Abstract
The poly(C)-binding proteins, αCPs, comprise a set of highly conserved KH-domain factors that participate in mRNA stabilization and translational controls in developmental and viral systems. Two prominent models of αCP function link these controls to late stages of erythroid differentiation: translational silencing of 15-lipoxygenase (Lox) mRNA and stabilization of α-globin mRNA. These two controls are mediated via association of αCPs with structurally related C-rich 3′-untranslated region elements: the differentiation control elements (DICE) in Lox mRNA and the pyrimidine-rich motifs in α-globin mRNA. In the present report a set of mRNA translation and stability assays are used to determine how these two αCP-containing complexes, related in structure and position, mediate distinct posttranscriptional controls. While the previously reported translational silencing by the DICE is not evident in our studies, we find that the two determinants mediate similar levels of mRNA stabilization in erythroid cells. In both cases this stabilization is sensitive to interference by a nuclear-restricted αCP decoy but not by the same decoy restricted to the cytoplasm. These data support a general role for αCPs in stabilizing a subset of erythroid mRNAs. The findings also suggest that initial binding of αCP to target mRNAs occurs in the nucleus. Assembly of stabilizing mRNP complexes in the nucleus prior to export may maximize their impact on cytoplasmic events.
Posttranscriptional controls play a pivotal role in mammalian gene expression (10, 14, 33). These controls are mediated by sequence-specific interactions between a structure(s) in an mRNA and RNA-binding proteins. While some RNP complexes are highly specific to a particular control or mRNA target, others are more widely expressed and have the capacity to impact on multiple steps in gene expression. The association of disparate mRNAs in a cell with a single species of RNA binding protein may serve to coordinate critical posttranscriptional controls in cell differentiation and function (17).
αCPs, along with hnRNP K, comprise a subset of RNA-binding proteins that contain the 70-amino-acid KH RNA-binding domain (18, 20, 25, 35). This subset is characterized by a shared triple KH domain repeat structure and prominent poly(C)-binding specificity. The high-affinity RNA binding by these proteins is dependent on the presence of C-rich motifs in accessible secondary structures (38). hnRNP K is encoded at a single locus, its expression is ubiquitous, and it serves general packaging functions for nuclear RNAs (1, 8). αCPs are also widely distributed, are encoded by four dispersed loci, and comprise a set of closely related isoforms (21, 23). In contrast to the general role of hnRNP K, αCPs bind a defined set of mRNA targets (39) and appear to mediate a variety of specific gene control functions (24).
The two most intensively studied posttranscriptional controls mediated by αCPs are mRNA stabilization and translational modulation (24). The initial mRNA to be identified as an αCP stabilization target was human α-globin (hα-globin) mRNA. The identification of a C-rich stability motif in the 3′-untranslated region (UTR) of this mRNA (43) led to the identification of the corresponding RNP complex, the α-complex, that forms at this site (41). αCPs were identified as the primary constituents of this RNP complex (18). Subsequent studies revealed that additional mRNAs with long half-lives, such as those encoding β-globin (44), collagen (36), and tyrosine hydroxylase (30), have closely related, if not identical, αCP-containing 3′-UTR complexes. These findings have led to a model in which 3′-UTR-αCP complexes can serve as general determinants of mRNA stability (38).
A second reported role of αCP is in translational control. Remarkably, αCP binding has been reported to enhance translation in certain settings and to block ribosome loading in other settings. For example, αCPs markedly enhance translation of the polio mRNA by binding to a stem-loop structure (stem-loop IV) centrally located in the internal ribosomal entry site (IRES) (11). The mechanism of this translational enhancement remains to be fully defined. In contrast, αCP binding to a CU-rich differentiation control element (DICE) determinant in the 3′-UTR of 15-lipoxygenase (Lox) mRNA has been reported to result in mRNA sequestration and translational repression (29). hnRNP K may also contribute to this silencing activity (29), although αCP appears to be sufficient in experimental models (32). The mechanism involved in this translational repression by the DICE has been linked to a block in 60S joining at the AUG (28). The observation that αCPs can stabilize mRNA, enhance translation, or silence translation in distinct settings suggests that its functions are mediated by selective downstream interactions that feed into an assortment of mechanistic pathways.
The DICE is proposed to play a critical role in posttranscriptional control of erythroid gene expression (15, 37). This model states that Lox mRNA, once synthesized in the early erythroblast, is stored in a translationally inert form until it is activated at the terminal stage of reticulocyte maturation. This reversible silencing is attributed to binding of αCP1, and possibly hnRNP K, to the repeated CU-rich DICE sequence (29). This control has been modeled in transfected HeLa cells and in a rabbit reticulocyte lysate (RRL) in vitro translation system by linking the DICE to a luciferase reporter (29). The number of DICE repeats varies from 3 to 10 among 15-Lox mRNAs in a variety of mammalian species. It has been demonstrated that a single DICE is inactive, while the activity of the DICE can be effectively modeled by linking a minimum of two DICE repeats (DICE-2R) to a fLuc reporter (Luc-2R) (29).
Based on prior studies (see above), binding of αCP to 3′-UTR pyrimidine-rich (PR) elements can silence translation (15-Lox mRNA) or stabilize the bound mRNA (α-globin mRNA). Evidence suggests that these are nonoverlapping functions; although the DICE shares significant sequence similarity with the PR element in α-globin mRNA, the latter structure has been reported to lack translational silencing activity (29). In the present report we attempt to investigate the basis for the functional differences between these two αCP complexes. Surprisingly, our analysis of the DICE-2R and DICE-8R determinants fails to reveal translational control over the linked fLuc reporter either in vitro or in vivo. However, we did observe that the DICE-2R can stabilize mRNA and that this stabilization activity is equivalent to that mediated by the human α-globin PR determinant. Blockade of αCP function with RNA decoys confirmed that the mRNA stabilization functions of both the Lox DICE-2R and hα-globin PR reflect interactions with αCP. Of note, this αCP decoy effect appears to be mediated by blockade of α-complex formation in the nuclear compartment. These data suggest that αCPs play a role in coordinating stabilization of a subset of erythroid mRNAs and point to a functional link between assembly of αCP mRNP complexes in the nucleus with their impact on cytoplasmic events.
MATERIALS AND METHODS
Plasmids.
fLuc-2R and fLuc-2Rm plasmids were kind gifts from M. W. Hentze (29); the sequences of the two plasmids were confirmed upon receipt. These plasmids have a simian virus 40 early promoter and a T7 promoter 5′ of the Luc gene, allowing them to be used for both cell transfection studies and for in vitro transcription. pRLuc31 reporter plasmid was a kind gift from R. Andino at the University of California, San Francisco (12). pRL-TK (encoding Renilla luciferase as an internal control [rLuc]) was purchased from Promega. fLuc-8R and fLuc-8Rm plasmids were generated by replacing the SacI-BamHI fragments from fLuc-2R with DICE-8R (5′-SacI-[CCCCACCCTCTTCCCCAAG]8-BamHI-3′) and DICE-8Rm (5′-SacI-[CCCCAAGAGAGACCCCAAG]8-BamHI-3′), respectively. pTet-α2R and pTet-α2Rm plasmids were generated by insertion of a 38-bp DICE-2R (5′-CCCCACCCTCTTCCCCAAGCCCCACCCTCTTCCCCAAG-3) or DICE-2Rmut (5′-CCCCAAGAGAGACCCCAAGCCCCAAGAGAGACCCCAAG-3′) fragment in the place of the 42-bp pyrimidine-rich protection region of the α-globin 3′-UTR (PR) in parent plasmid pTet-αWT. The underlined sequences are the regions of the DICE that are mutated in mut 3′-UTR.
In vitro transcription and translation.
In vitro translation of fLuc-2R, fLuc-2Rm, fLuc-8R, and fLuc-8Rm reporters was assayed using the TNT T7 Quick Coupled Transcription/Translation system (Promega). Equal amounts of the fLuc-2R/8R and fLuc-2Rm/8Rm vectors were added to the system, along with a fixed amount of pRL-TK (where indicated) to 11 μl of reaction mixture (Promega). The levels of the fLuc and the rLuc vectors were used at concentrations that were in the linear range of reporter detection. Recombinant αCP1 and hnRNP K proteins were added to the reaction mixture individually or together in three concentrations (50 ng, 250 ng, and 500 ng). Bovine serum albumin was used at the same concentrations as a negative control. Firefly and Renilla luciferase activities were determined using a dual-luciferase reporter assay system (Promega). In separate studies (RNA decoy studies described below and data not shown), capped fLuc-2R/8R and fLuc-2Rm/8Rm mRNAs were synthesized in vitro (MegashortScript kit; Ambion) from the corresponding linearized plasmid templates, quantified by UV absorption, and checked for concentration and structure by glyoxal agarose electrophoresis, and equal amounts of each transcript (in the linear range of translational activity) were added to Red Nova lysate (RRL; Novagen) along with a fixed quantity of Renilla reporter (where needed). For decoy studies, the in vitro-transcribed VA1, VA1-R7α1 RNA, or dC17 DNA in the indicated amount was added to the RRL along with the mRNA of interest.
RNA-EMSA.
An RNA-electrophoresis mobility shift assay (RNA-EMSA) and supershift assays were carried out as described previously (41). 32P-labeled RNA probes [32P]α2R, [32P]α2Rm, and [32P]αWT (α-globin) 3′-UTR were transcribed in vitro with T7 RNA polymerase (MaxiScript MegashortScript kit; Ambion). Unlabeled RNA competitors (VA1 and VA1-R7α1) were synthesized with the MagashortScript kit (Ambion). The labeled probe (3 × 105 cpm per reaction mixture) was incubated with 15 μg of HeLa cell cytoplasmic extract or 5 μl of RRL (Novagen) at room temperature for 30 min in 25 μl binding buffer (10 mM Tris-HCl, pH 7.4, 150 mM KCl, 1.5 mM MgCl2, and 0.5 mM dithiothreitol). Unbound RNAs were degraded with RNase T1 (1 U/μl) for 10 min at room temperature. Heparin (final concentration of 1 mg/ml) was added to each reaction mixture prior to loading. The terminated reactions were run in a 5% native polyacrylamide gel in 0.5% Tris-borate-EDTA buffer at 10 V/cm2. Supershift assays were performed with polyclonal rabbit antisera against αCP1 (FF1 antibody) (3). Antibodies were added to reaction mixtures during incubation at a concentration of 1 μg/30 μl.
Western blot analysis.
Cytoplasmic extracts of HeLa cells transfected with pCDNA3.1-αCP1 or pCDNA3.1 (empty plasmid) were resolved in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and blotted to a nitrocellulose filter (Protran; Schleicher & Schuell) in Tris-CAPS buffer (Bio-Rad). Rabbit polyclonal anti-αCP1 antibody FF1 (1:5,000) or polyclonal anti-c-myc antibody (1:1,000; Santa Cruz Biotechnology) as well as anti-rabbit or anti-mouse antibody coupled to peroxidase (1:5,000; Amersham) were used to detect αCP1 protein by enhanced chemiluminescence (Lumi-Light; Roche).
Cell transfection and analysis of luciferase activity.
HeLa cells were grown in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 1× antibiotic-antimycotic (Invitrogen) and replated to 60-mm dishes 1 day before transfection. One μg of fLuc-2R or fLuc-2Rmut DNA, 0.5 μg of pRL-TK (encoding Renilla luciferase as an internal control), and 4.5 μg of carrier DNA or the indicated amount of pcDNA3.1-αCP1 plasmid DNA were cotransfected into HeLa cells with liposomal reagent Mirus Trans-IT (Fisher). After 36 h of incubation, the cells were washed twice with cold phosphate-buffered saline and harvested by scraping into 500 μl of lysis buffer (passive lysis buffer; Promega) followed by one freeze-thaw cycle and tested for luciferase activity (dual luciferase reporter assay system; Promega).
Determination of mRNA half-lives: cell transfection and RPA.
MEL/tTA cell transfection and RNA stability analysis in an RNase protection assay (RPA) were performed as described previously (19). MEL/tTA cells were grown in minimal essential medium supplemented with 10% fetal bovine serum, 1× antibiotic-antimycotic (Invitrogen), and 500 ng/ml tetracycline and split 1 day before transfection. MEL/tTA cells were electroporated with 2 μg pTet-hα2R, pTet-hα2Rm, or pTet-hαWT DNA and 18 μg carrier, decoy, or decoy control DNA. The transfected cells were incubated overnight in complete minimal essential medium as described above with 100 ng/ml tetracycline. A 4-h transcriptional pulse was then induced by transferring the cells to Tet(-) medium, after which tetracycline was added back to the medium at 500 ng/ml. Total RNA was purified at the indicated time points after readdition of tetracycline. α2R, α2Rm, and αWT mRNAs were quantified by RPA with a [32P]CTP-labeled 244-nucleotide antisense hα-globin RNA probe. A mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antisense RNA probe was used as an internal control (template from Ambion). The protected RNA fragments were resolved on a 6% polyacrylamide gel containing 8 M urea. Quantification was performed with PhosphorImage 1.1 software. The α2R, α2Rm, and αWT RNA concentrations at each time point were normalized to the internal control RNA and to cell number to account for cell proliferation during the study. The data were plotted against a logarithmic scale, and half-life was determined by establishing the time to 50% loss of mRNA. The half-lives of each mRNA from each of several individual experiments (n) were determined, and the P values of critical comparisons were calculated by using Student's t test. The relevant P values are noted in the text.
RESULTS
The DICE-2R unit fails to silence translation both in vitro and in vivo.
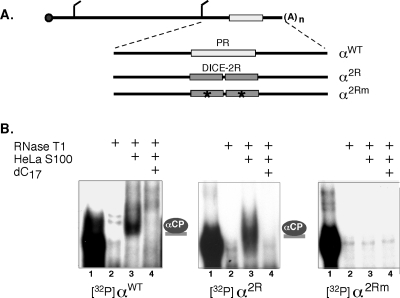
In prior studies, translational repression by the Lox DICE determinant was modeled in transfected HeLa cells and by in vitro translation in RRL (29). Our aim was to use these assays to explore the mechanistic basis for the ability of the DICE, but not the α-globin PR, to mediate translational repression. The initial studies were carried out by HeLa transfection. We first assessed whether αCPs in HeLa extracts could bind directly to the Lox DICE and whether the generated αCP RNP complex resembled that formed on the hα-globin PR motif. To simplify this comparison and normalize for surrounding sequence effects, RNP formation on the two elements was studied in an identical sequence context. Thus, complex formation was assessed for the wild-type α-globin 3′-UTR containing the native PR motif and a derivative sequence in which the PR element was replaced with a two-copy DICE unit, DICE-2R (Fig. 1A). DICE-2R has been shown by others to be sufficient to mediate translational control on a linked firefly luciferase open reading frame ORF (ORF; fLuc-2R) in transfected HeLa cells (29). As a control, a mutant DICE sequence (DICE-2Rm) that ablates the reported translational repression was inserted at the same position (see Materials and Methods). RNA-EMSA demonstrated that the DICE-2R RNA forms an RNP complex in HeLa S100 extract (Fig. 1B, middle panel). The electrophoretic migration of the complex and its sensitivity to poly(C) competition are remarkably similar to the α-complex that assembles on the native α-globin mRNA 3′-UTR (Fig. 1B, left panel, and data not shown). This α-complex has been previously demonstrated to comprise a single αCP bound to the α-globin 3′-UTR PR determinant (3), as is shown schematically to the right of the gel. The DICE-2Rm fails to form a comparable complex (Fig. 1B, right panel). More extensive studies demonstrated that, as is the case for the α-globin PR, the poly(C) sensitivity of the DICE complex is specific, as there was no comparable inhibition of complex formation by other homoribopolymers or unrelated RNAs (Fig. 1B and data not shown). These data confirm that the DICE-2R motif can serve as a direct binding target for αCPs endogenous to HeLa cells.
FIG. 1.
The DICE-2R motif binds to αCPs endogenous to HeLa cells. (A) Diagram of α2R and α2Rm reporter mRNAs. The human α-globin mRNA is diagrammed at the top with a 7meG cap (solid circle), poly(A) tail [(A)n], and AUG initiation and UAA termination codons. The PR stability element of wild-type α-globin (αWT) mRNA (open rectangle) was replaced by two repeats of the DICE (DICE-2R; shaded rectangles) derived from 15-Lox mRNA (α2R) or with a set of derivative DICEs containing inactivating multinucleotide substitutions (stars) (α2Rm) (see Materials and Methods). (B) RNA-EMSA demonstrating in vitro binding of the α2R 3′-UTR to proteins in HeLa cell extract. Each of three 32P-labeled probes, αWT, α2R, or α2Rm, was analyzed for RNP complex formation with proteins from a HeLa S100 extract (left, center, and right panels, respectively). The 32P-labeled probes are shown in lanes 1, and their RNase sensitivities in the absence of extract are shown in lanes 2. An RNase-resistant RNP complex (lanes 3) assembles on the αWT and α2R probes but not the α2Rm probe. This complex is sensitive to competition with dC17 (lanes 4), and its position is marked by a binary α-complex (3) shown to the right of the respective autoradiographs.
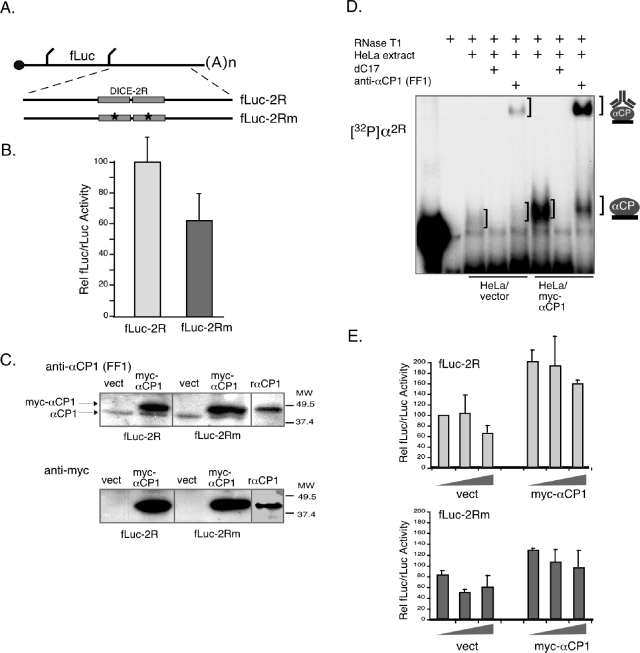
The function of the DICE was next addressed using a set of fLuc expression vectors in which the DICE-2R or the DICE-2Rm was placed in the 3′-UTR (fLuc-2R and fLuc-2Rm plasmids were kind gifts of M. Hentze) (Fig. 2A). The two reporter plasmids, fLuc-2R and fLuc-2Rm, were transfected into HeLa cells, and their expression was monitored. Surprisingly, analysis of equimolar transfections of fLuc-2R and fLuc-2Rm plasmids failed to reveal silencing of fLuc-2R gene expression (Fig. 2B). In fact, contrary to predictions, fLuc-2R expression was consistently higher than that of fLuc-2Rm. The same result is observed in MEL cell transfection studies (data not shown). These data indicate that the 3′-UTR DICE-2R does not repress fLuc expression in the context of abundant endogenous αCP in either HeLa cells or in the erythroid MEL cells (Fig. 2B and data not shown).
FIG. 2.
Expression of a luciferase reporter linked to the 3′-UTR DICE-2R (fLuc-2R) determinant fails to be repressed by αCPs endogenous to HeLa cells and is unaffected by overexpression of exogenous αCP1. (A) Diagram of the fLuc-2R and fLuc-2Rm mRNAs. Diagram details are as for Fig. 1. (B) The DICE-2R determinant fails to repress expression of an fLuc reporter in transfected HeLa cells. Equimolar amounts of plasmids encoding the fLuc-2R or fLuc-2Rm mRNAs were transfected into HeLa cells along with an internal control rLuc reporter. The ratio of fLuc and rLuc activities for 12 independent studies is represented on the histogram. The means and standard deviations are shown. (C) Overexpression of myc-tagged αCP1 in HeLa cells cotransfected with fLuc-2R or fLuc-2Rm expression vector. Top panel: Western blots of the cells cotransfected with the indicated vectors (empty vector [vect] and myc-epitope-tagged αCP1 vector [myc-αCP1]). The membranes were probed with monospecific antibody to αCP1 (anti-αCP1; lab designation FF1 (3). This antibody detects both endogenous αCP1 and myc-αCP1 (light lower and dark upper bands, respectively). Recombinant myc-tagged αCP1 (rαCP1) was included in the analysis (right lane) as a positive control. Lower panel: Western blots from a parallel gel were probed with anti-myc antibody to specifically detect the epitope-tagged exogenous αCP1 protein. Molecular weight markers are indicated to the right of the gel figure. (D) RNA-EMSA analysis demonstrating increased levels of α-complex formation in HeLa cells cotransfected with the myc-αCP1 expression vector. The identity of the sample in each lane is as indicated by the legend above the autoradiograph. The comparison is of extract isolated from HeLa cells transfected with the empty vector to HeLa cells transfected with the myc-αCP1 expression cassette. The positions of the α-complex and the supershifted α-complex are indicated to the right of the gel. (E) Expression of fLuc-2R fails to be repressed in HeLa cells and is unresponsive to elevated levels of exogenous αCP1. HeLa cells were cotransfected with fLuc-2R or fLuc-2Rm expression vectors along with either empty vector (vect) or the same vector containing the myc-αCP1 expression cassette (myc-αCP1). The graded input of expression vector used in the transfections (see Materials and Methods) is indicated by the wedge. The data represent five studies.
The apparent absence of a repressive effect of the DICE-2R in transfected HeLa cells was further tested by coexpressing fLuc-2R along with a plasmid encoding myc-tagged αCP1. The αCP1 isoform has been previously reported to mediate DICE-dependent translational repression (29). Western analysis confirmed a substantial increase in αCP1 over the endogenous levels (Fig. 2C), and a corresponding increase in RNA-binding activity was detected by EMSA (Fig. 2D). Remarkably, the increased level of αCP1 failed to repress fLuc-2R gene expression (Fig. 2E), and expression of the fLuc-2R was greater than the matched fLuc-2Rm even in the presence of elevated αCP1 levels. Thus, αCP1 endogenous to the HeLa cells fails to repress expression of a DICE-linked reporter, and significant boosting of αCP1 levels in the cell does not alter this result.
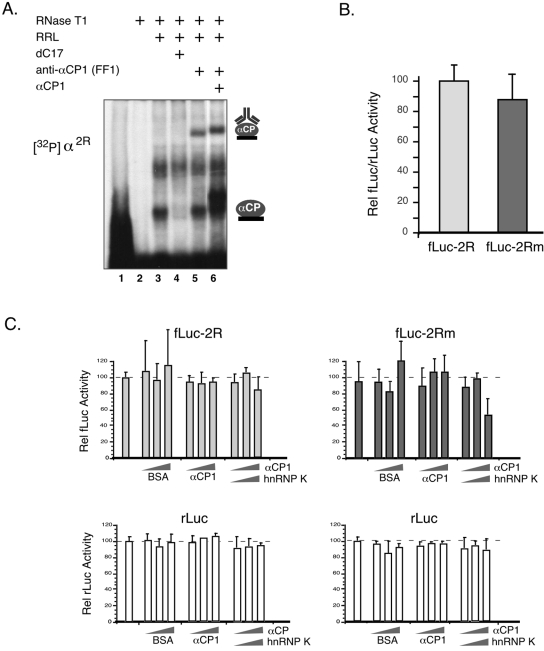
To extend the DICE analysis and more specifically focus on translational effects, we carried out a series of in vitro studies in RRL extract. Endogenous αCP activity is readily detected in the RRL by RNA-EMSA (Fig. 3A), and supershift analysis specifically identifies αCP1 in these complexes (Fig. 3A, lane 5). Addition of equal amounts of fLuc-2R and fLuc-2Rm plasmids to a coupled transcription/translation RRL (see Materials and Methods) fails to reveal a selective repression of the fLuc-2R expression in the presence of the endogenous αCP proteins (Fig. 3B). The only reproducible observation from multiple (>20) independent trials is a slightly higher expression of fLuc-2R compared to fLuc-2Rm. Addition of recombinant αCP1 with validated DICE-binding activity (Fig. 3A, lane 6) also fails to repress fLuc-2R expression (Fig. 3C). The coaddition of hnRNP K, a second poly(C)-binding protein reported to synergize with αCP1 in mediating DICE repression in this system (29), is also without appreciable effect on the expression of the fLuc-2R reporter (Fig. 3C). Finally, the in vitro translations were repeated by adding to the RRL equal amounts of capped synthetic fLuc-2R and fLuc-2Rm mRNAs rather than by using the coupled transcription/translation system. The mRNAs were added to the assay mixtures in the linear range of translational activity. As was the finding with the coupled transcription/translation system, we found a lower activity from fLuc-2Rm than with fLuc-2R mRNA (data not shown; results were indistinguishable from those in Fig. 3B). Thus, the in vitro studies using two different approaches failed to detect a repressive effect of the wild-type DICE-2R element when linked to the Luc reporter.
FIG. 3.
In vitro translation of fLuc-2R mRNA is not repressed by αCPs endogenous to the RRL, and the DICE-2R does not respond to addition of recombinant αCP1 or hnRNP K to the system. (A) RNA-EMSA demonstrating in vitro binding of the α2R 3′-UTR to αCP endogenous to RRL. The probe (lane 1) and the probe digested with RNase T1 (lane 2) are shown. The RNase-resistant RNP α-complex (lane 3) is sensitive to the competitor dC17 (lane 4) and is enhanced by the addition of recombinant αCP1 (lane 6). The complex is supershifted with the anti-αCP1 antibody FF1 (lanes 5 and 6). The recombinant αCP1 contains His6 and myc epitope tags, accounting for its slightly retarded mobility on the gel. (B) Comparison of fLuc-2R and fLuc-2Rm mRNA translation in RRL. Equimolar amounts of the two plasmids were separately added to RRL along with a fixed amount of rLuc plasmid as an internal control. The data represent the results from 15 independent studies. There is no significant difference between translational activities of the two mRNAs. (C) The translation of fLuc-2R is not repressed by the addition of increasing amounts of recombinant αCP1, or the combined addition of recombinant αCP1 and hnRNP K (upper panel). The translation of the fLuc-2R mRNA is shown in the left panels and that of the fLuc-2Rm mRNA is shown in the right panels. Translation of Renilla luciferase (rLuc) mRNA added to each translation mix as an internal control is shown in the corresponding lower panels. The translation mixtures were supplemented with increasing amounts of recombinant αCP1 or αCP1 plus hnRNP K. In a parallel set of studies, an equivalent amount of bovine serum albumin (BSA) was added to serve as a control. The data from five studies are represented by the histograms; means and standard deviations are shown for each condition.
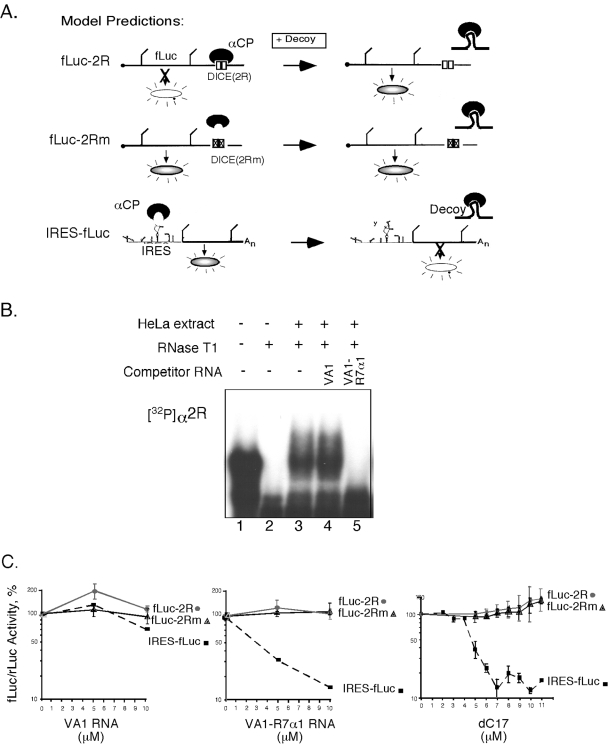
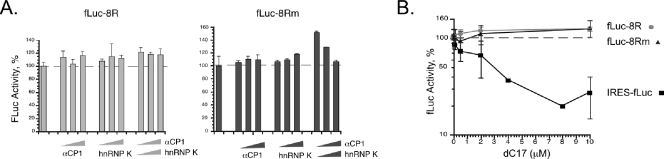
A reciprocal study was next attempted to detect DICE activity. In this study αCP function endogenous to the RRL was blocked by a high-affinity αCP decoy RNA (22). The rationale for the study and the predicted outcomes, based on previous reports, are summarized in Fig. 4A. A synthetic decoy RNA aptamer, R7α1, generated by SELEX (38), was embedded within a VA1 RNA framework (22). The highly structured VA1 RNA serves as an effective and stable RNA framework for the decoy cassette (for details, see reference 16). The binding activity of the αCP decoy RNA was validated in vitro by EMSA (Fig. 4B), and its biologic activity was validated by demonstrating an effective blockade of poliovirus IRES function (IRES-fLuc) (Fig. 4C). However, the predicted increase (i.e., derepression) of fLuc-2R but not fLuc-2Rm mRNA translation by αCP blockade (Fig. 4A, top and middle diagrams) was not observed (Fig. 4C). The same lack of effect was seen when a second highly effective αCP decoy, dC17, was used (Fig. 4C). Thus, the analyses of DICE-2R activity in transfected HeLa cells and in RRL under native conditions failed to reveal translational repression activity, and this activity was not revealed by a specific increase or blockade of αCP activity.
FIG. 4.
A high-affinity αCP decoy fails to enhance translation of a DICE-containing mRNA. (A) Models summarizing the predicted impact of an αCP decoy on the in vitro translation of the fLuc-2R, fLuc-2Rm, and IRES-fLuc (encoded by pRLuc31) mRNAs. Based on prior studies (29), blockade of αCP activity is predicted to derepress translation of the fLuc-2R mRNA. In contrast, the mutant DICE, 2Rm, cannot bind αCP and should be unresponsive to the αCP decoy. The poliovirus IRES-driven translation is dependent on αCP, and addition of the αCP decoy to the system is predicted to block efficient translation of a linked reporter fLuc ORF. (B) RNA-EMSA demonstrating that α-complex formation on the DICE-2R is blocked in vitro by the presence of the VA1-R7α1 decoy RNA. (C) Blockade of αCP activity with RNA decoys fails to alter translational activity of an fLuc ORF linked to the DICE-2R element. fLuc-2R and fLuc-2Rm mRNA translation is comparable in the presence of a nondecoy RNA (VA1 RNA). The addition of the SELEX decoy RNA, VA1-R7α1, markedly inhibits IRES function but fails to enhance translation of the fLuc-2R mRNA. The same marked inhibition of IRES function and lack of impact on the DICE-linked reporter is noted when a second αCP decoy, dC17, is added to the RRL.
The DICE-8R unit fails to silence translation in vitro.
Although the DICE-2R has been reported to be sufficient for silencing translation both in vitro (29, 32) and in vivo (29), we failed to observe this effect (Fig. 2 to 4). To extend this analysis of DICE function, we multimerized the DICE from 2R to 8R. This DICE-8R unit has been shown to have a maximal binding affinity to αCP1 when studied in a comparison of multimer repeats (32). The translation of fLuc-8R and fLuc-8Rm was analyzed in the in vitro translation RRL system as described above for the fLuc-2R study. Addition of equal amounts of fLuc-8R and fLuc-8Rm plasmids to a coupled transcription/translation RRL failed to reveal a selective repression of the fLuc-8R expression in the presence of the endogenous αCP proteins. In fact, the expression of fLuc-8R in this setting was substantially (fourfold) higher than the fLuc-8Rm (data not shown). This higher translational activity of fLuc-8R versus fLuc-8Rm is consistent with, and more exaggerated than, our observations comparing the translation of fLuc-2R with fLuc-2Rm mRNAs (Fig. 2B and E and 3B). In addition, the supplementation of the extract with recombinant αCP1, hnRNP K, or the combination of both proteins failed to selectively repress fLuc-8R expression (Fig. 5A). In addition, the presence of increasing levels of dC17, which effectively blocks the poliovirus IRES function (IRES-fLuc) (Fig. 5B), does not selectively stimulate (i.e., derepress) fLuc-8R mRNA translation. The entire set of experiments described above using the coupled transcription/translation system was then repeated in an in vitro translation in RRL of equal amounts of capped synthetic fLuc-8R and fLuc-8Rm mRNAs. The mRNAs were added to the assay mixtures in the linear range of translational activity. We again observed a higher translational activity from fLuc-8R mRNA compared with fLuc-8Rm mRNA (data not shown; results were indistinguishable from those in Fig. 5) and no significant impact of alterations in αCP levels on fLuc-8R expression.
FIG. 5.
In vitro translation of fLuc-8R does not respond to alterations in the levels or availability of the DICE-binding proteins αCP and hnRNP K. (A) Translation of fLuc-8R mRNA is not repressed by the addition of recombinant αCP1, recombinant hnRNP K, or the combined addition of recombinant αCP1 and hnRNP K (left panel) to the translation extract. The translations of the fLuc-8R and fLuc-8Rm mRNAs were carried out in reticulocyte extracts supplemented with increasing amounts of recombinant αCP1, hnRNP K, or αCP1 plus hnRNP K as indicated by the wedge below the histogram. The data represent four independent studies; means and standard deviations are shown for each of the translation conditions. (B) Blockade of αCP activity with dC17 fails to alter translational activity of fLuc-8R mRNA. Addition of dC17 to the RRL markedly inhibits translation of fLuc under the control of the αCP-dependent poliovirus IRES function (IRES-fLuc). There is no apparent impact of the same decoy effect on the DICE-linked reporter fLuc-8R. The data represent the results from three independent studies; means and standard deviations are shown for each of the translation conditions.
The DICE-2R motif acts as an mRNA stabilization determinant.
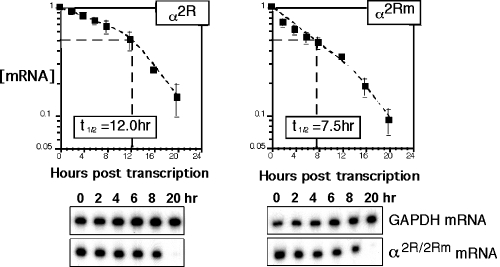
Interaction of αCP with the hα-globin mRNA 3′-UTR PR element contributes significantly to mRNA stability (18, 19, 41, 42). The structural similarity of the Lox DICE to the α-globin PR suggested that the DICE might share an mRNA stabilization function. To assess this model of DICE function, the PR was deleted from α-globin mRNA and replaced by the DICE-2R motif or with its mutant 2Rm counterpart. Deletion of the PR, or its replacement with random sequences, has been previously shown to decrease the hα-globin mRNA half-life in transfected MEL cells from 11 h to 7.5 h (19). Each of the hα-globin gene constructs was inserted into a Tet-off expression vector and separately transfected into the MEL/tTA cell line (19). The genes were activated for 4 h, and the decay of the newly synthesized mRNA was then followed over time by quantitative RPA (see Materials and Methods). The half-life of α2R mRNA (t1/2 of 12 h) is comparable to the wild-type α-globin mRNA (t1/2 of 11 h) (19) (also see below). Mutation of the DICE decreased the half-life of α2Rm mRNA to 7.5 h (Fig. 6) (P = 1.1 × 10−6), a value equivalent to that of an α-globin mRNA lacking the PR determinant (19). These data suggest that DICE-2R is fully efficient in mRNA stabilization. In contrast, the same experiments performed with C127/tTA cells, a nonerythroid Tet-off cell line (19), failed to show destabilization of α2Rm mRNA (data not shown). These results parallel and extend the previously reported erythroid specificity of PR-mediated α-globin mRNA stabilization (reference 19 and unpublished data). Thus, the DICE-2R appears to have an erythroid-specific mRNA stabilization profile quite similar to that of the α-globin PR determinant.
FIG. 6.
The DICE-2R motif stabilizes a human α-globin mRNA lacking its cognate PR determinant. The C-rich PR stabilization determinant in the αWT 3′-UTR was replaced with the DICE-2R determinant (α2R) or mutant DICE-2Rm determinant (α2Rm) (Fig. 1A). The two corresponding genes were placed under transcriptional control of a Tet-controlled promoter. Each construct was transfected into MEL/tTA cells followed by a 4-h transcriptional pulse and subsequent chase. RPA detected the remaining mRNA at each time point. The α2R or α2Rm signals were normalized to the internal mGAPDH control and to cell number (see Materials and Methods). The computed half-life is shown in the box, and a representative gel is shown under the decay curve. The data points for the α2R and α2Rm mRNAs represent the results from nine and seven independent studies, respectively.
mRNA stabilization by the DICE and PR depends on αCP activity in the nucleus.
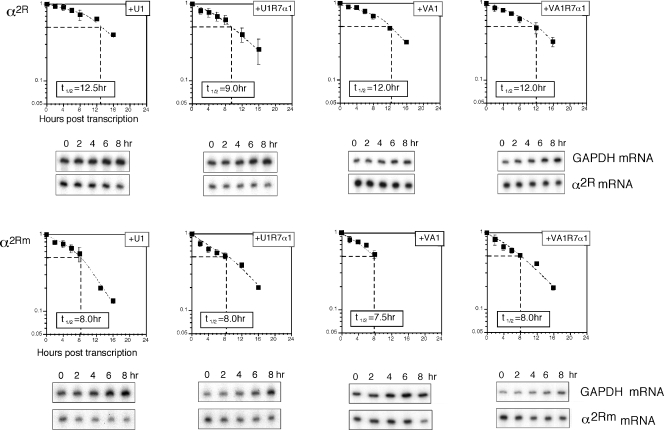
The analysis of PR and DICE indicated that they are capable of mediating comparable levels of mRNA stabilization in vivo. mRNA stabilization by PR has been shown in prior studies to be tightly linked to its binding of αCP. A corresponding dependence of DICE-mediated stabilization on αCPs was tested with a set of αCP decoys. Vectors that specifically express the VA1-R7α1 or U1-R7α1 decoy RNAs in the cytoplasm or nucleus, respectively, were used in separate studies. VA1-R7α1 generates the high-affinity αCP-binding RNA aptamer R7α1 within the framework of the adenoviral VA1 RNA (described above); the fusion VA1-R7α1 RNA is localized in the cytoplasm. In contrast, insertion of the same R7α1 aptamer within the U1 snRNA framework results in the expression of a fusion U1-R7α1 RNA in the nucleus (for details, see reference 22). The assumption going into the study was that the cytoplasmic decoy would have the most direct and substantial effect on mRNA stabilization by the two 3′-UTR determinants. Each decoy expression vector was cotransfected into MEL/tTA cells along with vectors expressing α2R or α2Rm under Tet control. Expression of the decoys was allowed to proceed for 24 h prior to the 4-h transcriptional pulse of the target mRNAs. The expression of the decoys in the appropriate compartment of the transfected cells was confirmed by reverse transcription-PCR (data not shown) (22). When cotransfected with U1-R7α1, the half-life of α2R mRNA is shortened from 12 h to 9.0 h (P = 0.006), while the control vector expressing the U1 framework RNA without the inserted decoy cassette has no appreciable effect (Fig. 7). Surprisingly, the cytoplasmic decoy, VA1-R7α1, fails to destabilize α2R mRNA. Destabilization of the α2R mRNA by the R7α1 decoy is consistent with the linkage of the DICE stabilization function to αCP binding. The specific utility of the nuclear as opposed to cytoplasmic decoy is unexpected and suggests the possibility that the critical interaction between DICE-2R and αCP occurs in the nucleus.
FIG. 7.
Stabilization of mRNA by DICE-2R is sensitive to nuclear αCP decoys. The α2R or α2Rm expression vector was separately transfected into MEL/tTA cells along with plasmids encoding the nuclear decoy U1-R7α1, its empty framework U1 RNA, the cytoplasmic decoy VA1-R7α1, or its corresponding empty framework VA1 RNA. The half-life of α2R or α2Rm mRNA under each treatment was derived from the decay curve plotted from the RPA data. The computed half-life is shown in the box, and representative RPA gels of 8-h chase studies are displayed below each of the corresponding graphs. The data represent results from five to seven independent 8-h or 16-h chase studies.
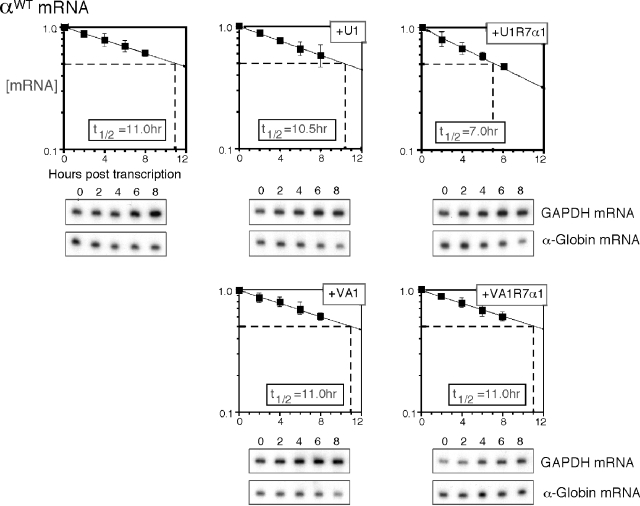
To further explore the specificity of the nuclear decoy, the above studies were repeated with the αWT-globin mRNA. MEL/tTA cells were cotransfected with U1-R7α1 or VA1-R7α1 expression vectors along with the αWT globin gene (Fig. 8). The half-life of α-globin mRNA in the presence of nuclear αCP decoy U1-R7α1 was shortened from 11 h to 7.0 h (P = 9.1 × 10−5). In contrast, the cytoplasmic VA1-R7α1 decoy had no appreciable effect. These data indicate that the selective impact of nuclear decoys on mRNA turnover is shared between the Lox DICE and the α-globin PR stability determinants. Since mRNA turnover during the 16-hour chase period is a cytoplasmic event, these observations led us to propose that nuclear events impact on the cytoplasmic function of αCP mRNP complexes.
FIG. 8.
Wild-type α-globin mRNA is selectively destabilized by nuclear αCP decoys. The αWT expression vector was cotransfected with a plasmid encoding the nuclear decoy U1-R7α1, its framework U1 RNA, the cytoplasmic decoy VA1-R7α1, or its framework VA1 RNA. The half-life of αWT mRNA under each treatment (value in box) was derived from the decay curve plotted from the RPA data. The data represent results from five to eight independent studies; representative RPA gels are shown below the respective graphs.
DISCUSSION
Terminal differentiation of mammalian erythroblasts takes place in cells that are undergoing a staged process of global transcriptional shutdown, chromatin condensation, and nuclear extrusion. As such, much of the gene expression profile must be established and coordinated via posttranscriptional controls (40). mRNAs that are important for continuous or scheduled expression late in the process of erythroid differentiation must be selectively preserved over a period of several days. The stability of the α- and β-globin mRNAs is supported by the binding of αCPs to the C-rich determinants in the 3′-UTR (18, 19, 44). Mutations in the hα-globin mRNA that destroy its stability result in a dramatic loss of globin protein production; such mutations underlie the most common cause of nondeletion α-thalassemia in the human population (4-6, 42). A number of mRNAs in addition to globin mRNAs must also be preserved in the differentiating erythroid cells to program the final steps in red cell maturation (e.g., 15-Lox and carbonic anhydrase). Thus, mechanisms that are involved in the stabilization of globin mRNAs may be shared with other mRNAs to coordinate and regulate late-stage erythroid cell differentiation and function.
In addition to stability control, the ability of a cell to reversibly control translation allows for a dynamic alteration in the gene expression profile in a transcriptionally silent environment. The most clearly defined translational controls of this sort have been described in early embryos and reflect specific 3′ adenylation and subsequent translational activation of target mRNAs (7, 34). The canonical cytoplasmic polyadenylation elements comprise a set of related UA-rich structures in the 3′-UTRs of the target mRNAs (9, 26, 27). The 15-lipoxygenase mRNA, as reported in the literature, represents a distinct example of translational control. Lox mRNA is transcribed in the early erythroblast, stored in an inert state, and then translationally activated in the late reticulocyte stage, where it produces an enzyme necessary for clearance of mitochondrial membranes from the maturing red blood cell (15, 31, 37). Translational control of Lox mRNA is attributed to a 19-bp pyrimidine-rich DICE repeat element (see the introduction) that is conserved among mammalian species. The 19-bp DICE monomer consists of C-patches interrupted at conserved sites by purines (32). Studies indicate that a single DICE repeat is insufficient for activity, while two or more are able to mediate translational control (32). A single molecule of αCP1 is proposed to bind to each of the repeat units, and this binding appears to have cooperative kinetics when two or more units are present (32). Such αCP-αCP interactions may be critical to the translational control pathway (13). It is implicit in this scenario of translational control that LOX mRNA must be maintained intact throughout 4 to 6 days of erythroblast differentiation in order to be available for reactivation in the reticulocyte. While translational silencing has been attributed to the DICE, the basis for the stable maintenance of the mRNA has not been explored.
Thus, two distinct categories of posttranscriptional controls, mRNA stabilization and translational silencing, are linked to programming gene expression in the differentiating erythroblast. The widely expressed poly(C)-binding protein αCP has been reported to be involved in both of these processes. The DICE and the C-rich PR element in the α-globin 3′-UTR share significant similarity in that they are both composed of C-rich motifs that target αCP binding. It is thus of clear interest to determine the basis for their distinct functions.
The DICE-2R motif is not sufficient for translational silencing.
As an initial step to compare and define the functional differences between the DICE and PR determinants, we attempted to recapitulate the DICE-mediated translational control model. The translational silencing activity of the DICE has been modeled by placing the DICE-2R unit 3′ of an fLuc reporter and expressing this mRNA in transfected HeLa cells (29). In the present study, the expression of fLuc-2R mRNA was compared in transfected HeLa cells to that of a derivative mRNA in which the DICE motif was mutated to block its function (fLuc-2Rm). The expectation was that expression of fLuc activity would be suppressed by insertion of the DICE-2R to the fLuc mRNA and that this repression would be relieved by the 2Rm mutation. Surprisingly, in multiple attempts, the fLuc-2R reporter was expressed as well as, or better than, fLuc-2Rm (Fig. 2B). Thus, although αCP proteins are expressed at high levels in HeLa cells and these proteins actively bind to the DICE-2R elements (Fig. 1B and 4B), they do not appear to silence fLuc-2R mRNA translation. The same comparisons of the functional and inactive DICE-2R motifs were repeated in a context of elevated αCP1 levels. αCP1 binding has been specifically linked to DICE translational repression (29). Again, the results were counter to the anticipated response. A subsequent set of in vitro studies supported these negative results. Linkage of the 2R motif to the fLuc mRNA failed to repress in vitro translation in RRL either in the context of the endogenous αCPs or in the presence of added recombinant αCP1 (Fig. 3C). In all cases the translation of the fLuc-2R mRNA was somewhat higher rather than lower than the fLUC-2Rm mRNA (Fig. 2 and 3). Increasing the combined levels of αCP1 and hnRNP K also failed to silence the translation of fLuc-2R compared to fLuc-2Rmut mRNA. Finally, a reciprocal approach was attempted in which the activity of αCP in the RRL was blocked by introducing two independent, high-affinity decoy RNAs (Fig. 4A). These decoys sequester αCP and theoretically should selectively enhance (i.e., derepress) fLuc-2R mRNA translation. As shown in Fig. 4C, the SELEX decoy, VA1-R7α1, and dC17 resulted in the expected suppression of poliovirus IRES function. However, neither decoy had the expected derepressive effect on translation of the fLuc-2R mRNA, nor did either demonstrate a differential effect on the translation of the fLuc mRNA linked to the active versus the mutant DICE motif (DICE-2R versus DICE-2Rm). Thus, DICE-2R failed to mediate appreciable translational control on the fLuc reporter either in vitro or in vivo in the context of ambient αCPs and in situations in which αCP activity was specifically augmented or blocked.
While it is possible that unappreciated differences in experimental approaches account for the apparent discrepancies between the presently reported translation assays and prior studies of DICE function (29), the nature of these differences remains undefined. We used the DICE-2R determinant in our studies. The existence of 10 copies of the DICE in the native rabbit 15-Lox mRNA may reflect a requirement for a higher number of DICE repeats to repress translation under physiological settings or under specific experimental conditions. However, the lower number of DICE repeats in the 15-Lox 3′-UTR of other mammalian species (three in humans) argues against this model, as do prior studies that reported that two copies (DICE-2R) were sufficient in a variety of experimental settings (28, 29, 32). When we increased the DICEs in the 3′-UTR of fLuc reporter to eight copies (fLuc-8R) and repeated the in vitro translation study, we were still unable to detect evidence for translational control. The fLuc-8R translation was neither repressed by the addition of recombinant αCP1 and/or hnRNP K nor stimulated by competing off αCP1 and hnRNP K by dC17. The present translational analyses of DICE-2R and DICE-8R lead us to conclude that the DICEs do not mediate reproducible translational silencing in these experimental systems.
The DICE-2R motif is sufficient for the mRNA stabilization function.
Human α-globin mRNA is stabilized by the 3′-UTR C-rich PR determinant. The longevity of Lox mRNA in erythroid cells and the common pyrimidine-rich and C-rich structures in the 3′-UTRs of the α-globin and Lox mRNA determinants suggested a common function. To determine whether the DICE might represent an mRNA stability element, it was inserted in the α-globin mRNA in place of the PR determinant (Fig. 1A). Remarkably, the replacement of the PR by the DICE-2R results in an mRNA with stability equivalent to that of the wild-type hα-globin mRNA (Fig. 6). In contrast, parallel replacement with the DICE-2Rm fails to mediate stabilization of the α-globin mRNA. These data suggest that the DICE-2R and PR determinants can stabilize mRNA to the same extent. Recent studies have identified a large subset of mRNAs in erythroid cells that are bound in vivo by αCPs (39). To what extent the stabilization function of αCPs is more generally exerted in this setting can now be addressed.
mRNA stabilization by the DICE-2R determinant is dependent on nuclear αCP activity.
The DICE-2R element is able to confer full stabilization to the hα-globin mRNA lacking its cognate PR stability determinant. The linkage of mRNA stabilization to αCP was tested by blocking αCP function in trans with a set of decoy RNAs. The U1-R7α1 vector expresses high levels of the R7α1 aptamer in the nucleus, while VA1-R7α1 packages the same R7α1 aptamer in a cytoplasmic (VA1 RNA) delivery framework (22). The U1-R7α1 and VA1-R7α1 decoy RNAs bind αCP with equivalent affinities (Fig. 4B and data not shown). Each RNA decoy was coexpressed with α2R in MEL/tTA cells. It was assumed that if DICE-mediated stabilization reflected αCP function, the presence of the decoy would destabilize the target α2R mRNA. While this was the observed effect, it was surprising to find that the effect was only seen with the nuclear, but not the cytoplasmic, decoy (Fig. 7). To evaluate whether these results were peculiar to the function of the DICE-2R, the same set of decoys were expressed along with wild-type α-globin mRNA. The data revealed that α-globin mRNA is also selectively destabilized by the nuclear, but not the cytoplasmic, αCP decoy (Fig. 8). These results support a model in which the αCP complex critical to cytoplasmic mRNA stability is assembled in the nucleus. This model is supported by recent observations that the major αCP isoforms, αCP1, αCP2, and αCP2KL, contain novel nuclear localization signals and appear to shuttle between the nuclear and cytoplasmic compartments (2). The decoy RNA appears to be most effective in blocking the initial nuclear assembly of the α-complex as opposed to displacing αCP from the cytoplasmic RNP complex.
The possibility that αCPs load on α-globin transcripts in the nucleus prior to cytoplasmic export brings up the question of whether αCPs have direct nuclear functions that might complement their role in cytoplasmic regulation. In support of this model, αCPs have been found to associate with splicing factors in vivo, and αCP1 is specifically enriched in nuclear speckles (2). Recent studies have further revealed that αCP is associated with prespliced hα-globin mRNA and may play a direct role in nuclear processing events (X. Ji et al., submitted for publication). Thus, while αCP-mediated mRNA stabilization is a cytoplasmic event, transcript processing and/or RNP assembly in the nucleus may be dependent on the same, or a closely related, αCP-containing complex(es). Coordination of nuclear and cytoplasmic events mediated by αCP complexes may be critical to robust expression of the α-globin transcripts. The details of this linkage and whether it can be generalized to other mRNAs targeted by αCP can now be explored.
Acknowledgments
This work was supported by NIH funding from grants R37-MERIT HL 65449 and PO1-CA72765. We also acknowledge the generous support of the Doris Duke Foundation.
REFERENCES
- 1.Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein, multiple processes. Bioessays 26:629-638. [DOI] [PubMed] [Google Scholar]
- 2.Chkheidze, A. N., and S. A. Liebhaber. 2003. A novel set of nuclear localization signals determine distributions of the αCP RNA-binding proteins. Mol. Cell. Biol. 23:8405-8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chkheidze, A. N., D. L. Lyakhov, A. V. Makeyev, J. Morales, J. Kong, and S. A. Liebhaber. 1999. Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein αCP. Mol. Cell. Biol. 19:4572-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clegg, J. B., D. J. Weatherall, I. Contopolou-Griva, K. Caroutsos, P. Poungouras, and H. Tsevrenis. 1974. Haemoglobin Icaria, a new chain-termination mutant with causes alpha-thalassaemia. Nature 251:245-247. [DOI] [PubMed] [Google Scholar]
- 5.Clegg, J. B., D. J. Weatherall, and P. F. Milner. 1971. Haemoglobin Constant Spring: a chain termination mutant? Nature 234:337-340. [DOI] [PubMed] [Google Scholar]
- 6.De Jong, W. W., P. Meera Khan, and L. F. Bernini. 1975. Hemoglobin Koya Dora: high frequency of a chain termination mutant. Am. J. Hum. Genet. 27:81-90. [PMC free article] [PubMed] [Google Scholar]
- 7.de Moor, C. H., and J. D. Richter. 2001. Translational control in vertebrate development. Int. Rev. Cytol. 203:567-608. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 9.Fox, C. A., M. D. Sheets, and M. P. Wickens. 1989. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 3:2151-2162. [DOI] [PubMed] [Google Scholar]
- 10.Frischmeyer, P. A., and H. C. Dietz. 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8:1893-1900. [DOI] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 14.Hazelrigg, T. 1998. The destinies and destinations of RNAs. Cell 95:451-460. [DOI] [PubMed] [Google Scholar]
- 15.Hohne, M., B. J. Thiele, S. Prehn, E. Giessmann, B. Nack, and S. M. Rapoport. 1988. Activation of translationally inactive lipoxygenase mRNP particles from rabbit reticulocytes. Biomed. Biochim. Acta 47:75-78. [PubMed] [Google Scholar]
- 16.Ilves, H., C. Barske, U. Junker, E. Bohnlein, and G. Veres. 1996. Retroviral vectors designed for targeted expression of RNA polymerase III-driven transcripts: a comparative study. Gene 171:203-208. [DOI] [PubMed] [Google Scholar]
- 17.Keene, J. D., and P. J. Lager. 2005. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 13:327-337. [DOI] [PubMed] [Google Scholar]
- 18.Kiledjian, M., X. Wang, and S. A. Liebhaber. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 14:4357-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, J., X. Ji, and S. A. Liebhaber. 2003. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell. Biol. 23:1125-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leffers, H., K. Dejgaard, and J. E. Celis. 1995. Characterisation of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur. J. Biochem. 230:447-453. [PubMed] [Google Scholar]
- 21.Makeyev, A. V., A. N. Chkheidze, and S. A. Liebhaber. 1999. A set of highly conserved RNA-binding proteins, αCP-1 and αCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol. Chem. 274:24849-24857. [DOI] [PubMed] [Google Scholar]
- 22.Makeyev, A. V., D. L. Eastmond, and S. A. Liebhaber. 2002. Targeting a KH-domain protein with RNA decoys. RNA 8:1160-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makeyev, A. V., and S. A. Liebhaber. 2000. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics 67:301-316. [DOI] [PubMed] [Google Scholar]
- 24.Makeyev, A. V., and S. A. Liebhaber. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matunis, M. J., W. M. Michael, and G. Dreyfuss. 1992. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGrew, L. L., E. Dworkin-Rastl, M. B. Dworkin, and J. D. Richter. 1989. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 3:803-815. [DOI] [PubMed] [Google Scholar]
- 27.McGrew, L. L., and J. D. Richter. 1990. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 9:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostareck, D. H., A. Ostareck-Lederer, I. N. Shatsky, and M. W. Hentze. 2001. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104:281-290. [DOI] [PubMed] [Google Scholar]
- 29.Ostareck, D. H., A. Ostareck-Lederer, M. Wilm, B. J. Thiele, M. Mann, and M. W. Hentze. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89:597-606. [DOI] [PubMed] [Google Scholar]
- 30.Paulding, W. R., and M. F. Czyzyk-Krzeska. 1999. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J. Biol. Chem. 274:2532-2538. [DOI] [PubMed] [Google Scholar]
- 31.Rapoport, S. M., and T. Schewe. 1986. The maturational breakdown of mitochondria in reticulocytes. Biochim. Biophys. Acta 864:471-495. [DOI] [PubMed] [Google Scholar]
- 32.Reimann, I., A. Huth, H. Thiele, and B. J. Thiele. 2002. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J. Mol. Biol. 315:965-974. [DOI] [PubMed] [Google Scholar]
- 33.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seydoux, G. 1996. Mechanisms of translational control in early development. Curr. Opin. Genet. Dev. 6:555-561. [DOI] [PubMed] [Google Scholar]
- 35.Siomi, H., M. J. Matunis, W. M. Michael, and G. Dreyfuss. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21:1193-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefanovic, B., C. Hellerbrand, M. Holcik, M. Briendl, S. Aliebhaber, and D. A. Brenner. 1997. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiele, B. J., H. Andree, M. Hohne, and S. M. Rapoport. 1981. Regulation of the synthesis of lipoxygenase in erythroid cells. Acta Biol. Med. Ger. 40:597-602. [PubMed] [Google Scholar]
- 38.Thisted, T., D. L. Lyakhov, and S. A. Liebhaber. 2001. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and αCP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem. 276:17484-17496. [DOI] [PubMed] [Google Scholar]
- 39.Waggoner, S. A., and S. A. Liebhaber. 2003. Identification of mRNAs associated with αCP2-containing RNP complexes. Mol. Cell. Biol. 23:7055-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waggoner, S. A., and S. A. Liebhaber. 2003. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. 228:387-395. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., M. Kiledjian, I. M. Weiss, and S. A. Liebhaber. 1995. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol. 15:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss, I. M., and S. A. Liebhaber. 1994. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol. Cell. Biol. 14:8123-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, I. M., and S. A. Liebhaber. 1995. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol. Cell. Biol. 15:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, J., and J. E. Russell. 2001. Structural and functional analysis of an mRNP complex that mediates the high stability of human beta-globin mRNA. Mol. Cell. Biol. 21:5879-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]