Abstract
The MafA transcription factor is both critical to islet β-cell function and has a unique pancreatic cell-type-specific expression pattern. To localize the potential transcriptional regulatory region(s) involved in directing expression to the β cell, areas of identity within the 5′ flanking region of the mouse, human, and rat mafA genes were found between nucleotides −9389 and −9194, −8426 and −8293, −8118 and −7750, −6622 and −6441, −6217 and −6031, and −250 and +56 relative to the transcription start site. The identity between species was greater than 75%, with the highest found between bp −8118 and −7750 (∼94%, termed region 3). Region 3 was the only upstream mammalian conserved region found in chicken mafA (88% identity). In addition, region 3 uniquely displayed β-cell-specific activity in cell-line-based reporter assays. Important regulators of β-cell formation and function, PDX-1, FoxA2, and Nkx2.2, were shown to specifically bind to region 3 in vivo using the chromatin immunoprecipitation assay. Mutational and functional analyses demonstrated that FoxA2 (bp −7943 to −7910), Nkx2.2 (bp −7771 to −7746), and PDX-1 (bp −8087 to −8063) mediated region 3 activation. Consistent with a role in transcription, small interfering RNA-mediated knockdown of PDX-1 led to decreased mafA mRNA production in INS-1-derived β-cell lines (832/13 and 832/3), while MafA expression was undetected in the pancreatic epithelium of Nkx2.2 null animals. These results suggest that β-cell-type-specific mafA transcription is principally controlled by region 3-acting transcription factors that are essential in the formation of functional β cells.
Insulin is an essential hormonal regulator of glucose, fatty acid, and amino acid metabolism in the liver, adipose tissue, muscle, and brain and is exclusively produced in the β cells of the islet of Langerhans. The cis-acting elements controlling β-cell-selective insulin transcription are located within conserved 5′ flanking sequences located from bp −340 to −91 upstream of the transcription initiation site. Several key control elements have been identified within this region, including those activated by the binding of PDX-1 (32, 37, 38), BETA2 (28), Pax6 (42), and MafA (17, 25, 34), respectively.
Gene ablation experiments in mice have shown that PDX-1 (15, 31), BETA2 (27), and Pax6 (3, 42, 46) are critical for pancreatic development. Thus, PDX-1 null mice are apancreatic, while islet β-cell-conditional knockouts develop diabetes in part due to decreased insulin expression (1). In contrast, BETA2 and Pax6 affect relatively late steps in pancreatic development through their specific actions on islet cell formation: Pax6−/−, reduced number of islet α and β cells (3, 42, 46); BETA2−/−, reduced islet β>α>δ cells (27). Furthermore, dysfunctional heterozygous mutations in PDX-1 (45), BETA2 (23), and PAX6 (50) cause diabetes in humans, presumably due to reduced expression of target genes required for proper islet cell function.
In contrast to the other islet-enriched transcriptional regulators of the insulin gene, MafA−/− mice do not appear to manifest a defect in islet cell development, although they are diabetic as adults due to an inability to properly regulate insulin secretion (51). The MafA expression pattern in the pancreas is also unusual, as no other characterized activator factor is expressed exclusively in insulin+ cells or induced at the onset of the secondary and principal wave of islet β-cell formation during embryogenesis (24). Arguably, MafA may be an even better marker for properly functioning β cells than insulin, as this hormone is produced in dysfunctional cells during development (35, 36, 49) and in the adult (18). Interestingly, heterozygous MafA mutant mice are also diabetic (51), a property previously only found for PDX-1 (1).
Because MafA displays novel islet β-cell expression and functional properties, we sought to identify sequences within mammalian mafA that confer β-cell-specific expression. Six potential conserved control regions were found within 25 kb upstream of the transcription initiation site in the mouse, human, and rat. Functional analyses of each demonstrated that only one, termed region 3 (bp −8118 to −7750), mediated β-cell-selective expression in transfected cell lines. The islet-enriched PDX-1, FoxA2, and Nkx2.2 regulatory factors were found to bind in vitro and in vivo to conserved region 3 sequences, a process shown to be important for activation in β cells. These results strongly suggest that region 3 plays a unique role in defining the specific expression pattern of mafA in the pancreas.
MATERIALS AND METHODS
DNA sequence analysis.
Mouse (GenBank NT 039621), rat (GenBank NW 047780), and human (GenBank NT 023684) mafA sequence alignment was performed on approximately 25 kb of DNA upstream and downstream of the translation start site. The VISTA alignment program (26) was used in the analysis, with a window size of 500 bp and a minimal sequence identity of 50%. Potential transcription factor binding sites within conserved chicken (GenBank NW 060306), human, mouse, and rat region 3 were found using the ClustalW application in MacVector 7.0 (Oxford Molecular) and the TRANSFAC program (11).
RNase protection assay.
The mouse mafA bp −35 to +106 promoter fragment was generated by PCR and subcloned into the TOPO TA vector (Invitrogen). A radiolabeled antisense RNA probe was generated using the MAXIscript kit (Ambion) and added to either 10 μg of total RNA from βTC3 cells or 10 μg of yeast control RNA. The RNase protection assay was performed using the RPA III kit (Ambion) according to the manufacturer's instructions. The exact location of the mafA transcription start site was determined from a DNA sequencing reaction run in parallel on the gel.
Transfection constructs.
Mouse mafA 5′ flanking deletion DNAs were obtained by homologous recombination from the bacterial artificial chromosome RP22-260L4 (Children's Hospital Oakland Research Institute) and inserted into the pSVOAPL2L luciferase (LUC) expression vector (22). The bacterial recombination methods were performed as previously described (21). The following primers were used to amplify the 5′ homology arms: R1-6, −10393 (5′-GGACCCCTCTGCCAGGCTTTGTCC-3′) to −10194 (5′-ATCTCAAAATCATCCATGGTGCCA-3′); Δ1-2, −8292 (5′-CAGGCCCCCAGGAGCCCTCGCACC-3′) to −8119 (5′-TGGTCTTCAAGGCAGCCGCTGCTG-3′); Δ1-3, −7033 (5′-CTCCCCAACACCTCAGTTTCCCAT-3′) to −6834 (5′-CAGTGGAGGAGGGCCACGCGGCCC-3′). The 3′ homology arm was amplified using primers from bp +57 (5′-CGGGCGGGAGAGCCCGGAGCGCGG-3′) to +230 (5′-CTGTGCTCAGGGGACGCCGCCGGC-3′). Mouse mafA sequences spanning nucleotides −9940 to −4217 and −510 to +239 were obtained from RP22-260L4, with bp −9940 to −6615, −6615 to −4217, and −5777 to −4217 subcloned directly upstream of the herpes simplex virus minimal thymidine kinase (Tk) promoter region in the chloramphenicol acetyltransferase (CAT) expression vector pTk(An) (13). The bp −510 to +239 promoter fragment was cloned into pSVOAPL2L.
Individual conserved and nonconserved mouse mafA constructs were constructed in pTk(An) by PCR using the following primers: region 1, −9413 (5′-CCGTGTGTAGCCATAGTACCCACTT-3′) to −9163 (5′-AGTGATCTGTCACCTGAGTACGTGG-3′); region 2, −8428 (5′-AGCCCCTTCCTTGGAGCAGGAAGAA-3′) to −8295 (5′-GCCCAGTCTCAGTCTGTCCTCCTGG-3′; region 3, −8120 (5′-CACCCCAGCGAGGGCTGATTTAATT-3′) to −7750 (5′-AGCAAGCACTTCAGTGTGCTCAGTG-3′); region 4, −6624 (5′-TGCAATGGCTTGATGCTTGACCTTT-3′) to −6443 (5′-CCAGCTGGGCTCAGTGGCAGGCTGC-3′); region 5, −6244 (5′-ATCCATCAGGGTGCCATGTGAGGTT-3′) to −6000 (5′-GCCATACTCTTGCCTCCCTTATATG-3′); −3785 (5′-ACTAATGGTGCCTGGATTATAGGC-3′) to −3286 (5′-AGAAGTTCAAGGTCACCCTCAGCTAC-3′); −1779 (5′-AGAGAACTGAAACACTGGCTCTCC-3′) to −1280 (5′-ACAAGGTGGAAAAAGTTTGAAAGG-3′). Noncomplementary transversional mutations (G to T; C to A) were produced in mouse region 3:pTk using the QuikChange mutagenesis kit (Stratagene) with the following oligonucleotides (mutated bases are underlined): FoxA2 mutant, ACGACCCTTTCTGTACCACTTTTACAGCTCTCTG (−7943 to −7910); Nkx2.2 mutant, TGAGCACACTGCCTGTCTTGCTGGAG (−7771 to −7746); PDX-1 mutant, CCCATTTCTGTGCCGTTGTTCTGGA (−8087 to −8063). Restriction enzyme digestion and DNA sequencing were used to verify each construct.
Transient transfections.
Monolayer cultures of pancreatic islet β (βTC3, MIN6, INS-1 [lines 832/3 and 832/13]) and non-β (NIH 3T3) cells were maintained as described previously (7, 12). The INS-1-derived cell lines are functionally similar, except 832/3 cells express five times more Nkx6.1 mRNA than 832/13 cells (43). Cells were transfected using the Lipofectamine reagent (Invitrogen Life Technologies) with 1 μg of mafA:pTk and 1 μg of Rous sarcoma virus enhancer-driven luciferase (pRSV-LUC) (2 μg total). Cellular extracts were collected after 40 to 48 h, and LUC (Promega, Madison, WI) and CAT (29) assays were performed. The pRSV-LUC activity was used to normalize the CAT activity from mafA:pTk. The firefly LUC activity from mafA:pSVOAPL2L (0.5 μg) was normalized to the Renilla luciferase activity of the cotransfected pHRLTk (22) (10 ng) control. Each experiment was performed on several separate occasions with at least two independently prepared plasmid preparations.
Electrophoretic mobility shift assays.
Nuclear extract from βTC3 cells was prepared as described previously (44). The binding analysis was performed with double-stranded DNA oligonucleotides to mouse region 3 sequences. The probe and competitor sequences were as follows (mutated bases are underlined): wild-type FoxA2, −7943 ATTAACGACCCTTTCTGTAAACATTTTACAGCTCTCTG −7910; mutant, ACGACCCTTTCTGTACCACTTTTACAGCTCTCTG; wild-type Nkx2.2, −7771 ATTATGAGCACACTGAAGTGCTTGCTGGAG −7746; mutant, TGAGCACACTGCCTGTCTTGCTGGAG; wild-type PDX-1, −8087 ATTACCCATTTCTGTTAATTTGTTCTGGA −8063; mutant, CCCATTTCTGTGCCGTTGTTCTGGA. The binding reactions were performed at 4°C (20-μl final volume) with 10 μg of nuclear protein and 400 fmol of polynucleotide kinase-labeled 32P-radiolabeled probe in a buffer containing 20 mM HEPES (pH 7.9), 10% glycerol, 20 mM KCl, 50 mM NaCl, 1 mM dithiothreitol, and 1 μg of poly(dI-dC). Competition analysis was performed with a molar excess of unlabeled competitor to labeled probe (wild type, 5- to 50-fold; mutant, 25- to 50-fold). Antibody supershift analyses were performed by preincubating nuclear extract protein with specific FoxA2 (Santa Cruz Biotechnology), Nkx2.2 (Developmental Studies Hybridoma Bank), and PDX-1 (Chris Wright, Vanderbilt University) antibodies for 20 min at 4°C prior to addition of the radiolabeled probe. Samples were electrophoresed on 6% nondenaturing polyacrylamide gels at 150 V for 2 h in 1× TGE buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA). Gels were then dried and visualized by autoradiography.
Chromatin immunoprecipitation assays.
βTC3 cells (∼0.5 × 108 to 1.0 × 108) were formaldehyde cross-linked, and the sonicated chromatin-DNA complexes were isolated as previously described (7). Anti-FoxA2, anti-Nkx2.2, anti-PDX-1, normal rabbit immunoglobulin G (IgG; Santa Cruz Biotechnology), or no antibody was added to the sonicated chromatin, and the antibody-protein-DNA complexes were isolated by incubation with A/G-agarose (Santa Cruz Biotechnology). PCR was performed on 1/10 of the purified immunoprecipitated DNA using Ready-to-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, N.J.) with 15 pmol of each of the following mouse primers: region 3, −8120 (5′-CACCCCAGCGAGGGCTGATTTAATT-3′) to −7750 (5′-AGCAAGCACTTCAGTGTGCTCAGTG-3′); phosphoenolpyruvate carboxykinase (PEPCK), −434 (5′-GAGTGACACCTCACAGCTGTGG-3′) to −96 (5′-GGCAGGCCTTTGGATCATAGCC-3′). The PCR products were confirmed by sequencing with amplified products resolved in a 1.4% agarose gel in TAE buffer and then visualized by ethidium bromide staining. Each experiment was repeated at least three times.
Recombinant adenoviruses, treatment of cells, and RNA purification.
Adenoviruses containing small interfering RNA (siRNA) sequences specific for rat pdx-1 or a random siRNA sequence were prepared as described previously (4, 43). INS-1-derived cell lines (832/13 or 832/3) were plated into 6-cm dishes at 5 × 105 per dish and maintained overnight. Cells were then treated with the various siRNA-containing adenoviruses for 18 h at a titer of 20 infectious units/cell. RNA was harvested 96 h later and purified by a column-based method (RNeasy; Qiagen), and 500 ng was used as a template for cDNA synthesis using the iScript system (Bio-Rad).
RT-PCR analysis.
The mafA mRNA level was quantified by real-time PCR (RT-PCR) with SYBR green PCR chemistry at a final primer concentration of 100 nM (forward, 5′-AGGAGGAGGTCATCCGACTG-3′; reverse, 5′-CTTCTCGCTCTCCAGAATGTG-3′). Prevalidated primer and probe sets based on Taqman chemistry (Applied Biosystems) were used to quantify pdx-1 mRNA (no. Rn00755591_m1; Applied Biosystems) and ribosomal 18S (no. 4308329; Applied Biosystems) levels. Triplicate reactions from five independent RNA samples were carried out in a final volume of 25 μl containing 10 ng of cDNA template.
Mice and immunohistochemistry.
Staged embryos (embryonic day 18.5 [E18.5]) were obtained from Nkx2.2 mutant animals (47). The day of vaginal plug discovery was designated stage E0.5. Immunofluorescence and confocal image analyses were performed on paraffin sections as described previously (25). The primary antibodies used were rabbit anti-MafA (1:1,000, BL658; Bethyl Laboratories, Montgomery, TX) and goat anti-ghrelin (Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were Cy3- or Cy5-conjugated donkey anti-goat and anti-rabbit IgG (1:500; Jackson ImmonoResearch, West Grove, PA). Fluorescent images were captured with a Zeiss LSM 510 confocal microscope using an optical depth of 1 μm. Nuclear counterstaining was performed using YoPro1 (Molecular Probes, Eugene, OR).
RESULTS
Analysis of the flanking regions of mammalian mafA.
As conservation between species often highlights important functional sequences, a bioinformatics approach was used to reveal identity around mammalian mafA. This strategy has been used to identify and characterize other pancreatic transcriptional regulatory domains, including amylase (10), insulin (33), pdx-1 (8, 9), and pax6 (19). Six highly conserved regions were found within 25 kb of noncoding 5′ flanking sequence from human, mouse, and rat mafA, although little to no sequence identity was found within a comparable region 3′ to the mafA coding exon.
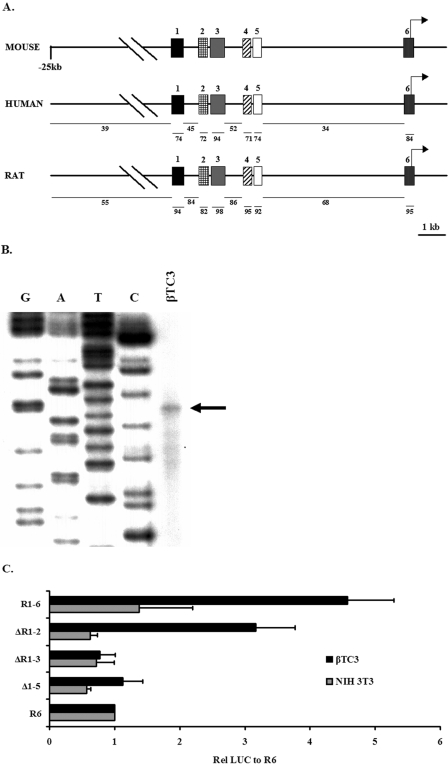
All 5′ conserved regions had greater than 70% identity and were labeled 1 through 6 in relation to their 5′ to 3′ position to the gene (Fig. 1A). Region 3 was the largest and most conserved, with >94% identity over 370 bp. An RNase protection assay was performed with βTC3 cellular RNA to determine the transcription start of mouse mafA (Fig. 1B), allowing assignment of the precise nucleotide location of regions 1 (i.e., −9389 to −9194), 2 (−8426 to −8293), 3 (−8118 to −7750), 4 (−6622 to −6441), 5 (−6217 to −6031), and 6 (−250 to +56) relative to the transcription start site. One initiation site at 407 bp upstream of the translational start site was identified in region 6, with a conserved consensus TATAA site found 22 bp upstream.
FIG. 1.
Analysis of the effects of 5′ flanking deletions spanning the conserved sequence domains of mammalian mafA on β-cell-line-selective expression. (A) Diagram of the human, mouse, and rat 5′ flanking region. The percent identity of the human and rat to mouse is indicated with bars below the locus diagram. Mouse regions 1 (bp −9389 to −9194), 2 (bp −8426 to −8293), 3 (bp −8118 to −7750), 4 (bp −6622 to −6441), 5 (bp −6217 to −6031), and 6 (bp −250 to +56) are numbered relative to the transcription start site determined in panel B. (B) Total RNA from βTC3 cells was hybridized with the mouse antisense mafA bp −35 to +106 probe and then treated with RNase. The arrow in the autoradiograph denotes the mafA RNase protected band, the exact size of which was determined using the M13mp18 plasmid sequencing ladder produced with the M13-40 primer. The same initiation site was determined independently using a bp −35 to +339 probe (data not shown). (C) The 5′ flanking deletion mutants were transfected into β (βTC3) and non-β cells (NIH 3T3). The normalized activity of each construct was presented relative to region 6:pSVOAPL2L ± the standard error of the mean.
To broadly define the transcriptional regulatory properties of the conserved mafA regions, a series of 5′ deletion constructs were constructed and analyzed in transfected β (βTC-3) and non-β (NIH 3T3) cells. The region 1 to 6 and region 3 to 6 spanning reporters were fourfold more active in β cells than the proximal promoter region 6, which displayed no cell-type-specific activity (Fig. 1C). Cell-selective stimulation was also lost upon deletion of bp −10393 to −7033, consistent with regions 1, 2, and/or 3 being important in β-cell-specific expression of mafA.
Region 3 alone confers islet β-cell-line-selective expression.
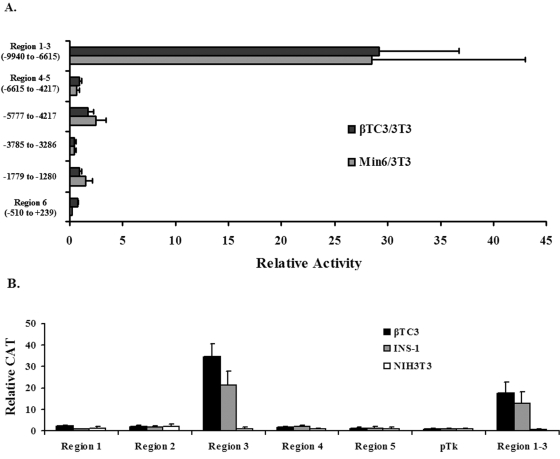
To more precisely define the sequences necessary for cell-specific activity, conserved and nonconserved sequences within 10 kb upstream of the mouse mafA promoter were subcloned into the Tk minimal promoter driven expression plasmid, pTk(An), and transfected into β and non-β cell lines. The mafA construct spanning regions 1 to 3 was selectively activated in a β-cell-specific manner in this heterologous system (Fig. 2A), whereas little or no cell-specific stimulation was observed from regions 4 to 5, region 6, or nonconserved sequence-driven constructs. These results further supported a role for sequences within the area of regions 1, 2, and/or 3 in mafA-mediated activation.
FIG. 2.
Region 3 imparts β-cell-line-specific activation. (A) Mouse mafA:pTk constructs driven by regions 1 to 3 (bp −9940 to −6615), regions 4 and 5 (bp −6615 to −4217), region 6 (bp −510 to +239), and nonconserved (bp −5777 to −4217, bp −3785 to −3286, and bp −1779 to −1280) sequences were transfected into βTC3, MIN6, and NIH 3T3 cells. The ratio of the normalized activity of mafA:pTk to pTk and mafA:pSVOAPL2L to pSVOAPL2L were calculated for each cell line. Results are presented as the relative activity of each construct ± the standard error of the mean in β cells divided by the NIH 3T3 cell activity. (B) Mouse region 1, 2, 3, 4, 5, and 1 to 3 mafA:pTk constructs were transfected into βTC3, INS-1 (832/3), and NIH 3T3 cells. The ratio of the normalized activity of mafA:pTk to pTk was calculated for each cell line. Results are presented as the relative activity of each construct ± the standard error of the mean.
To directly determine how region 1, 2, or 3 was involved in activation, the ability of each to independently mediate pTk activity was analyzed. Strikingly, region 3 was uniquely activated in β cells, with a stimulation level comparable to regions 1 to 3 (Fig. 2B). In contrast, region 1 or 2 was as inactive as region 4 or 5 in β or non-β cells. Taken together, these results strongly suggest that region 3 is sufficient to support pancreatic β-cell-selective transcription.
Region 3 contains PDX-1, FoxA2, and Nkx2.2 binding sites.
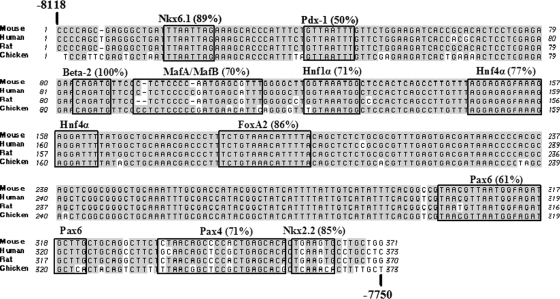
The identity within region 3 between rat, mouse, and human mafA was higher than the other mafA conserved regions, which ranged from 74 to 94% (Fig. 1A). In fact, region 3 was also highly conserved in chicken mafA, with 88% identity to the human sequence (Fig. 3), while there was no significant likeness to region 1, 2, 4, 5, or 6 (<40% identity to mouse mafA). The 94% identity level of region 3 between mice and humans was much greater than in other known islet β-cell-selective control regions, like insulin (63% between bp −350 to −90 of human and mouse I or mouse II) (22) and pdx-1 (78% between bp −2139 to −1958 [area II] of human and mouse) (8). As chicken mafA is also expressed in the pancreas (20), these results provided further support for region 3 playing an essential role in regulating tissue-specific expression.
FIG. 3.
Sequence identity within region 3 between mouse, human, rat, and chicken mafA. Mouse, human, rat, and chicken mafA region 3 sequences were aligned in MacVector 7.0 using the ClustalW application. The shaded sequences are conserved between all species. The numbering is relative to the mouse mafA transcription start site. The potential Nkx6.1 (−8101 to −8094), PDX-1 (−8078 to −8071), BETA2 (−8036 to −8031), MafA and/or MafB (−8026 to −8008), Hnf-1α (−8000 to −7992), Hnf-4α (−7973 to −7955), FoxA2 (−7934 to −7920), Pax6 (−7818 to −7797), Pax4 (−7784 to −7764), and Nkx2.2 (−7762 to −7757) regulatory factor binding sites identified by TRANSFAC database analysis of mouse region 3 is shown, with the percent identity to the factor consensus binding site denoted. The identity to the Nkx2.2 consensus site was reduced in the human and chicken gene to 73% and 43%, respectively.
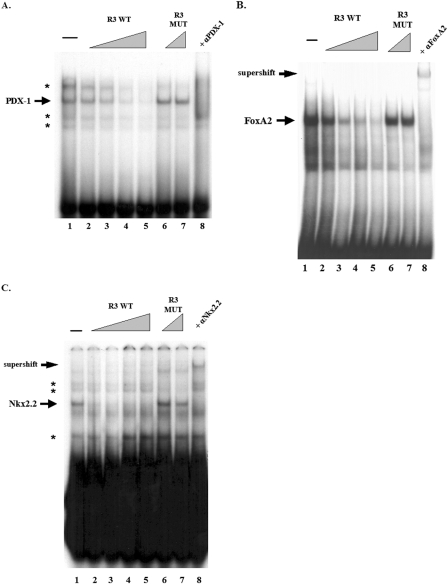
The TRANSFAC database program was used to localize potential islet-enriched transcription factor binding sites in region 3. Since MafA is selectively expressed in β cells within the pancreas (24) and essential to their function (51), potential regulatory sites for factors involved in these processes were expected (Fig. 3). To specifically determine if PDX-1, FoxA2, and Nkx2.2 actually bound to mafA sequences, gel mobility shift assays were first performed with βTC-3 nuclear extract using mouse nucleotide −8087 to −8063 (i.e., for PDX-1), −7943 to −7910 (FoxA2), and −7771 to −7746 (Nkx2.2) probes. Both antibody addition and wild-type and mutant site competition experiments demonstrated that PDX-1, FoxA2, and Nkx2.2 were capable of specifically binding in vitro to region 3 (Fig. 4).
FIG. 4.
Identification of FoxA2, Nkx2.2 and PDX-1 binding sites in region 3. Sequences spanning −8087 to −8063 (PDX-1) (A), −7943 to −7910 (FoxA2) (B), and −7771 to −7746 (Nkx2.2) (C) were used in gel shift binding assays with βTC3 nuclear extract. The specificity of protein-DNA complex formation was determined by competition with a 5- to 50-fold molar excess of unlabeled wild-type competitor (R3 WT, lanes 2 to 5) or a 25- to 50-fold molar excess of binding mutant competitor (R3 MUT, lanes 6 and 7). In addition, preincubation of nuclear extract with anti-PDX-1 (A), anti-FoxA2 (B), or anti-Nkx2.2 (C) antibodies were found to influence the formation (anti-PDX-1 [αPDX-1]) or mobility (anti-FoxA2 [αFoxA2] and anti-Nkx2.2 [αNkx2.2]) of the factor-specific complex (lane 8). Arrowheads indicate the specific binding and supershifted complexes, with the asterisks denoting the nonspecific complexes.
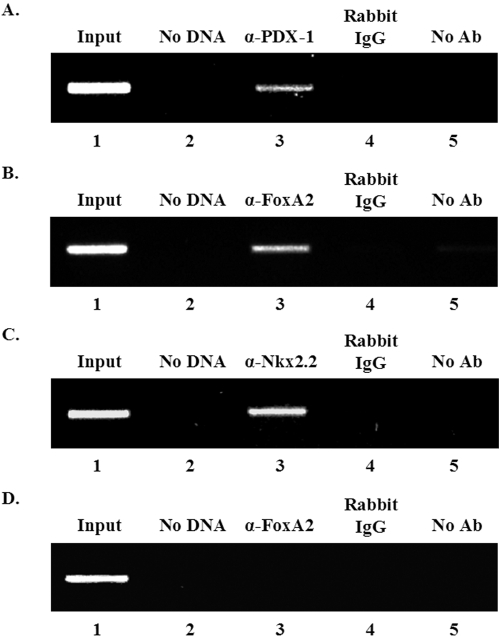
The chromatin immunoprecipitation assay was next used to examine whether PDX-1, FoxA2, and Nkx2.2 bound to region 3 within the context of endogenous mafA. Region 3 sequences were selectively amplified by PCR from formaldehyde cross-linked β-cell chromatin precipitated with antibodies specific to FoxA2, Nkx2.2, and PDX-1 but not from chromatin treated with rabbit IgG or in the absence of antibody (Fig. 5). In contrast, the anti-FoxA2, anti-Nkx2.2, and anti-PDX-1 antisera did not immunoprecipitate promoter sequences from the PEPCK gene, which is not transcribed in β cells (Fig. 5D; also data not shown). These results strongly suggest that FoxA2, Nkx2.2, and PDX-1 bind to region 3 in β cells.
FIG. 5.
FoxA2, Nkx2.2, and PDX-1 bind to region 3 of endogenous mafA in β cells. Formaldehyde cross-linked chromatin from βTC3 cells was incubated with antibodies specific to PDX-1 (lane 3) (A), FoxA2 (lane 3) (B), and Nkx2.2 (lane 3) (C). Immunoprecipitated DNA was analyzed by PCR using primers specific to region 3 (A to C) and PEPCK (D). As controls, PCRs were performed with total input DNA (1/100 dilution, all panels, lane 1), no DNA (all panels, lane 2), DNA immunoprecipitated with rabbit IgG (all panels, lane 4), and DNA that was precipitated in the absence of antibody (all panels, lane 5). Only the anti-FoxA2 (α-FoxA2) results are shown with PEPCK (D), although the same pattern was found with the anti-Nkx2.2 (α-Nkx2.2) and anti-PDX-1 (α-PDX-1) antibody precipitates (data not shown). Each experiment was repeated on three separate occasions.
PDX-1, FoxA2, and Nkx2.2 activate region 3-driven expression in β cells.
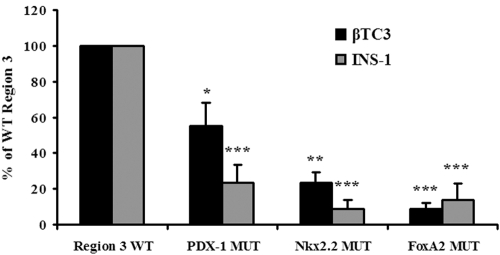
To understand how PDX-1, FoxA2, and Nkx2.2 regulate region 3-driven expression, mutations that specifically disrupt transcription factor binding were incorporated into the region 3:pTk construct. The FoxA2 site mutant led to the largest decrease from the wild type in transfected β cells, although region 3:pTk activity was also substantially decreased in the Nkx2.2 and PDX-1 site mutants (Fig. 6). These results indicate that FoxA2, Nkx2.2, and PDX-1 function as positive regulators of mafA expression in β cells.
FIG. 6.
FoxA2, Nkx2.2, and PDX-1 binding stimulate region 3 activity in β cell lines. βTC3 and INS-1 (832/13) cells were transfected with the wild type (WT) and PDX-1, FoxA2, or Nkx2.2 binding site mutant (MUT) versions of region 3:pTk. Normalized region 3:pTk activity ± standard error of the mean of each mutant is presented as a percentage of the region 3 WT activity. Asterisks denote that there was a statistically significant decrease between PDX-1, Nkx2.2, FoxA2, and the WT in a Student t test: *, P < 0.05; **, P < 0.005; ***, P < 0.001. The data were compiled from the results from at least six independently performed transfections.
Endogenous mafA expression is compromised in the absence of PDX-1 and Nkx2.2.
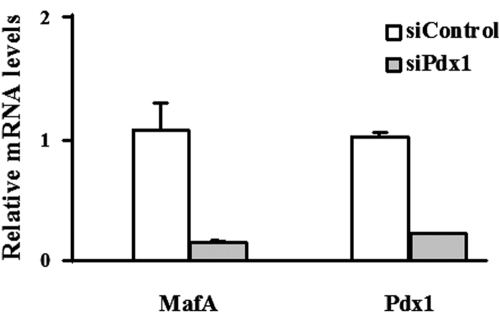
To determine how limiting PDX-1 levels influenced mafA expression, INS-1 (832/13) cells were treated with a recombinant adenovirus containing an siRNA specific for PDX-1. Under these conditions, basal and glucose-stimulated insulin secretion is compromised (43). In addition, we found that mafA mRNA levels were also reduced by suppression of PDX-1 synthesis in these cells (Fig. 7). Similar findings were obtained in an independent INS-1-derived cell line, 832/3 (data not shown).
FIG. 7.
mafA mRNA levels are decreased by siRNA-mediated knockdown of pdx-1. INS-1 (832/13) cells were treated with recombinant adenoviruses expressing siControl or siPDX-1. The effects on pdx-1 and mafA mRNA were analyzed by RT-PCR. The mafA and pdx-1 values are normalized to those for siControl-treated cells and presented as means ± standard errors of the means. Each experiment was performed in triplicate and repeated five times.
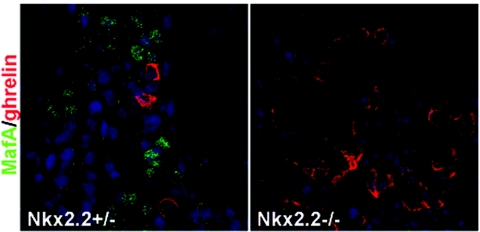
MafA expression was next analyzed in Nkx2.2-deficient animals. Disruption of the Nkx2.2 gene leads to a loss in β-cell neogenesis during the secondary transition, resulting in hyperglycemia and profoundly increased numbers of ghrelin-producing islet ɛ cells (39, 47). Furthermore, PDX-1 levels are decreased in null mice, and there is no detectable expression of Nkx6.1, Pax6, or GLUT2. MafA was also not detected in the pancreatic epithelium of Nkx2.2−/− mice (Fig. 8). Collectively, these results demonstrate that mafA expression is compromised upon reducing PDX-1 and Nkx2.2.
FIG. 8.
MafA is not present in Nkx2.2−/− pancreata. Sections of E18.5 wild-type and Nkx2.2 mutant mice were stained with anti-MafA (green) and anti-ghrelin (red). MafA was not detected in the mutant nor in wild-type ghrelin+ cells. Nuclei were stained blue.
DISCUSSION
The present study provides insight into the mechanisms directing mafA transcription to islet β cells. Expression of this key regulator is novel in relation to other islet transcription factors in being produced exclusively in insulin+ cells of the developing and adult pancreas. Six areas of identity were found within the 5′ flanking region of mouse, rat, and human mafA. However, only region 3, which is located between nucleotides −8118 and −7750 in mouse mafA, was capable of independently driving pancreatic β-cell-line-selective reporter gene expression. In addition, region 3 was the most highly conserved mafA sequence domain found between these mammalian genes, with a level of identity greater than the β-cell-specific control domains of pdx-1 and insulin. These results suggested that the factors acting upon region 3 specifically direct mafA transcription to β cells. Analysis of potential binding sites within this region indicates that many factors important to β-cell formation and function may be involved in regulation. Among these, evidence was provided for PDX-1, FoxA2, and Nkx2.2 having a direct role in region 3 activation.
MafA expression has been most extensively studied in birds, where it is found in cells of the nervous system, lens, and pancreas (20). Although MafA is expressed in the same tissues, differences may exist at the cellular level between birds and mammals (16, 20). As a consequence, it was quite interesting that region 3 was the only conserved 5′ flanking sequence domain retained in chicken mafA, whose position was further upstream than in mammals (i.e., between bp −14312 and −13942 versus bp −8525 and −8157 [mouse] relative to the start of translation). In contrast, chicken pdx-1 retains 3 of the 4 mammalian conserved 5′ flanking domains (i.e., areas I [bp −2839 to 2520 in the mouse gene], III [bp −1939 to 1664], and IV [bp −6200 to −5670]). Of these, areas I, II, and IV can all independently direct β-cell-line-selective expression, with at least areas I and II mediating mammalian islet cell-selective expression in vivo (40, 48).
Our mutational and functional data strongly suggest that region 3 is the only conserved domain upstream of mafA that is capable of independently directing β-cell-specific transcription. Several potential sites for factors associated with β-cell development and function were found in region 3 (Fig. 3), with our biochemical data directly associating PDX-1, FoxA2, and Nkx2.2 in activation. Further support for PDX-1 and Nkx2.2 in regulation was provided via our demonstration that mafA expression was reduced in response to suppression of either of these factors. Potential binding sites for other islet-enriched transcription factors were found within region 3, some of which are also found in nonpancreatic tissues associated with mafA transcription (e.g., nervous system [BETA2 and Nkx6.1] and lens [MafB and Pax6]). Regrettably, β-cell-specific expression is unlikely to be defined by these potential regulators or PDX-1, FoxA2, and Nkx2.2 due to their broad expression pattern during pancreas development (14). Perhaps the exception is the closely related MafB protein, whose expression precedes MafA and is essentially only found in insulin+ and glucagon+ cells during development (2).
The Nkx2.2 binding site mutant profoundly reduced region 3 activity, which is consistent with the complete loss of MafA in the Nkx2.2 null animals (Fig. 8). Because Nkx2.2 binds to region 3 in β cells and activates mafA-driven expression, the combined data strongly suggest an important role in mafA expression in vivo. Alternatively, since the loss of β cells in the Nkx2.2 mutant reflects an endocrine cell fate switch to ghrelin-producing ɛ cells (39), the absence of MafA may be only indirectly related to Nkx2.2 control. Experiments to remove Nkx2.2 during pancreatic development and in adults are planned with recently generated Nkx2.2loxP/loxP mice to further address the significance of this regulator in mafA expression in vivo.
Previously, it had been shown that a large Maf protein activates pdx-1 expression through binding to conserved sequences in the mammalian-specific area II control domain (41). Here we present evidence that PDX-1 regulates mafA expression through region 3. These data indicate that PDX-1 and MafA might work together in a positive feedback loop in the developing pancreas and/or mature islet β cell. This mode of regulation has been demonstrated for other transcription factors important in liver and pancreatic β-cell function. As an example, the maturity-onset diabetes of the young (MODY) factors HNF-1α (MODY3) and HNF-4α (MODY1) form a positive feedback loop in these tissues (5, 6, 30). It has been hypothesized that MODY is caused by disrupting this feedback loop and thus influencing the expression of target genes involved in glycemic control in heterozygous HNF-1α or HNF-4α mutant individuals (6). Interestingly, the phenotype of the β-cell-specific PDX-1 (1) and MafA (51) knockouts are similar, as both have decreased β-cell numbers, compromised β-cell function, and altered islet architecture. Their coregulatory nature is also suggested by the finding that hyperglycemia is only found in heterozygous mutant mice of PDX-1 and MafA and not in other islet-enriched regulators that cause diabetes in humans. If so, MafB may be regulating early pdx-1 expression in the islet and thus initiating the feedback loop, as this factor is produced in first and second wave insulin+ cells during pancreatic organogenesis (2).
The unique ability of region 3 to direct β-cell-line-specific reporter gene expression indicates that the transcription factors important for its activation are also essential in making the mafA locus competent for transcription. Our observations strongly suggest that the programs controlling mafA transcription during pancreatic differentiation and in adult β cells are principally defined by cell-enriched regulators, like FoxA2, Nkx2.2, and PDX-1. Because of MafA's unusual islet expression pattern and significance in β-cell function, we believe that understanding how region 3-mediated activation is controlled will provide insight into the role that characterized (and likely novel) transcription factors play in regulating mafA transcription and pancreas development normally and may help to identify heritable defects that cause insulin deficiency and diabetes.
Acknowledgments
The goat anti-PDX-1 antibody was kindly provided by C. V. E. Wright, and EL250 cells were provided by D. Mortlock.
This work was supported by grants from the National Institutes of Health (DK50203 to R.S., U19 DK061248 to L.S., and U01-DK-56047 to C.B.N.), the Juvenile Diabetes Research Foundation (postdoctoral grant no. 10-2006-5 to I.A.), and the Vanderbilt Molecular Endocrinology Training Program (5T32 DK07563 to J.R.). Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (Public Health Service grant P60 DK20593).
REFERENCES
- 1.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, and H. Edlund. 1998. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artner, I., J. Le Lay, Y. Hang, L. Elghazi, J. C. Schisler, E. Henderson, B. Sosa-Pineda, and R. Stein. 2006. MafB: an activator of the glucagon gene expressed in developing islet {alpha}- and {beta}-cells. Diabetes 55:297-304. [DOI] [PubMed] [Google Scholar]
- 3.Ashery-Padan, R., X. Zhou, T. Marquardt, P. Herrera, L. Toube, A. Berry, and P. Gruss. 2004. Conditional inactivation of Pax6 in the pancreas causes early onset of diabetes. Dev. Biol. 269:479-488. [DOI] [PubMed] [Google Scholar]
- 4.Bain, J. R., J. C. Schisler, K. Takeuchi, C. B. Newgard, and T. C. Becker. 2004. An adenovirus vector for efficient RNA interference-mediated suppression of target genes in insulinoma cells and pancreatic islets of langerhans. Diabetes 53:2190-2194. [DOI] [PubMed] [Google Scholar]
- 5.Boj, S. F., M. Parrizas, M. A. Maestro, and J. Ferrer. 2001. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc. Natl. Acad. Sci. USA 98:14481-14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrer, J. 2002. A genetic switch in pancreatic beta-cells: implications for differentiation and haploinsufficiency. Diabetes 51:2355-2362. [DOI] [PubMed] [Google Scholar]
- 7.Gerrish, K., M. A. Cissell, and R. Stein. 2001. The role of hepatic nuclear factor 1 alpha and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 276:47775-47784. [DOI] [PubMed] [Google Scholar]
- 8.Gerrish, K., M. Gannon, D. Shih, E. Henderson, M. Stoffel, C. V. Wright, and R. Stein. 2000. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J. Biol. Chem. 275:3485-3492. [DOI] [PubMed] [Google Scholar]
- 9.Gerrish, K., J. C. Van Velkinburgh, and R. Stein. 2004. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol. Endocrinol. 18:533-548. [DOI] [PubMed] [Google Scholar]
- 10.Gumucio, D. L., K. Wiebauer, R. M. Caldwell, L. C. Samuelson, and M. H. Meisler. 1988. Concerted evolution of human amylase genes. Mol. Cell. Biol. 8:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinemeyer, T., X. Chen, H. Karas, A. E. Kel, O. V. Kel, I. Liebich, T. Meinhardt, I. Reuter, F. Schacherer, and E. Wingender. 1999. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 27:318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohmeier, H. E., H. Mulder, G. Chen, R. Henkel-Rieger, M. Prentki, and C. B. Newgard. 2000. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49:424-430. [DOI] [PubMed] [Google Scholar]
- 13.Jacoby, D. B., N. D. Zilz, and H. C. Towle. 1989. Sequences within the 5′-flanking region of the S14 gene confer responsiveness to glucose in primary hepatocytes. J. Biol. Chem. 264:17623-17626. [PubMed] [Google Scholar]
- 14.Jensen, J. 2004. Gene regulatory factors in pancreatic development. Dev. Dyn. 229:176-200. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606-609. [DOI] [PubMed] [Google Scholar]
- 16.Kajihara, M., H. Sone, M. Amemiya, Y. Katoh, M. Isogai, H. Shimano, N. Yamada, and S. Takahashi. 2003. Mouse MafA, homologue of zebrafish somite Maf 1, contributes to the specific transcriptional activity through the insulin promoter. Biochem. Biophys. Res. Commun. 312:831-842. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka, K., S. I. Han, S. Shioda, M. Hirai, M. Nishizawa, and H. Handa. 2002. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 277:49903-49910. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura, Y. I., T. Kitamura, J. P. Kruse, J. C. Raum, R. Stein, W. Gu, and D. Accili. 2005. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2:153-163. [DOI] [PubMed] [Google Scholar]
- 19.Kleinjan, D. A., A. Seawright, A. J. Childs, and V. van Heyningen. 2004. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev. Biol. 265:462-477. [DOI] [PubMed] [Google Scholar]
- 20.Lecoin, L., K. Sii-Felice, C. Pouponnot, A. Eychene, and M. P. Felder-Schmittbuhl. 2004. Comparison of maf gene expression patterns during chick embryo development. Gene Expr. Patterns 4:35-46. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 22.Le Lay, J., T. A. Matsuoka, E. Henderson, and R. Stein. 2004. Identification of a novel PDX-1 binding site in the human insulin gene enhancer. J. Biol. Chem. 279:22228-22235. [DOI] [PubMed] [Google Scholar]
- 23.Malecki, M. T., U. S. Jhala, A. Antonellis, L. Fields, A. Doria, T. Orban, M. Saad, J. H. Warram, M. Montminy, and A. S. Krolewski. 1999. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 23:323-328. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka, T. A., I. Artner, E. Henderson, A. Means, M. Sander, and R. Stein. 2004. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. USA 101:2930-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuoka, T. A., L. Zhao, I. Artner, H. W. Jarrett, D. Friedman, A. Means, and R. Stein. 2003. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol. Cell. Biol. 23:6049-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayor, C., M. Brudno, J. R. Schwartz, A. Poliakov, E. M. Rubin, K. A. Frazer, L. S. Pachter, and I. Dubchak. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046-1047. [DOI] [PubMed] [Google Scholar]
- 27.Naya, F. J., H. P. Huang, Y. Qiu, H. Mutoh, F. J. DeMayo, A. B. Leiter, and M. J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 11:2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naya, F. J., C. M. Stellrecht, and M. J. Tsai. 1995. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9:1009-1019. [DOI] [PubMed] [Google Scholar]
- 29.Nordeen, S. K., P. P. Green III, and D. M. Fowlkes. 1987. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA 6:173-178. [DOI] [PubMed] [Google Scholar]
- 30.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Offield, M. F., T. L. Jetton, P. A. Labosky, M. Ray, R. W. Stein, M. A. Magnuson, B. L. Hogan, and C. V. Wright. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122:983-995. [DOI] [PubMed] [Google Scholar]
- 32.Ohlsson, H., K. Karlsson, and T. Edlund. 1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohneda, K., H. Ee, and M. German. 2000. Regulation of insulin gene transcription. Semin. Cell Dev. Biol. 11:227-233. [DOI] [PubMed] [Google Scholar]
- 34.Olbrot, M., J. Rud, L. G. Moss, and A. Sharma. 2002. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. USA 99:6737-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oster, A., J. Jensen, P. Serup, P. Galante, O. D. Madsen, and L. I. Larsson. 1998. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1). J. Histochem. Cytochem. 46:707-715. [DOI] [PubMed] [Google Scholar]
- 36.Pang, K., C. Mukonoweshuro, and G. G. Wong. 1994. Beta cells arise from glucose transporter type 2 (Glut2)-expressing epithelial cells of the developing rat pancreas. Proc. Natl. Acad. Sci. USA 91:9559-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peshavaria, M., L. Gamer, E. Henderson, G. Teitelman, C. V. Wright, and R. Stein. 1994. XIHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol. Endocrinol. 8:806-816. [DOI] [PubMed] [Google Scholar]
- 38.Petersen, H. V., P. Serup, J. Leonard, B. K. Michelsen, and O. D. Madsen. 1994. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc. Natl. Acad. Sci. USA 91:10465-10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prado, C. L., A. E. Pugh-Bernard, L. Elghazi, B. Sosa-Pineda, and L. Sussel. 2004. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. USA 101:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samaras, S. E., M. A. Cissell, K. Gerrish, C. V. Wright, M. Gannon, and R. Stein. 2002. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in pancreatic beta cells: role for hepatocyte nuclear factor 3 beta and Pax6. Mol. Cell. Biol. 22:4702-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samaras, S. E., L. Zhao, A. Means, E. Henderson, T. A. Matsuoka, and R. Stein. 2003. The islet beta cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J. Biol. Chem. 278:12263-12270. [DOI] [PubMed] [Google Scholar]
- 42.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11:1662-1673. [DOI] [PubMed] [Google Scholar]
- 43.Schisler, J. C., P. B. Jensen, D. G. Taylor, T. C. Becker, F. K. Knop, S. Takekawa, M. German, G. C. Weir, D. Lu, R. G. Mirmira, and C. B. Newgard. 2005. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc. Natl. Acad. Sci. USA 102:7297-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoffers, D. A., J. Ferrer, W. L. Clarke, and J. F. Habener. 1997. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 17:138-139. [DOI] [PubMed] [Google Scholar]
- 46.St-Onge, L., B. Sosa-Pineda, K. Chowdhury, A. Mansouri, and P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature 387:406-409. [DOI] [PubMed] [Google Scholar]
- 47.Sussel, L., J. Kalamaras, D. J. Hartigan-O'Connor, J. J. Meneses, R. A. Pedersen, J. L. Rubenstein, and M. S. German. 1998. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125:2213-2221. [DOI] [PubMed] [Google Scholar]
- 48.Van Velkinburgh, J. C., S. E. Samaras, K. Gerrish, I. Artner, and R. Stein. 2005. Interactions between areas I and II direct pdx-1 expression specifically to islet cell types of the mature and developing pancreas. J. Biol. Chem. 280:38438-38444. [DOI] [PubMed] [Google Scholar]
- 49.Wilson, M. E., J. A. Kalamaras, and M. S. German. 2002. Expression pattern of IAPP and prohormone convertase 1/3 reveals a distinctive set of endocrine cells in the embryonic pancreas. Mech. Dev. 115:171-176. [DOI] [PubMed] [Google Scholar]
- 50.Yasuda, T., Y. Kajimoto, Y. Fujitani, H. Watada, S. Yamamoto, T. Watarai, Y. Umayahara, M. Matsuhisa, S. Gorogawa, Y. Kuwayama, Y. Tano, Y. Yamasaki, and M. Hori. 2002. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes 51:224-230. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, C., T. Moriguchi, M. Kajihara, R. Esaki, A. Harada, H. Shimohata, H. Oishi, M. Hamada, N. Morito, K. Hasegawa, T. Kudo, J. D. Engel, M. Yamamoto, and S. Takahashi. 2005. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 25:4969-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]