Abstract
Glycine receptors are ligand-gated chloride channels that mediate inhibitory neurotransmission in the adult nervous system. During development, glycine receptor alpha 2 (GlyRα2) is expressed in the retina, in the spinal cord, and throughout the brain. Within the cortex, GlyRα2 is expressed in immature cells and these receptors have been shown to be active and excitatory. In the developing retina, inhibition of glycine receptor activity prevents proper rod photoreceptor development. These data suggest that GlyRα2, the developmentally expressed glycine receptor, may play an important role in neuronal development. We have generated mice with a targeted deletion of glycine receptor alpha 2 (Glra2). Although these mice lack expression of GlyRα2, no gross morphological or molecular alterations were observed in the nervous system. In addition, the cerebral cortex does not appear to require glycine receptor activity for proper development, as Glra2 knockout mice did not show any electrophysiological responses to glycine.
In the adult nervous system, inhibitory neurotransmission is mediated by γ-aminobutyric acid (GABA) and glycine. Glycine receptors are ligand-gated chloride channels that are composed of pentamers of alpha and beta subunits. Four alpha subunits (α1, α 2, α3, and α4) and one beta (β) subunit are present in the murine genome. In the mature nervous system, glycine receptors are heteropentamers composed of alpha subunits and beta subunits. Glycine receptors were initially described as being composed of three alpha subunits, which contained the ligand binding domain, and two beta subunits (5, 13). However, a recent study provides evidence that glycine receptor heteropentamers are composed of two alpha subunits and three beta subunits, with both the alpha and beta subunits contributing to the ligand binding domain (10). Homopentamers of alpha subunits also have been identified, and these forms were shown to be expressed extrasynaptically (28, 30). In the mature nervous system, glycine receptors are expressed at high levels in the spinal cord and brain stem and at lower levels in the thalamus and hypothalamus (reviewed in reference 17). The α1 and β subunits of the glycine receptor are expressed at very low levels embryonically, with increasing expression from birth to 2 weeks after birth and maintenance of these high levels thereafter (18). GlyRα3 expression mirrors GlyRα1 expression, although at lower levels (18). In contrast, GlyRα2 expression is high and widespread in the embryonic nervous system and decreases in the mature brain (18).
Defects in glycine receptor function in humans, mice, and cattle lead to startle disease phenotypes. These disorders are characterized by transient muscle rigidity in response to unexpected stimuli. Human startle disease, or hyperekplexia, has been genetically linked to glycine receptor function. Several dominant mutations have been identified in both the glycine receptor alpha 1 (Glra1) and beta (Glrb) loci (reviewed in reference 17). These mutations lead to either a reduction in levels of glycine receptors or impairment of glycine receptor chloride conductance. Two recessive mutations also have been identified in Glra1 that, when homozygous, lead to complete loss of GlyRα1 expression (4, 25). Interestingly, these mutations are no more severe than the dominant point mutations.
In addition to human disease, there are three naturally occurring mouse startle disease mutants: spastic, spasmodic, and oscillator. The spastic mutant contains a transposon insertion in Glrb, which results in aberrant splicing and a severe reduction in GlyRβ protein levels (11, 21). Spasmodic mice harbor a point mutation in Glra1 that results in a sixfold decrease in glycine sensitivity (26, 27). The oscillator mutant harbors a frameshift mutation in Glra1 that results in a drastic reduction in GlyRα1 cell surface expression (12). This is the only glycine receptor mutation that leads to a lethal phenotype. Each of these three mouse mutants appears phenotypically normal up to 2 weeks after birth. Phenotypic startle responses appear at 3 weeks, corresponding to the developmental switch in subunit expression from GlyRα2 to GlyRα1, leading to the suggestion that developmental GlyRα2 expression is responsible for the delay in phenotypic onset (2).
Ligand binding to glycine receptors induces channel pore opening, followed by chloride ion diffusion through the pore. In mature adult neurons, this results in chloride ion influx and hyperpolarization of the membrane. In contrast, during embryonic development, the chloride ion concentration is higher intracellularly than in the extracellular environment. Therefore, glycine receptor activation embryonically leads to chloride ion efflux from the cell and calcium ion influx (8). Embryonically, both GlyRα2 homopentamers and GlyRα2 and GlyRβ heteropentamers are expressed (1, 14). GlyRα2 is expressed in the cortex before functional synapses are formed (18). Flint et al. have shown that these extrasynaptic glycine receptors are excitatory (8). The functional implications of the developmental physiology of glycine receptors have not been established.
Previously, we showed that transient knockdown of GlyRα2 at postnatal day 0 (P0) with short hairpin RNA in the rat retina inhibits the production of rod photoreceptors (34). At P0, ∼85% of the cells that exit mitosis become rod photoreceptors (24). As GlyRα2 is expressed throughout the central nervous system during development, we wanted to determine whether it is involved more generally in neuronal development. In order to further examine the role of GlyRα2, we generated mice that have a targeted deletion within the Glra2 genomic locus that results in a complete loss of GlyRα2 expression. Glra2−/− mice are viable and fertile. Morphological and molecular analyses of developing and adult retinae and brains reveal no profound phenotype. In the retina and spinal cord, expression of other glycine receptor subunits may compensate for the lack of a profound phenotype. However, electrophysiology of the embryonic cortex showed no responses to glycine in Glra2−/− animals.
MATERIALS AND METHODS
Generation of Glra2 knockout (KO) mice.
Mice were housed and cared for under the guidelines set up by Harvard University in compliance with federal standards.
Bacterial artificial chromosome (BAC) filters containing 129/Sv mouse genomic DNA inserts were purchased from Genome Systems Inc. and hybridized with a probe encoding exons 6 and 7 of mouse glycine receptor alpha 2 (GlyRα2). BAC end sequencing and Southern blot restriction mapping revealed a 160-kb insert that contained genomic DNA that included GlyRα2 exons 3 to 8. In order to replace exons 6 and 7 with an FLP recombination target (FRT)-flanked neomycin cassette within the BAC, the methodology for bacterial recombination developed by Stewart et al. was employed (22). A PGK-neo cassette was PCR amplified with primers that added 62-bp homology arms to each side of the PGK-neo cassette. The sequences of these homology arms are TACATTTTGCCTTTTCTCAGAAAAGATACATTGTTTGTTACTACTTCTTT and CGGAAGTGTGCTTTCGACAAGACCTTTGGAAAGCCTCTGAGGTCACCTG. The amplified PGK-neo cassette with the homology arms was electroporated into bacteria containing the BAC and recombination machinery. Homologous recombination within the BAC resulted in the deletion of a 2.3-kb fragment containing both exons 6 and 7 and replacement with the PGK-neo cassette. The vector sequence was removed from the BAC by cleavage with NotI, and the modified BAC insert was purified. This fragment was electroporated into 129/SvJ embryonic stem (ES) cells. Homologously recombined ES cell clones were analyzed by PCR and Southern blotting for the presence of Glra2 exon 9 and the absence of Glra2 exons 6 and 7. The Glra2 exon 6/7 probe was generated and sequence verified by PCR amplification with the primers TATTGCACAAAGCATTACAACA and CATTTTACCTTCCTATTTTATTTTG.
Homologously recombined ES cells were injected into blastocysts to generate chimeric animals. F1 animals were genotyped by Southern blot analysis with probe 1 (Fig. 1). Probe 1 was generated by PCR amplification of genomic DNA with primers TCATTGGGGTTCTTTGCCTTAG and AATGAGCCTTGGTTTGTCTTATTG. The resultant PCR product was subcloned into pBluescript and sequence verified.
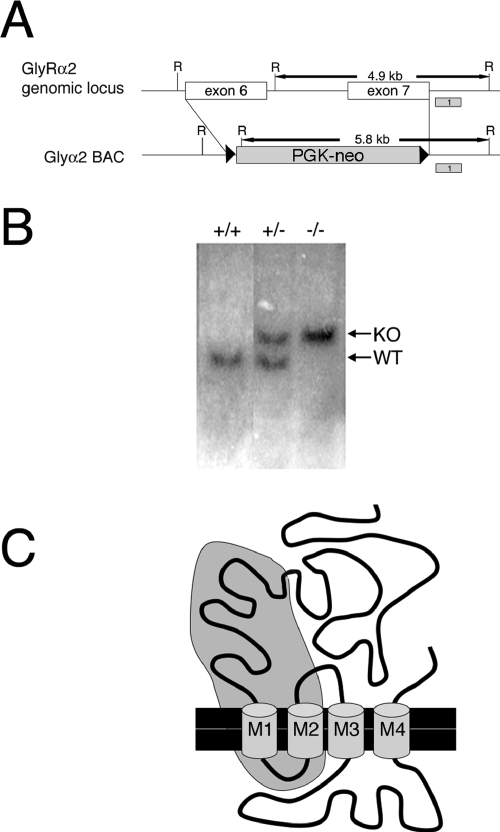
FIG. 1.
Targeted mutation at the Glra2 locus. (A) Strategy for targeted deletion of Glra2 exons 6 and 7. Homologous recombination within a BAC containing exons 6 and 7 was used to replace exons 6 and 7 and the intervening intron with a PGK-neo cassette flanked by FRT sites. Shaded boxes labeled 1 represent probes used in Southern blot assays to screen for homologous recombination and deletion of exons 6 and 7. Wild-type (WT) genomic DNA digested with EcoRI and probed with probe 1 yields a fragment of 4.9 kb. In KO alleles, an endogenous EcoRI site present in the intron between exons 6 and 7 is deleted, yielding a 5.8-kb fragment when probed with probe 1. R, EcoRI. (B) Southern blot analysis of genomic DNA isolated from mouse tails. Southern blot assays of EcoRI-digested DNA probed with probe 1 presents a 4.9-kb band for the wild-type allele and a 3.5-kb band for the deletion-containing allele. (C) Diagram of GlyRα2 protein showing its four transmembrane domains (M1 to M4) with the approximate region of the protein encoded by exons 6 and 7 shaded in gray. Alternate splicing of exon 5 to exon 8 would result in a premature stop codon four amino acids into exon 8.
Brain slice preparation.
Timed pregnant Glra2 KO mice and control C57/BL6 mice (Taconic, New York) at embryonic day 17 (E17) were used for all experiments, except where indicated otherwise. Mice were anesthetized (with ketamine at 90 mg/kg and xylazine at 10 mg/kg), and embryos were removed for further dissection. Following rapid decapitation, the brain was removed in chilled artificial cerebrospinal fluid (ACSF; 125 mM NaCl, 5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgSO4, 2 mM CaCl2, 25 mM NaHCO3, 25 mM glucose, pH 7.4, 310 mosM/liter). The dissected brain was placed in 4% low-melting-point agarose (Fisher Scientific) in ASCF. Agarose was cooled on ice and allowed to solidify, and the embedded brain was sliced into coronal sections (400 μm) in ice-cold ACSF with a VT100S vibrating-blade microtome (Leica, Nussloch, Germany). Slices were allowed to recover at room temperature in oxygenated (95% O2 and 5% CO2) ACSF. They were subsequently used for electrophysiological recording or loaded for calcium imaging.
Calcium imaging.
Brain slices were bath loaded with the acetoxymethyl ester form of the calcium indicator dye fluo-3 (10 to 15 μM; Molecular Probes, Eugene, OR). Loading was performed in the dark at room temperature for 1 to 3 h. Loaded slices were placed in an imaging chamber on the stage of an upright compound microscope (BX50-WI; Olympus, Tokyo, Japan) and continuously perfused with oxygenated ACSF. Epifluorescence imaging of fluo-3 was performed with a 100-W mercury light source and a low-light charge-coupled device camera (300-T; Dage-MTI, Michigan City, IN). For fluo-3 imaging, we used the following fluorescence filters (Chroma Technology, Brattleboro, VT): excitation filter, 480 ± 20 nm; dichroic mirror, 505 nm long pass; emission filter, 535 ± 25 nm. Cells were imaged with a 10× water immersion objective, and photobleaching was minimized by controlling a shutter positioned in the excitation light path (Uniblitz S25; Vincent Associates, Rochester, NY). Time-lapse images were acquired every 2 s by the Scion Image program on a Macintosh G3 computer equipped with a video frame grabber (Scion LG-3; Scion Corp., Frederick, MD). Fluorescence changes were measured in selected cells by Scion Image.
Pharmacological agents.
Glycine, taurine (Sigma, St. Louis, MO), and GABA (RBI, Natick, MA) were prepared as stock solutions in double-distilled H2O and diluted in ACSF to final concentration. All drugs were applied focally with the DAD-12 Superfusion System (ALA Scientific Instruments, Westbury, NY).
Electrophysiology.
Whole-cell patch clamp recordings were performed with an EPC-9 patch clamp amplifier (HEKA Electronic, Lambrecht, Germany). Data were acquired with HEKA Pulse v. 8.0 software (HEKA Electronic, Lambrecht, Germany). Borosilicate glass (Warner Instrument Corp., Hamden, CT) electrode pipettes (5 to 8 MΩ) were filled with 130 mM CsCl, 2 mM CaCl2, 10 mM HEPES, and 11 mM EGTA (pH = 7.4 at 25°C, 265 to 275 mosM/liter). Unless otherwise indicated, neurons were voltage clamped at −60 mV).
In situ hybridization and immunofluorescence.
Littermates were decapitated, and their heads were immediately fixed in 4% paraformaldehyde overnight at 4oC. Tissues were washed two times in phosphate-buffered saline (PBS), equilibrated in 30% sucrose in PBS overnight, equilibrated in a 1:1 mixture of 30% sucrose in PBS-optimal cutting temperature embedding medium, and embedded in optimal cutting temperature medium. Section in situ hybridizations were performed on retinal cryosections (20 μm) as previously described (6). Probes used were transcribed from the following DNA clones: Glra2 (full length) (34), taurine-upregulated gene 1 (TUG1; accession no. AI842058), GABA(A) subunit 6 (accession no. AI841957), Btg2 (B-cell translocation gene 2; accession no. BF464976), inhibitor of differentiation 2 (Id2) (accession no. AI848408), Notch1 (accession no. BE952133), and Nrl (neural retina leucine zipper protein; accession no. BF464350).
For immunofluorescent staining, retinal cryosections (20 μm thick) were blocked with 2% goat serum-2% donkey serum-0.1% Triton X-100 for 1 h at room temperature. Slides were incubated in primary antibody overnight at 4°C. The primary antibodies used were anti-rhodopsin Rho4D2 (mouse monoclonal, 1:250) (20), anti-cone arrestin (rabbit polyclonal, 1:100, Zymed), and anti-neurofilament 200 (mouse monoclonal; Sigma). After several washes (PBS, 0.1% Triton X-100), slides were incubated in goat anti-mouse or goat anti-rabbit Cy3 or Cy5 (1:200; Jackson Immunoresearch Laboratories, Inc.) for 1 to 2 h at room temperature. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and washed several times. Hematoxylin-eosin staining was performed on paraffin sections. Images were taken on a Nikon Eclipse E1000 microscope with a Leica DC200 digital camera.
RESULTS
Generation of GlyRα2 KO mice.
In order to generate a Glra2 targeting construct, a BAC containing exons 3 to 8 of the Glra2 locus was identified through hybridization of a BAC library (Genome Systems) with a probe containing exons 6 and 7 of GlyRα2. BAC end sequencing revealed that the BAC contained ∼160 kb of sequence, 56 kb upstream of exon 6 and 103 kb downstream of exon 7. Bacterial recombination was utilized to replace exons 6 and 7 in the BAC with an FRT-flanked PGK-neomycin cassette by the system developed by Stewart et al. (22). This targeting construct was used to delete exons 6 and 7 in ES cells. As the Glra2 locus is present on the X chromosome and the ES cells used contained only one X chromosome, correctly targeted ES cells were identified by the loss of exons 6 and 7 by Southern blotting and PCR analysis. One correctly targeted ES cell clone out of 390 clones screened was identified. A higher frequency of homologous recombination was expected because of the large homology arms of this construct (33), suggesting that the genomic location of Glra2 may be refractory to recombination. Blastocyst injection of this positive ES clone generated 22 founder mice. Five of these lines were further expanded for analysis. Genomic deletion of exons 6 and 7 in mice was confirmed by Southern blotting with a probe 3′ to the deleted region and within an EcoRI fragment. This probe recognizes a 4.9-kb fragment in the wild-type allele and a 5.8-kb fragment in the KO allele (Fig. 1A and B).
Exons 6 and 7 encode two transmembrane domains of GlyRα2 and a large portion of its ligand binding domain (Fig. 1C). Alternative splicing of exon 5 to exon 8 would result in an immediate premature stop codon and should lead to nonsense-mediated degradation of the mRNA (7). In order to confirm the lack of GlyRα2 mRNA, reverse transcription-PCR, Northern blotting, and in situ hybridization analyses were performed with P0 wild-type and Glra2−/− brain RNAs (Fig. 2).
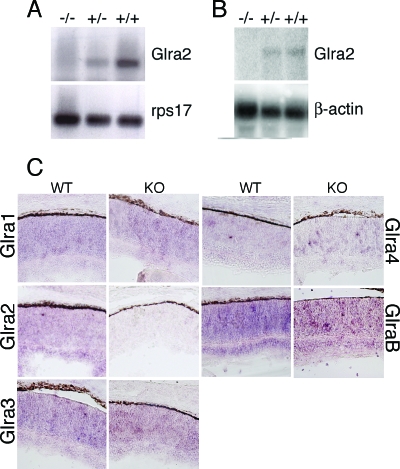
FIG. 2.
Glycine receptor expression in animals with Glra2 disrupted. (A) Reverse transcription-PCR analysis for Glra2 with RNA isolated from P0 mouse brain from wild-type (+/+), heterozygous (+/−), or homozygous Glra2 KO (−/−) littermates with primers specific for Glra2 or ribosomal protein 17 (rps17). (B) Northern analyses were performed with total RNA (10 μg) isolated from P0 mouse brain. The Glra2 probe was a 32P-labeled, full-length GlyRα2 cDNA. A probe for β-actin was used as a loading control. (C) In situ hybridization analysis of glycine receptor subunits in wild-type (WT) or Glra2 KO retinae at P0. The probes used recognized RNA species encoding glycine receptors alpha 1 (Glra1), alpha 2 (Glra2), alpha 3 (Glra3), alpha 4 (Glra4), and beta (GlrB).
Glycine receptor KO animals exhibit normal spinal cord development.
Glra2−/− animals were born in normal ratios and were healthy and fertile. No gross behavioral or morphological phenotypes were apparent in newborn or adult mice, as they suckled normally, had a normal gait, and bred and ate as their wild-type littermates did.
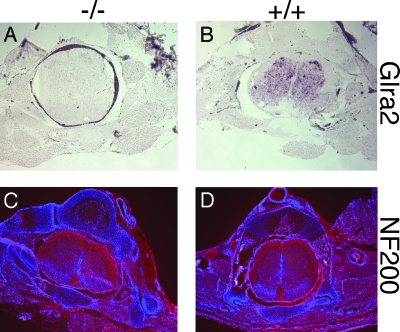
GlyRα2 is the predominant subunit expressed in the developing spinal cord (1, 18). Although Glra2−/− animals do not express GlyRα2 RNA in the P0 spinal cord (Fig. 3A and B), the developmental morphology of the spinal cord appeared normal; the diameter and dell density within the spinal cord were unchanged in Glra2−/− and wild-type littermates throughout the anterior-posterior axis (Fig. 3C and D).
FIG. 3.
Analysis of P0 spinal cords from wild-type and Glra2 KO animals. (A) In situ hybridization for Glra2 RNA in cryosections of P0 spinal cord from wild-type (+/+) and Glra2 KO (−/−) animals. (B) Immunofluorescent staining for neurofilament 200 (red) and nuclei counterstained with DAPI (blue) in P0 spinal cord sections.
Glycine receptor activity is not necessary for normal cortical development.
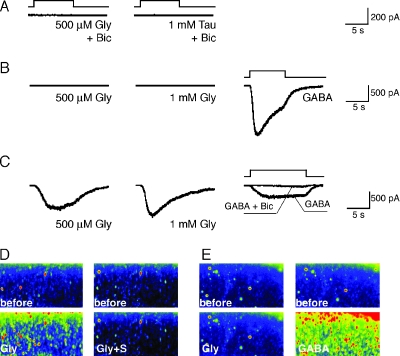
GlyRα2 is expressed and active in the developing cerebral cortex (8, 18). Patch clamp recordings of neonatal wild-type neurons show that these cells respond to the application of glycine and taurine by activating endogenously expressed glycine receptors (8). In addition, wild-type neonatal (8) and late embryonic cortical brain slices loaded with a calcium indicator showed an increase in intracellular calcium levels in an entire field of imaged cortical neurons in response to glycine application (Fig. 4D). This glycine-evoked calcium influx was reversibly inhibited by preincubation with strychnine, a specific inhibitor of glycine receptors (Fig. 4D). In contrast, standard whole-cell patch clamp recordings of individual cortical neurons in brain slices from Glra2−/− animals did not show responses to a low (500 μM) or a high (1 mM) concentration of glycine or taurine in the presence or absence of bicuculline at E17 (Fig. 4A and B). The results obtained with individual neurons were further confirmed by calcium imaging of a population of neurons in Glra2−/− cortical slices (Fig. 4E). However, Glra2−/− neurons did respond normally to GABA (30 μM) (Fig. 4B), and that response induced a calcium increase in slices loaded with a calcium indicator (Fig. 4E). At postnatal day 7, glycine responses were recovered in Glra2−/− cortical cells (Fig. 4C), presumably because of neonatal expression of other glycine receptor subunits.
FIG. 4.
Electrophysiological and calcium imaging responses to glycine (Gly), taurine (Tau), and GABA recorded in cortical pyramidal neurons. (A, B) Single-cell patch clamp recordings of E17 Glra2 KO mice. Current responses recorded in upper-layer pyramidal neurons (n = 5/5) demonstrate that cortical neurons do not respond to 500 μM glycine or 1 mM taurine in the presence (A) or absence (B) of 50 μM bicuculline (Bic). However, these neurons exhibit normal GABA (30 μM) responses (B). (C) Recordings of P7 Glra2 KO neurons (n = 7/7) demonstrate normal responses to glycine and GABA. GABA responses are abolished in the presence of 50 μM bicuculline. (D, E) Calcium imaging of E17 coronal cortical slices from wild-type control (D) and Glra2 KO (E) animals. Calcium influx is induced in response to 500 μM glycine (Gly) in wild-type (D) but not Glra2 KO (E) slices (n = 2). Preincubation with strychnine (Gly+S) inhibits glycine-induced calcium influx (D). Normal calcium influx in response to GABA is maintained in Glra2 KO slices. before, before stimulation.
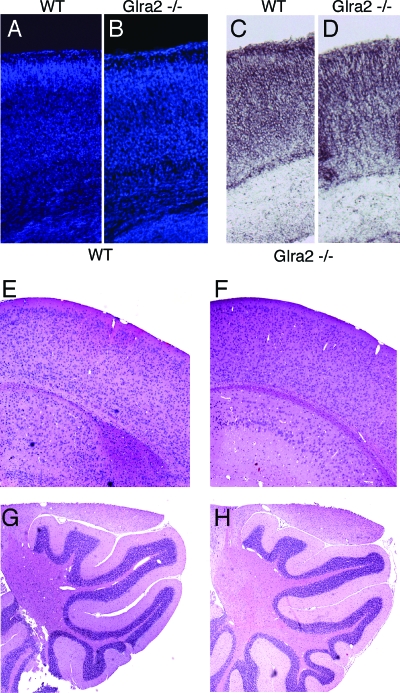
In spite of a lack of developmental electrophysiological responses to glycine and taurine in the cortex, wild-type and Glra2−/− cortexes at P0 (Fig. 5A to D) and adult ages (Fig. 5E and F) were morphologically indistinguishable. At P0, there appeared to be no obvious change in the expression pattern of Notch1, Id2, or GABA(A) subunit 6 (Fig. 5C and D and data not shown). In addition, the cerebella of Glra2−/− animals also appeared normal by hematoxylin-eosin staining (Fig. 5G and H).
FIG. 5.
Analysis of adult and neonatal cortexes from Glra2 KO animals. (A to D) Coronal sections of wild-type (WT) (A, C) and Glra2 mutant (B, D) P0 cortexes. Sections were stained with DAPI (A, B) or by in situ hybridization with a probe for GABA(A) receptor alpha 6 (C, D). (E to H) Hematoxylin-eosin staining of sections from adult (3-month-old) wild-type cortex (E) or cerebellum (G) or Glra2 mutant cortex (F) or cerebellum (H).
Retinal development and function appear normal in Glra2−/− animals.
GlyRα2 is expressed in the developing rodent retina (34). Section in situ hybridization of P0 retinae from wild-type and Glra2−/− animals revealed no detectable morphological or molecular difference (Fig. 2C). In addition to GlyRα2, other glycine receptor subunits also are expressed in the P0 retina. However, no differences in α1, α3, α4, or β expression could be detected between wild-type and Glra2 KO retinae (Fig. 2C).
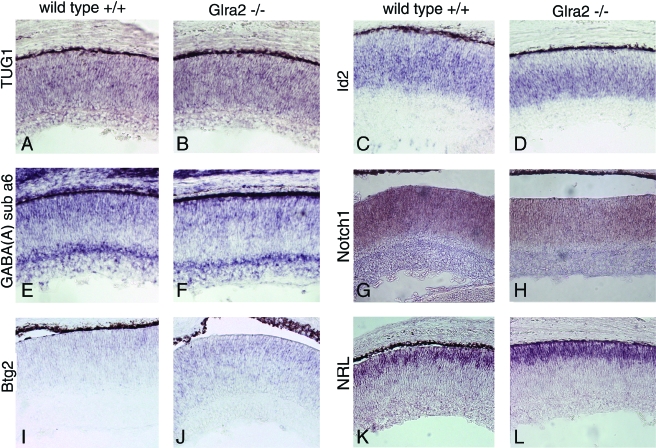
At P0, the majority of retinal cells are mitotic progenitor cells or postmitotic undifferentiated cells, with smaller numbers of differentiated ganglion, amacrine, and cone photoreceptor cells. Notch1, Id2, Btg2, TUG1, and GABA(A) subunit 6 are each expressed in subsets of progenitor and/or precursor cells. Nrl is expressed in photoreceptor precursor cells (19). The expression of these genes appeared unchanged in Glra2−/− retinae (Fig. 6).
FIG. 6.
Expression of markers in wild-type and Glra2 mutant retinae at P0. In situ hybridization for TUG1 (A, B), Id2 (C, D), GABA(A) alpha 6 (E, F), Notch1 (G, H), Btg2 (I, J), and Nrl (K, L) in wild-type (A, C, E, G, I, K) or Glra2 KO (B, D, F, H, J, L) P0 retinae.
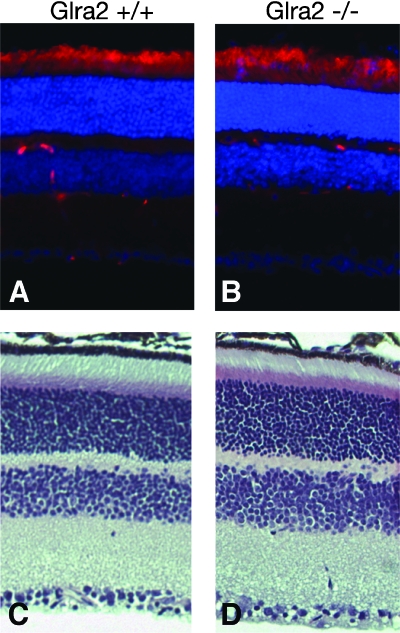
Hematoxylin-eosin staining of adult retinae from wild-type and Glra2−/− littermates showed no obvious morphological difference; outer nuclear and plexiform layers, inner nuclear and plexiform layers, and ganglion cell layers were present and of similar thicknesses and cell densities (Fig. 7). In addition, the inner and outer segments of photoreceptors appeared normal. Electroretinogram analyses revealed no appreciable difference between adult retinae from wild-type and Glra2−/− animals (B. Pawlyk and T. Li, personal communication).
FIG. 7.
Analysis of retinae of adult wild-type and Glra2 KO animals. Retinal sections of adult (3-month-old) wild-type (A, C) or Glra2 KO (B, D) animals are shown. (A, B) Retinal sections were immunostained for rhodopsin (RHO4D2, red), and nuclei were counterstained with DAPI (blue). (C, D) Hematoxylin-eosin staining of adult retinal sections.
DISCUSSION
Glycine receptors mediate inhibitory neurotransmission in the mature brain stem and spinal cord. However, glycine receptors, particularly the α2 subunit, also are expressed throughout the developing nervous system (18). During development, functionally active glycine receptors are expressed on migrating and differentiating cortical neurons (8). In addition, a ligand for these receptors, taurine, is present at abundant levels in the developing cortex (29). In contrast to glycine receptors expressed in the adult, activation of these developmentally expressed receptors results in excitation of immature neuronal cells (8). Taken together, these observations raise the question of the functional role of glycine receptors in neuronal development.
Additional evidence that supports a developmental role for glycine receptors came from our studies of retinal development. Acute knockdown of GlyRα2 mRNA levels in P0 rodent retinal cells resulted in the inhibition of production of rod photoreceptor cells, which are the most abundant cell type produced at P0 (34). However, retinae from Glra2−/− mice appear normal at P0 and in the adult. Nrl expression at P0 and rhodopsin expression in the adult were identical in KO and wild-type littermates, suggesting that rod photoreceptors are produced normally in both. Acute knockdown of GlyRα2 may preclude compensatory mechanisms that are employed in Glra2−/− animals. Glycine receptor alpha subunits exhibit 80 to 90% amino acid similarity to one another, and the α1, α2, α3, and β subunits are expressed in the retina. Compensatory mechanisms between alpha subunits have been proposed in other glycine receptor mutant animals. Oscillator mice harbor a frameshift mutation in Glra1 that completely eliminates alpha 1 expression. Studies of alpha subunit expression in oscillator mice suggest that other alpha subunits are upregulated to compensate for the loss of GlyRα1 in the retina and spinal cord (9, 12, 31).
Although the lack of a retinal phenotype and spinal cord may be attributable to compensation by other alpha subunits, the absence of a cortical phenotype cannot. Glra2−/− embryonic cortical neurons did not exhibit an electrophysiological response to glycine or taurine, showing that the allele generated here is a null allele and that no other functional glycine receptors are present embryonically to compensate for the loss of GlyRα2. However, Glra2−/− cortical neurons did respond to GABA. Applications of GABA and glycine are both excitatory in the embryonic cortex, as binding of these ligands to their respective receptors results in opening of the chloride-permeable channel (8, 15). In both cases, since embryonic neurons have a higher intracellular chloride concentration, the channel opening results in chloride ions flowing out of the cell and the subsequent depolarization leads to an influx of calcium ions. Thus, it is possible that GABA activity may compensate for the loss of glycinergic activity in the Glra2−/− embryonic cortex sufficiently to bridge the gap of glycine receptor inactivity until the neonatal expression of other glycine subunits. In fact, GABA(A) receptor protein expression has been shown to be upregulated in spastic mice, which contain a mutation in GlrB, and in myoclonic cattle which has been linked to a nonsense mutation in Glra1 (3, 16, 23, 32). In addition, humans suffering from hyperplexia, a human disease linked to loss of glycine receptor function, are treated with benzodiazepines such as clonazepam (23). The effectiveness of clonazepam is proposed to be due to its potentiation of GABA(A) receptors.
One possibility that this Glra2−/− mouse raises is that, in spite of the widespread expression of glycine receptors and their ligands embryonically, glycine receptor signaling may not be critical for neuronal development. In the future, interbreeding of Glra2−/− mice with previously established Glra1, Glra3, and Glrb null mice, and perhaps with GABA receptor-deficient animals, could provide valuable insights into the roles of glycine receptors in the nervous system.
Acknowledgments
We thank J. Munson and J. Trimarchi for technical assistance and B. Pawlyk and T. Li for providing electroretinogram data.
The mice described in this study were generated within the Mental Retardation Research Center (MRRC)/Children's Hospital Gene Manipulation Facility (NIHP30-HD 18655) and the Brigham and Women's Hospital Transgenic Mouse Facility. This work was supported by funds from NIH (RO1EY009676). C.L.C. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Becker, C. M., W. Hoch, and H. Betz. 1988. Glycine receptor heterogeneity in rat spinal cord during postnatal development. EMBO J. 7:3717-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, C. M., V. Schmieden, P. Tarroni, U. Strasser, and H. Betz. 1992. Isoform-selective deficit of glycine receptors in the mouse mutant spastic. Neuron 8:283-289. [DOI] [PubMed] [Google Scholar]
- 3.Biscoe, T. J., J. P. Fry, and C. Rickets. 1984. Changes in benzodiazepine receptor binding as seen autoradiographically in the central nervous system of the spastic mouse. J. Physiol. 352:509-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., R. G. Weber, B. Saul, M. von Knebel Doeberitz, C. Grond-Ginsbach, K. Kellerman, H. M. Meinck, and C. M. Becker. 1996. A GLRA1 null mutation in recessive hyperekplexia challenges the functional role of glycine receptors. Am. J. Hum. Genet. 58:989-997. [PMC free article] [PubMed] [Google Scholar]
- 5.Burzomato, V., P. J. Groot-Kormelink, L. G. Sivilotti, and M. Beato. 2003. Stoichiometry of recombinant heteromeric glycine receptors revealed by a pore-lining region point mutation. Recept. Channels 9:353-361. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. M., and C. L. Cepko. 2002. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development 129:5363-5375. [DOI] [PubMed] [Google Scholar]
- 7.Conti, E., and E. Izaurralde. 2005. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17:316-325. [DOI] [PubMed] [Google Scholar]
- 8.Flint, A. C., X. Liu, and A. R. Kriegstein. 1998. Nonsynaptic glycine receptor activation during early neocortical development. Neuron 20:43-53. [DOI] [PubMed] [Google Scholar]
- 9.Graham, B. A., P. R. Schofield, P. Sah, and R. J. Callister. 2003. Altered inhibitory synaptic transmission in superficial dorsal horn neurones in spastic and oscillator mice. J. Physiol. 551:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grudzinska, J., R. Schemm, S. Haeger, A. Nicke, G. Schmalzing, H. Betz, and B. Laube. 2005. The beta subunit determines the ligand binding properties of synaptic glycine receptors. Neuron 45:727-739. [DOI] [PubMed] [Google Scholar]
- 11.Kingsmore, S. F., B. Giros, D. Suh, M. Bieniarz, M. G. Caron, and M. F. Seldin. 1994. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat. Genet. 7:136-141. [DOI] [PubMed] [Google Scholar]
- 12.Kling, C., M. Koch, B. Saul, and C. M. Becker. 1997. The frameshift mutation oscillator (Glra1spd-ot) produces a complete loss of glycine receptor alpha1-polypeptide in mouse central nervous system. Neuroscience 78:411-417. [DOI] [PubMed] [Google Scholar]
- 13.Langosch, D., L. Thomas, and H. Betz. 1988. Conserved quaternary structure of ligand-gated ion channels: the postsynaptic glycine receptor is a pentamer. Proc. Natl. Acad. Sci. USA 85:7394-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Legendre, P. 2001. The glycinergic inhibitory synapse. Cell. Mol. Life Sci. 58:760-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LoTurco, J. J., D. F. Owens, M. J. Heath, M. B. Davis, and A. R. Kriegstein. 1995. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron 15:1287-1298. [DOI] [PubMed] [Google Scholar]
- 16.Lummis, S. C., A. L. Gundlach, G. A. Johnston, P. A. Harper, and P. R. Dodd. 1990. Increased gamma-aminobutyric acid receptor function in the cerebral cortex of myoclonic calves with an hereditary deficit in glycine/strychnine receptors. J. Neurochem. 55:421-426. [DOI] [PubMed] [Google Scholar]
- 17.Lynch, J. W. 2004. Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev. 84:1051-1095. [DOI] [PubMed] [Google Scholar]
- 18.Malosio, M. L., B. Marqueze-Pouey, J. Kuhse, and H. Betz. 1991. Widespread expression of glycine receptor subunit mRNAs in the adult and developing rat brain. EMBO J. 10:2401-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mears, A. J., M. Kondo, P. K. Swain, Y. Takada, R. A. Bush, T. L. Saunders, P. A. Sieving, and A. Swaroop. 2001. Nrl is required for rod photoreceptor development. Nat. Genet. 29:447-452. [DOI] [PubMed] [Google Scholar]
- 20.Molday, R. S., and D. MacKenzie. 1983. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry 22:653-660. [DOI] [PubMed] [Google Scholar]
- 21.Mulhardt, C., M. Fischer, P. Gass, D. Simon-Chazottes, J. L. Guenet, J. Kuhse, H. Betz, and C. M. Becker. 1994. The spastic mouse: aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron 13:1003-1015. [DOI] [PubMed] [Google Scholar]
- 22.Muyrers, J. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce, K. D., C. A. Handford, R. Morris, B. Vafa, J. A. Dennis, P. J. Healy, and P. R. Schofield. 2001. A nonsense mutation in the alpha1 subunit of the inhibitory glycine receptor associated with bovine myoclonus. Mol. Cell. Neurosci. 17:354-363. [DOI] [PubMed] [Google Scholar]
- 24.Rapaport, D. H., L. L. Wong, E. D. Wood, D. Yasumura, and M. M. LaVail. 2004. Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474:304-324. [DOI] [PubMed] [Google Scholar]
- 25.Rees, M. I., T. M. Lewis, B. Vafa, C. Ferrie, P. Corry, F. Muntoni, H. Jungbluth, J. B. Stephenson, M. Kerr, R. G. Snell, P. R. Schofield, and M. J. Owen. 2001. Compound heterozygosity and nonsense mutations in the α1-subunit of the inhibitory glycine receptor in hyperekplexia. Hum. Genet. 109:267-270. [DOI] [PubMed] [Google Scholar]
- 26.Ryan, S. G., M. S. Buckwalter, J. W. Lynch, C. A. Handford, L. Segura, R. Shiang, J. J. Wasmuth, S. A. Camper, P. Schofield, and P. O'Connell. 1994. A missense mutation in the gene encoding the α1 subunit of the inhibitory glycine receptor in the spasmodic mouse. Nat. Genet. 7:131-135. [DOI] [PubMed] [Google Scholar]
- 27.Saul, B., V. Schmieden, C. Kling, C. Mulhardt, P. Gass, J. Kuhse, and C. M. Becker. 1994. Point mutation of glycine receptor α1 subunit in the spasmodic mouse affects agonist responses. FEBS Lett. 350:71-76. [DOI] [PubMed] [Google Scholar]
- 28.Singer, J. H., and A. J. Berger. 2000. Development of inhibitory synaptic transmission to motoneurons. Brain Res. Bull. 53:553-560. [DOI] [PubMed] [Google Scholar]
- 29.Sturman, J. A. 1993. Taurine in development. Physiol. Rev. 73:119-147. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, T., A. Momiyama, K. Hirai, F. Hishinuma, and H. Akagi. 1992. Functional correlation of fetal and adult forms of glycine receptors with developmental changes in inhibitory synaptic receptor channels. Neuron 9:1155-1161. [DOI] [PubMed] [Google Scholar]
- 31.Wassle, H., P. Koulen, J. H. Brandstatter, E. L. Fletcher, and C. M. Becker. 1998. Glycine and GABA receptors in the mammalian retina. Vision Res. 38:1411-1430. [DOI] [PubMed] [Google Scholar]
- 32.White, W. F., and A. H. Heller. 1982. Glycine receptor alteration in the mutant mouse spastic. Nature 298:655-657. [DOI] [PubMed] [Google Scholar]
- 33.Yang, Y., and B. Seed. 2003. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat. Biotechnol. 21:447-451. [DOI] [PubMed] [Google Scholar]
- 34.Young, T. L., and C. L. Cepko. 2004. A role for ligand-gated ion channels in rod photoreceptor development. Neuron 41:867-879. [DOI] [PubMed] [Google Scholar]