Abstract
The interaction of Prep1 and Pbx homeodomain transcription factors regulates their activity, nuclear localization, and likely, function in development. To understand the in vivo role of Prep1, we have analyzed an embryonic lethal hypomorphic mutant mouse (Prep1i/i). Prep1i/i embryos die at embryonic day 17.5 (E17.5) to birth with an overall organ hypoplasia, severe anemia, impaired angiogenesis, and eye anomalies, particularly in the lens and retina. The anemia correlates with delayed differentiation of erythroid progenitors and may be, at least in part, responsible for intrauterine death. At E14.5, Prep1 is present in fetal liver (FL) cMyb-positive cells, whose deficiency causes a marked hematopoietic phenotype. Prep1 is also localized to FL endothelial progenitors, consistent with the observed angiogenic phenotype. Likewise, at the same gestational day, Prep1 is present in the eye cells that bear Pax6, implicated in eye development. The levels of cMyb and Pax6 in FL and in the retina, respectively, are significantly decreased in Prep1i/i embryos, consistent with the hematopoietic and eye phenotypes. Concomitantly, Prep1 deficiency results in the overall decrease of protein levels of its related family member Meis1 and its partners Pbx1 and Pbx2. As both Prep1 and Meis interact with Pbx, the overall Prep1/Meis-Pbx DNA-binding activity is strongly reduced in whole Prep1i/i embryos and their organs. Our data indicate that Prep1 is an essential gene that acts upstream of and within a Pbx-Meis network that regulates multiple aspects of embryonic development.
Biochemical studies have shown that Prep1 (also known as pKnox1), a member of the TALE class of homeodomain proteins (8, 27), is an important regulator of Pbx activity (4-6, 13, 16, 34). Pbx genes, in turn, play a central role in development and organogenesis. The Pbx family comprises four genes in mammals which are differentially expressed during embryonic development and in the adult (18, 38). Pbx1-deficient mice exhibit an embryonic lethal phenotype, characterized by homeotic transformation of elements of the second branchial arch and by defective organogenesis affecting the spleen, pancreas, kidney, and organs of the caudal pharyngeal pouches (26, 33, 44, 46). Pbx1-deficient embryos are also affected in their definitive hematopoiesis (14) and are unable to induce splenic cell fate specification during early embryogenesis (7). On the other hand, the lack of Pbx3 causes lethality at birth (P0) by respiratory failure (41), while Pbx2-deficient mice show no evident phenotype, most likely due to compensation by other family members (47).
Pbx activity is regulated by the TALE proteins Prep1, Prep2, Meis1, Meis2, and Meis3 (4, 6, 8, 10, 19, 21, 27, 49), which form transcriptionally active complexes with Pbx, important during embryonic development (4, 10, 18, 27, 28, 43, 53, 54). DNA-bound Meis/Prep-Pbx complexes, in turn, bind to and modify the activity of other proteins, like anterior clustered or nonclustered Hox proteins, such as Hoxb1, Hox11, Pdx1, and other transcription factors like MyoD (2, 7, 17, 25, 42 50). Hence, they control expression of numerous genes, including Hoxb2, Hoxb1, Hoxa3, Hox11, and glucagon (7, 17, 22, 25, 34, 42, 43), that are required for development and organogenesis. In zebra fish embryos, Meis1 overexpression increases the stability of Pbx (53, 54). Likewise, in mammalian cells, overexpression of Prep1 increases the stability of Pbx1 and Pbx2 by preventing their proteasomal degradation (31). On the other hand, down-regulation of prep1.1 in zebra fish causes an overall reduction of all Pbx proteins (13).
Meis1-deficient mice exhibit an embryonic lethal phenotype (embryonic day 13.5 [E13.5] to 14.5) with major defects in hematopoiesis, angiogenesis, and eye formation (1, 23), while Meis2 (Mrg2) appears to be involved in controlling chick limb outgrowth (9, 36). In Xenopus laevis, Meis1b regulates hindbrain gene expression (32), while ectopic expression of Meis caudalizes neural cell fates (43). Importantly, the expression levels of other TALE proteins have not been assessed in Meis1-deficient models.
Mammals have two Prep genes, Prep1 and Prep2 (4, 19, 21), while zebra fish have three, prep1.1, prep1.2, and prep2 (13). Down-regulation with morpholino antisense oligonucleotides of the prep1.1 gene in zebra fish causes an embryonic lethal phenotype with extensive brain apoptosis, loss of hindbrain rhombomeric segmentation, lack of cartilage differentiation of neural crest cells, pericardial edema, and lack of fins (13). In mice, a null Prep1 mutation results in early lethality (E7.5) (L.C. Fernandez, N. Jenkins, N. Copeland, and F. Blasi, unpublished data), precluding a study of the Prep1 role(s) in later developmental processes.
An insertion of a retroviral vector in the first intron of the Prep1 gene (Prep1i/i) results in a hypomorphic mutation that exhibits variable penetrance and expressivity. Most Prep1i/i embryos die between E17.5 and P0 (see below), although about 1/4 of these escape embryonic lethality. The mice escaping embryonic lethality show T-cell development anomalies (40). In this paper, we show that the Prep1i/i embryonic phenotype recapitulates, at least in part, the Meis1 and Pbx1 phenotypes. Indeed, erythropoiesis and angiogenesis are impaired, with liver hypoplasia, decreased hematocrit, anemia, and delayed erythroid differentiation together with a decrease in capillary formation. Moreover, much like in Meis1 mutants, Prep1i/i embryos also display major eye anomalies. Finally, Prep1i/i embryos exhibit decreased levels of Pbx1, Pbx2, and Meis1 proteins as well as decreased expression of cMyb and Pax6, consistent with the hematopoietic and eye phenotype, respectively. Our data highlight a novel hierarchy wherein Prep1 acts upstream in the network regulating hematopoiesis (and specifically erythropoiesis), angiogenesis, and eye development by controlling the levels of Pbx and Meis proteins.
MATERIALS AND METHODS
Antibodies.
Anti-Pbx1b and -Pbx2 antibodies were kindly donated by Michael Cleary, anti-Meis1 was donated by Miguel Torres, and an anti-Meis1 antiserum was donated by A. M. Buchberg. Pan-Pbx antibodies recognizing all of the Pbx splice variants were a kind gift of H. Poepperl. The anti-Prep1 polyclonal antibodies were previously described (5). An anti-Prep1 monoclonal antibody (Upstate Biotechnology, Upstate House, Dundee, United Kingdom) was also used in some experiments.
Prep1 targeting.
Prep1 targeted mice were generated by gene trapping by Lexikon Genetics, Inc. (The Woodlands, Texas) using a clone isolated from a library of embryonic stem cells (129/SvEvBrd strain) randomly targeted with a retroviral vector (VICTR45) (55). Prep1+/i mice were obtained in the C57BL/6-SV129 strain. Heterozygous mice were backcrossed with wild-type (wt) C57BL/6 for up to 9 generations. All animal handling conformed to regulations of the Ethics Committee on Animal Use of H. S. Raffaele (IACUC permission number 207).
Genotyping of Prep1i/i mice.
Southern Blot analysis of EcoRI-digested total DNA from tail biopsy specimens or yolk sacs employed a 132-bp double-stranded Prep1 cDNA probe prepared from full-length Prep-1 cDNA with the forward primer 5′-ATGATGGCGACACAGACGCTAAGTATA-3′ and reverse primer 5′-GGGGTCTGAGACTCGATGGGAGGAGGACTC-3′.
The PCR genotyping strategy employed oligonucleotides Prep-R1 and LTR2 (sequences provided below) that amplify a 230-bp fragment in the disrupted allele, while the Prep-F1-Prep-R1 couple amplifies a 300-bp fragment of the wild-type allele. Sequences of oligonucleotides are as follows: Prep-F1, 5′-CCAAGGGCAGTAAGAGAAGCTCTGGAG-3′; Prep-R1 5′-GGAGTGCCAACCATGTTAAGAAGAAGTCCC-3′; LTR2, 5′-CAAAATGGCGTTACTTAAGCTAGCTTGCC-3′.
Nuclear extract preparation and immunoblotting analysis.
For Electrophoretic Mobility Shift assays (EMSA) and immunoblotting assays, nuclear (or total) extracts were prepared from dissected mouse embryos and organs at the indicated stages as described previously (3).
EMSA.
EMSA were performed with the O-1, Sp1, b2-PH, and b2-PM-PH oligonucleotides, as described previously (3, 4, 17). The oligonucleotide sequences are as follows: O-1, 5′-CACCTGAGAGTGACAGAAGGAGGCAGGGAG-3′; b2-PH, 5′-GGGGCTAAGATTGATCGCCTC-3′; b2-PM-PH, 5′-GGAGCTGTCAGGGGGCTAAGATTGATCGCCTCA-3′; Sp1, 5′-GATCGATCGGGGCGGGGCGATC-3′.
mRNA extraction and QT-PCR from mouse embryos.
For quantitative PCR (QT-PCR), total RNA was extracted from single E10.5 embryos with the TRIZOL reagent (Life Technologies) and the guanidine isothiocyanate method (11). The Taqman gene expression assay (Applied Biosystems, Foster City, CA) was used with predesigned, gene-specific Taqman probe and primer sets and the ABI-Prism 7900HT sequence detection system (Applied Biosystems). The data are standardized to the level of 18S rRNA.
Hematocrit determination.
For hematocrit determination, peripheral blood obtained by cardiac puncture from E16.5 embryos was centrifuged in 32 by 0.8 mm Na-heparinized capillary tubes (Hirschmann Laborgerate, Ebenstadt, Germany).
Flow cytometry.
Ter119 and CD71 antibodies (Pharmingen, San Diego, CA) were used to analyze erythroid subpopulations in fetal liver (FL) (57).
BFU-E assays.
Colony assays were carried out by incubating 50,000 Prep1+/+, Prep1+/i, or Prep1i/i FL single-cell suspensions in 1 ml of methylcellulose enriched with erythropoietin (Methocult GF M-3434; Stem Cell Technologies, Vancouver, Canada) in triplicate. The growth of erythroid colonies was quantitated after 10 days (35).
Cultures of allantois.
Allantoids from E7.5 to 7.75 embryos were cultured for 18 to 20 h, fixed in 4% paraformaldehyde for 30 min, washed, and permeabilized for 30 min as described elsewhere (45). The cultures were stained with a rat anti-mouse PECAM/CD31 (clone Mec 13.3; Pharmingen, San Diego, Calif.) monoclonal antibody, revealed with Cy3 donkey anti-rat secondary antibodies (1:300), and washed, and images were acquired by confocal fluorescence microscopy. The vessel density was blindly measured as the percentage of CD31-stained pixels in identical areas of the central portion of each allantois using Pixel-Counter software (courtesy of M. Mazzieri).
Immunohistochemistry.
For immunohistochemistry, E10.5 or E14.5 embryos were fixed, dehydrated, and embedded in paraffin. Deparaffinized sections (7 μm) were incubated with antibody overnight at 4°C, then incubated with biotinylated secondary anti-mouse, -rat, or -rabbit immunoglobulin G (Vectastain ABC kit, Vector Laboratories, Inc.), and detected with the DAB substrate kit for peroxidase or the Vector red alkaline phosphatase substrate kit I (Vector Laboratories, Inc.).
RESULTS
Prep1i/i mice exhibit an embryonic lethal phenotype with variable penetrance.
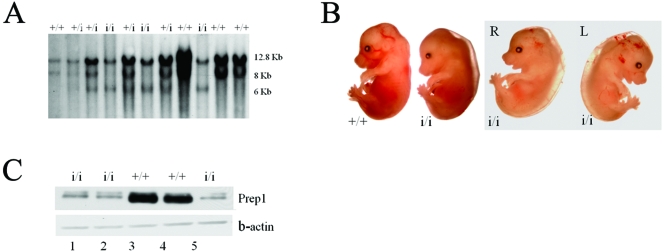
Prep1i/i mice were generated at Lexicon Genetics with the VICTR45 enhancer trap strategy (55, 56). The insertion occurred at nucleotide 849 of intron 1 located within the 5′ untranslated region (2.8 kb from the ATG) (not shown). We call this mutation Prep1i (for insertion). The insertion of VICTR45 provides, in addition to the two wt 12.8- and 8.0-kb EcoRI bands, a third 6.0-kb band in Southern blot analysis (Fig. 1A).
FIG. 1.
Prep1i/i phenotype. (A) Southern blotting analysis of EcoRI-digested DNAs from the progeny obtained by crossing F1 Prep1i/+ × Prep1i/+. (B) Gross morphology of Prep1i/i embryos. The two rightmost panels show the same embryo viewed from both sides (R and L), exhibiting edema, pallor, smaller size, small liver spot and hemorrhaging. (C) Nuclear extracts prepared from E14.5 embryonic brains of 5 littermate embryos (2 wt and 3 Prep1i/i, as indicated) were immunoblotted with monoclonal anti-Prep1 and anti-beta-actin antibodies. Lane 1, 2, and 5 are extracts from Prep1i/i embryos; lanes 3 and 4 are extracts from wt embryos.
The phenotype of the first heterozygous cross generated Prep1i/i pups with a frequency slightly lower (20%) than expected (not shown). The phenotype of the offspring of Prep1+/i heterozygous crosses stabilized at the fourth backcross with wt C57BL/6. Here we report data obtained with mice backcrossed 7 to 9 times. The percentage of Prep1i/i mice born from heterozygous crosses was 6.3% (Table 1). These surviving Prep1i/i embryos lived for at least 15 months (not shown). Table 1 also shows that most Prep1i/i embryos died between E17.5 and P0, since homozygous Prep1i/i embryos were about 22% at E14.5 to E17.5, but only 6% were born (P0). The most frequently visible phenotypes (Fig. 1B) in E15.5 embryos were a general organ hypoplasia, pallor (75%), edema (67%), smaller body size (75%), and smaller liver spot (71%). Eye abnormalities (61%, see below) and hemorrhages (13%) were also seen. We conclude that the Prep1i/i mutation causes a pleiotropic embryonic lethal phenotype with variable penetrance.
TABLE 1.
Timed pregnancy analysis of Prep1i/i embryosa
| Embryonic stage | nb | No. of crosses | No. (%) of embryos or pups with genotype
|
||
|---|---|---|---|---|---|
| +/+ | +/i | i/i | |||
| E14.5 | 200 | 15 | 52 (26) | 104 (52) | 44 (22) |
| E15.5-17.5 | 194 | 26 | 51 (25.4) | 100 (50.5) | 43 (21.4) |
| P0 | 221 | 34 | 73 (33.0) | 132 (59.7) | 14 (6.3), 2 (0.9)c |
The mice employed in these crosses had been backcrossed from 7 to 9 generations with wt C57BL/6.
Numbers of embryos or newborn pups analyzed.
Two mice were born alive but died within the first day.
RNA and protein analyses show that Prep1i/i is a hypomorphic mutation.
Comparison by quantitative PCR of RNA from E10.5 wt and Prep1i/i embryos showed that 1.5% of Prep1 mRNA was still present in 6 different Prep1i/i embryos analyzed (an arbitrary 100% value was given to wt littermates) (not shown). The adopted gene trap strategy is reported to be leaky in about 4% of the mouse lines generated (56). The presence of Prep1 mRNA suggests that the Prep1i/i embryos may produce low levels of protein. In fact, E14.5 embryonic brain nuclear extracts also showed low levels of residual Prep1 protein in Prep1i/i embryos (Fig. 1C, lanes 1, 2, and 5). Similar results were obtained with whole embryos, other embryonic organs, and cultured embryonic fibroblasts (not shown). Densitometric analysis (not shown) of many data demonstrated that Prep1i/i embryos still exhibited 3 to 10% of wt levels of Prep1 protein, resulting in a Prep1 mutation that was not null but hypomorphic. Interestingly, the severity of the phenotype (in particular, pallor and edema) was higher in those embryos that expressed less Prep1. Indeed, in Fig. 1C, the Prep1i/i embryo with the lowest residual level of Prep1 protein (lane 5) corresponded to a more severe phenotype than that of the embryo analyzed in lane 1 (not shown). This observation has been reproduced in all of the experiments reported below.
Prep1i/i embryonic phenotype.
In agreement with the striking pallor, edema, and smaller liver spot in the majority of embryos, in most E16.5 Prep1i/i embryos, blood smear analyses showed a profound anemia and a relative increase of nucleated cells (not shown), which resulted in a very low hematocrit (13% versus 37% of wt littermates) (Table 2). To test whether this phenotype may be due to a delayed or abnormal differentiation of the erythroid lineage in the FL, we analyzed the expression of Ter119 and CD71 by flow cytometry. This approach identifies five erythroid progenitor subpopulations at differentiation stages from proerythroblasts to reticulocytes (57). In Table 3, the R1 subpopulation (CD71med, Ter119−) includes the earliest progenitors, proerythroblasts and early basophilic erythroblasts fall into R2 (CD71high, Ter119low), early and late basophilic erythroblasts in R3 (CD71high, Ter119high), and chromatophilic and ortochromatic erythroblasts in R4 (CD71med, Ter119high) (57). The analysis of E15.5 FL (13 wt or heterozygous versus 10 Prep1i/i) showed, with respect to wt, a general decrease in total FL cells, an increase in early progenitor subpopulations R2 and R3, and a similar decrease in the more differentiated R4 subpopulation (Table 3). Although not large, the differences observed in R2, R3, and R4 were statistically significant. At E16.5, the differences were more marked, although there was more variability (Table 3). The total number of FL cells was lower in Prep1i/i than in the wt. In wt embryos, the majority of more differentiated erythroid progenitors was in the R3 and R4 subpopulations. In the case of the three Prep1i/i embryos, the results were more variable from embryo to embryo, and hence, we present the data for the individual embryos in Table 3. In one embryo, almost all erythroid cells were retained in the R1 subpopulation (i.e., the earliest erythroid progenitors). In a second embryo, about half of the erythroid cells were in the R1 subpopulation (early progenitors) and the other half were in R4. In the third Prep1i/i embryo, instead, most erythroid cells were present in the R4 subpopulation, indicating that they had reached a rather advanced stage of differentiation. Thus, at least two of three Prep1i/i embryos exhibit a variable degree of delay in erythroid differentiation, which may be correlated with different levels of residual Prep1protein. It is interesting that the greatest differences were observed at E16.5, i.e., when a major hematocrit decrease becomes evident (Table 3).
TABLE 2.
Hematocrit analysis of E16.5 Prep1i/i embryos
| Genotype | n | Hematocrit (%) |
|---|---|---|
| +/+ or +/i | 12 | 37 ± 3.9 |
| i/i | 5a | 13 ± 4.1 |
In one additional embryo, the amount of blood was not enough to determine the hematocrit.
TABLE 3.
Analysis of the erythroid subpopulations in E15.5 and E16.5 wt and Prep1i/i fetal liver cellsa
| Stage | No. of embryos | Genotype(s) | % Erythroid differentiation for subpopulationb:
|
Total no. of FL cells (106) | |||
|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | ||||
| E15.5 | 13 | +/+, +/i | 3.48 ± 0.56 | 3.8 ± 1.15 | 54.7 ± 5.44 | 32.5 ± 4.18 | 46.3 ± 9.73 |
| E15.5 | 10 | i/i | 3.15 ± 0.63 | 5.1 ± 1.9 | 59.1 ± 6.1 | 27.9 ± 4.8 | 33.1 ± 5.3 |
| E16.5 | 6 | +/+, +/i | 4.6 ± 2.2 | 2.19 ± 0.9 | 45.1 ± 24.3 | 42.2 ± 24.7 | 71.6 ± 7.4 |
| E16.5 | 1 | i/i | 96.2 | 0.02 | 0 | 0.19 | 30.0 |
| E16.5 | 1 | i/i | 44.9 | 1.2 | 2.6 | 43.3 | 4.0 |
| E16.5 | 1 | i/i | 4.9 | 1.74 | 9.2 | 79.5 | 57.0 |
The table shows the data averaged from three E15.5 litters and from a single E16.5 litter generated by crossing Prep1 heterozygous mice. The mean values and standard deviations are shown for the E15.5 embryos, while only the wt and heterozygous embryos are averaged for the E16.5 embryos. For E16.5 Prep1i/i embryos, the data for each individual are shown. For this reason, no P values are shown for the E16.5 embryos. However, it is evident that 2 of 3 Prep1i/i embryos show enrichment in the least differentiated R1 subpopulation. P values: R1, 0.1; R2, 0.029; R3, 0.04; R4, 0.012; total, 0.00045.
Erythroid differentiation was monitored by measuring the expression of CD71 and Ter119 by flow cytometry. The various subpopulations are classified as follows: R1 (CD71med-Ter119−/lo), earliest progenitors; R2 (CD71hi-Ter119−/lo), proerythroblasts and early basophilic erythroblasts; R3 (CD71hi-Ter119hi), early and late basophilic erythroblasts; R4 (CD71med-Ter119hi), chromatophilic and orthochromatic erythroblasts (57).
To further analyze the efficiency of Prep1i/i FL erythroid progenitors, we measured the erythroid colony-forming activity using an erythroid-selective, erythropoietin-enriched, methylcellulose medium (35). While in 4 heterozygous embryos the FL cells produced at least 6,000 BFU-E colonies/FL (average of 10,100 ± 4,500), the two Prep1i/i embryonic samples produced about 1,000 colonies/FL in one case and less than 100 colonies/FL in the other (Table 4). A similar decrease in BFU-E-forming ability of Prep1i/i FL cells in erythroid-selective medium has been reproduced several times. Overall, the data demonstrate a deficient colony forming efficiency of Prep1i/i FL cells and confirm a requirement for Prep1 in erythroid development.
TABLE 4.
BFU-E colony-forming potential of E14.5 FL cells from wt, heterozygous, and homozygous Prep1i/i mice
Number of embryos employed.
FL cells (50,000) were plated in erythropoietin-supplemented methylcellulose, and BFU-E colonies were counted after 10 days. The data are the averages of results from at least three plates per each individual embryo and are expressed as numbers of colonies per FL. The average ± standard deviation for Prep1 heterozygous (+/i) mice is 10,100 ± 4,500. P value (+/i versus i/i), 0.02.
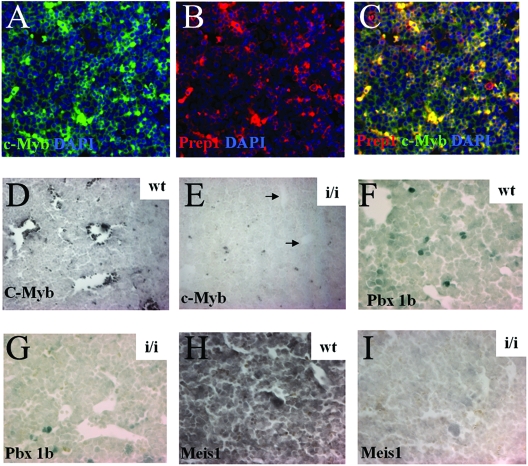
Subsequently, we analyzed expression of Prep1 in the FL using immunohistochemistry and immunofluorescence. As one of the genes which is required for normal erythropoiesis is cMyb (15), we tested for the coexpression of Prep1 and cMyb in wt FL cells. As shown in Fig. 2A to C, immunofluorescence demonstrated that the proteins encoded by those genes colocalized in the nucleus of a subset of FL cells. We therefore tested whether cMyb-positive cells were affected in the FL of Prep1i/i embryos. We found that the number of cMyb-positive cells was profoundly decreased in Prep1i/i embryos (Fig. 2D and E). Since Pbx1b and Meis1 are also essential in hematopoietic development (1, 14, 23), we analyzed Pbx1b and Meis1 protein levels in FL cells by immunohistochemistry and found a profound decrease of Pbx1b- and Meis1-positive cells in Prep1i/i FL (Fig. 2F to I).
FIG. 2.
Presence of Prep1, Pbx1b, Meis1, and cMyb in wt and Prep1i/i FL. (A to C) Immunofluorescence analysis of E14.5 wt FL sections. Triple staining with anti-cMyb and anti-Prep antibodies and 4′,6′-diamidino-2-phenylindole (DAPI) to stain nuclei is shown. The antibodies used are indicated. Magnification (A to C), ×20. In panel C, colocalization of cMyb and Prep1 appears as a yellow color. (D to I) Immunohistochemistry of E14.5 wt and Prep1i/i FL, developed with the DAB kit. (D and E) Anti-cMyb antibodies; (F and G) anti-Pbx1b antibodies; (H and I) anti-Meis1 antibodies. Magnification, ×20.
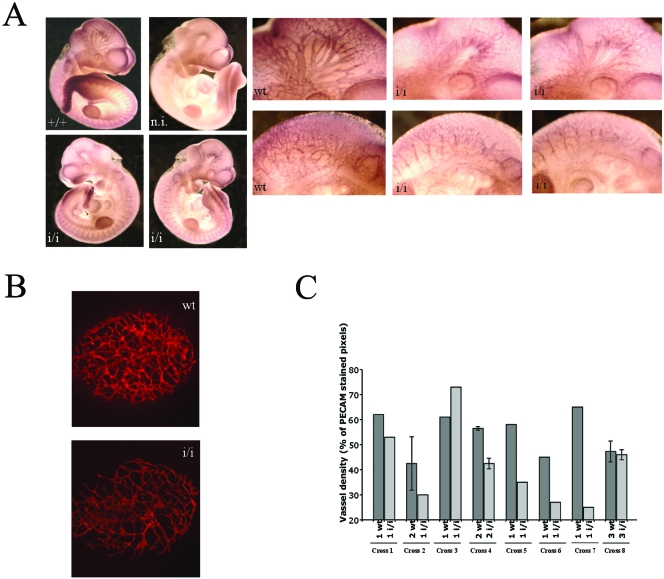
Visible hemorrhages were detected in Prep1i/i embryos, although only in 13% of the samples. Whole-mount immunostaining of E10.5 embryos with an antibody to the endothelial marker CD31 (PECAM) revealed a decreased vascular tree at the level of both the head and intersomitic vessels (Fig. 3A). We also examined embryonic angiogenesis by culturing allantois preparations of 23 E7.5 to 7.75 (12 wt and 11 Prep1i/i) embryos using CD31 immunofluorescence. A decrease in the number of capillaries (CD31-positive area) and of the microvasculature complexity was observed in most (8/11) Prep1i/i allantois specimens (Fig. 3B and C). Only in 3 of the 11 pairs of wt and Prep1i/i embryos analyzed were no differences observed. Furthermore, immunofluorescence microscopy showed the coexpression of Prep1 in a rather large percentage of CD31-positive cells in wt FL (not shown).
FIG. 3.
Prep1i/i embryos exhibit angiogenesis defects. (A) Whole-mount CD31 (PECAM) immunofluorescence on E10.5 embryos. The genotype is shown in each panel. A nonimmune (n.i.) serum gave essentially no staining (not shown). A set of close-up pictures is inserted. The top row shows details of the head region; the bottom row the intersomitic area. (B) Immunofluorescence on E7.5 wt and Prep1i/i allantois cultured for 18 h and stained with anti-CD31 antibodies. (C) Vessel density (percentage of CD31-stained pixels) in 11 different litters containing wt and Prep1i/i embryos. At the bottom of each histogram, the symbols indicate the numbers of wt and Prep1i/i littermates from different crosses.
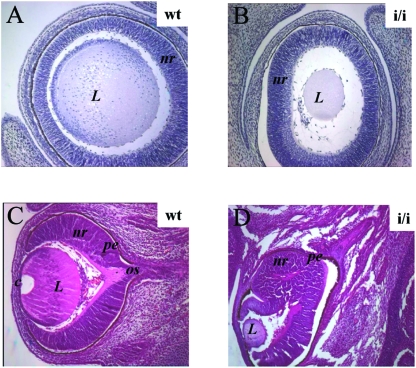
Prep1i/i embryos also showed ocular abnormalities (Table 5). Of 18 E14.5 embryos analyzed, 61.5% of them displayed eye abnormalities affecting different structures of the developing eye with different penetrance, as judged by histological examination. In 40% of all Prep1i/i embryos, we observed a reduction of the lens size (see example in Fig. 4A to D): in 23%, abnormalities of the neural retina (Fig. 4C and D), and in 14%, the eye was still encased deep within the head (data not shown).
TABLE 5.
Frequency of eye phenotypes in E14.5 Prep1i/i embryosa
| Phenotype | % of embryos with:
|
|||
|---|---|---|---|---|
| Overall eye phenotype | Lens size reduction | Retinal epithelium | Position of the eye | |
| Abnormal | 61.5 | 45 | 23 | 14 |
| Normal | 38.5 | 41 | 63 | 81 |
The data refer to a total of 18 Prep1i/i embryos, for a total of 36 eyes examined. The position is considered abnormal when the eye is not visible at the gross morphology analysis.
FIG. 4.
Eye defects in E14.5 Prep1i/i embryos. Comparison of hematoxylin (A and B) or hematoxylin-eosin (C and D) staining of wt and Prep1i/i embryonic eyes (indicated). Notice the reduction of the lens size (A and B) and anomalies and duplication of the retinal epithelium (D). L, lens; nr, neural retina; pe, pigmented retinal epithelium; c, cornea; os, optic stalk. Magnification, ×10 (A and B); ×4 (C and D).
We then tested Prep1 levels in the developing eye structures using immunohistochemistry. At E14.5, Prep1 immunoreactivity was prominent in the neural retina in all the different cell layers (Fig. 5A). Low Prep1 staining was observed in Prep1i/i eyes (Fig. 5D). Control immunostaining with actin antibodies showed the presence of similar numbers and types of cells in both wt and Prep1i/i embryos (Fig. 5C and E). Prep1 immunoreactivity was also detected in the lens epithelium and was drastically reduced in Prep1i/i embryos (Fig. 5F and G).
FIG. 5.
Prep1 and Pax6 are colocalized in wt embryonic eye structures, and Pax6 is present at lower levels in Prep1i/i embryos. Immunohistochemistry with anti-Prep1 (A, B, D, F, G, and I), antiactin (C and E), anti-Pax6 (H, J, and K), and anti-Meis1 (L to O) antibodies in wt and Prep1i/i embryonic eye sections (genotype and antibody used are indicated). Comparison of plates H and I shows that Prep1 and Pax6 colocalize, while plates J and K show lower levels of Pax6 in Prep1i/i eye structures. ch, choroids; c, cornea; ce, corneal epithelium; gc, ganglion cell layer; in, inner nuclear layer; i, iris/ciliar body; L, lens; le, lens epithelium; lf, lens fiber cells; nr, neural retina; os, optic stalk; on, outer nuclear layer; pe, pigmented retinal epithelium; rc, rod and cone photoreceptor cell layer; s, sclera. All panels were developed by the alkaline phosphatase reaction, except panels H and L to O. Magnification, ×60 (A); ×20 (B to E and L to O); ×40 (F, G); ×10 (H to K).
As Pax6 is a factor essential for lens development (20, 29, 48, 52), we analyzed Pax6 levels in wt and Prep1i/i embryos. Pax6 was present in the same cells of the neural retina and lens as Prep1 (compare Fig. 5H and I), but its levels were drastically reduced in Prep1i/i embryos (Fig. 5J and K). Pax6 levels were clearly reduced in the iris, ciliar body, corneal epithelium, cornea, and lens epithelium in Prep1i/i embryos.
As Meis1 homeoprotein has been implicated in the direct regulation of Pax6 during vertebrate lens morphogenesis (58), we tested Meis1 levels in E14.5 developing eyes of wt and Prep1i/i embryos by immunohistochemistry. Meis1 protein was high in the neural retina and low in the lens epithelium (Fig. 5L and N) of wt embryos, while almost totally absent in Prep1i/i embryos (Fig. 5 M and O). Lower levels of Meis1 in the lens versus retina have been previously reported (23). As a result, the decrease of Meis1 in the Prep1i/i neural retina was more evident than in the lens.
Prep1 deficiency almost abolishes Pbx-dependent DNA-binding activity and decreases Pbx and Meis1 protein levels.
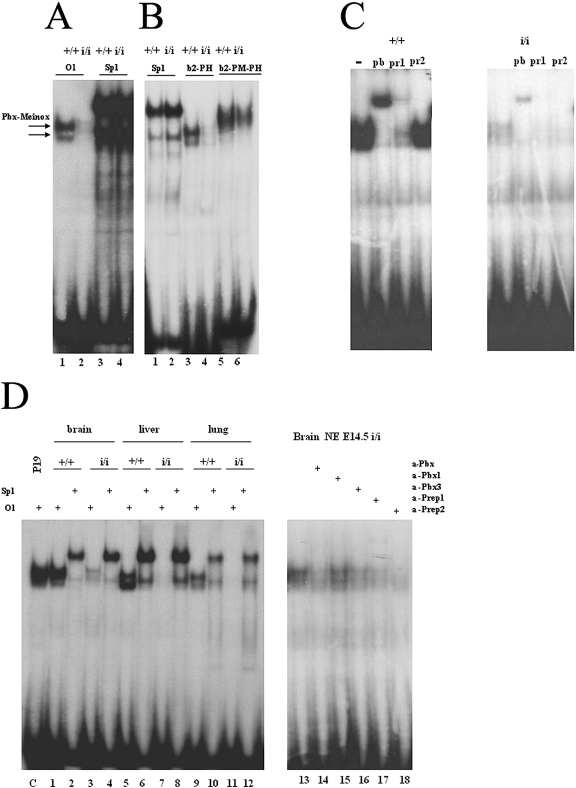
Pbx proteins bind DNA upon dimerization with Prep and Meis proteins (4, 10). However, nothing is known about the relative contribution of individual Prep or Meis proteins in vivo. In fact, Pbx proteins can dimerize with either Prep or Meis proteins, producing dimers with similar DNA-binding properties. We therefore exploited Prep1i/i embryos to test the contribution of Prep1 to the overall DNA binding activity of Pbx proteins, using EMSA. Nuclear extracts of whole E10.5 Prep1i/i embryos showed almost no DNA binding to oligonucleotide O1 (Fig. 6A), which can bind both Prep1-Pbx and Meis-Pbx dimers (3, 5). The binding to the control Sp1 oligonucleotide, specific for the ubiquitous Sp1 transcription factor (24), was unchanged. Prep1i/i extracts also displayed no or weak binding to oligonucleotides that, in addition, also recognize Pbx-Hox dimers (oligo b2-PH) or Prep/Meis-Pbx-Hox ternary complexes (b2-PM-PH) (16, 17, 25) (Fig. 6B). We then used antibodies to verify the contribution of Prep1 to the above binding activities. In wt as well as Prep1i/i extracts, a supershift was observed with anti-Pan-Pbx or inhibition with specific anti-Prep1 antibodies, indicating that the measured activity was mostly due to Prep1-Pbx dimers (Fig. 6C); no effect was observed with anti-Prep2 antibodies. Thus, Prep1 represents the most abundant, among Prep-Meis family members, DNA binding partner for Pbx. The results shown in Fig. 6A and B also confirm the presence of residual, functional Prep1 in Prep1i/i extracts. When we analyzed extracts from Prep1i/i E14.5 embryonic liver, a site of active hematopoiesis, essentially no DNA-binding activity was observed (Fig. 6D, lanes 5 and 7). In brain (Fig. 6D, lanes 1 and 3) and lung (Fig. 6D, lanes 9 and 11) extracts, a similar result was observed. Competition with specific antibodies revealed that the residual binding activity in Prep1i/i brain nuclear extracts was due to Prep1, Prep2, Pbx1, and Pbx3 (Fig. 6D, lanes 17, 18, 14, and 16, respectively). We conclude that Prep1 is the most abundant partner of Pbx in embryonic DNA-binding activity and that this is profoundly decreased in Prep1i/i embryos.
FIG. 6.
Decreased Pbx DNA-binding activity in Prep1i/i embryos. (A) Electrophoretic mobility shift analysis of the DNA-binding activity of nuclear extracts from an E10.5 total embryo with labeled O1 (specific for Prep1/Meis-Pbx dimers) and Sp1 (control) oligonucleotides. On the top of the panels, the genotype of the embryo is indicated. The two arrows to the left indicate the migration of Pbxa and Pbxb-Prep/Meis complexes. (B) Same as in panel A, with labeled oligonucleotide Sp1 (control), b2-PH (binding both Pbx-Hoxb1 and Prep1/Meis-Pbx dimeric complexes), and b2-PM-PH (binding also Meis/Prep1-Pbx-Hoxb1 ternary complexes). TC indicates the migration of the ternary complex. (C) Identification of the nature of the binding activity by EMSA in the presence of anti-Prep1 (pr1), anti-panPbx (pb), and anti-Prep2 (pr2) antibodies. Comparison of nuclear extracts from E10.5 wt and Prep1i/i embryos. The oligonucleotide used was O1. (D) Left panel, analysis by EMSA on nuclear extracts obtained from embryonic brain (lanes 1 to 4), liver (lanes 5 to 8), and lung (lanes 9 to 12). The genotype is indicated on the top of the panel. Right panel, analysis by EMSA of the residual binding activity of nuclear extracts obtained from a Prep1i/i brain in the presence of different antibodies (+), as indicated on the top of the panel. α, anti.
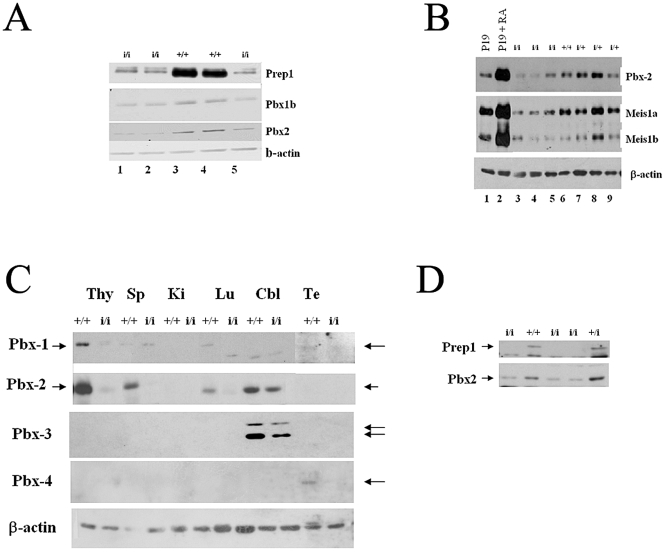
We also used immunoblotting to analyze the levels of Pbx and Meis proteins in tissues from Prep1i/i embryos. An immunoblot performed on E14.5 brain nuclear extracts from the same litter tested for Prep1 in Fig. 1C showed a concomitant decrease of Pbx2 and Pbx1b in Prep1i/i embryonic brains (Fig. 7A). In another litter, a reduction of Pbx2 and Meis1 was observed in an E14.5 embryonic brain (Fig. 7B), while a general reduction of all Pbx proteins was observed in nuclear extracts of several E10.5 and E11.5 whole Prep1i/i embryos (data not shown). Furthermore, in the rare adult Prep1i/i mice, Pbx1a was reduced in the lung and testis, Pbx2 in spleen and lung, Pbx3a and 3b in the cerebellum, and Pbx4 in the testis, as revealed by immunoblotting (Fig. 7C). Finally, a blot of an E14.5 FL showed not only the reduction of Prep1 but also the concomitant reduction of Pbx2 (Fig. 7D).
FIG. 7.
Prep1i/i embryonic and adult organs show a decrease of Pbx1b, Pbx2, and Meis1 proteins. (A) Immunoblotting analysis of the same filter shown in Fig. 1C: nuclear extracts from the E14.5 embryonic brain of 5 littermate embryos (2 wt and 3 Prep1i/i, as indicated) tested with monoclonal anti-Pbx1b, anti-Pbx2, and anti-beta-actin antibodies. Lanes 1, 2, and 5 contain extracts from Prep1i/i embryos; lanes 3 and 4 contain extracts from wt embryos. (B) Immunoblotting analysis of brain extracts from 1 wt, 3 heterozygous, and 3 Prep1i/i embryos, using anti-Pbx2 and anti-Meis1 antibodies. (C) Immunoblotting analysis of nuclear extracts obtained from organs, indicated at the top of the panel, of an adult Prep1i/i mouse, tested with anti-Pbx1, anti-Pbx2, anti-Pbx3, anti-Pbx4, and antiactin antibodies. Cbl, cerebellum; Lu, lung; Ki, kidney; Sp, spleen; Te, testis; Thy, thymus. (D) Immunoblotting analysis of E14.5 liver extracts from 1 wt, 1 heterozygous, and 3 Prep1i/i embryos, using anti-Pbx2 (α-Pbx2) and anti-Prep1 (α-Prep1) antibodies.
These results agree with the immunohistochemical data showing decreased levels of Pbx1b and Meis1 in the fetal liver (Fig. 2D to I) and eye (Fig. 5L to O) and in the total embryo (not shown). Therefore, we conclude that the deficiency of Prep1 in the hematopoietic liver and other organs is accompanied by a decrease of Pbx and Meis1 proteins. In agreement with these data, we have recently shown that in adult Prep1i/i mice, Pbx2 (the most abundant Pbx) disappears from the thymus (40).
MRNA levels of cMyb, some Pbx, and some Meis genes are reduced in Prep1i/i embryos.
Quantitative PCR on E10.5 total embryonic RNA demonstrated a statistically significant reduction of several mRNAs (Table 6). Interestingly, while the levels of Pbx1, Pbx2, and Meis1 proteins were decreased in Prep1i/i embryos (Fig. 7), the levels of their mRNAs did not change. These data are in keeping with the absence of Pbx2 proteins from the thymus of adult Prep1i/i mice in the presence of unaltered levels of Pbx2 mRNA (40). However, the levels of Pbx3, Pbx4, Meis2, and Meis3 mRNAs were significantly reduced. Finally, the level of cMyb mRNA was also significantly decreased (Table 6), supporting the immunohistochemistry of Prep1i/i FL (Fig. 2). In conclusion, these data indicate that Prep1 is required for the expression of related TALE proteins and of cMyb.
TABLE 6.
QT-PCR analysis of mRNA from wt versus Prep-1i/i E10.5 embryosa
| Gene | No. of embryos | mRNA level (% of wt)b | SD | P valuec |
|---|---|---|---|---|
| Prep1 | 5 | 1.5 | 0.004 | 0.0000007 |
| Meis1 | 5 | 58.6 | 19.9 | 0.27 |
| Meis2 | 3 | 56.3 | 0.6 | 0.001 |
| Meis3 | 5 | 58.6 | 7.7 | 0.001 |
| Pbx1 | 5 | 79.4 | 23.4 | 0.23 |
| Pbx2 | 5 | 105 | 19.4 | 0.39 |
| Pbx3 | 5 | 56.4 | 12 | 0.009 |
| Pbx4 | 5 | 64.4 | 23.3 | 0.09 |
| cMyb | 5 | 43.8 | 18 | 0.01 |
The table pools together two experiments conducted with two different litters.
The data are standardized to that of the 18S rRNA. A value of 100% is attributed to the mean value obtained for wt embryos.
P values in boldface type indicate statistical significance.
DISCUSSION
Prep1 is required for embryonal development.
The hypomorphic Prep1i/i mutation caused embryonic lethality with variable penetrance, as only about 1/4 of the homozygous Prep1i/i embryos survived pregnancy (Table 1). Death of most embryos occurred between E17.5 and P0, but a small percentage survived the pregnancy and lived a normal-length life (40). This variability in the Prep1i/i phenotype may be due to differences in residual Prep1 protein in different embryos. Among the observed embryonic phenotypes, hematopoietic abnormalities exhibited a very high penetrance (about 70%) and variable expressivity, with a reduced hematocrit and anemia. The anemia might be, at least in part, the cause of embryonic death. Frequent angiogenesis and eye defects were also observed in Prep1i/i embryos.
The hematopoietic phenotype consisted of a dramatic decrease in the number of circulating erythrocytes and a delay in erythroid differentiation. Indeed, E15.5 and E16.5 Prep1i/i FL contained more erythroid progenitors and fewer differentiated cells (Table 3). Deficiency in erythroid progenitors was also shown by the measurement of erythropoietin-dependent colony formation, which uncovered a dramatic deficiency in Prep1i/i FL (Table 4). The presence of Prep1 in FL hematopoietic cells, as shown by its colocalization with cMyb (Fig. 2), and Sca1 (not shown), is consistent with the observed phenotype. FL from Prep1i/i embryos exhibited a drastic decrease of cMyb-positive cells. However, a few cells still exhibited apparently normal levels of cMyb (Fig. 2D and E). This finding is in agreement with the observation that cMyb and Prep1 were colocalized in most but not all FL cells (Fig. 2). Nonetheless, the overall decrease of cMyb can, at least in part, explain the erythroid phenotype (15).
Prep1i/i mice which escape embryonic lethality show a defect in T-cell development, with a decreased number of circulating CD4+ and CD8+ T cells, increased apoptosis, decreased proliferation of double-positive thymocytes, and anomalies in αβ and γδ T-cell receptor expression, a phenotype reproduced in wt mice transplanted with Prep1i/i FL cells (40). Present data show that, in addition to the lymphoid lineage, Prep1 is also required for the proper development of the erythroid lineage. Whether the lymphoid and erythroid phenotypes derive from anomalies in common stem cell progenitors or from the concomitant roles of Prep1 in different hematopoietic lineages remains to be elucidated.
Angiogenesis was also impaired in Prep1i/i embryos. Indeed, E7.5 to 7.75 Prep1i/i allantois preparations and E10.5 whole embryos showed reduced, thinner, and less-organized capillaries (Fig. 3). These data suggest, therefore, that angiogenic precursors may also be affected by Prep1 deficiency. In fact, Prep1 is present in endothelial precursors, where it colocalizes with CD31 and c-Kit (data not shown) in E14.5 FL. Furthermore, the finding of a decreased microvasculature in Prep1i/i allantois cultures indicates that Prep1i/i embryos have an intrinsic angiogenic defect, which does not simply reflect a decrease in circulating blood cells, and thus is independent from the hematopoietic phenotype.
Another frequent phenotype of Prep1i/i embryos involved eye development (Table 5). In some cases, the eye was not detectable but was found deep inside the head. In most cases, the size of the lens was strongly reduced, similar to the phenotype of Pax6-deficient mice, where no lens induction and anomalies of the neural retina have been reported (48, 52). Prep1 is present in E14.5 neural retina, cornea, and lens epithelium and specifically colocalized with Pax6. Interestingly, Pax6 levels were drastically reduced in the Prep1i/i neural retina, cornea, iris, and lens epithelium (Fig. 5J and K). Pax6 down-regulation may have a critical role in determining the eye phenotype of Prep1i/i embryos, since Pax6 is essential for oculogenesis (20, 29, 48, 52). As Prep1i/i embryos exhibit overall lower levels of Meis1 protein, the Prep1i/i ocular phenotype might be due to reduced Meis1 expression. Previous biochemical and genetic data demonstrated that Meis1 directly regulates Pax6 expression during vertebrate lens morphogenesis (58). Furthermore, the specific 107-bp minimal lens lineage enhancer element contains a functionally essential Meis1 binding site (which potentially could also be a Prep1 site) that directs expression in the prospective mouse lens (58). At this point, our data do not allow us to conclude whether Prep1 directly regulates Pax6 expression in the lens, cornea, and/or neural retina or whether it does so by controlling the levels of Meis1 (Fig. 8). As Prep1 and Meis act by dimerizing with Pbx proteins, it is possible that Pbx also participates in the regulation of Pax6 expression and eye development. While no information is available in the mouse, in Xenopus laevis Pbx has been shown to be required for lens development (39). It is worth noting that the angiogenic, hematopoietic, and eye phenotypes have also been reported in Meis1-deficient embryos (1, 23).
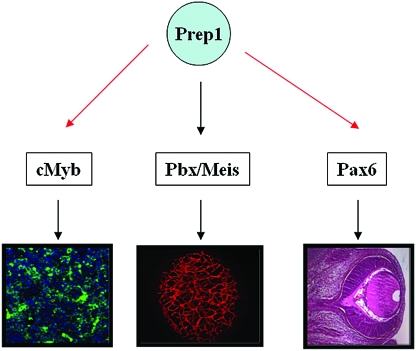
FIG. 8.
Establishment of a hierarchical role for Prep1 within the TALE protein network in embryogenesis. Prep1 is required for normal hematopoiesis, angiogenesis, and oculogenesis, as illustrated. The scheme depicts the upstream role of Prep1 as it controls Pbx and Meis TALE class homeoproteins and their target genes. Such genes become “effectors” within specific developmental processes: cMyb for erythropoiesis and Pax6 for eye development. Black arrows indicate direct control; red arrows indicate hierarchical control, whose direct or indirect nature remains to be established.
Molecular basis for the Prep1i/i phenotype.
Prep1 deficiency affects the expression of both TALE class partners Pbx and Meis. In fact, a reduction of Pbx1, Pbx2, and Meis1 proteins was directly demonstrated by immunoblotting and agrees overall with the deficient DNA-binding activities of both whole embryos and embryonic organ extracts (Fig. 6, 7). The reduction of Pbx and Meis gene expression in Prep1i/i embryos (Fig. 7) is in agreement with the decrease of Pbx1b-positive and Meis1-positive cells in FL of Prep1i/i embryos (Fig. 2). These results are of particular interest, since both Meis1 and Pbx1 are required for embryonic hematopoiesis (1, 14, 23). Indeed, Pbx1-deficient embryos also exhibit striking defects in erythroid colony formation (14). Therefore, it could be envisaged that the hematopoietic phenotype exhibited by the Prep1i/i embryo is mediated by the decrease of both Meis1 and Pbx1/Pbx2. Even though the Pbx2-deficient mouse has no evident phenotype (47), the absence of Pbx2 protein may be relevant, since in the context of a general decrease of all Pbx proteins in Prep1i/i embryos, the lack of Pbx2 would functionally uncover Pbx1 deficiency in hematopoietic cells.
The decrease of cMyb-positive cells in Prep1i/i embryos is also consistent with the reported hematopoietic abnormality, as cMyb-deficient embryos fail to produce all of the hematopoietic lineages (12, 15, 30, 37, 51). However, at present, it is not clear whether the Prep1i/i hematopoietic phenotype derives from a direct or indirect effect of Prep1 on cMyb gene expression.
Likewise, the decrease of Pax6 in the eye of Prep1i/i embryos appears to be, at least in part, the cause of the reported eye defect. Understanding whether the Prep1i/i eye abnormality is a direct effect of the regulation of Pax6 gene expression by Prep1 will require further experiments. Nonetheless, the decrease of Pax6 levels in the developing eye of Prep1i/i embryos is well in keeping with the established requirement of Pax6 in eye formation (20, 48, 52).
The Prep1 role in mouse development is epistatic to that of Pbx and Meis genes.
The decrease not only of Prep1 but also of Pbx and Meis proteins in Prep1i/i embryos almost abolishes the DNA-binding activity of Meis/Prep-Pbx dimer-specific target sequences. Prep1 deficiency causes a reduction of related family members and TALE partners, such as Pbx1, Pbx2, and Meis1. As the mRNA levels of these proteins is not affected in a statistically significant manner (Table 6), their reduction appears to be at the posttranscriptional level. Likewise, in zebra fish prep1.1 down-regulation reduces the levels of all Pbx proteins (13). Furthermore, in mammalian cells in culture, Prep1 overexpression does not affect Pbx1 and Pbx2 mRNA levels but increases the stability of Pbx1b and Pbx2 by preventing their proteasomal degradation (31). In light of these results, it is likely that, in the absence of Prep1, Pbx proteins are not protected from proteasomal degradation. However, Prep1 deficiency results in a decrease of Pbx3, Pbx4, Meis2, and Meis3 mRNAs in whole E10.5 embryos (Table 6). Thus, Prep1 not only forms transcriptional complexes with Pbx but also hierarchically controls the expression of all Pbx and Meis genes. It will be interesting to analyze whether the levels of Hox genes is also affected in Prep1i/i embryos, as described for zebra fish (13).
We conclude that Prep1 is a master gene that is required for hematopoietic, angiogenic, and eye development, as well as other developmental functions, by controlling the levels of Pbx and Meis TALE proteins and their target genes (Fig. 8). Many of the phenotypes observed in Prep1i/i embryos may be mediated by the concomitant loss of Meis and Pbx partners, therefore resulting in defects closely resembling those of Pbx1 and Meis1 null embryos. Nonetheless, Prep1 undoubtedly also exerts unique functions in vertebrate development, as demonstrated by the presence of unique abnormalities in Prep1i/i versus either Meis- or Pbx-deficient embryos. For example, the lack of Meis1 mainly causes a defect in megakaryocyte production, while in Prep1i/i embryos, erythroid cells are primarily affected. In addition, Prep1 null embryos exhibit very early embryonic lethality (Fernandez et al., unpublished data), while the Pbx and Meis mutants survive in utero until late gestation. Therefore, our studies establish that Prep1, while controlling other TALE partners, also plays unique, selective roles in vertebrate development.
Acknowledgments
We are very grateful to C. Camaschella, G. Cossu, E. Dejana, and A. Brendolan for helpful discussions and to Heike Poepperl, Michael E. Cleary, Mark Featherstone, Miguel Torres, and Arthur Buchberg for gifts of monoclonal antibodies. Many thanks go to IFOM Services for QT-PCR (Laura Tizzoni and Sara Volorio), to Marco Chiaravalli and Davide Moi for mouse husbandry and genotyping, to Cesare Covino (Alembic, HSR) for confocal microscopy, and to Massimo Mazzieri for software generation.
This work was supported by grants from the Telethon Foundation Onlus (GGP02031), the Italian Ministry of University and Research (COFIN 2002-2005), and the Italian Association for Cancer Research (AIRC). The experiments performed by N.M. were supported by an AIRC grant to Massimo Crippa. L.S. acknowledges grants from the National Institutes of Health (HD43997) and from the March of Dimes and Birth Defects Foundation (6-FY03-071).
REFERENCES
- 1.Azcoitia, V., M. Aracil, A. Carlos-Martinez, and M. Torres. 2005. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev. Biol. 280:307-320. [DOI] [PubMed] [Google Scholar]
- 2.Berkes, C., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 3.Berthelsen, J., J. Vandekerkhove, and F. Blasi. 1996. Purification and characterization of UEF3, a novel factor involved in the regulation of the urokinase and other AP-1 controlled promoters. J. Biol. Chem. 271:822-3830. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen, J., V. Zappavigna, E. Ferretti, F. Mavilio, and F. Blasi. 1998. Prep1, a novel partner of Pbx proteins, modifies Pbx-Hox protein cooperativity. EMBO J. 17:1434-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthelsen, J., V. Zappavigna, F. Mavilio, and F. Blasi. 1998. Prep1, a novel functional partner of Pbx proteins. EMBO J. 17:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brendolan, A., E. Ferretti, V. Salsi, K. Moses, S. Quaggin, F. Blasi, M. L. Cleary, and L. Selleri. 2005. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 132:3113-3126. [DOI] [PubMed] [Google Scholar]
- 8.Burglin, T. R. 2005. Homeodomain proteins, p. 179-222. In R. A. Meyers (ed.), Encyclopedia of molecular cell biology and molecular medicine, vol. 6. Wiley-VCH, Hoboken, N.J. [Google Scholar]
- 9.Capdevila, J., T. Tsukui, C. Rodriquez Esteban, V. Zappavigna, and J. C. Izpisua Belmonte. 1999. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol. Cell 4:839-849. [DOI] [PubMed] [Google Scholar]
- 10.Chang, C. P., Y. Jacobs, T. Nakamura, N. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. Meis proteins are major in vivo DNA binding partners for wt but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 12.Creavin, T. 2005. Hematopoiesis: cMyb enters centre stage. Drug Discov. Today 10:381-382. [DOI] [PubMed] [Google Scholar]
- 13.De Florian, G., N. Tiso, E. Ferretti, F. Blasi, M. Bortolussi, and F. Argenton. 2004. Prep1.1 has essential and unique genetic functions in hindbrain development and neural crest cells differentiation. Development 131:613-627. [DOI] [PubMed] [Google Scholar]
- 14.Dimartino, J., L. Selleri, D. Traver, M. Firpo, J. Rhee, R. Warnke, S. O'Gorman, I. L. Weissman, and M. L. Cleary. 2001. The Hox cofactor and protoncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood 98:618-626. [DOI] [PubMed] [Google Scholar]
- 15.Emambokus, N., A. Vegiopoulos, B. Harman, E. Jenkinson, G. Anderson, and J. Frampton. 2003. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of cMyb. EMBO J. 22:4478-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferretti, E., F. Cambronero, S. Tumpel, L. Weidemann, F. Blasi, and R. Krumlauf. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: Complex interplay between Prep1-Pbx1-Hoxb1 binding sites. Mol. Cell. Biol. 25:8541-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti, E., H. Marshall, H. Pöpperl, M. Maconochie, R. Krumlauf, and F. Blasi. 2000. A complex site including both Pbx-Hox and Prep-Meis-responsive elements and binding a retinoic acid-inducible ternary Hoxb1-Pbx-Prep1 complex is required for Hoxb2 rhombomere 4 expression. Development 127:155-166. [DOI] [PubMed] [Google Scholar]
- 18.Ferretti, E., H. Schulz, D. Talarico, F. Blasi, and J. Berthelsen. 1999. The Pbx-regulating protein Prep1 is present in a Pbx-complexed form throughout mouse embryogenesis. Mech. Dev. 83:53-64. [DOI] [PubMed] [Google Scholar]
- 19.Fognani, C., C. Kilstrup-Jensen, E. Ferretti, V. Zappavigna, and F. Blasi. 2002. Human PREP-2, a novel interactor of PBX proto-oncogene, defines a novel sub-family of TALE homeodomain transcription factors. Nucleic Acids Res. 30:2043-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehring, W. J., and K. Ikeo. 1999. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 15:371-377. [DOI] [PubMed] [Google Scholar]
- 21.Haller, K., I. Rambaldi, E. Nagy Kovacs, E. Daniels, and M. S. Featherstone. 2002. Prep2: cloning and expression of a new Prep family member. Dev. Dyn. 225:358-364. [DOI] [PubMed] [Google Scholar]
- 22.Herzig, S., L. Fuzesi, and W. Knepel. 2000. Heterodimeric Pbx-Prep1 homeodomain protein binding to the glucagon gene restricting transcription in a cell type dependent manner. J. Biol. Chem. 275:27989-27999. [DOI] [PubMed] [Google Scholar]
- 23.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibanez-Tallon, I., C. Ferrai, E. Longobardi, I. Facetti, F. Blasi, and M. Crippa. 2002. Binding of Sp1 to the proximal promoter links constitutive expression of the human uPA gene and metastatic potential of PC3 cells. Blood 100:3325-3332. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, Y., Schnabel, C. A., and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. K., L. Selleri, J. S. Lee, Y. Jacobs, and M. L. Cleary. 2002. Defective pancreas development and function in mice deficient for Pbx1. Nat. Genet. 30:430-435. [DOI] [PubMed] [Google Scholar]
- 27.Knoepfler, P. S., K. R. Calvo, H. Chen, S. E. Antonarakis, and M. P. Kamps. 1997. Meis 1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. USA 94:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurant, E., C.-Y. Pai, R. Sharf, N. Halachmi, Y. H. Sun, and A. Salzberg. 1998. Dorsothonal/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning the embryonic PNS. Development 125:1037-1048. [DOI] [PubMed] [Google Scholar]
- 29.Lang, R. A. 2004. Pathways regulating lens induction in the mouse. Int. J. Dev. Biol. 48:783-791. [DOI] [PubMed] [Google Scholar]
- 30.Lieu, Y. K., A. Kumar, A. G. Pajerowski, T. J. Rogers, and E. P. Reddy. 2004. Requirement of c-myb in T cell development and in mature T cell function. Proc. Natl. Acad. Sci. USA 101:14853-14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longobardi, E., and F. Blasi. 2003. Overexpression of PREP1 leads to a functionally relevant increase of PBX2 by preventing its degradation. J. Biol. Chem. 278:39235-39241. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, R., A. Ishimura, K. Mood, E. K. Park, A. M. Buchberg, and I. O. Daar. 2002. Xpbx1b and Xmeis1b play a collaborative role in hindbrain and neural crest gene expression in Xenopus embryos. Proc. Natl. Acad. Sci. USA 99:5448-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manley, N., L. Selleri, A. Brendolan, J. Gordon, and M. L. Cleary. 2004. Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev. Biol. 47:301-312. [DOI] [PubMed] [Google Scholar]
- 34.Manzanares, M., S. Bel-Vialar, L. Ariza-McNaughton, E. Ferretti, H. Marshall, M. M. Maconochie, F. Blasi, and R. Krumlauf. 2001. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain. Development 128:3595-3607. [DOI] [PubMed] [Google Scholar]
- 35.May, C., S. Rivella, A. Chadburn, and M. Sadelain. 2002. Successful treatment of murine-thalassemia intermedia by transfer of the human-globin gene. Blood 99:1902-1908. [DOI] [PubMed] [Google Scholar]
- 36.Mercader, M., E. M. Tanake, and M. Torres. 2005. Proximodistal identity during vertebrate limb development is regulated by Meis homeodomain proteins. Development 132:4131-4142. [DOI] [PubMed] [Google Scholar]
- 37.Metcalf, D., M. R. Carpinelli, C. Hyland, S. Mifsud, L. Dirago, N. A. Nicola, D. J. Hilton, and W. S. Alexander. 2005. Anomalous megakaryocytopoiesis in mice with mutations in the c-Myb gene. Blood 105:3480-3487. [DOI] [PubMed] [Google Scholar]
- 38.Monica, K., N. Galili, J. Nourse, D. Saltman, and M. L. Cleary. 1993. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol. Cell. Biol. 11:6149-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan, R., J. Sohal, M. Paleja, and R. Pettengell. 2004. Pbx genes are required in Xenopus lens development. Int. J. Dev. Biol. 48:623-627. [DOI] [PubMed] [Google Scholar]
- 40.Penkov, D., P. Di Rosa, L. Fernandez Diaz, V. Basso, E. Ferretti, F. Grassi, A. Mondino, and F. Blasi. 2005. Involvement of Prep1 in the αβ T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol. Cell. Biol. 25:10768-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee, J. W., A. Arata, L. Selleri, Y. Jacobs, S. Arata, H. Onimaru, and M. L. Cleary. 2004. Pbx3 deficiency results in central hypoventilation. Am. J. Pathol. 165:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryoo, H. D., T. Marty, F. Casares, M. Affolter, and R. S. Mann. 1999. Regulation of Hox target genes by a DNA bound homothorax/Hox/extradenticle complex. Development 126:5137-5148. [DOI] [PubMed] [Google Scholar]
- 43.Salzberg, A., S. Elias, N. Nachaliel, L. Bonstien, C. Henig, and D. Frank. 1999. A Meis family protein caudalizes neural cell fate in Xenopus laevis embryos. Mech. Dev. 80:3-13. [DOI] [PubMed] [Google Scholar]
- 44.Schnabel, C., R. E. Godin, and M. L. Cleary. 2003. Pbx1 regulates nephrogenesis and ureteric branching in the developing kidney. Dev. Biol. 254:262-276. [DOI] [PubMed] [Google Scholar]
- 45.Scott, W. 2004. Angiogenesis assays. Methods Mol. Med. 88:239-246. [DOI] [PubMed] [Google Scholar]
- 46.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. E. Cheah, J. L. R. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128:3543-3557. [DOI] [PubMed] [Google Scholar]
- 47.Selleri, L., J. DiMartino, J. van Deursen, A. Brendolan, M. Sanyal, E. Boon, T. Capellini, K. S. Smith, J. Rhee, H. Popperl, G. Grosveld, and M. L. Cleary. 2004. The TALE homeodomain protein Pbx2 is not essential for development and long-term survival. Mol. Cell. Biol. 24:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson, T. I., and D. J. Pryce. 2002. Pax 6; a pleiotropic player in development. Bioessays 24:1041-1051. [DOI] [PubMed] [Google Scholar]
- 49.Steelman, S., J. J. Moskow, K. Muzynski, C. North, T. Druck, J. C. Montgomery, K. Huebner, I. O. Daar, and A. M. Buchberg. 1997. Identification of a conserved family of Meis1-related homeobox genes. Genome Res. 7:142-156. [DOI] [PubMed] [Google Scholar]
- 50.Swift, G. H., Y. Liu, S. D. Rose, L. J. Bischof, S. Steelman, A. M. Buchberg, C. V. Wright, and R. J. MacDonald. 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol. Cell. Biol. 9:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, M. D., C. S. Kremer, K. S. Ravichandran, K. Rajewsky, and T. P. Bender. 2005. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity 23:275-286. [DOI] [PubMed] [Google Scholar]
- 52.Treisman, J. E. 2004. How to make an eye. Development 131:3823-3827. [DOI] [PubMed] [Google Scholar]
- 53.Vlachakis, N., S.-K. Choe, and C. G. Sagerstrom. 2001. Meis3 synergizes with Pbx4 and Hoxb1 in promoting hindbrain fates in the zebrafish. Development 128:1299-1312. [DOI] [PubMed] [Google Scholar]
- 54.Waskiewicz, A. J., H. A. Rikhof, R. E. Hernandez, and C. B. Moens. 2001. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128:4139-4151. [DOI] [PubMed] [Google Scholar]
- 55.Zambrowicz, B. P., G. A. Friedrich, E. C. Buxton, S. L. Lilleberg, C. Person, and A. T. Sands. 1998. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392:608-611. [DOI] [PubMed] [Google Scholar]
- 56.Zambrowicz, B. P., A. Abuin, R. Ramirez-Solis, L. J. Richter, J. Piggott, H. BeltrandelRio, E. C. Buxton, J. Edwards, R. A. Finch, C. J. Friddle, A. Gupta, G. Hansen, Y. Hu, W. Huang, C. Jaing, B. W. Key, Jr., P. Kipp, B. Kohlhauff, Z. Q. Ma, D. Markesich, R. Payne, D. G. Potter, N. Qian, J. Shaw, J. Schrick, Z. Z. Shi, M. J. Sparks, I. Van Sligtenhorst, P. Vogel, W. Walke, N. Xu, Q. Zhu, C. Person, and A. T. Sands. 2003. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc. Natl. Acad. Sci. USA 100:14109-14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, J., M. Socolovsky, A. W. Gross, and H. F. Lodish. 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood 102:3938-3946. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, X., A. Friedman, S. Heaney, P. Purcell, and R. L. Maas. 2002. Meis1 homeoprotein directly regulates Pax 6 during vertebrate lens morphogenesis. Genes Dev. 16:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]