Abstract
Random assortment within mammalian genomes juxtaposes genes with distinct expression profiles. This organization, along with the prevalence of long-range regulatory controls, generates a potential for aberrant transcriptional interactions. The human CD79b/GH locus contains six tightly linked genes with three mutually exclusive tissue specificities and interdigitated control elements. One consequence of this compact organization is that the pituitarycell-specific transcriptional events that activate hGH-N also trigger ectopic activation of CD79b. However, the B-cell-specific events that activate CD79b do not trigger reciprocal activation of hGH-N. Here we utilized DNase I hypersensitive site mapping, chromatin immunoprecipitation, and transgenic models to explore the basis for this asymmetric relationship. The results reveal tissue-specific patterns of chromatin structures and transcriptional controls at the CD79b/GH locus in B cells distinct from those in the pituitary gland and placenta. These three unique transcriptional environments suggest a set of corresponding gene expression pathways and transcriptional interactions that are likely to be found juxtaposed at multiple sites within the eukaryotic genome.
The random assortment of structural loci in higher eukaryotic genomes brings with it a need to functionally “insulate” genes that are closely packed and yet must maintain distinct tissue and developmental specificities. While such insulation can be mediated by fixed chromatin structures (2, 21), not all unwanted transcriptional interactions can be blocked in this simple linear manner. For example, genes in close proximity can be under the control of remote control elements that are interdigitated and/or situated within chromatin domains or transcription units of neighboring genes (30). Thus, establishing and maintaining specificity of gene expression in the context of varied locus organizations is likely to demand an array of mechanistic solutions. In certain cases, problems of transcriptional insulation may not be fully resolved and may result in gene expression in the “wrong” tissue (5). Such ectopic expression or “leakiness” within the transcriptome may be the price that the cell pays for the random distribution of its genetic elements.
The hGH multigene cluster represents an informative model for the study of developmental control (17, 27). This cluster, located on chromosome 17q22-24, encompasses five growth hormone paralogs (Fig. 1A). hGH-N is expressed in pituitary somatotrope cells; the four remaining genes, hCS-L, hCS-A, hGH-V, and hCS-B, are expressed in the syncytiotrophoblast (STB) layer of the placental villi (6, 22). Activation of the pituitary and the placental genes is dependent on distinct actions of a 5′-distal locus control region (LCR). This hGH LCR is comprised of five DNase I hypersensitive sites (HS) (Fig. 1A). The pituitary cell-specific HSI and HSII, located −14.5 and −15.5 kb 5′ to the cluster, are sufficient to drive high-level hGH-N expression in pituitary somatotropes (17). Selective inactivation of HSI results in a dramatic loss of hGH-N expression (15). HSIV, located at −30 kb, forms exclusively in STB chromatin, but a specific role for this HS has not yet been demonstrated. HSIII and HSV are shared by placental and pituitary chromatin. The CD79b gene, which encodes the signal transduction subunit of the B-cell antigen receptor (26), is located between the hGH cluster and the hGH LCR (Fig. 1A) (3). The CD79b protein first appears at the pre-B-cell stage, and its expression is maintained throughout all stages of B-cell differentiation (4, 14). Remarkably, CD79b mRNA is also abundant in pituitary somatotropes (5). This pituitary CD79b mRNA appears to be structurally the same as that in B cells, yet it fails to produce appreciable protein. The mechanism of this posttranscriptional control remains undefined.
FIG. 1.
hCD79b/GH locus and expression of CD79b in human B-cell lines. A. The hCD79b/GH locus on human chromosome 17q22-24. Vertical arrows indicate the positions of DNase I HS previously identified in chromatin of pituitary somatotropes and placental STB. HSI and HSII are pituitary gland specific, and HSIV is placenta specific. The horizontal arrows indicate the direction of transcription from each gene. The corresponding tissue specificities are noted. B. CD79b expression in developmental-stage human B-cell lines. RNA samples from the K562 human erythroleukemia cell line and from a series of established human B-cell lines were analyzed by Northern blotting. The identity of each B-cell line and its reported stage of differentiation along the B-cell lineage is noted above the respective lanes. The membrane was hybridized with 32P-cDNA probes to human CD79b mRNA and to mouse ribosomal protein 32 (mRPL32) mRNA as a loading control. The expected positions of the two mRNA signals are noted to the left of the autoradiograph.
Transcription of CD79b in pituitary cells has been attributed to a “bystander” pathway of gene activation. That is to say, this ectopic expression of CD79b is triggered by its position within the 32-kb pituitary cell-specific domain of acetylated chromatin that links the hGH LCR and hGH-N in this tissue (5, 15). In contrast, acetylation at this locus in placental chromatin is restricted to the HSIII-HSV region and to the placental genes, leaving the region between the LCR and the cluster unmodified (19). In this way, CD79b is excluded from “activated” regions of modified chromatin and remains unexpressed in the placental STB nuclei (5). Thus, chromatin structures in pituitary and placenta tissue can account for selective “bystander” activation of CD79b in the pituitary cells by hGH regulatory sequences. In the present report, we explore the structure of the CD79b transcriptional environment in B cells. The aim of these studies is to understand how the corresponding B-cell-specific determinants and chromatin modifications interface with the adjacent hGH cluster to result in the overall pattern of transcriptional interactions within this multigene cluster.
MATERIALS AND METHODS
Cell lines and purification of B cells from mouse spleen.
The following cell lines were used in this study: K562 (erythroleukemia cell line); JM1 and REH (pro-B-cell lines); Nalm6 and 697 (pre-B-cell lines); CA46, 15A, and 25A (mature B-cell lines); U266, CRL9068, CRL1621, and CRL1484 (plasma B-cell lines); and JEG3 (human choriocarinoma cell line). Several of the cell lines have specific ATCC designations: JM1 is designated CRL10423; REH, CRL8286; CA46, CRL1648; and U266, TIB196. B-cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin; JEG3 cells were cultured in minimal essential medium. All cells were grown at 37°C and 5% CO2 and were maintained at a maximum cell density of 1 × 106 cells/ml.
To purify mouse lymphocyte cells, spleens removed from anesthetized mice were dispersed through a 30-μm nylon mesh to generate a single-cell suspension. Lymphocytes were isolated by Ficoll gradient using Ficoll-Plaque plus (Amersham Biosciences, Piscataway, NJ) by centrifugation at 650 × g for 30 min without braking. After centrifugation, the cells at the interface between Ficoll and the aqueous phase were collected and washed twice with phosphate-buffered saline (PBS).
For primary B-cell chromatin immunoprecipitation (ChIP) assays, lymphocytes were further purified by negative selection using MACS CD43 (Lys48) microbeads (Militenyi Biotec, Germany). B cells were magnetically enriched (excluding pro-B cells) using MACS LD depletion columns and Mini-MACS cell sorting kits (Militenyi Biotec, Germany) according to the manufacturer's instructions.
Transgenic constructs and generation of transgenic mouse lines.
All transgenic mouse lines used in this study are listed in Table 1 (also see the diagram in Fig. 3A). The hGH/P1 line (87-kb hCD79b/GH transgene), −1.3CD79b (5.6-kb hCD79b transgene), and −0.2CD79b (4.6-kb hCD79b transgene) have been previously reported (5, 27). −0.5CD79b contains a 4.9-kb genomic fragment generated by BspMI and EcoRI digestions of a 5.6-kb hCD79b genomic DNA fragment (5). This fragment was gel isolated, extracted with a QIAquick gel extraction column (QIAGEN, Germany), purified through an Elutip D column (Schleicher & Schuell, Germany), and adjusted to a concentration of 2 ng/μl in TE buffer (10 mM Tris-HCl [pH 7.6], 0.1 mM Na2EDTA) prior to microinjections into fertilized mouse oocytes (C57BL/6 × SJL). A BAC genome clone encompassing the hCD79b/GH locus was screened by Invitrogen Life Technologies using colony hybridization with TCAM and SCNex13 probes corresponding to TCAM1 and SCN4A genes, respectively. A clone of 148.3 kb was identified (clone #535D15). A 123-kb insert was released by NotI digestion. The NotI sites are present in exon 1 of the SCN4A gene and between TCAM1 and SMARCD2 genes. The resulting released BAC (named CD/hGH BAC) insert was separated by pulsed-field gel electrophoresis (Bio-Rad, Hercules, CA) and purified from an excised slice of the agarose gel by digestion with 1 U/100 μl β-agarase I (New England Biolabs, Beverly, MA) followed by phenol-chloroform extraction. This purified 123-kb DNA fragment was used for microinjection as described above. All transgenic mouse founders were generated by routine procedures by the University of Pennsylvania Transgenic and Chimeric Mouse Core Facility. Positive founders were identified by Southern blot analyses of NcoI-digested tail DNA. Transgene integrity was intact in all reported lines. Transgene copy numbers were quantified by PhosphorImager (Storm 840 PhosphorImager; Molecular Dynamics, Sunnyvale, CA) analyses of Southern blots. The hCD79b transgene signal was normalized to the hybridization signal from the endogenous mouse ζ-globin gene on the same filter.
TABLE 1.
Transgenic mouse line constructs, lines, and copy numbers
| Transgene | Construct size (kb) | Line designation | Copy no. |
|---|---|---|---|
| CD/hGH BAC | 123 | 1210B | 2 |
| 1252E | 24 | ||
| 1253A | 17 | ||
| 1254D | 3 | ||
| 1255B | 2 | ||
| hGH/P1 | 87 | 811D | 4 |
| −8.0CD79b | 12 | 989D | 7 |
| 1002D | 8 | ||
| 1047C | 3 | ||
| 1047E | 4 | ||
| 1194F | 5 | ||
| −1.3CD79b | 5.6 | 1014A | 3 |
| 1014F | 2 | ||
| 1092C | 3 | ||
| 1092Fa | 5 | ||
| 1092Fb | 10 | ||
| −0.5CD79b | 4.9 | 1261E | 4 |
| 1262A | 12 | ||
| 1265E | 9 | ||
| 1265F | 3 | ||
| 1266H | 7 | ||
| −0.2CD79b | 4.6 | 1290A | 5 |
| 1290B | 3 | ||
| 1290D | 2 | ||
| 1290K | 2 | ||
| 1294E | 10 |
FIG. 3.
hCD79b mRNA levels in splenic lymphocytes of transgenic mice. A. Transgenes are depicted. The map of the CD79b gene and its 5′-flanking region indicates the positions of B-cell-specific HS (black arrows) and the two HS specific to the pituitary gland (HSI and HSII; white arrows) for reference. A CpG hypomethylated region of DNA specific to B cells is shown (stippled circle). The four CD79b transgenes are indicated on the map along with their respective sizes. The name of each of these transgenes corresponds to the length of the 5′-flanking region in each case. B. Quantitation of human versus mouse CD79b mRNAs. hCD79b mRNA and endogenous mCD79b mRNA were reverse transcribed (RT) coamplified from B cells purified from the spleens of the indicated transgenic mice. The coamplification was accomplished using a set of conserved sequence primers (mhIgβ forward and reverse; Table 2; white horizontal arrows). The PCR product spans exons 5 and 6, permitting size spliced RNA products. The cDNAs corresponding to human and mouse mRNAs were differentiated by restriction enzyme cleavage. Digestion of the 5′ 32P end-labeled cDNA products with SfcI (S) will exclusively generate the hCD79b cDNA product (95 bp), whereas digestion with HinfI (H) exclusively generates the mCD79b cDNA product (63 bp). A diagram of the predicted fragment migration on an analytic gel is represented. C to G. hCD79b mRNA expression in B cells of each CD79b transgenic mouse line. For each transgene, five independent lines with unique transgene insertion sites were analyzed. Signals corresponding to the human and mouse CD79b mRNAs were determined according to the scheme depicted in panel B. The SfcI (S) and HinfI (H) digestion products were quantified, and the human/mouse mRNA ratio was divided by the transgene copy number (indicated below each pair of lanes). The results of hCD79b mRNA expression per transgene copy were expressed as a percentage of a single endogenous mCD79b gene (also shown below each pair of lanes). H. Summary of CD79b mRNA expression in B cells of the six transgenic mouse lines. The hCD79b expression percentages derived from analysis of each of the transgenic lines (C to G) are displayed on the semilog plot. The value from each line, shown as a diamond, reflects the average of two or more independent assays. The data from the hGH/P1 lines have been previously reported (5) and are included to facilitate comparisons.
Northern blot analysis.
Total RNAs were extracted with RNA-Bee (Tel-Test, Freindswood, TX) from cultured cells and mouse tissues. A 20-μg aliquot of each RNA sample was separated on a 1.2% agarose gel containing 2.2 M formaldehyde in 1× morpholinepropanesulfonic acid and transferred to a Zetaprobe membrane (Bio-Rad, Hercules, CA). The membrane was probed with 32P-labeled hCD79b cDNA at 65°C overnight in hybridization solution (0.5 M NaHPO4 [pH 7.2], 0.1 mM EDTA, 1% bovine serum albumin, 7% sodium dodecyl sulfate [SDS]) and then washed (1× SSC, 0.1% SDS, 0.5× SSC, 0.1% SDS, and finally 0.1× SSC, 0.1% SDS [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at room temperature to 65°C, and signals were detected by exposure to a PhosphorImager screen.
Isolation of intact nuclei from B-cell lines and DNase I hypersensitive site mapping.
Cells from the various B-cell lines were collected, washed with PBS, and resuspended at 108 cells/ml in ice-cold sucrose buffer I (0.32 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 10 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.05% Triton X-100). The cell suspensions were centrifuged at 500 × g for 5 min at 4°C and washed with RB buffer (100 mM NaCl, 50 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 0.1 mM PMSF, 5 mM sodium butyrate). For DNase I mapping, the nuclei were resuspended again at 108 cells/ml of RB buffer, and a baseline (at 0 s) aliquot was withdrawn. RB buffer containing 3 mM CaCl2 and 300 U of DNase I (Invitrogen, Carlsbad, CA) per ml was added to yield final concentrations of 1 mM CaCl2 and 100 U of DNase I per ml. The sample was incubated at 37°C. Aliquots were withdrawn at the indicated times and EDTA was added to 50 mM, followed by digestion of the nuclei with 120 μg of proteinase K per ml in 0.8 M NaCl, 0.5% SDS, extraction with phenol and chloroform, and a final ethanol precipitation terminating each reaction. DNase I-digested DNA samples were subsequently digested with restriction enzymes and transferred to Zetaprobe membranes (Bio-Rad, Hercules, CA) for hybridization. Hybridization probes were synthesized by PCR amplification using primer sets (Prb1 and Prb2) from human genomic DNA (Table 2).
TABLE 2.
Primer sets used for ChIP, HS mapping, and coamplified reverse transcription-PCR assays
| Name | Forward | Reverse | Location (kb)a |
|---|---|---|---|
| P7 | GCTCATCAAGATCATTGGCA | AATTTGAGAACTACGGG | −25.2 |
| HSV | CTTGCCCAGTCCTCACACTT | CTGAGGCTTCTGTCCTCCTT | −18.8 |
| HSIV | TGCCTCTACGTGGACATCTC | TATCAGCAGAGAGTGCACAA | −16.5 |
| P6 | CTGGGTGGCGTAGAGATG | GACCCACGTTGTCGTAGTTG | −13.4 |
| P5 | GCCTCAAAACCTGATTGG | GGAGATCTCTGAGGCTGG | −7.88 |
| HSI/II | CCAAGCCTTTCCCAGTTATAC | GATCTTGGCCTAGGCCTC | −1.70 |
| CDp | GGACAGGTGCCTATTTCGCTC | GACCCCAAACCCGTGACAAC | −0.40 |
| P3 | CTTCCCCCAGAAGACTGA | GGAGGAAAACGTGTAACTGC | +0.08 |
| P2 | TGCTCAGACCAGCCTATGCA | TCAACAGGAAGTGGAGCACA | +3.90 |
| P1 | GATTACAAGCGCCCACTACC | GAGAGAATAAGCCAGGAGGTG | +7.90 |
| GHNp | CAGGGCTATGGGAGGAAGAGCTTG | CTTCTCTCCCACTGTTGCCC | +12.7 |
| Prb1 | AGCTCTCGTTCTGGCCTTCTTGGT | GAGTCCATTTGCGGGCTCTGGAC | +2.58 |
| Prb2 | TCTCCTTCCTCATCGTGGTCAAC | ATTCAAAGCCCTCCTCCCTCACTC | −13.7 |
| mhGAPDH | GCCAAAAGGGTCATCATCTC | CTGCTTCACCACCTTCTTGA | |
| mhIgβ | GGAGGAAGATCACACCT | ATCCCCAGAGAACTCC | —b |
The first ATG of hCD79b is located at +1 kb.
—, forward on exon 5 (+2.8 kb), reverse on exon 6 (+3.2 kb).
Coamplified reverse transcription-PCR endonuclease cleavage assay.
To quantify human CD79b mRNA expression in each B-cell preparation, reverse transcription, PCR, and restriction enzyme digestion methods were performed as previously described (5). All assays were carried out on lymphocytes purified from transgenic mouse spleens (as described above). In all cases, 0.5 μg of total RNA, extracted from tissues or cell lines using RNA-Bee (Tel-Test Inc., Freindswood, TX), was reverse transcribed with an oligo(dT) primer in the presence of avian myeloblastosis virus reverse transcriptase. Human and mouse CD79b mRNAs were then coamplified using a primer set corresponding to regions perfectly conserved between hCD79b and mCD79b (mhIgβ forward and reverse; Table 2). The forward primer was 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. This primer set spans intron 5 to distinguish cDNA from amplified genomic DNA. PCR products at 25 cycles were digested; HinfI specifically cleaves mCD79b cDNA, and SfcI specifically cleaves hCD79b cDNA. Fragments were separated on 6% polyacrylamide gels, and bands were quantified by PhosphorImager analyses. The ratios of hCD79b to mCD79b cDNA products were normalized to the transgene copy numbers, and the final hCD79b mRNA expression value was calculated as a percentage of the expression from a single endogenous mCD79b gene. All PCR results were confirmed to be within the linear range of amplification.
Chromatin immunoprecipitation (ChIP) assay for modified histones.
ChIP studies were performed with human B-cell lines (1484 and U266), a human erythroid cell line (K562), and tissues isolated from hGH/P1 mice (line 811D) (27). Preparation of unfixed chromatin and the ChIP assay were carried out as described previously (8). Briefly, 0.3 mg nuclei was digested with 25 U of micrococcal nuclease at 37°C for 6 min in 1 ml 50 mM NaCl, 20 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 1 mM CaCl2, 10 mM sodium butyrate, 0.1 mM PMSF. The reaction was stopped by the addition of Na2EDTA to a final concentration of 0.5 mM, and salt-soluble chromatin was isolated as described previously (13). Soluble chromatin was concentrated using a Microcon centrifugal filter (Amicon Inc., Bedford, PA), and 250 μg of this chromatin input was incubated with 10 μl each of the antibodies to acetylated H3 and H4 (Upstate Biotechnologies, Lake Placid, NY), or in the absence of antibody, in a total volume of 500 μl. Protein A-Sepharose (Amersham Pharmacia Biosciences, Piscataway, NJ) precipitates were generated and washed, and DNA was purified from the pellets (bound) as described previously (23). DNA samples from the input and bound fractions were amplified by PCR using various primer sets listed in Table 2. Parallel dilutions of 1/300, 1/600, and 1/1,200 were made from 15% of the input sample and from the entire immunoprecipitated sample pellet. Aliquots containing a final concentration of 0.16%, 0.08%, and 0.04% of each immunoprecipitated sample were analyzed by PCR to confirm that each assay was within the linear range of amplification. Each product was analyzed by electrophoresis on 1% agarose gels followed by Southern blotting. The blots were hybridized with 1 × 106 to 2 × 106 cpm/ml random primer-labeled probe at 65°C overnight. The membranes were subsequently washed at 60°C in 0.1% SDS and 0.5× SSC. Signals were quantified by PhosphorImager analysis. Each ratio was normalized to the comparable signal detected at the ubiquitously expressed GAPDH locus. For the histone H3 acetylation and H4 acetylation ChIP assays, two independent assays were performed from different preparations of nuclear extracts. For the histone H3 K4 dimethylation ChIP assay, a single experiment was performed.
RESULTS
B-cell-specific DNase I HS are established at the CD79b locus.
The chromatin structure at the hCD79b/GH locus was analyzed in a series of B-cell lines representing different stages of differentiation. Consistent with their B-cell characteristics, each line expresses CD79b mRNA (Fig. 1B). However, the levels of CD79b mRNA differ significantly among these lines and fail to correlate with the stage of B-cell differentiation. A subset of these lines representing various levels of CD79b expression was chosen for HS mapping. Control studies were carried out on K562 erythroleukemia cells and a human choriocarcinoma cell line (JEG3).
HS mapping surveyed two adjacent EcoRI restriction fragments spanning the CD79b gene and 31 kb of 5′-flanking sequences (Fig. 2). HS mapping of the 12.5-kb EcoRI fragment containing the human CD79b gene, the contiguous 8.0 kb of 5′-flanking sequences, and 850 bp of 3′-flanking sequences revealed two HS (HSB1 and HSa; Fig. 2A). HSa mapped within CD79b intron 1 and was detected in all cells studied, irrespective of cell type or CD79b expression. HSB1 mapped to the CD79b promoter and formed specifically in B-cell chromatin. DNase I analysis of the adjacent 5′ EcoRI fragment, 23.1 kb in length, identified three HS (Fig. 2B). Two of these HS corresponded to the previously reported HSIII and HSV of the hGH LCR (17). HSIII was detected in all cell types tested, including pituitary, placenta, B, and erythroid cells, whereas HSV was observed in B-cell lines and pituitary, placenta, and JEG3 cells but not in K562 cells (Fig. 2B and data not shown). A novel HSB2 was also identified in this region. This HS was exclusive to B cells and mapped 14 kb 5′ to the CD79b promoter. HSB2 intensity roughly paralleled the levels of CD79b mRNA in the various B-cell lines. In summary, the DNase I mapping identified three novel HS at the human CD79b/GH locus; a constitutive HSa within the CD79b gene, a B-cell-specific HS mapping to the CD79b promoter, and a second B-cell-specific HS located 14 kb 5′ of the CD79b gene (Fig. 2C).
FIG. 2.
Identification of a B-cell-specific DNase I HS at the hCD79b/GH locus. A. Two DNase I HS are detected within and immediately 5′ of the CD79b gene; HSB1 is B cell specific. Chromatin samples from the B-cell lines were subjected to DNase I HS mapping. For the left panel, chromatin preparations from six human B-cell lines representing various developmental stages (697, REH, 25A, 9068, 1484, and 1621) were analyzed. The relative level of CD79b mRNA in each cell line, as determined from Northern blot analysis (Fig. 1 and data not shown), is indicated above the corresponding lanes (strong, ++; intermediate, +; low, +/−). The first lane contains a DNA size marker (M). The analyzed EcoRI fragment is 12.5 kb (marked by a black dot). Sub-bands generated by graded DNase I digestions (times of 0 s, 10 s, and 1 min) are indicated by the horizontal arrows to the right of the autoradiograph (HSB1 and HSa). The band at 8 kb was present irrespective of DNase I digestion (time 0) and was considered to be a cross-hybridizing band of unknown origin. For the right panel, DNase I HS mapping of chromatin from a human choriocarcinoma cell line (JEG3) and the erythroleukemia cell line (K562) were compared to a pre-B-cell line (Nalm6). HSB1 was only detectable in Nalm6 cells, whereas HSa was present in all three lines. The diagram below the blots summarizes the strategy and results of the HS mapping studies depicted in the blots. The relevant EcoRI restriction enzyme sites, the probe (Prb1; Table 2; filled box below the CD79b gene), fragment sizes, and positions of HSB1 and HSa, as well as pituitary cell-specific HSI and HSII for reference, are shown within the 12.5-kb EcoRI fragment. B. Three DNase I HS are detected in the remote 5′-flanking region of CD79b; HSB2 is B cell specific. Chromatin mapping and labeling of the autoradiograph are as described for panel A. The 23.1-kb EcoRI fragment (marked by a black dot) studied with probe Prb2 is located immediately 5′ to the fragments depicted in panel A. The DNase I sub-bands generated from this fragment are HSV, HSIII, and HSB2 (positions indicated at the right of each autoradiograph). The left panel shows DNase I mapping of the human B-cell lines; positions of HSV, HSIII, and HSB2 are indicated at the right. The right panel shows DNase I mapping of two nonlymphoid lines (JEG3 and K562) and a B-cell line (CA46). HSB2 was formed in the B-cell line but not in JEG3 or K562. The band below the 23.1-kb master band in panel B is a nonspecific, cross-hybridizing band. The diagram at the bottom summarizes the mapping experimental scheme and results as described in the legend to panel A. The hybridization probe (Prb2) is indicated by the filled rectangle. C. Localization of B-cell-specific HS in the hCD79b/GH locus. Arrows (open) above the map indicate B-cell-specific HS and the HSa identified in the current report. Arrows (closed) summarize HS identified in prior studies. HSI and HSII are specific to pituitary chromatin, HSIV is specific to placental chromatin, and HSIII and HSV are constitutive.
hCD79b expression is under local control.
Expression of the hGH cluster in pituitary and placenta tissue is dependent on a remote LCR (Fig. 1A). Identification of HSB2 suggested that CD79b might be under similar remote regulatory control. The full set of HS mapping data (Fig. 2C) was consistent with a model in which the pituitary tissue-specific HSI and HSII, the B-cell-specific HSB2, and the placenta tissue-specific HSIV might each act in conjunction with the shared HSV and HSIII to selectively activate their respective target genes. To test this model, sequences necessary for hCD79b expression were mapped in vivo using a series of mouse transgenic models (Fig. 3A). For each transgene, five independent mouse lines were established. For each line, the transgene copy number was determined by Southern blotting (Table 1 and data not shown), and the level of hCD79b mRNA expression in the transgenic B cells was quantified in two or more mice relative to endogenous mouse CD79b (mCD79b) mRNA (Fig. 3B). The ratio of hCD79b mRNA to mCD79b mRNA in each line was divided by the respective transgene copy number. In this manner, hCD79b transgene expression was calculated as a percentage of the expression from a single endogenous mCD79b gene (see Materials and Methods for details).
Prior analyses had revealed that the hGH/P1 transgene supports robust hCD79b expression in mouse B cells at levels of 25 to 42% of a single endogenous mCD79b gene (5). To extend this baseline, we established five lines carrying a 123-kb BAC transgene encompassing CD79b with more extensive flanking regions (details will be reported separately) (Fig. 3A). CD79b mRNA was expressed from this CD/hGH BAC transgene at levels similar to that observed in the hGH/P1 lines (10 to 53% of the endogenous mCD79b locus; Fig. 3C). With these two large transgenes as standards, we next assessed expression from a set of transgenes that isolated CD79b from its remote sequences. The first transgene, −8.0CD79b, contained the CD79b gene along with 8 kb of 5′-flanking sequences. This transgene specifically excluded HSV, HSIII, and HSB2 as well as much of the 3′-flanking region (Fig. 3A). hCD79b was expressed in the splenic B cells of all five −8.0CD79b lines. The mean expression was indistinguishable from that of the larger transgenes, although there was slightly more prominent variation in expression values (6% to 49% of the endogenous mouse locus) (Fig. 3D and H). Two additional hCD79b transgenes were generated that shared a 3′ end with −8.0CD79b (Fig. 3A). The −1.3CD79b transgene excluded the pituitary cell-specific HSI and HSII, and the −0.5CD79b transgene further excluded a B-cell-specific hypomethylated CpG island (Fig. 3A and unpublished data). The −1.3CD79b transgene was robustly expressed in all five lines. Its expression per copy was 10 to 30% of that of the endogenous mCD79b gene (Fig. 3E and H). The −0.5CD79b transgene maintained a similar level of expression, ranging from 16 to 19% of that of the endogenous gene (Fig. 3F and H). Thus, the five transgenes, CD/hGH BAC, hGH/P1, −8.0CD79b, −1.3CD79b, and −0.5CD79b, expressed mean levels of CD79b mRNA that were within a twofold range (17 to 27%). These data indicated that a complement of cis-acting elements necessary for full activation and expression of CD79b is in close juxtaposition to the structural gene.
A final hCD79b transgene was generated that contained only a 200-bp minimal promoter region (Fig. 3A). It should be noted that the CD79b gene utilizes a GC-rich, TATA-less promoter, and for this reason it has a cluster of multiple transcription initiation sites (5, 28). The expression of the −0.2CD79b transgene (Fig. 3G and H) was remarkable in three respects. First, its level of expression, 4.6% relative to that of the endogenous mCD79b gene, was substantially below that of the other five transgenes. Second, despite its weak expression, the −0.2CD79b transgene retained site-of-integration independence and copy number-dependent expression, with four of five lines demonstrating essentially identical levels (4%) of CD79b mRNA per gene copy. Finally, this transgene retained B-cell specificity (Fig. 4). Thus, determinants sufficient to establish an autonomous and B-cell-specific chromatin locus are encompassed within the CD79 gene or in its immediate 200 bp of 5′-flanking sequences. An additional element(s) necessary for enhancement of expression to full levels is located between −200 and −500 of the transcription start site.
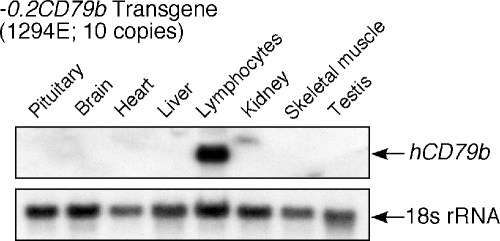
FIG. 4.
Human CD79b transgene containing the 0.2-kb minimal promoter (−0.2CD79b) maintains B-cell specificity. Total RNAs purified from the indicated tissues of a −0.2CD79b transgenic mouse (1294E) were analyzed by Northern blot hybridization. A human-specific CD79b probe was used to detect hCD79b mRNA. The level of mouse 18S rRNA served as a loading and transfer control.
Histone modifications at the hCD79b/GH locus in B-cell chromatin are tightly restricted to the CD79b gene.
Transgenic studies indicate that the major determinant(s) of CD79b transcription in B cells is located within, or in close proximity to, the structural gene (Fig. 3 and 4). To explore the basis for this control, we mapped histone modifications at the hCD79b/GH locus in B-cell chromatin. Three histone modifications that are consistently associated with active chromatin domains were assessed by ChIP: acetylated histone H3 (H3ac), acetylated histone H4 (H4ac), and histone H3 dimethylated at the lysine 4 position (H3K4me2). ChIP-enriched DNA was surveyed at 11 sites across the hCD79b/GH locus by semiquantitative PCR (Fig. 5A; Table 2). Each value was calculated as the ratio of DNA in the antibody-bound chromatin to that in the input sample. The values were further normalized to levels of enrichment for the corresponding histone modification at the constitutively active glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) (see Materials and Methods).
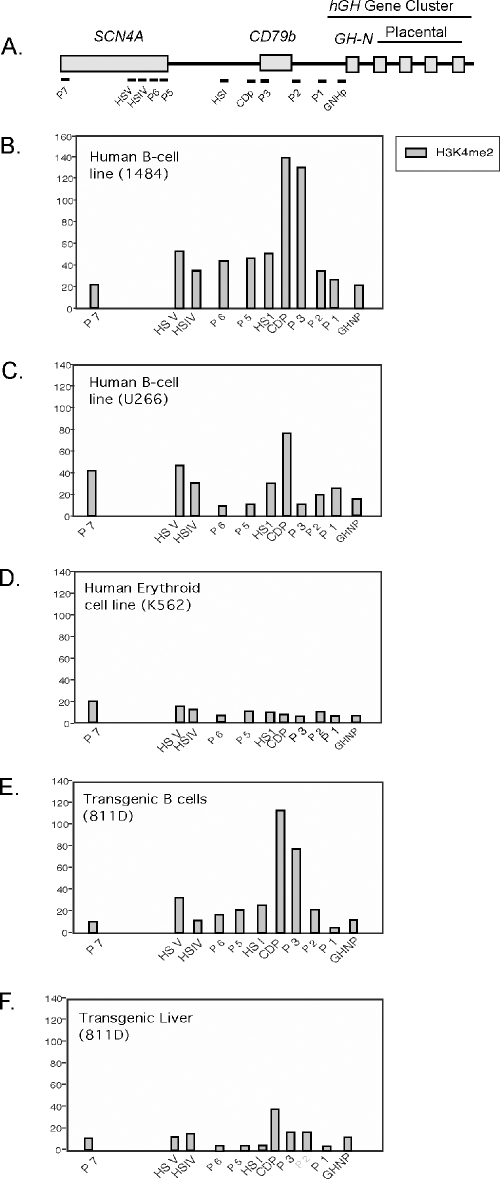
FIG. 5.
Histone acetylation profiles at the hCD79b/GH locus in B-cell chromatin are predominantly localized to the area of CD79b. A. Diagram of the hCD79b/GH locus. The position of each amplimer set used in the chromatin immunoprecipitation (ChIP) analysis is indicated below the diagram (labeled dashes). B. Patterns of histone H3 and H4 acetylation throughout the human hCD79b/GH region in a B-cell line expressing high levels of CD79b mRNA. Chromatin from line 1484 was used as a representative high-expressing line (Fig. 1B). DNAs isolated from anti-acetyl H3 and anti-acetyl H4 ChIPs were amplified with the indicated primer sets and normalized (described in Materials and Methods). Each histone modification value was calculated as the ratio of DNA in the antibody-bound chromatin to that in the input sample. Mean histone H3 acetylation (dark bars) and histone H4 acetylation (light bars) are plotted. The amplimer regions analyzed are indicated underneath each pair of bars. The means ± standard deviations are representations of a minimum of two independent assays. C. Patterns of histone H3 and H3 acetylation in a plasma cell line with low levels of CD79b. U266 was selected as a representative B-cell line expressing trace levels of CD79b mRNA (Fig. 1B). Analyses and labeling are as described for panel B. D. Patterns of histone H3 and H4 acetylation in an erythroid cell line. The K562 erythroleukemia line does not express CD79b mRNA (Fig. 1B). E. Patterns of histone H3 and H4 acetylation in splenic B cells expressing hCD79b from an hGH/P1 transgenic mouse (line 811D). F. Patterns of histone H3 and H4 acetylation in liver tissue from the same hGH/P1 transgenic mouse used in the experiment depicted in panel E (line 811D).
Acetylated histones H3 and H4 were mapped in a B-cell line with high CD79b expression (1484), a B-cell line with trace CD79b expression (U266), and an erythroid line lacking CD79b mRNA (K562) (Fig. 1B). Line 1484 chromatin contained a prominent peak of H3 and H4 acetylation centered on the CD79b promoter (amplimer CDP) and its immediate flanking regions (amplimers HSI and P3; Fig. 5B). A slight enrichment for acetylated H3 and H4 was also noted at HSV (Fig. 5B). The U266 B-cell line lacked significant enrichment for H3 acetylation throughout the locus, but it had a prominent peak of H4 acetylation at HSV and a minor peak at the CD79b promoter (Fig. 5C). The K562 cell chromatin lacked H3 and H4 acetylation throughout the locus (Fig. 5D). A second set of ChIP studies was carried out on the primary splenic B cells and primary hepatocytes of an hGH/P1 transgenic mouse (line 811D). The B cells contained an isolated peak of histone H3 and H4 acetylation coincident with the CD79b promoter and an additional moderate peak over HSV (Fig. 5E). Hepatocyte chromatin from the same mouse revealed peaks of H4 acetylation at the CD79b promoter (probe CDP) and at HSV; there was no evidence of H3 acetylation in the hepatic sample (Fig. 5F). Thus, within the resolution of the assay, H3/H4 acetylation in CD79b-expressing B cells was limited to the CD79b promoter. The comparison of the two B-cell lines further established a positive correlation between levels of CD79b expression and local acetylation.
Levels of H3K4me2 were determined by ChIP analysis at the hCD79b/GH locus (Fig. 6). A prominent peak of H3K4me2 was observed at the CD79b promoter (probes CDP and P3) in the high-expressing 1484 cell line (Fig. 6B). A low level of modification also appeared to be present between CD79b and HSV in these cells. In contrast, the region 3′ of the gene remained unmodified. In the U266 cells, which contain only trace levels of CD79b mRNA, a modest peak of modification was identified at the CD79b promoter and at HSV (Fig. 6C). K562 cell chromatin lacked modification throughout the surveyed region (Fig. 6D). The primary B cells from an hGH/P1 transgenic mouse had a strong H3K4me2 peak at the CD79b promoter (probes CDP and P3) and minimal enrichment at HSV (Fig. 6E). Hepatocytes from the same line had low levels of modification at the CD79b promoter (probe CDP) and lacked significant enrichment elsewhere. As was the case in the H3 and H4 acetylation ChIP study, there was no evidence for 3′ extension of H3K4me2 modification within the hGH cluster. The peak of H3K4me2 modification at two adjacent sites in the two expressing cell sources (1484 and transgenic B cells) were mutually confirming. Comparisons among the high- and low-expressing B-cell lines and the nonexpressing K562 cells revealed a direct relationship between gene activation and H3K4 methylation at the CD79b promoter. Analysis of primary B cells and hepatocytes from the hGH/P1 mouse extended these observations. The significance of the H4 acetylation and H3K4 dimethylation at the CD79b promoter in the hepatocytes remains unclear. Overall, these results reveal that histone modifications associated with chromatin activation in B cells are limited to the immediate vicinity of the hCD79b gene and fully exclude the adjacent hGH cluster.
FIG. 6.
Histone H3 lysine 4 dimethylation (H3K4me2) profiles at the hCD79b/GH locus. Experimental details are the same as those described in the legend to Fig. 5, except that anti-H3K4me2 was utilized for immunoprecipitation. The histogram represents a single assay in each case.
DISCUSSION
Approximately 20,000 to 25,000 genes are present in the human genome (16). These genes, which for the most part are randomly distributed, are differentially regulated by sets of cis-acting determinants, including enhancers, silencers, insulators, and boundary elements. Since these elements are frequently remote from target promoters, there is a clear potential for genes with distinct expression profiles to cross paths and interfere with each other's functions (11). Remote regulatory elements can initiate and regulate gene expression via targeted alterations in chromatin structure; these modifications often constitute critical steps in complex developmental pathways. Thus, defining interactions of tightly linked genes necessitates combined structural and functional mapping of chromatin structures in developmentally valid model systems.
The hCD79b/GH locus presents an informative model for analysis of chromatin structures and their developmental regulation. This locus contains six closely packed genes with three distinct tissue specificities: pituitary, placenta, and B cell. In addition, the striated muscle-specific SCN4A gene is juxtaposed 5′ of these genes, and a testis-specific TCAM gene is juxtaposed 3′ of the locus (Fig. 6). HS formation and histone modifications at this locus have been previously mapped in pituitary and placental chromatin (7, 15, 17, 19). In the pituitary tissue, the hGH LCR establishes a 32-kb acetylated chromatin domain that directly links the LCR with the target hGH-N gene promoter (15). In the placenta tissue, the LCR determinants HSIII-HSV and the target placental genes are each individually acetylated and are separated by a 28-kb region of unmodified chromatin (8, 19). Of note, the CD79b gene is located between the hGH cluster and its LCR. As such, it is situated in the “activated” domain in pituitary chromatin and in a region of hypoacetylated histones in placental chromatin. Significantly, CD79b is robustly transcribed in the pituitary tissue and is inactive in the placenta (5). The “bystander” activation of CD79b in the pituitary tissue is independent of B-cell-specific transcription factors, and as such it appears to represent a mechanistically independent and epigenetically driven pathway of gene activation (5). Thus, the specific configurations of chromatin modifications at the hCD79b/GH locus in the pituitary and placenta tissues predict divergent pathways of LCR-mediated transcriptional activation and have distinct effects on the adjacent CD79b gene.
To further explore transcriptional interactions at the hCD79b/GH locus, we have mapped its transcriptional determinants and chromatin structures in B cells. HS mapping, which has the capability to scan for critical chromatin determinants (10, 12), identified a set of B-cell-restricted sites, HSB1 and HSB2, and a constitutive site, HSa (Fig. 2). The B-cell specificity of HSB1 and HSB2 suggested that they play a role in CD79b expression. The functional importance of these sites was tested in a series of transgenic mouse lines. The initial studies assessed the expression of hCD79b from transgenes that encompass the entire locus on 123-kb or 87-kb genomic fragments. These levels were then compared to a series of smaller transgenes that isolate the CD79b gene with limited flanking regions (Fig. 3). Three major observations resulted from these studies. First, CD79b expression was maintained within a twofold mean range for all but the smallest transgene, −0.2CD79b. These comparisons led us to conclude that the region between −200 bp and −500 bp upstream from the CD79b gene contains one or more determinants that enhance CD79b expression. Second, the copy number dependence and site-of-integration independence of CD79b expression was maintained in all transgenes, including −0.2CD79b. Whereas we did observe as much as an eightfold variation of expression in some of the transgenes, the remarkable consistency of expression for the −0.5CD79b and −0.2CD79b transgenes and the maintenance of B-cell specificity in the −0.2CD79b transgene (Fig. 4) indicated that an element(s) sufficient to establish an autonomous chromatin domain in B cells is located within or in very close juxtaposition to the hCD79b gene itself. Third, these studies effectively eliminated HSB2 as a critical determinant of CD79b expression. Exclusion of this HS from the CD79b transgene had no appreciable impact on expression levels, and the copy number dependence and B-cell specificity was maintained in its absence. This data set leads us to conclude that control over CD79b is local as opposed to long range, and that the two HS retained in the −0.2CD79b transgene, the B-cell-specific HSB1 and the constitutive HSa, may be sufficient to establish an autonomous B-cell chromatin domain.
Although the −0.2CD79b transgene contained elements sufficient to establish an autonomous chromatin domain, its level of expression was significantly below that of the transgenes with more extensive 5′-flanking regions. As noted above, this comparison suggests that one or more determinants residing in the region between the 5′ termini of −0.2CD79b and −0.5CD79b have transcription-enhancing activity. Prior analyses by others using in vitro and cell transfection approaches (28) have identified a putative B-cell-specific activation element, BCS (for “B29 conserved sequence”), in this region. This BCS can bind to the multifunctional transcription factor Ying Yang 1 (YY1) in vitro (25). YY1, a zinc finger, GL1-Kruppel family member, can function as either a silencer or an enhancer through interaction with a variety of chromatin-modifying proteins (1, 9, 20). However, the role of this factor in CD79b gene expression has not been further evaluated. Previous studies using cell transfection models have also suggested that the minimal promoter of the hCD79b gene, extending to −0.2 kb, exhibits B-cell-specific activity. This minimal promoter region contains EBF, Oct1-2, PU.1 (Ets family), Ikaros, and Sp1 transcription factor motifs (28). Ikaros is essential for normal development and proliferation of lymphoid cells and exists as an integral component of chromatin-remodeling complexes (18, 24). PU.1, a member of the Ets family, is critical for B-lymphocyte development and interacts with the chromatin-modifying enzymes CBP and p300 (29). The activity of these factors may correlate with HSB1 formation and function and may contribute to the autonomous chromatin domain established by the −0.2CD79b transgene. However, these studies in transfected B cells also suggested that the minimal promoter (−193 bp) has full transcriptional activity, and extension of the 5′-flanking region to −469 bp resulted in a prominent loss of activity (28). These data, which led the authors to propose that repressor determinants are located between −193 and −496, are inconsistent with our transgenic studies. Indeed, we detect enhancer activity in this region based on comparison of the −0.2CD79b versus −0.5CD79b transgenes. Thus, the preceding cell transfection studies, while useful in identifying potential cis-trans interactions at the CD79b promoter, may reflect functional activities that are not in operation during in vivo B-cell differentiation in the intact animal.
The analysis of histone modifications at the hCD79b/GH cluster in B cells (Fig. 5 and 6) revealed that histone H3 and H4 acetylation and H3K4 methylation are predominantly limited to the immediate vicinity of the CD79b promoter. This stands in marked contrast to the distribution of histone modifications at this locus in pituitary and placental chromatin. Therefore, three distinct patterns of chromatin modification are established at the hCD79b/GH locus (Fig. 7). These patterns correspond to three specific gene expression profiles. In the pituitary cell, an extensive domain of histone acetylation links the LCR with the target hGH-N promoter. This pituitary domain, which may be established by linear “spreading” of histone acetyltransferase complexes, encompasses the intervening CD79b gene and results in bystander gene activation. Consistent with this model of bystander activation and its dependence on HSI action, hCD79b is robustly transcribed in hGH/P1 and −8.0CD79b transgenic mouse pituitary glands but not in the −1.3CD79b, −0.2CD79b, and hGH/P1(ΔHSI) transgenic mouse pituitary glands (5). In the placenta, histone acetylation is limited to the LCR and to the placental genes. These modified regions are separated by an extensive segment of unmodified chromatin containing the pituitary cell-specific LCR determinants HSI and HSIII, the CD79b gene, and the hGH-N gene. Previous studies have demonstrated that hCD79b, which is excluded from the modified region, is not expressed in the placenta of humans and transgenic mice (5). The discontinuous pattern of histone modification in the placenta predicts a long-distance interaction, or “looping,” between the LCR and the four placental genes to activate expression. Finally, the activating modifications in B-cell chromatin are tightly restricted to the CD79b gene; these modifications exclude all adjacent genes in the region. These data, taken together, reveal a remarkable complexity of chromatin structures at a single locus. These structures impact the expression of neighboring genes in an asymmetric and tissue-specific fashion. How common these sorts of interactions are and how well they represent other multispecificity loci can now be further explored.
FIG. 7.
Comparison of chromatin structures and gene activation pathways at the hCD79b/GH locus in pituitary tissue, placenta tissue, and B cells. The hCD79b/GH locus is diagrammed with the set of DNase I HS present in each tissue. Transcriptional activities are denoted by angled arrows at the indicated promoters. The “bystander” transcription of CD79b in the pituitary gland is indicated. Regions of H3 and H4 hyperacetylation are indicated by the shaded ovals below the diagrams. Proposed pathways of gene activation are indicated as long-range “spreading” (pituitary), “looping” (placenta), or “local” (B cell).
. . . . .
Acknowledgments
We thank Jean Richa, Pei Fu He, and Kathleen Moosbrugger of the University of Pennsylvania Transgenic & Chimeric Mouse Facility (NIH P30DK50306, NIH P30DK19525, and NIH P30CA16520) for generating transgenic founders for the study.
This work was supported by NIH R01 HD25147 and NIH HD046737 (N.E.C. and S.A.L.) and by postdoctoral fellowship 5013-03 from the Leukemia and Lymphoma Society (I.C.).
REFERENCES
- 1.Baumeister, P., S. Luo, W. C. Skarnes, G. Sui, E. Sato, Y. Shi, and A. S. Lee. 2005. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell. Biol. 25:4529-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, A. C., A. G. West, and G. Felsenfeld. 1997. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 3.Bennani-Baiti, I. M., N. E. Cooke, and S. A. Liebhaber. 1998. Physical linkage of the human growth hormone gene cluster and the CD79b(Igβ/B29) gene. Genomics 48:258-264. [DOI] [PubMed] [Google Scholar]
- 4.Benschop, R. J., and J. C. Cambier. 1999. B cell development: signal transduction by antigen receptors and their surrogates. Curr. Opin. Immunol. 11:143-151. [DOI] [PubMed] [Google Scholar]
- 5.Cajiao, I., A. Zhang, E. J. Yoo, N. E. Cooke, and S. A. Liebhaber. 2004. Bystander gene activation by a locus control region. EMBO J. 23:3854-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, E. Y., Y.-C. Liao, D. H. Smith, H. A. Barrera-Saldana, R. E. Gelinas, and P. H. Seeburg. 1989. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics 4:179-197. [DOI] [PubMed] [Google Scholar]
- 7.Elefant, F., S. A. Liebhaber, and N. E. Cooke. 2000. Targeted recruitment and spreading of histone acetyltransferase activity by a locus control region. J. Biol. Chem. 275:13827-13834. [DOI] [PubMed] [Google Scholar]
- 8.Elefant, F., Y. Su, S. A. Liebhaber, and N. E. Cooke. 2000. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19:6814-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliassen, K. A., A. Baldwin, and E. M. Sikorski. 1998. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol. Cell. Biol. 18:7106-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenfeld, G. 1996. Chromatin unfolds. Cell 86:13-19. [DOI] [PubMed] [Google Scholar]
- 11.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 12.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitivity sites in chromatin. Annu. Rev. Biochem. 57:159-197. [DOI] [PubMed] [Google Scholar]
- 13.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 13:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermanson, G. G., D. Eisenberg, P. W. Kincade, and R. Wall. 1988. B29: a member of the immunoglobulin gene superfamily exclusively expressed on B-lineage cells. Proc. Natl. Acad. Sci. USA 85:6890-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 16.International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931-945. [DOI] [PubMed] [Google Scholar]
- 17.Jones, B. K., B. R. Monks, S. A. Liebhaber, and N. E. Cooke. 1995. The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15:7010-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, J., S. Sif, B. Hones, A. Jackson, J. Loipally, E. Hiller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, R. Kingston, and K. Georgopoulos. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10:345-355. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, A. P., S. A. Liebhaber, and N. E. Cooke. 2004. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol. Endocrinol. 18:1018-1032. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. S., K. M. Galvin, and Y. Shi. 1993. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Sp1. Proc. Natl. Acad. Sci. USA 90:6145-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, A., and A. Murrill. 2004. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 14:R284-R286. [DOI] [PubMed] [Google Scholar]
- 22.MacLeod, J. N., A. K. Lee, S. A. Liebhaber, and N. E. Cooke. 1992. Developmental control and alternative splicing of the placentally expressed transcripts from the human growth hormone gene cluster. J. Biol. Chem. 267:14219-14226. [PubMed] [Google Scholar]
- 23.O'Neill, L. P., and B. M. Turner. 1996. Immunoprecipitation of chromatin. Methods Enzymol. 274:189-197. [DOI] [PubMed] [Google Scholar]
- 24.O'Niell, D. W., S. S. Schoetz, R. A. Lopez, M. Castle, L. Rabinoxitz, E. Shor, D. Krawchuk, M. G. Goll, M. Renz, H. Seelig, S. Han, R. H. Seong, S. D. Park, T. Agalioti, N. Munshi, D. Thanos, G. Erdjument-Bromage, P. Tempst, and A. Bank. 2000. An Ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol. Cell. Biol. 20:7572-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrone, L., S. E. Henson, R. Wall, and C. S. Malone. 2004. A conserved sequence upstream of the B29 (Igβ, CD79b) gene interacts with YY1. Mol. Biol. Rep. 31:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez, M., Z. Misulovin, A. L. Burkhardt, S. Mahajan, T. Costa, R. Franke, J. B. Bolen, and M. Nussenzweig. 1993. Signal transduction by immunoglobulin is mediated through Igα and Igβ. J. Exp. Med. 178:1049-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su, Y., S. A. Liebhaber, and N. E. Cooke. 2000. The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J. Biol. Chem. 275:7902-7909. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, A. A., W. J. Wood, M. J. Gilly, M. A. Damore, S. A. Omori, and R. Wall. 1996. The promoter and 5′-flanking sequences controlling human B29 gene expression. Blood 87:666-673. [PubMed] [Google Scholar]
- 29.Yang, C., L. H. Shapiro, M. Rivera, A. Kumar, and P. K. Brindle. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusufzai, T. M., and G. Felsenfeld. 2004. The 5′-HS4 chicken β-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 101:8620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]