluted 20-fold with T4 DNA ligase buffer (New England Biolabs, Inc.) and 1% Triton X-100 and incubated for 1 h at 37°C. The DNA was ligated using 1,000 units T4 DNA ligase (New England Biolabs, Inc.) for 4 h at 16°C followed by 30 min at room temperature. Proteinase K (10 μg/ml; GIBCO, Maryland) was added to the ligation mixtures and incubated overnight at 65°C to reverse the cross-links. The DNA was extracted with phenol-chloroform and precipitated with ethanol. The purified DNA (100 ng for untransfected cells and 300 ng for transfected cells) was used as a PCR template with the following primer pair sets: P1, 5′-ATGGCATCTCTCAACTTGTTCA, and P2, 5′-CCTGGCTATCATCACATTACCTTC-3′; P3, GGAGGAGGACAGTGACTAGC, and P4, GGTGCTTCCAGAATGCTTCC. Additional control primers used included P5, CTCCACGACTCTCTATGAAAGACAAATC, and P6, GGTATCTTGATTTCCTCCTTCC. Twenty-five to 30 PCR cycles were performed, with each cycle including 30 s at 95°C, 30 s at 60°C, and 45 s at 68°C. The PCR products were analyzed on 1.5% agarose gels. Each 3C assay was performed at least three times with independent cross-linked samples. Similar results were obtained for each experiment. The 3C assay product was sequenced and found to represent the predicted junction. Some 3C products were quantitated by scanning gels and using the “Quantity One” software package for imaging and analyzing one-dimensional electrophoresis gels (Bio-Rad, Inc., version 4.2). The results are expressed as the averages from three experiments with standard deviations.
ChIP assays.
ChIP assays were performed as previously described (10). The anti-RelA/p65 (C-20, sc-372), anti-CARM1, anti-Sp1, anti-dimethyl-H3-Arg17, and anti-H3-lysine-14 antibodies were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). The anti-H3-lysine-K18 was either from Upstate Biotechnology, Inc., or Abcam, Inc. (Cambridge, United Kingdom). The following primer sets were used to amplify the distal regulatory region and proximal regulatory region of the MCP-1 gene, respectively: distal, FWDChip2, 5′-TTTCCACGCTCTTATCCTACTCTGC, and RVChip2, 5′-TTGTCTGTTTCCCTCTCACTTCAC; proximal ProxFW2, 5′-TACCAAATTCCAACCCACAGTTTC, and ProxRv2, 5′-GAGAGCTGGCTTCAGTGAGAG.
RNA extraction and RT-PCR.
Total RNA was isolated from cells using the RNAeasy kit (QIAGEN, Valencia, CA). cDNA synthesis by SuperScript II reverse transcriptase (Invitrogen, Inc., Carlsbad, CA) was performed using the GeneAmp RNA PCR kit (PerkinElmer, Boston, MA) according to the manufacturer's instructions. Two micrograms of total RNA was used per sample, and each reaction mixture contained a parallel control with no reverse transcriptase added. One-twentieth of the reverse transcriptase reactions were analyzed by real-time PCR as described previously (42). The reactions were subjected to 40 cycles of PCR amplification in a Bio-Rad, Inc., iCycler and detection system. All results were normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. The sequence of primers used in reverse transcription (RT)-PCR or real-time PCR of the MCP-1 gene were published previously (36). At least three independent experiments were performed, and results were averaged.
siRNA knockdown experiments.
SMART pool siRNAs for mouse CBP and p300 were purchased (Dharmacon RNA Technologies) to knock down the expression of CBP and p300 genes, respectively. NIH 3T3 cells were transfected using TransIT-TKO transfection reagent following the manufacturer's instructions (Mirus, Inc., Madison, WI). TNF was added at 500 U/ml for 2 h after 48 h of incubation of 50 nmol of the respective siRNAs. The samples were prepared in parallel either for western, RT-PCR, ChIP, or 3C assay as indicated.
Western blot analysis.
Whole-cell lysates were prepared by lysing cells with 2× SDS-polyacrylamide gel electrophoresis loading buffer (Bio-Rad, Inc.). Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The membrane was blocked with 10% nonfat dry milk and probed with either anti-CARM1 (Upstate Biotechnology, Inc.), anti-RelA/p65 (C-20, sc-372), anti-p300 (sc-585), anti-CBP (sc-369), anti-Sp1 (Sc-39), or anti-GAPDH antibodies (Santa Cruz Biotechnology, California) for 1 h. Anit-RelA/p65 antibodies from Abcam, Inc. (12145-30), were also used. The membranes were rinsed twice with phosphate-buffered saline containing 0.1% Tween 20 and incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibody. Membranes were extensively washed with phosphate-buffered saline containing 0.1% Tween 20 and incubated with enhanced chemiluminescence substrate (Amersham Life Sciences, Arlington Heights, IL) for 1 min and placed on film.
RESULTS
TNF induces an interaction linking a distal NF-κB enhancer to a proximal Sp1-regulated element.
MCP-1 is regulated by two distinct modules (Fig. 1A). The distal regulatory region (DRR) containing two NF-κB sites is required for TNF responsiveness (25, 50). The proximal regulatory region (PRR) encodes an essential GC box that binds Sp1 (49, 50) and is also required for TNF-mediated induction. Importantly, Sp1 binding does not occur until the gene is activated by TNF, suggesting that the distal and proximal regions interact in some manner (12, 50). To explore this possibility, the 3C assay (22, 34, 57, 58) was developed for the MCP-1 gene (Fig. 1A) using NIH 3T3 cells, which are highly inducible for MCP-1 expression (25, 48-50). In this assay, nuclei from formaldehyde-treated cells were subjected to NcoI digestion. Following enzyme inactivation and intramolecular DNA ligation, the cross-links are reversed and the presence of the unique ligation junction, which is indicative of an interaction between the distal and proximal regions, was assessed by PCR and termed a 3C product (21, 34, 57-59).
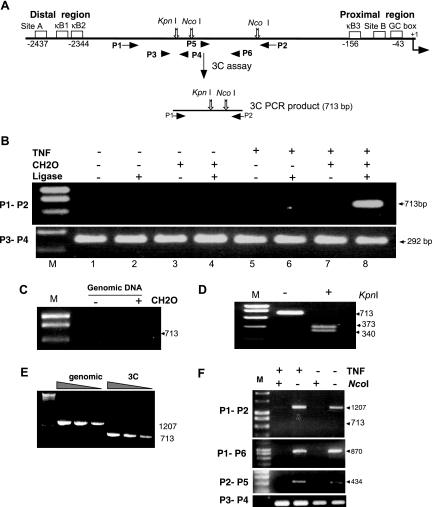
FIG. 1.
An interaction between the distal and proximal regulatory regions of the MCP-1 gene forms following TNF stimulation. (A) A schematic representation of the 3C assay and the architecture of the MCP-1 promoter with cis regulatory sites with selected restriction sites and primer positions (P1 through P4) is shown. In the 3C assay, cells were fixed with formaldehyde and chromatin was isolated and digested with NcoI. Following inactivation of the restriction enzyme, the sample was diluted and subjected to DNA ligation such that only intramolecular ligations would be preferred. The cross-links were then reversed, and the DNA was purified. PCR primers P1 and P2 were used to detect the formation of the novel ligated 3C product. Primers P3 and P4 were used to verify that similar levels of MCP-1 DNA were present in the assay and that the DNA was amplifiable. Products from the 3C assay were examined on agarose gels stained with ethidium bromide. (B) NIH 3T3 cells treated in the absence (−) (lanes 1 to 4) or presence (+) (lanes 5 to 8) of TNF for 2 h were processed in the above 3C assay with and without formaldehyde (CH2O) or DNA ligase, as indicated, to control for specificity of the protocol. M, DNA marker. (C) Purified mouse genomic DNA was processed in the 3C assay and demonstrates that the unique PCR product cannot form with naked DNA templates. (D) KpnI cleaves the PCR product generated by the 3C assay, producing the anticipated DNA fragments as shown on the agarose gel. (E) PCR amplification of the 3C product and the non-3C product display similar relative efficiencies. PCR products generated from undigested genomic DNA (1,207 bp) and DNA from a 3C assay (713 bp) were quantitated by fluorometry and used as templates in PCRs to determine if there were major differences in their PCR efficiency. DNA templates (40 pg, 8 pg, and 1.6 pg) were amplified by primers P1 and P2 under identical PCR conditions. (F) Chromatin isolated from TNF-treated or control cells fixed with formaldehyde was subjected to NcoI digestion as indicated. Following deproteination, the DNA was purified and analyzed by PCR using the indicated primers. Because the ligation step of the 3C assay was not performed, P1/P2, P1/P6, and P2/P5 PCR products can only be observed when NcoI was omitted from the reaction, indicating that the chromatin is accessible to restriction digestion even in the absence of TNF.
To test the efficacy of this system, NIH 3T3 cells were left untreated or treated with TNF for 2 h and the 3C assay was conducted (Fig. 1B) in the presence and absence of formaldehyde and/or DNA ligase. A 3C product was observed only following TNF induction, and detection of this product required the addition of formaldehyde and DNA ligase. Because a 3C product was detected only during TNF induction and not in uninduced cells, it was likely that random ligation events were not occurring. However, a number of critical controls were carried out to verify that the system was in fact working properly. First, a series of PCRs demonstrated that each of the treated samples contained similar levels of amplifiable MCP-1 DNA (Fig. 1A and B, primers P3 and P4). Second, the interaction did not occur when the 3C assay was carried out with purified genomic DNA (Fig. 1C). Third, the PCR product generated can be cleaved with KpnI, providing the exact-sized fragments produced from the predicted ligation reaction (Fig. 1D). Sequencing of the 3C product confirmed the exact sequence predicted by the ligation of the adjoining NcoI fragments.
One additional possibility was that the absence of a 3C product might be due to the inability of the restriction enzyme, NcoI, to digest the chromatin templates of the TNF control samples. To test if the region was being digested during the 3C assay, chromatin from formaldehyde-cross-linked cells was isolated and restriction digested with NcoI or left undigested. Following deproteination, the samples were PCR amplified with primers P1/P2 as in the 3C assay. Primers P1 and P2 can amplify naked genomic DNA with or without the intervening NcoI fragment with similar relative efficiencies (Fig. 1E). Irrespective of TNF treatment, chromatin samples digested with NcoI failed to produce the full-length 1,207-bp product (Fig. 1F), demonstrating that the chromatin is accessible to NcoI digestion and that digestion is relatively efficient. Additionally, PCR amplification across each NcoI site only occurred in the absence of NcoI, indicating that both sites were accessible in the absence of TNF (Fig. 1F). Thus, the 3C assay is functional in this system and can provide information addressing potential interactions between the proximal and distal regulatory regions of the MCP-1 gene.
NF-κB and Sp1 are required for distal/proximal region interactions.
Previously, Sp1 and the RelA, interacting at the PRR and DRR, respectively, were found to be essential for TNF-mediated activation of MCP-1 (12, 48, 49). In genes where NF-κB and Sp1 are both present at a proximal regulatory region, they have been found to cooperatively bind DNA (14, 45). Additionally RelA and Sp1 have been found to interact in the absence of DNA (45). Such results suggest that the interactions shown by the 3C assay may be dependent on these proteins. To determine if this is indeed the case, the 3C assay was conducted on MEF lines deficient for RelA or Sp1 and compared to a wild-type MEF line. The results demonstrated that 3C product formation required both of these factors (Fig. 2A), as the 3C product was detected in wild-type MEFs but not in either RelA- or Sp1-deficient cells. Western blot analysis shows that the RelA- and Sp1-deficient cells each express wild-type levels of Sp1 and RelA, respectively (Fig. 2B). Coupled with the previous observations that the sequences between the PRR and DRR are dispensable for MCP-1 regulation (48), the present data suggest that the interactions observed in the 3C assay are mediated by the factors bound at the DRR and PRR and possibly by RelA and Sp1.
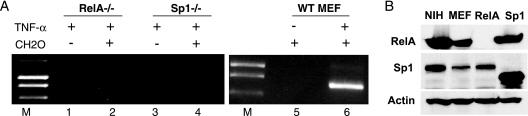
FIG. 2.
RelA and Sp1 are required for generation of the 3C product. (A) Murine cell lines deficient for RelA or Sp1 were compared to a wild-type MEF line for their ability to form the MCP-1 3C product. Cells were treated with TNF as indicated (+, present; −, absent), and formaldehyde was excluded in some experiments to control for nonspecific events. Only the wild-type (WT MEF) cells were able to generate a 3C assay product, suggesting that the 3C product is mediated by the factors bound to the DRR and PRR and that interactions between these regions occur in response to TNF. (B) Western blots for the presence of RelA, Sp1, or actin were conducted on the indicated cell lines. The lower migrating Sp1 band in the Sp1−/− cells represents the N-terminal portion of Sp1 that is expressed in the knockout line. As described by Marin et al., this mutant is inactive in DNA binding and transactivation (38).
3C product formation is coincident with MCP-1 expression.
If DRR and PRR interactions play a significant role in the regulation of MCP-1 transcription, such interactions and gene expression should occur with similar kinetics. Cells were stimulated with TNF over a 2-h time course (Fig. 3). MCP-1 mRNA was detected as quickly as 15 min after TNF exposure and was expressed throughout the time course (Fig. 3A). Analysis of similarly treated cells revealed that the 3C product could be detected as early as 15 min following TNF treatment (Fig. 3B and C). The intensity of the 3C signal increased to a maximum at 60 min and displayed nearly similar kinetics as the accumulation of MCP-1 RNA steady-state levels, demonstrating that 3C product and distal/proximal region interactions coincided with expression, suggesting that such interactions may be a prerequisite for expression.
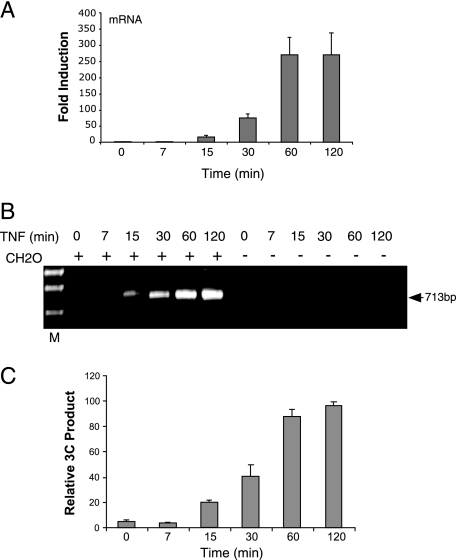
FIG. 3.
MCP-1 mRNA accumulation and distal/proximal region interactions are coincident events. (A) MCP-1 mRNA is detectable in the cytoplasm after 15 min of TNF stimulation and increases over the first hour of the time course. Real-time RT-PCR was carried out on RNA isolated from NIH 3T3 cells treated with 500 units of TNF for the indicated time. The results from three independent assays were averaged following normalization to the levels of GAPDH mRNA detected by real-time RT-PCR. (B) The 3C product was not detected until 15 min of TNF stimulation. NIH 3T3 cells treated with TNF for the indicated time were assayed using the 3C assay described above. The presence (+) of the formaldehyde (CH2O) was required for generation of the 3C product. (C) The 3C product increased in intensity over the first hour of the time course and was present at 2 h. Images from stained agarose gels, representing data from three independent sample sets, were quantitated and averaged as described in Materials and Methods. The mean data are plotted with their standard deviations.
The Rel homology domain of RelA is required for MCP-1 expression and distal/proximal region interactions.
To determine the domains of RelA that may mediate the observed interactions between the DRR and PRR, a series of deletion constructions targeting critical regions of RelA were generated and transfected into RelA-deficient MEFs along with or without an MCP-1 promoter reporter construction (Fig. 4A). The endogenous locus-derived MCP-1 mRNA (Fig. 4B) or the activity of a cotransfected reporter gene (Fig. 4C) was assayed. While important for maximal expression of the MCP-1 reporter construct, deletions encompassing the bulk of the C-terminal domain of RelA, including the transcriptional activation domain, were not essential for activity in this system (Fig. 4B and C). The C-terminal half of RelA (amino acids 310 to 550) was inactive. The endogenous locus was slightly more sensitive to the absence of the C-terminal region of RelA but did display >30% of wild-type RelA activity in its absence (Fig. 4B, RelA1-309). The Rel homology domain was essential and is the region known to interact with Sp1.
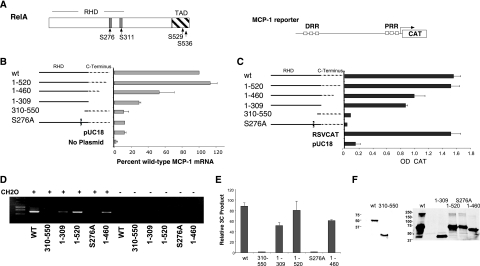
FIG. 4.
The Rel homology domain and serine 276 of RelA are required for expression and distal/proximal region interactions. A series of RelA mutants (A) were transfected into RelA-deficient cells and tested for their ability to drive expression of the endogenous gene (B) or an MCP-1 promoter CAT reporter gene (C) and for their ability to form the 3C product on the endogenous MCP-1 locus (D). TNF was not added to these assays, as overexpression of RelA is sufficient to bypass sequestration in the cytoplasm (48). All RelA expression constructions that contained a wild-type (WT) Rel homology domain were able to drive MCP-1 expression and generate a 3C product. (A) A schematic of the MCP-1 reporter gene construct (pJECAT2.6) and of the RelA p65 domain structure is shown. RHD, rel homology domain; TAD, transcriptional activation domain. Phosphorylatable serine residues shown to be important for RelA activity are indicated. (B) Real-time RT-PCR of MCP-1 mRNA levels from RelA-deficient cells transiently transfected as above with the indicated plasmids were normalized to the amount of GAPDH mRNA and plotted as the percentage of the wild-type RelA transfectant. The averages of results from three independent transfections are shown. (C) RelA-deficient cells transfected with the indicated RelA expression vector and the MCP-1 reporter. CAT values, derived from an enzyme-linked immunosorbent assay for CAT protein, normalized to the expression of an alkaline phosphatase expression vector were averaged from three experiments. (D) The 3C assay was carried out on the indicated plasmids transfected into the RelA-deficient cells as above. (E) Images from stained agarose gels representing three independent assays of the experiment shown in panel D were quantitated and plotted as described in Materials and Methods. (F) Western blots performed on cells transfected with the RelA mutant series show similar levels of expression between wild-type and mutant proteins. Different antibodies were used due to the loss of epitopes in some of the mutants (left blot, sc-372 [Santa Cruz, Inc.]; right blot, 12145-30 [Abcam, Inc.]).
To test whether mutations in RelA that affect the ability to recruit CBP or p300 would alter expression of MCP-1 in response to TNF, the single amino acid substitution mutant S276A was assayed as above. S276 phosphorylation in response to TNF is associated with CBP binding and RelA function (29, 64, 65). In both the reporter and endogenous MCP-1 mRNA assays, the S276A mutation showed only background levels of induction of MCP-1 (Fig. 4B and C).
The above mutants were also tested for their ability to mediate interactions between the distal and proximal regions (Fig. 4D and E). All constructions that were able to induce MCP-1 gene expression were able to generate a 3C assay product. Importantly, no 3C product was observed in cells complemented with the S276A mutation, suggesting that interaction with CBP and/or p300 is likely to be important for MCP-1 expression and the distal/proximal region interactions. This conclusion is supported by the fact that the adenovirus E1a protein can interfere with MCP-1 expression (12). E1a can sequester CBP/p300 from acting on non-E1a-regulated genes (26, 44). Moreover, because RelA mutants that lacked the TAD were modestly functional, the results suggest that interactions with CBP/p300 are sufficient to provide a transactivation signal, albeit not a complete signal. Western blots of the transfected mutant series showed that each of the RelA mutants expressed similar levels of protein (Fig. 4F).
CBP and p300 are critical to interactions between the proximal and distal regulatory regions.
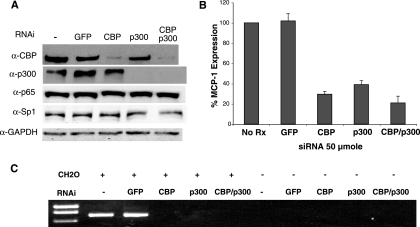
To demonstrate a direct role for CBP and p300 in the regulation of MCP-1 expression, SMART pool siRNAs (Dharmacon, Inc.) to these transcripts were used to knock down the expression of these factors in NIH 3T3 cells. Following transfection of siRNAs, Western blot assays were conducted to examine the level of CBP and p300 protein (Fig. 5A). The results showed that protein levels for each factor were reduced by greater than 90% by their respective siRNA pools but not by an siRNA to the green fluorescence protein (GFP). Importantly, knockdown of CBP or p300 did not affect each other, nor did any of the siRNAs tested affect the steady-state protein levels of RelA, Sp1, or GAPDH, suggesting that the siRNAs were specific to their targets. In a parallel experiment, siRNA-transfected cells were treated with TNF and examined for their ability to stimulate MCP-1 mRNA expression. The results showed that MCP-1 mRNA was reduced to 32 and 39% of wild-type levels when CBP and p300 siRNAs were used, respectively (Fig. 5B). Thus, each of these factors is required for expression. When siRNA pools to both CBP and p300 were used, the ability of TNF to induce MCP-1 mRNA was reduced to 21% of the control (Fig. 5B). This value was significantly lower than the single siRNAs (P < 0.041), indicating an additive effect.
FIG. 5.
CBP and p300 are required for MCP-1 expression and distal/proximal region interaction. siRNAs to CBP, p300, or both were transfected into NIH 3T3 cells. Cells were incubated for 48 h and then treated with TNF for 2 h. (A) Western blot analysis of the transfected cells using antibodies to CBP (α-CBP), p300 (α-p300), RelA (α-p65), Sp1 (α-Sp1), and GAPDH (α-GAPDH), demonstrating specificity and efficacy of the siRNA. (B) In similar siRNA transfections, MCP-1 expression was measured by real-time RT-PCR. (C) The 3C assay was performed on the transfected cells treated with TNF as above. All siRNA transfections and assays were performed at least three times with identical results. +, present; −, absent.
Also tested in parallel with the above siRNA transfections was the ability of distal and proximal regulatory regions to interact in cells with reduced levels of CBP and/or p300. Indeed, knockdown of CBP, p300, or both results in the inability of the cells to form the regulatory region interaction (Fig. 5C). Control siRNA to GFP- or mock-treated cells were able to form the 3C product under the conditions tested. These results place CBP and p300 in the pathway for MCP-1 gene activation. Because RelA binding is necessary for CBP and p300 recruitment to NF-κB dependent genes, it is likely that CBP and p300 recruitment follow the binding of NF-κB to the distal regulatory region.
CARM1 is necessary for expression but not distal/proximal region interactions.
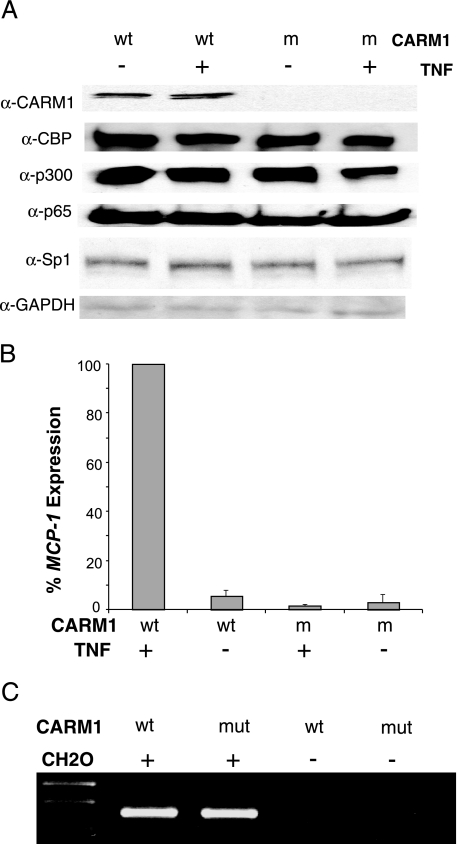
The arginine methyltransferase CARM1 is known to interact with and modify CBP and p300 (1, 18, 60) and was recently found to regulate TNF induction of MCP-1 (18). To examine the role of CARM1 in this system, CARM1-deficient MEFs were compared to wild-type MEFs. To determine if CARM1 cells have a deficiency in the factors necessary for activation in this system, immunoblots for CBP, p300, Sp1, and RelA were performed. The results showed that CARM1-deficient cells lacked CARM1 but had wild-type (or near wild-type) levels of CBP, p300, Sp1, and RelA (Fig. 6A). As anticipated, CARM1-deficient cells were not inducible for MCP-1 expression by TNF (Fig. 6B). Intriguingly, when tested, a 3C product was detected from CARM1−/− cells in response to TNF. Thus, CARM1 is required for gene activation but not distal/proximal region interactions. These data therefore suggest that recruitment of CARM1 follows the recruitment of CBP/p300 and is required for a downstream activation event.
FIG. 6.
CARM1 is required after the distal and proximal regions interact. CARM1-deficient MEFs were used to assay its role in MCP-1 activation. (A) Immunoblot analysis confirms that CARM1 protein is absent in the mutant cells and that CBP, p300, RelA (p65), Sp1, and GAPDH protein levels are unaffected. α, anti. (B) CARM1-deficient cells fail to activate MCP-1 in response to TNF. Real-time RT-PCR was conducted on RNA samples that were isolated from wild-type (wt) and CARM1-deficient (m) MEFs following no TNF stimulation (−) or 2 h of TNF stimulation (+). Real-time RT-PCR values were normalized to the amount of GAPDH mRNA in the samples and plotted with respect to the wild-type sample induced with TNF. The average of three independent experiments is shown. (C) An interaction forms between the proximal and distal regulatory regions regardless of whether CARM1 is present. The 3C assay as described above was performed on wild-type and CARM1-deficient (mut) MEFs. All samples in this assay were derived from cells treated with TNF. All experiments were performed three times with identical results.
CBP/p300-mediated histone modifications are enhanced at the proximal and distal regulatory regions, but CARM1 modifications are only present at the distal region.
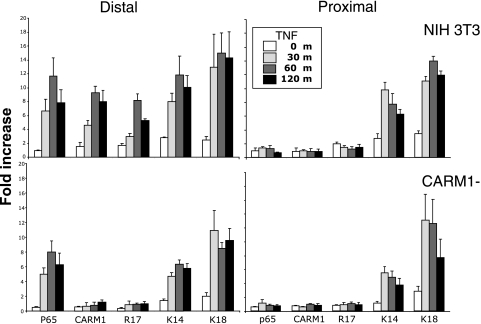
Previous analysis of the MCP-1 upstream sequences showed that, following TNF treatment, an increase in histone H3 acetylation occurred at the distal and proximal regulatory regions (12). The HAT activites of CBP and p300 are known to acetylate histone H3 at lysine 14 and 18 (1, 20). To determine if known CBP and p300 modifications occurred in the MCP-1 regulatory regions, ChIP assays using antibodies specific to these modifications were conducted following a 2-h time course of TNF treatment (Fig. 7). In addition, the binding of RelA to each region was assayed. As expected, in a TNF-dependent manner, RelA associated with the distal but not proximal regulatory region. ChIP for histone H3 K14 and K18 found TNF-dependent increases (maximum of 10- to 16-fold) at both regulatory regions. The binding of RelA and the increase in histone H3 K14 and K18 acetylation also occurred in CARM1-deficient cells, suggesting that the CARM1 deficiency does not affect the H3 K14 and K18 HAT activities associated with this system (Fig. 7).
FIG. 7.
CARM1 binds to and modifies the distal regulatory region in a time course that follows CBP/p300 histone modifications. ChIP assays coupled with real-time PCR for the distal and proximal regulatory regions were conducted with antibodies to p65, CARM1, and to the indicated histone modifications following TNF treatment of NIH 3T3 and CARM1-deficient cells for the indicated time. The data from three separate chromatin preparations and assays were averaged and plotted as relative increases over an irrelevant antibody control.
Because CBP and p300 appear to be functioning in the CARM1−/− background, this may suggest that the role of CARM1 is at the proximal regulatory region. To determine if this was the case, ChIP for CARM1 and its associated modification as determined by reactivity to the histone H3 R17 methylation-specific antibody was conducted. CARM1 was found associated with the distal but not the proximal regulatory region (Fig. 7). The binding of CARM1 reached full occupancy after 1 h. Moreover, the activity of CARM1 was associated with the distal regulatory region. This activity is strongly linked to CARM1, as it was absent in CARM1−/− cells (Fig. 7) (M. Bedford, M. D. Anderson Cancer Center, personal communication). The fact that modifications induced by CBP and p300 (H3 K14 and K18), which increase the affinity of CARM1 for histone H3, reach their maximum at 30 min suggests that CARM1 is recruited after these modifications are in place.
CBP/p300 are required for Sp1 access to the PRR.
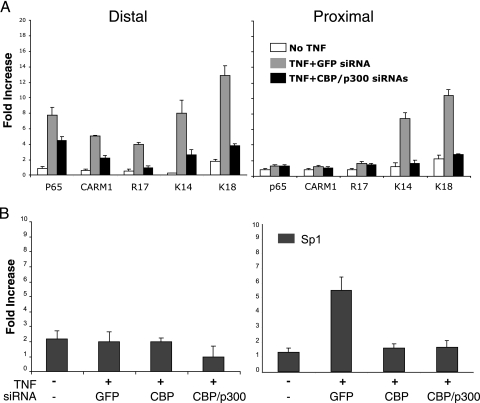
The above results suggest that CBP/p300 recruitment was required for the histone modifications observed at both the DRR and PRR. To determine the role of CBP and p300 on the above histone modifications in vivo, ChIP assays on NIH 3T3 cells transiently transfected with siRNAs to GFP (control) or CBP and p300 were conducted. In comparison to non-TNF-treated cells, TNF-treated GFP siRNA-transfected cells produced similar results (Fig. 7 and 8A). Importantly, siRNA-transfected, TNF-treated cells displayed robust reductions of histone H3 K14 and K18 acetylation at both the distal and proximal regulatory regions, suggesting that these coactivators were indeed responsible for these modifications in an in vivo system (Fig. 8A). The binding of CARM1 was also reduced, as was the CARM1-specific modification. RelA binding was slightly reduced as well, suggesting that CBP/p300 also plays a role in stabilizing the binding of RelA to the DRR.
FIG. 8.
CBP/p300 control distal and proximal regulatory region acetylation, CARM1 binding, and access to the proximal regulatory region. (A) siRNA to CBP/p300 in NIH 3T3 cells but not GFP reduce histone H3 K14 and K18 acetylation and CARM1 binding and its associated activity as indicated by H3 R17 methylation. (B) siRNAs to CBP or CBP/p300 prevent the binding of Sp1 to the proximal regulatory region. ChIP assays using the indicated antibodies were conducted on NIH 3T3 cells treated with the siRNAs as described in the legend to Fig. 5. The data from three separate chromatin preparations and assays were averaged and plotted as relative increases over an irrelevant antibody control.
The data thus far suggest a model where CBP/p300 recruited to the DRR controls access to the PRR and Sp1 binding. One prediction of this model is that Sp1 binding to the PRR should only occur when CBP/p300 are present. To test this, the above siRNA experiment (Fig. 8B) was repeated and ChIP was performed for Sp1 binding to the DRR and PRR. As expected, Sp1 did not bind the DRR. However, at the PRR, Sp1 binding was only detected in the TNF-treated, GFP control, siRNA-transfected cells and no Sp1 was detected at the PRR in either CBP or CBP/p300 knockdown cells. These results demonstrated that CBP/p300 activity is critical for Sp1 binding activity to the PRR and suggest that one role for CBP/p300 in this system is to control the accessibility of the PPR.
DISCUSSION
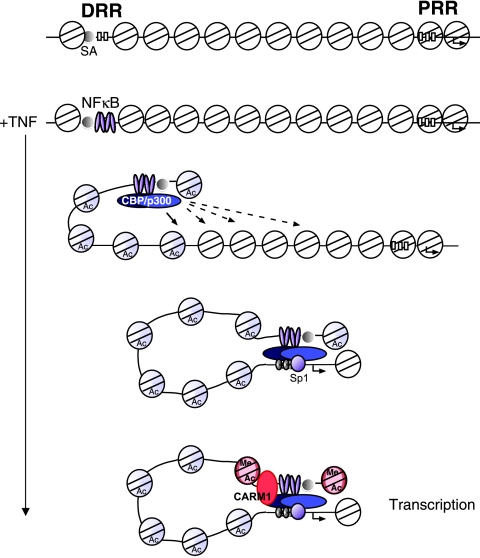
Here, we demonstrate an ordered sequence of transcription regulatory events that modulate the expression of the MCP-1 gene by TNF (Fig. 9). Induction of MCP-1 begins with the translocation of NF-κB into the nucleus and to the MCP-1 DRR. Our model predicts that the DRR is accessible to NF-κB binding due to the constitutive factor occupancy of site A located upstream of the two κB binding sites (31, 50). Once bound, NF-κB recruits CBP and p300, which acetylates the distal region histones. Previously, we showed that the sequences between the DRR and PRR (termed middle) as well as the PRR become acetylated in a RelA-dependent manner (12). Acetylation of the proximal region increases the accessibility of the PRR for binding by Sp1. This is likely then followed by the formation of a stable interaction between the two regulatory regions. CARM1 is recruited after CBP and p300, most likely due to the modifications of the distal regulatory region histones, as suggested from previous reports (1, 20). CARM1-dependent methylation of the distal regulatory region then occurs. Each of these steps is required for MCP-1 transcription. The formation of the interactions between the PRR and DRR likely allows the additional coactivator functions of CBP and p300 to be used in activating transcription.
FIG. 9.
Model of MCP-1 induction by TNF. A schematic is shown of the uninduced state of the MCP-1 gene followed by the predicted events that occur after TNF induction. Blue shaded nucleosomes indicate those that have been modified by CBP/p300. We suggest that CBP/p300 nucleosome modifications occur until an it reaches the PRR. Once the PRR nucleosomes have been modified, the region is accessible to Sp1, allowing a direct interaction with the DRR. CARM1 is then recruited and provides a signal to activate transcription of the gene. Red shaded nucleosomes indicate those that have been modified by CARM1 and CBP/p300. DRR, distal regulatory region; PRR, proximal regulatory region; SA, site A—the constitutively bound region.
The current results provide a mechanistic role for how an inducible enhancer may function. The recruitment of the activation factor, RelA, to a region where it can bind initiates a process by which HATs are recruited. The HATs then serve to open up the local chromatin structure until a promoter-proximal, cis-acting element is accessible for the binding of a constitutively expressed transcription initiation factor such as Sp1. Once this factor binds, a stable interaction between the enhancer and the promoter region is formed. As mentioned above, our previous data showing that the “middle” region was also acetylated in a RelA-dependent manner during activation by TNF provides support for this model. In this model, interactions between an enhancer and promoter-proximal element would maintain an open/accessible chromatin structure at the promoter for the recruitment of the general transcription machinery.
The current model provides a mechanism that ensures that a gene that is off remains in that state until the correct activation signal is received. The off state is maintained by an inaccessible chromatin structure at the promoter region. Separation of the enhancer and proximal regulatory region allows the gene to maintain two distinct chromatin structures: an accessible structure that can bind the activating transcription factor and initiate the series of events required for gene expression and a closed structure to prevent the random binding of RNA polymerase or a factor like Sp1 that can recruit RNA polymerase. Moreover, by using HATs to control the accessibility, the system can be rapidly reversed as well. It is intriguing that the phosphorylation state of RelA can control whether it associates with HATs or histone deacetylases (64), enabling the possibility that NF-κB can also shut down this system by the recruitment of a histone deacetylase.
One interesting aspect of the data was the fact that both CBP and p300 were both required for the regulatory region interactions and maximal expression. These factors are often associated with interchangeable roles as coactivators, yet here they are both required (30). The data suggest two possible explanations. The first is that each of these factors is performing a unique and essential function to MCP-1 regulation. The fact that knockout animals deficient for each of these factors display slightly different phenotypes suggests that each has its own function (30, 32, 62). The double knockdown experiment showed consistently lower levels of expression, which is in agreement with this conclusion. This was also true for the ChIP assay (data not shown). Alternatively, as suggested by the similarities of these factors and their roles (30, 32, 62), the levels of CBP and p300 are limiting, and removing one may be sufficient to reduce the amount of available CBP/p300 coactivator in the system such that distal/proximal region interactions or the detection thereof is not efficient and therefore not observed. Irrespective of which is the case, the role of CBP/p300 in this system is more than the scaffold/adapter described for enhanceosome complexes, such as the beta interferon gene (40), with the major difference being that CBP/p300 are responsible for chromatin modifications at both the distal and proximal elements. Like a traditional enhanceosome, it is possible that CBP/p300 also directly mediates the interactions that are observed in the 3C assay; however, it is also possible that Sp1 and RelA directly mediate these interactions.
It was also interesting that knockdown of CBP/p300, while ablating the ability to detect the 3C product, did not ablate the ability to detect some MCP-1 mRNA by real-time PCR. This could be due to a number of reasons. The first is the relative sensitivities of the assays. RT-PCR is one of the most sensitive molecular assays available, whereas the 3C assay relies on a number of inefficient molecular reactions. Thus, the remaining CBP/p300 after knockdown may have been sufficient to allow expression of MCP-1 but not enough to allow detection of the 3C product. A second possibility is that the observed mRNA levels represent those that occur in the presence of TNF and NF-κB but in the absence of CBP/p300, suggesting that the role of CBP/p300 is also to participate in transactivation of the system. Alternatively, this level of expression could be the result of the recruitment of additional coactivators by NF-κB to the system that are not involved in the interactions between the distal and proximal regions.
The binding and role of CARM1 in this system appear to be complex. Because CARM1 was required for expression but not for distal/proximal region interactions, we initially predicted that CARM1 would associate with the proximal regulatory region. The opposite result occurred. CARM1 was found at the distal regulatory region, and its associated modifications were detected only in the distal regulatory region, despite the fact that both the proximal and distal regions displayed histone modifications that were reported to have increased affinity for CARM1 binding. This begs the question of what CARM1 is doing in this system. As an arginine methyltransferase, CARM1 is likely acting to methylate nucleosomes or other proteins that are recruited. CARM1 has been found to methylate CBP and p300 at several sites, altering its ability to associate with other factors or coactivators such as CREB or GRIP1 (16, 35, 60). While the above modifications function negatively with respect to the factors with which they interact, it is possible that other CARM1 modifications of CBP/p300 lead to activation roles. CARM1 could be methylating the other DNA-binding proteins in this 3C complex, including RelA or Sp1. The data suggest that CARM1 mediates the observed histone H3 R17 modifications, which are associated with increased transcriptional activation (1, 8, 18, 56). Yet, the conundrum with this activation-associated modification as well as others of the “histone code” is how does it lead to activation? Perhaps this modification serves to recruit components of the general transcription machinery. By placing such modifications on the distal regulatory region, this may ensure that the distal region controls RNA polymerase recruitment, leaving the role of the proximal regulatory region to stabilize the interaction and target the polymerase to the initiation site of transcription.
Acknowledgments
We are grateful for the comments of D. Reines, J. Lucchesi, S. Warren, P. Wade, X. Chen, A. Long, P. Majumder, C. Moran, and M. Stallcup on the contents on the manuscript.
This work was supported by a Public Health Service grant (CA96810) from the National Institutes of Health.
REFERENCES
- 1.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 2.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 3.Baeuerle, P. A., and D. Baltimore. 1996. NF-kappa B: ten years after. Cell 87:13-20. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini, M., and P. Loetscher. 2000. Chemokines in inflammation and immunity. Immunol. Today 21:418-420. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin, A. S. 1996. The NF-kB and IkB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-681. [DOI] [PubMed] [Google Scholar]
- 6.Banerji, J., L. Olson, and W. Schaffner. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33:729-740. [DOI] [PubMed] [Google Scholar]
- 7.Banerji, J., S. Rusconi, and W. Schaffner. 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299-308. [DOI] [PubMed] [Google Scholar]
- 8.Bauer, U. M., S. Daujat, S. J. Nielsen, K. Nightingale, and T. Kouzarides. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-kB in preventing TNF-α-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 10.Beresford, G. W., and J. M. Boss. 2001. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2:652-657. [DOI] [PubMed] [Google Scholar]
- 11.Beutler, B., and A. Cerami. 1988. Tumor necrosis, cachexia, shock, and inflammation: a common mediator. Annu. Rev. Biochem. 57:505-518. [DOI] [PubMed] [Google Scholar]
- 12.Boekhoudt, G. H., Z. Guo, G. W. Beresford, and J. M. Boss. 2003. Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J. Immunol. 170:4139-4147. [DOI] [PubMed] [Google Scholar]
- 13.Carswell, E. A., L. J. Old, R. L. Kassel, S. Green, N. Fiore, and B. Williamson. 1975. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA 72:3666-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman, N. R., and N. D. Perkins. 2000. Inhibition of the RelA(p65) NF-kappaB subunit by Egr-1. J. Biol. Chem. 275:4719-4725. [DOI] [PubMed] [Google Scholar]
- 15.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 16.Chevillard-Briet, M., D. Trouche, and L. Vandel. 2002. Control of CBP co-activating activity by arginine methylation. EMBO J. 21:5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiao, P. J., S. Miyamoto, and I. M. Verma. 1994. Autoregulation of I kappa B alpha activity. Proc. Natl. Acad. Sci. USA 91:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covic, M., P. O. Hassa, S. Saccani, C. Buerki, N. I. Meier, C. Lombardi, R. Imhof, M. T. Bedford, G. Natoli, and M. O. Hottiger. 2005. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 24:85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Acquisto, F., M. J. May, and S. Ghosh. 2002. Inhibition of nuclear factor kappa B (NF-B): an emerging theme in anti-inflammatory therapies. Mol. Interv. 2:22-35. [DOI] [PubMed] [Google Scholar]
- 20.Daujat, S., U. M. Bauer, V. Shah, B. Turner, S. Berger, and T. Kouzarides. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12:2090-2097. [DOI] [PubMed] [Google Scholar]
- 21.Dekker, J. 2006. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3:17-21. [DOI] [PubMed] [Google Scholar]
- 22.Dekker, J., K. Rippe, M. Dekker, and N. Kleckner. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 23.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 24.Eivazova, E. R., and T. M. Aune. 2004. Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proc. Natl. Acad. Sci. USA 101:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freter, R. R., J. A. Alberta, G. Y. Hwang, A. L. Wrentmore, and C. D. Stiles. 1996. Platelet-derived growth factor induction of the intermediate-early gene MCP-1 is mediated by NF-kB and a 90-kDa phosphorylation coactivator. J. Biol. Chem. 271:17417-17424. [DOI] [PubMed] [Google Scholar]
- 26.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 28.Guo, Z., G. H. Boekhoudt, and J. M. Boss. 2003. Role of the intronic enhancer in tumor necrosis factor mediated induction of manganous superoxide dismutase. J. Biol. Chem. 278:23570-23578. [DOI] [PubMed] [Google Scholar]
- 29.Hayden, M. S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195-2224. [DOI] [PubMed] [Google Scholar]
- 30.Kalkhoven, E. 2004. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 68:1145-1155. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S. N., and J. M. Boss. 2000. Site A of the MCP-1 distal regulatory region functions as a transcriptional modulator through the transcription factor NF1. Mol. Immunol. 37:623-632. [DOI] [PubMed] [Google Scholar]
- 32.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 34.Lee, G. R., C. G. Spilianakis, and R. A. Flavell. 2005. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 6:42-48. [DOI] [PubMed] [Google Scholar]
- 35.Lee, Y. H., S. A. Coonrod, W. L. Kraus, M. A. Jelinek, and M. R. Stallcup. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. USA 102:3611-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung, T. H., A. Hoffmann, and D. Baltimore. 2004. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell 118:453-464. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Z., and W. T. Garrard. 2005. Long-range interactions between three transcriptional enhancers, active Vκ gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 25:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marin, M., A. Karis, P. Visser, F. Grosveld, and S. Philipsen. 1997. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell 89:619-628. [DOI] [PubMed] [Google Scholar]
- 39.May, M. J., and S. Ghosh. 1998. Signal transduction through NF-kappa B. Immunol. Today 19:80-88. [DOI] [PubMed] [Google Scholar]
- 40.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 41.Nadjar, A., V. Tridon, M. J. May, S. Ghosh, R. Dantzer, T. Amedee, and P. Parnet. 2005. NFkappaB activates in vivo the synthesis of inducible Cox-2 in the brain. J. Cereb. Blood Flow Metab. 25:1047-1059. [DOI] [PubMed] [Google Scholar]
- 42.Nagarajan, U. M., A. Bushey, and J. M. Boss. 2002. Modulation of gene expression by the MHC class II transactivator. J. Immunol. 169:5078-5088. [DOI] [PubMed] [Google Scholar]
- 43.Paal, K., P. A. Baeuerle, and M. L. Schmitz. 1997. Basal transcription factors TBP and TFIIB and the viral coactivator E1A 13S bind with distinct affinities and kinetics to the transactivation domain of NF-kappaB p65. Nucleic Acids Res. 25:1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker, S. F., L. K. Felzien, N. D. Perkins, M. J. Imperiale, and G. J. Nabel. 1997. Distinct domains of adenovirus E1A interact with specific cellular factors to differentially modulate human immunodeficiency virus transcription. J. Virol. 71:2004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perkins, N. D., A. B. Agranoff, E. Pascal, and G. J. Nabel. 1994. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol. Cell. Biol. 14:6570-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 47.Ping, D., G. Boekhoudt, and J. M. Boss. 1999. trans-Retinoic acid blocks platelet-derived growth factor-BB-induced expression of the murine monocyte chemoattractant-1 gene by blocking the assembly of a promoter proximal Sp1 binding site. J. Biol. Chem. 274:31909-31916. [DOI] [PubMed] [Google Scholar]
- 48.Ping, D., G. H. Boekhoudt, E. M. Rogers, and J. M. Boss. 1999. Nuclear factor kappa B p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J. Immunol. 162:727-734. [PubMed] [Google Scholar]
- 49.Ping, D., G. H. Boekhoudt, F. Zhang, A. Morris, S. Philipsen, S. T. Warren, and J. M. Boss. 2000. Sp1 binding is critical for promoter assembly and activation of the MCP-1 gene by tumor necrosis factor. J. Biol. Chem. 275:1708-1714. [DOI] [PubMed] [Google Scholar]
- 50.Ping, D., P. L. Jones, and J. M. Boss. 1996. TNF regulates the in vivo occupancy of both distal and proximal regulatory regions of the MCP-1/JE gene. Immunity 4:455-469. [DOI] [PubMed] [Google Scholar]
- 51.Riley, J. L., and J. M. Boss. 1993. Class II MHC transcriptional mutants are defective in higher order complex formation. J. Immunol. 151:6942-6953. [PubMed] [Google Scholar]
- 52.Rollins, B. J. 1997. Chemokines. Blood 90:909-928. [PubMed] [Google Scholar]
- 53.Rollins, B. J. 1991. JE/MCP-1: an early-response gene encodes a monocyte-specific cytokine. Cancer Cell 3:517-524. [PubMed] [Google Scholar]
- 54.Sassone-Corsi, P., R. Hen, E. Borrelli, T. Leff, and P. Chambon. 1983. Far upstream sequences are required for efficient transcription from the adenovirus-2 E1A transcription unit. Nucleic Acids Res. 11:8735-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spilianakis, C. G., and R. A. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 56.Stallcup, M. R., J. H. Kim, C. Teyssier, Y. H. Lee, H. Ma, and D. Chen. 2003. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J. Steroid Biochem. Mol. Biol. 85:139-145. [DOI] [PubMed] [Google Scholar]
- 57.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 58.Vakoc, C. R., D. L. Letting, N. Gheldof, T. Sawado, M. A. Bender, M. Groudine, M. J. Weiss, J. Dekker, and G. A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17:453-462. [DOI] [PubMed] [Google Scholar]
- 59.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19:631-642. [DOI] [PubMed] [Google Scholar]
- 60.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 61.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 62.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 63.Yoon, K., M. A. Thiede, and G. A. Rodan. 1988. Alkaline phophatase as a reporter enzyme. Gene 66:11-17. [DOI] [PubMed] [Google Scholar]
- 64.Zhong, H., M. J. May, E. Jimi, and S. Ghosh. 2002. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell 9:625-636. [DOI] [PubMed] [Google Scholar]
- 65.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]