Abstract
Studies have demonstrated cross talk between β-catenin and peroxisome proliferator-activated receptor γ (PPARγ) signaling pathways. Specifically, activation of PPARγ induces the proteasomal degradation of β-catenin in cells that express an adenomatous polyposis coli-containing destruction complex. In contrast, oncogenic β-catenin is resistant to such degradation and inhibits the expression of PPARγ target genes. In the present studies, we demonstrate a functional interaction between β-catenin and PPARγ that involves the T-cell factor (TCF)/lymphocyte enhancer factor (LEF) binding domain of β-catenin and a catenin binding domain (CBD) within PPARγ. Mutation of K312 and K435 in the TCF/LEF binding domain of an oncogenic β-catenin (S37A) significantly reduces its ability to interact with and inhibit the activity of PPARγ. Furthermore, these mutations render S37A β-catenin susceptible to proteasomal degradation in response to activation of PPARγ. Mutation of F372 within the CBD (helices 7 and 8) of PPARγ disrupts its binding to β-catenin and significantly reduces the ability of PPARγ to induce the proteasomal degradation of β-catenin. We suggest that in normal cells, PPARγ can function to suppress tumorigenesis and/or Wnt signaling by targeting phosphorylated β-catenin to the proteasome through a process involving its CBD. In contrast, oncogenic β-catenin resists proteasomal degradation by inhibiting PPARγ activity, which requires its TCF/LEF binding domain.
Peroxisome proliferator-activated receptor γ (PPARγ) is a nuclear receptor expressed in many tissues but predominantly found in adipose tissue, where it regulates the expression of a diverse array of genes involved in energy metabolism (13, 14, 26, 47, 54). It is also abundantly expressed in the gut, where, in combination with the coactivator Hic-5, it can regulate the differentiation of specialized epithelial cells (12). The transcriptional activity of PPARγ is regulated in part by binding to ligands which include derivatives of polyunsaturated fatty acids as well as the thiazolidinedione class of synthetic insulin sensitizers (25). The PPARγ protein consists of multiple domains, including a ligand-independent transactivation domain at the N terminus, two zinc fingers in the center of the molecule required for binding to DNA, and the ligand-binding domain at the C terminus which facilitates ligand-dependent transactivation as well as heterodimerization with retinoic acid X receptor alpha (RXRα) (24). Activation of PPARγ in a variety of cell types induces programs of gene expression that reflect the differentiation potential of each progenitor cell. For instance, its ectopic expression in mesenchyme-derived cells induces adipogenesis (49), whereas its expression in epithelium-derived cells stimulates the production of markers of epithelial differentiation/maturation, such as kruppel-like factor 4 and keratin 20 (12). Additionally, PPARγ is a potent inhibitor of cell proliferation through mechanisms that include induction of cyclin-dependent kinase inhibitors (i.e., p21CIP) and attenuation of E2F transcriptional activity (1, 34). It is also a suppressor of tumor cell growth (35), and consequently, investigators have considered whether synthetic PPARγ ligands are effective chemotherapeutic agents (17). In fact, Girnun and collaborators have provided evidence that PPARγ is capable of suppressing colon carcinogenesis by downregulating the oncogene β-catenin (16).
β-Catenin is a versatile protein initially identified as a component of cell adhesion complexes in epithelial cells, where it binds to cadherins to link extracellular anchors to the cytoskeleton (4, 5, 10, 56). Additionally, β-catenin functions as a coactivator of T-cell factor (TCF)/lymphocyte enhancer factor (LEF) transcription factors to facilitate the expression of genes regulated by the canonical Wnt signaling pathway (37, 53). Consequently, it serves a critical function during early development (7), but it is also a major contributing factor to the development of many tumors due to its ability to undergo sporadic mutation to an oncogene (41). In the absence of a Wnt signal, β-catenin exists within a cytoplasmic complex (β-catenin destruction complex) along with glycogen synthase kinase 3β (GSK3β), adenomatous polyposis coli (APC), and axin, where it is phosphorylated and targeted for degradation by the proteasome (42). Wnt signaling perturbs this destruction complex, leading to the accumulation of underphosphorylated β-catenin, which translocates to the nucleus to coactivate TCF/LEF-associated gene expression. β-Catenin consists principally of three domains: the N-terminal region of 134 amino acids, a central core domain of 550 amino acids, and a C terminus of 100 amino acids, which contains the transactivation domain (53). The regulated phosphorylation of β-catenin by GSK3β and casein kinase 1 occurs on amino acids S33, S37, T41, and S45, generating a recognition tag for ubiquitylation and subsequent proteasomal degradation (53). Most oncogenic forms of β-catenin have mutations in these phosphoacceptor sites; for instance, S37A β-catenin is expressed abundantly in several human carcinomas (33, 43). TCF/LEF family members, APC, axin, and cadherins all bind to the central core region of β-catenin, which contains 12 imperfect 42-amino-acid armadillo repeats. The crystal structure of the central region reveals that each repeat consists of three α helices, and together the 12 repeats form a superhelix containing a long positively charged groove (21). This structure appears to facilitate binding to the negatively charged β-catenin binding domains (CBD) within TCF/LEF and the other interacting proteins (18, 19, 22). In fact, recent studies have identified two lysines (charged buttons) within β-catenin, K312 and K435, in armadillo repeats 5 to 9, that form salt bridges with negatively charged glutamate or aspartate in the CBD of the interacting protein (19).
Recent studies suggest that β-catenin can interact with transcription factors other than those mediating the canonical Wnt signaling pathway. These interactions include the β-catenin-associated coactivation of the nuclear receptor liver receptor homologue 1 (LRH-1) (6) as well as repression of β-catenin activity by association with other nuclear receptor complexes, including those containing androgen receptor, retinoic acid receptor, or vitamin D receptor (50). The interaction between LRH-1 and β-catenin leads to the transcriptional activation of cyclin D1 and cyclin E and enhanced proliferation of gut epithelial cells (6). In fact, it appears that LRH-1 contributes to colon tumor formation through pathways that involve β-catenin (46). The other nuclear receptors that repress β-catenin activity appear to have the ability to suppress tumor formation in some target tissues (36). In some tumors, the transformed epithelial cells have escaped the tumor-suppressing activity of the nuclear receptor, which involves resistance of β-catenin to the repression process. In the case of PPARγ, Sarraf and collaborators suggested that it is a powerful tumor suppressor in the colon, since the loss of one allele of the PPARγ gene led to an increase in sensitivity to chemical carcinogenesis (45). Additional studies suggested that PPARγ downregulates the levels of β-catenin but only in cells that contain a functional APC molecule and an intact destruction complex (16). Further support for a role for PPARγ in suppressing the oncogenic activity of β-catenin comes from the work of Lu and collaborators (30), who show that the repression of β-catenin function in malignant cells by nonsteroidal anti-inflammatory drugs requires high-level expression of PPARγ. Our recent studies have demonstrated that activation of PPARγ in mesenchymal cells induces the proteasomal degradation of β-catenin, which depends on its phosphorylation at the N terminus by GSK3β (28), whereas expression of an oncogenic β-catenin (S37A) resists such a PPARγ-associated destabilization and inhibits the ability of PPARγ to induce target genes.
In the present study, we questioned whether there is a functional interaction between β-catenin and PPARγ that explains how these two effectors can affect each other's activities. The results show a direct interaction between β-catenin and PPARγ, which is facilitated by the region armadillo repeats 5 to 9 encompassing lysines 312 and 435 within β-catenin. In fact, mutation of these lysines to glutamic acid (K312E and K435E) renders the oncogenic S37A β-catenin unstable in the presence of an activated form of PPARγ. In addition, these mutations also prevent S37A β-catenin from inhibiting PPARγ target gene expression. Finally, we have identified a region within PPARγ that is highly homologous to the CBD in TCF/LEF and facilitates its interaction with β-catenin. Mutation of select amino acids within the CBD of PPARγ inhibits its ability to induce the degradation of β-catenin.
MATERIALS AND METHODS
Plasmids and stable cell lines.
pRevTRE-HA-WT-β-catenin and pRevTRE-HA-S37A-β-catenin were generated as previously described (28). pGEX-PPARγ was produced by subcloning the PPARγ coding region of the pBabe-PPARγ vector into the SalI site of pGEX-5X-3 vector (Amersham Biosciences). Site-directed mutations were introduced into the pRevTRE-HA-S37A-β-catenin and pGEX-PPARγ vectors using a QuikChange II XL site-directed mutagenesis kit (Stratagene) following the manufacturer's protocol. All mutations were confirmed by DNA sequencing. The resulting PPARγ mutant cDNAs were excised from the corresponding pGEX-PPARγ plasmids and subcloned into pBabe retrovirus. Tetracycline-responsive hemagglutinin (HA)-tagged mutant β-catenin stable lines were generated as previously described (28), while mutant PPARγ cell lines were generated by infecting Swiss fibroblasts with the corresponding pBabe-PPARγ vectors. The cell lines were maintained in culture as described previously (28) and induced to differentiate by exposure of a confluent population of cells to dexamethasone (1 μM), isobutylmethylxanthine (0.5 mM), insulin (1.67 μM), troglitazone (5 μM), and 20% fetal bovine serum.
Antibodies, Western blotting, and immunoprecipitation.
The following antibodies were purchased from the listed companies: anti-β-catenin and anti-cyclin D1 from BD Biosciences Transduction Laboratories; anti-PPARγ, anti-C/EBPα, antiactin, anti-glucocorticoid receptor interacting protein 1 (GRIP1)/transcriptional intermediary factor 2 (TIF2), anti-RXRα, anti-SRC-1, and anti-HA from Santa Cruz Biotechnology, Inc.; and anti-adiponectin/Acrp30 from Affinity BioReagents. Immunoblotting and immunoprecipitation were performed as previously described (27, 28).
Electrophoretic mobility shift assay and biotinylated oligonucleotide pull-down of nuclear protein complexes.
Electrophoretic mobility shift assays (EMSA) were performed as previously described (55) using an oligonucleotide corresponding to the ARE7/DR-1 site within the fatty acid binding protein 4 (FABP4) gene. The sense and antisense ARE7/DR-1 oligonucleotides were synthesized and biotinylated by Invitrogen. Fifty micrograms of nuclear extracts was precleared for 1 h with ImmunoPure immobilized streptavidin agarose beads (Pierce Biotech, Inc.) in the same buffer used for EMSA and then incubated with 2 μg biotinylated, annealed ARE7 oligonucleotides overnight at 4°C. Biotinylated ARE7/DR-1 oligonucleotide-bound proteins were pulled down by streptavidin agarose beads for 1 h at room temperature. The beads were washed with phosphate-buffered saline (pH 7.0) three times and boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 30 s. The bound proteins were then subjected to Western blot analysis following separation by SDS-PAGE.
GST pull-down assays.
Glutathione S-transferase (GST)-PPARγ wild-type and mutated fusion proteins were purified by growing BL21 bacteria transformed with pGEX-PPARγ plasmids following the instructions provided by Amersham Biosciences. For GST pull-down assays, nuclear extracts obtained from fibroblasts expressing HA-tagged S37A β-catenin were incubated with equal amounts of GST (control), wild-type (WT) GST-PPARγ, and mutated GST-PPARγ fusion proteins overnight at 4°C. The GST fusion proteins were allowed to interact with 30 μl glutathione-Sepharose 4B beads (Amersham Biosciences), and the bound proteins were identified by Western blot analysis.
Reporter gene assays.
Luciferase/Renilla assays were performed using the DLRII kit (Promega, Madison, WI) and a Luminoskan Ascent luminometer (Thermo Labsystems) as described previously (28).
RESULTS
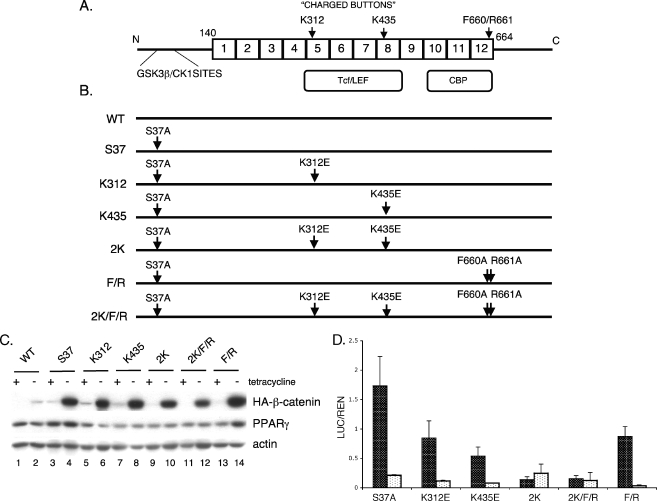
We previously demonstrated cross talk between PPARγ and β-catenin signaling during adipogenesis in which activation of PPARγ induced the degradation of β-catenin in the proteasome. Furthermore, expression of an oncogenic form of β-catenin (S37A) that activated canonical Wnt signaling was resistant to degradation and selectively inhibited PPARγ activity. To identify the molecular mechanisms regulating this process, we introduced a series of mutations within the parental S37A β-catenin molecule (Fig. 1) in order to effect a change in the activity of this protein by perturbing its association with known factors. Since S37A β-catenin activated Wnt signaling in addition to blocking PPARγ activity, we created mutations that likely affect binding to coactivators of the canonical Wnt signaling pathway, TCF/LEF and CBP/p300. In the former case, studies have shown that TCF/LEF binds to a positively charged groove of β-catenin in the region between armadillo (Arm) repeats 5 and 9 (19, 51). Two critical amino acids in this region are lysines at positions 312 and 435 that bind acidic residues in the extended region of the TCF CBD. Mutation of either of these lysines to glutamic acid (K312E or K435E) disrupts the association of β-catenin with TCF in in vitro immunoprecipitation “pull-down” assays (19). Furthermore, mutation of lysine 435 to alanine (K435A) significantly reduces the transactivation properties of β-catenin in a TCF/LEF reporter gene assay (51). On the basis of these observations, we generated mutants of the S37A molecule containing either K312E or K435E alone or together in the same construct (constructs K312, K435, and 2K in Fig. 1B) in order to selectively block the activation of β-catenin target gene expression. In the case of CBP/p300, studies have shown a direct binding to the Arm repeat region 10 to 12 of β-catenin, but there are no structural assays identifying the specific amino acids responsible for this interaction (20, 48). It appears, however, that the β-catenin binding site of inhibitor of β-catenin and TCF (ICAT) overlaps that of CBP/p300 and in so doing blocks p300-mediated activation of β-catenin target genes (11). The helix H1 of ICAT within the three-helix bundle domain is anchored to Arm repeat 12 by hydrophobic interactions with F660 of β-catenin and hydrogen bonds between ICAT E37 and β-catenin R661 (18). We questioned, therefore, whether mutation of the amino acids F660 and R661 would affect the transcriptional activity as well as the stability of S37A β-catenin. Consequently, we introduced two additional mutations corresponding to F660A and R661A into the parental S37A protein (F/R) as well as into the molecule containing both K312E and K435E (construct 2K/F/R in Fig. 1B).
FIG. 1.
The charged buttons (K312 and K435) of β-catenin are required to activate the canonical Wnt signaling. (A) Structure of β-catenin: GSK3β/CK1 sites are located in the N-terminal domain followed by 12 armadillo repeats (from amino acids [aa] 140 to 664). TCF/LEF and CBP binding domains span from repeats 5 to 8 and 10 to 12, respectively. Two charged buttons (K312 in repeat 5 and K435 in repeat 8) and F660/R661 (in repeat 12) are indicated. (B) Schematic representation of β-catenin mutants. S37 represents a mutant with serine 37 mutated to alanine (S37A). All of the other mutations as indicated were generated within the parental S37A β-catenin molecule (i.e., K312 represents lysine 312 mutated to glutamic acid [K312E]). (C) Expression of the ectopic mutant β-catenins. The stable cell lines expressing the WT or different mutated S37A β-catenins as well as PPARγ were cultured in the presence or absence of tetracycline for 5 days until confluent. Total cellular proteins were analyzed by Western blotting for HA-tagged β-catenin, PPARγ, and actin (loading control). (D) TCF/LEF reporter gene assay. The stable cell lines were cultured in the absence of tetracycline for 3 days, at which stage they were transfected with TOPFLASH (black) or FOPFLASH (white) firefly reporter plasmids along with Renilla luciferase plasmid for an additional 2 days. Then, the luciferase assay was performed as detailed in Materials and Methods. The same experiment was repeated at least three times. The final values (the ratio of luciferase to Renilla [LUC/REN]) and standard deviation (error bars) were calculated based on all repeats.
The charged buttons of β-catenin, Lys312 and Lys435, are required for activation of TCF/LEF reporter gene activity.
To investigate the interaction of β-catenin with PPARγ, we conditionally expressed the mutants of S37A β-catenin illustrated in Fig. 1B in Swiss fibroblasts that also express PPARγ (Swiss-Pγ cells) to generate a series of stable cell lines. Production of abundant quantities of each of these mutants can be achieved by culture of the cells in the absence of tetracycline as shown in Fig. 1C. As observed previously, the level of expression of the S37A protein is significantly higher than the control wild-type β-catenin (Fig. 1C, compare lane 4 with lane 2) due to its greater stability. Introduction of the additional mutations within the S37A molecule does not significantly alter its rate of degradation, at least under the culture conditions used in this experiment. We have also demonstrated that S37A β-catenin is capable of activating canonical Wnt signaling in Swiss cells expressing PPARγ based on its ability to transactivate a TCF-based reporter gene (TOPFLASH) (28). To examine the transactivating capabilities of each of the mutants, we transiently transfected the TOPFLASH vector and an appropriate control (FOPFLASH) into each of the corresponding cell lines. Figure 1D demonstrates that mutations at K435 significantly attenuate the transactivation properties of S37A β-catenin. In fact, the data showing the importance of K435 are consistent with earlier studies by von Kries et al. (51), who generated a collection of mutants within Arm repeats 3 to 8 in β-catenin to identify relevant TCF binding sites. In that study, mutation of K435 to alanine also reduced the ability of β-catenin to transactivate a TCF/LEF-based reporter gene. Those investigators, however, did not analyze K312. The data presented in Fig. 1D suggest that K435 plays a more prominent role in regulating TCF activity than K312, but when both lysines are mutated in the same S37A molecule (construct 2K or 2K/F/R), there is an almost complete obliteration of transactivity which is significantly greater than the reduction caused by K435 alone (compare K435E with 2K). It is of interest that mutation of the amino acids responsible for binding to ICAT (Fig. 1D, construct F/R) only modestly attenuates the ability of β-catenin to transactivate TCF/LEF.
The effect of the different β-catenin mutants on expression of TCF/LEF target genes and PPARγ activity.
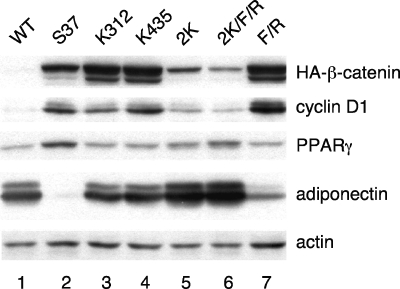
β-Catenin can activate the expression of a variety of target genes by interacting with different transcription factors. Consequently, analysis of TCF/LEF-based reporter genes in transient transfection assays provides only a limited insight into the function of the various β-catenin-transcription factor interactions. To address this concern, we analyzed the abilities of the different mutants to induce expression of a well-characterized TCF/LEF target gene, cyclin D1, in Swiss-Pγ fibroblasts. In the experiment shown in Fig. 2, the mutant cell lines were cultured in the absence of tetracycline to induce the expression of the corresponding β-catenins. The cells were also exposed to troglitazone along with a cocktail of adipogenic inducers in order to stimulate PPARγ activity. As observed previously, S37A protein significantly enhanced cyclin D1 production above the level expressed in the WT cells (Fig. 2, compare lane 2 with lane 1). The data show that S37A β-catenin with a single mutation at either K312 or K435 is able to activate cyclin D1 expression, whereas introduction of the double mutations (2K or 2K/F/R) completely abolishes this activity, which is consistent with the TOPFLASH reporter assay shown in Fig. 1D. It is of interest not only that the S37A β-catenin containing mutations in the ICAT binding site alone (Fig. 2, lane 7) retains the ability to induce cyclin D1 expression but also that it appears that the activity of this protein is significantly greater than that of the parental S37A protein. This may be due to the fact that ICAT, as an inhibitor of Wnt signaling, is incapable of binding to F660A/R661A S37A β-catenin and therefore not able to attenuate its activity.
FIG. 2.
Oncogenic β-catenin activates canonical Wnt signaling (cyclin D1) while inhibiting PPARγ target gene expression. The same cell lines as those used in Fig. 1D were cultured in the absence of tetracycline for 5 days until confluent, at which time they were treated with 5 μM troglitazone along with a cocktail of adipogenic inducers for 2 days as described in Materials and Methods. Total cellular proteins were analyzed by Western blotting for HA-β-catenin, PPARγ, cyclin D1, adiponectin, and actin.
We have previously shown that ectopic expression of S37A β-catenin in Swiss-Pγ fibroblasts blocks expression of select PPARγ target genes; most notably it blocks C/EBPα and adiponectin without affecting the expression of aP2/FABP4 (28). The data shown in Fig. 2 show that mutation of either K312 or K435 significantly attenuates this inhibitory action of S37A β-catenin on adiponectin expression (Fig. 2, compare lanes 3 and 4 with lane 2). This effect is amplified when both lysines are mutated in the same S37A molecule (Fig. 2, compare lanes 5 and 6 with lane 2). In contrast, S37A β-catenin containing mutations in the ICAT binding site (F/R) still retains the ability to inhibit the PPARγ-associated expression of adiponectin (Fig. 2, compare lane 7 with lane 1). Taken together, the data in Fig. 2 demonstrate that K312 and K435 contribute to the ability of oncogenic β-catenin (S37A) to inhibit PPARγ activity. They also suggest that this inhibitory action does not depend on cyclin D1 expression, since mutation of either K312 or K435 alone rescues the inhibition of PPARγ but has no significant effect on the induction of cyclin D1 by S37A β-catenin.
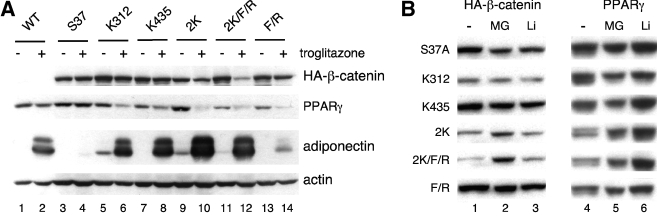
K312 and K435 contribute to the stability of oncogenic S37A β-catenin.
It is of interest that, in the experiment outlined in Fig. 2, the levels of abundance of the various mutant β-catenins differ somewhat from that of the parental S37A protein. One would have predicted that these mutant proteins would be as stable as S37A β-catenin, since they all exist within the parental S37A protein; however, the proteins containing mutations at K312 and K435 (Fig. 2, compare lanes 5 and 6 with lanes 2, 3, 4, and 7) appear to be unstable, since their abundance is manyfold lower than that of S37A β-catenin. As expected, the level of expression of WT β-catenin (Fig. 2, lane 1) is low because the protein is unstable due to its constant phosphorylation by GSK3β. The samples analyzed in Fig. 2 were extracted from cells exposed to troglitazone; therefore, it is possible that activation of PPARγ in these cells led to the targeted degradation of the proteins containing the two lysine mutations. To test this idea, we cultured the cell lines in the presence or absence of troglitazone and assessed the abundance of the mutant proteins on Western blots. Figure 3A, lanes 3 and 4, demonstrates that exposure of the cells to troglitazone for 5 days does not significantly affect the turnover of S37A β-catenin as previously reported. Inclusion of a single K312E or K435E mutation as well as the F660A and F661A mutations has no significant effect on the stability of the S37A molecule (Fig. 3A, compare lane 5 with lane 6, lane 7 with lane 8, and lane 13 with lane 14), whereas replacement of both lysines (K312 and K435) with glutamates renders S37A β-catenin unstable but only in cells exposed to troglitazone (Fig. 3A, compare lane 9 with lane 10 and lane 11 with lane 12). It is also important to point out that the ectopic PPARγ is unstable under conditions (i.e., coexpression with the double lysine mutants and plus troglitazone) that enhance its ability to induce adiponectin expression (for instance, compare lanes 10 and 12 with 9 and 11, respectively, in Fig. 3A). Similarly relevant is the observation that the oncogenic S37A β-catenin not only blocks the activity of PPARγ but also prevents its degradation in response to troglitazone (Fig. 3A, compare lanes 3 and 4). To assess whether activated PPARγ (plus troglitazone) was targeting the double lysine mutants to the proteasome, the Swiss-Pγ-β-catenin cell lines were cultured in the presence of troglitazone for 2 days and then treated with either MG132 (an inhibitor of the proteasome) or LiCl (GSK3 inhibitor) for 24 h or not treated. Figure 3B demonstrates that blocking the proteasome with MG132 results in a significant increase in the abundance of the β-catenin molecules containing the two lysine mutations and has no significant effect on the other mutants (compare lane 2 with lane 1). In addition, the degradation of the double lysine mutants appears to be independent of GSK3 activity, since LiCl has no effect on the abundance of any of the mutant forms of S37A β-catenin (Fig. 3B, compare lane 3 with lane 1). Analysis of the abundance of PPARγ in each of the corresponding cell lines serves to demonstrate the selective effect of MG132 on the double lysine mutants of β-catenin. It is interesting, however, that inhibition of GSK3 by LiCl appears to enhance the abundance of PPARγ without affecting S37A β-catenin, which suggests that GSK3 might have a role in controlling the turnover of PPARγ independently of β-catenin.
FIG. 3.
Lysine 312 and lysine 435 are required to maintain the stability of oncogenic S37A β-catenin. (A) The stable cell lines described in the legend to Fig. 2 were cultured in the absence of tetracycline until confluent and then treated with the adipogenic inducers for 7 days with (+) or without (−) troglitazone. Total cellular proteins were analyzed by Western blotting for HA-β-catenin, PPARγ, adiponectin, and actin. (B) The different β-catenin cell lines were cultured until confluent and then treated with 12.5 μM MG132 (MG) (Sigma), 30 mM LiCl (Li), or vehicle control (−) for 24 h in the presence of troglitazone and the adipogenic inducers as in panel A. The nuclear proteins were then analyzed by Western blotting for HA-β-catenin and PPARγ.
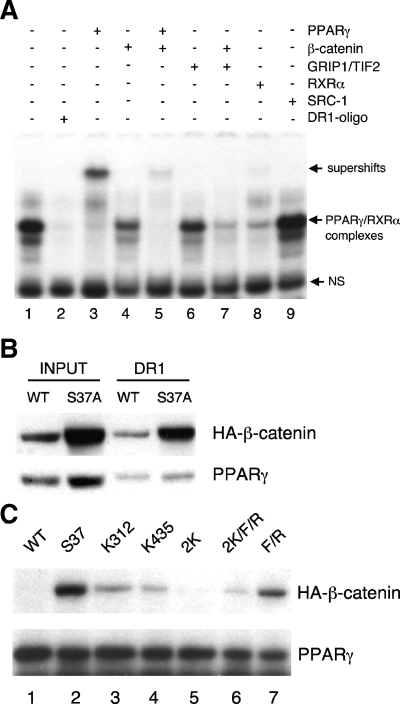
β-Catenin associates with PPARγ-DNA complexes.
A plausible mechanism by which oncogenic β-catenin (S37A) attenuates PPARγ activity is by associating with nuclear complexes containing PPARγ. To address this possibility, we analyzed the composition of complexes binding to a consensus PPAR response element (PPRE) using EMSA. Figure 4A shows the results of an EMSA generated by interacting the radiolabeled PPRE with nuclear proteins isolated from cells expressing S37A β-catenin and troglitazone-activated PPARγ. Fig. 4A, lane 1, shows the binding of two major complexes to the PPRE, but the faster-migrating species (labeled NS in Fig. 4A) appears to be nonspecific, since its binding is not disrupted by competition with excess unlabeled PPRE oligonucleotide (Fig. 4A, lane 2). Supershift analysis using antibodies against PPARγ and RXRα demonstrates that the major species corresponds, as expected, to PPARγ heterodimerized with RXRα (Fig. 4A, lanes 3 and 8). Of interest is the observation that incubation of an anti-β-catenin antibody with or without anti-PPARγ antibody significantly attenuates the binding of PPARγ to the PPRE (Fig. 4A, compare lane 4 with lane 1 and lane 5 with lane 3). Also of interest is the finding that an antibody against the coactivator TIF2 (anti-GRIP1/TIF2) attenuates the association of PPARγ with the PPRE (Fig. 4A, lane 6), and this effect is even greater in the presence of an anti-β-catenin antibody (Fig. 4A, lane 7). An antibody against SRC-1, another coactivator of PPARγ, had no effect on the binding of the PPRE to the PPARγ/RXRα complexes. Taken together, these assays suggest that PPARγ, as a heterodimer with RXRα, is in a complex with at least the GRIP1/TIF2 coactivator and the ectopically expressed S37A β-catenin. As confirmation of this association, we incubated a biotin-labeled PPRE oligonucleotide with nuclear extracts from cells expressing S37A β-catenin and PPARγ and pulled down the complexes with streptavidin beads. The Western blot of the PPRE-associated proteins is shown in Fig. 4B and reveals an association of both PPARγ and β-catenin with the PPRE.
FIG. 4.
Interaction of β-catenin with PPARγ and associated proteins. (A) Nuclear proteins from S37A β-catenin (S37A) cells were prepared as described in the legend to Fig. 3A in the presence of troglitazone and subjected to EMSA as outlined in Materials and Methods using the indicated antibodies to supershift or perturb the association of corresponding proteins with the PPRE oligonucleotide. (B) Nuclear extract proteins from WT and S37A β-catenin cells were incubated with biotinylated PPRE oligonucleotide as described in Materials and Methods. The complexes binding to the oligonucleotides were analyzed by Western blotting for HA-β-catenin and PPARγ. Equal amounts of proteins were analyzed on the two leftmost lanes for input. (C) Nuclear proteins extracted from the S37A β-catenin mutant cell lines exposed to troglitazone were immunoprecipitated with anti-PPARγ, and the complexes were subjected to Western blot analysis using anti-HA (β-catenin) and anti-PPARγ antibodies.
K312 and K435 are required for the interaction of β-catenin with PPARγ.
We next addressed whether the TCF/LEF binding site (K312 and K435) in β-catenin is required for the formation of PPARγ/β-catenin complexes. Figure 4C demonstrates that immunoprecipitation of complexes expressed in the Swiss-Pγ-β-catenin cell lines exposed to troglitazone with an anti-PPARγ antibody pulled down the ectopic parental S37A β-catenin. Mutation of K312 and/or K345 within S37A significantly reduces the extent of this pull-down (Fig. 4C, compare lane 3, 4, 5, or 6 with lane 2), whereas mutation of just the ICAT binding site (F660A and R661A) only partially affects the association of S37A β-catenin with PPARγ (Fig. 4C, lane 7). Immunoprecipitation of PPARγ from cells expressing a WT β-catenin shows undetectable amounts of a coprecipitating HA-β-catenin on the Western blot (Fig. 4C, lane 1) because the abundance of the ectopic β-catenin is very low due to its rapid rate of turnover. These data in Fig. 4C, taken together with those presented in Fig. 4A and B, provide strong evidence for an association of β-catenin with nuclear complexes containing PPARγ that appear to involve the same region of β-catenin required for binding to TCF/LEF.
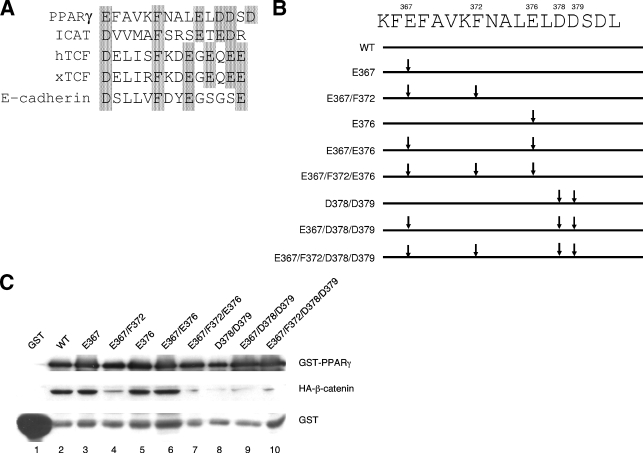
Identification of a domain in PPARγ facilitating its association with β-catenin.
The data presented above suggest that PPARγ might interact directly with the TCF/LEF binding site within β-catenin. We questioned, therefore, whether PPARγ contains a binding site for β-catenin similar to that present in TCF/LEF. In fact, Fig. 5A shows a region of PPARγ between amino acids 367 and 381 that is highly homologous to the β-catenin binding site in TCF/LEF (18, 19). To determine whether this region facilitates binding of PPARγ to β-catenin, we generated a series of GST-PPARγ fusion proteins in which highly conserved amino acids that we considered might be involved in the binding process were altered as shown in Fig. 5B. The GST-PPARγ mutants were then used to pull down nuclear proteins from Swiss cells expressing an ectopic S37A β-catenin. The Western blot shown in Fig. 5C demonstrates that the WT GST-PPARγ pulls down abundant amounts of S37A β-catenin compared to the undetectable amounts associating with GST alone (compare lane 2 and lane 1). These data are consistent with those shown in Fig. 4 and published reports by others (23, 30) confirming an interaction between PPARγ and β-catenin. Figure 5C demonstrates further that this interaction involves amino acids F372, D378, and D379, since mutation of each of these to an alanine significantly attenuates the ability of GST-PPARγ to pull down S37A β-catenin (compare lanes 4, 7, 8, 9, and 10 with lanes 2, 3, 5, and 6).
FIG. 5.
Phenylalanine 372 and aspartic acids 378 and 379 of PPARγ are required for its ability to bind to and induce the degradation of β-catenin. (A) Sequence alignment (ClustalX 1.8) of a putative catenin binding domain (aa 367 to 382) of PPARγ together with similar domains within ICAT, human and Xenopus TCFs, and E-cadherin. (B) Schematic representation of the WT and a series of mutated PPARγ-GST fusion proteins (only the catenin binding domain is shown). (C) GST-PPARγ pull-down of β-catenins. Nuclear proteins extracted from Swiss fibroblasts expressing S37A β-catenin were incubated with GST protein (lane 1), WT PPARγ-GST (lane 2), or mutated PPARγ-GST fusion proteins (lanes 3 to 10). The binding complexes were pulled down with glutathione Sepharose 4B beads and subjected to Western blot analysis of HA-β-catenin (anti-HA). One-twentieth the amount of each sample was used to analyze the presence of GST or GST-PPARγ fusion proteins on duplicate SDS-PAGE/Western blots using anti-GST and anti-PPARγ antibodies to verify equal input.
The PPARγ-associated degradation of normal β-catenin involves F372 within the CBD of PPARγ.
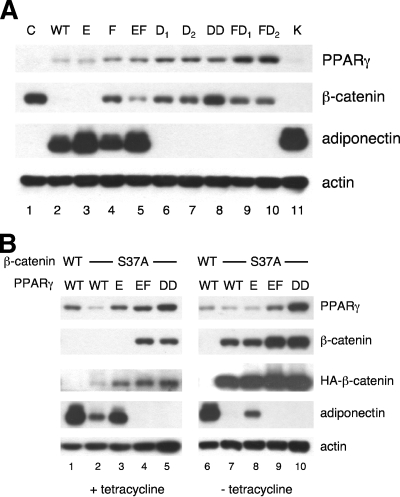
We next questioned whether this putative CBD within helices 7 and 8 is involved in any of the activities of PPARγ; consequently, we expressed select mutants of PPARγ corresponding to the mutations K365A (K), E367A (E), F372A (F), E367A/F372A (EF), D378A (D1), D379A (D2), D378A/D379A (D1D2), F372A/D378A (FD1), or F372A/D379A (FD2) as well as the wild-type protein in normal Swiss fibroblasts. The resulting Swiss-PPARγ cell lines were exposed to adipogenic inducers including troglitazone for 4 days, and total cell proteins were harvested for Western blot analysis. The data presented in Fig. 6A show that WT, E367A, and K365A PPARγ are capable of activating the degradation of the endogenous β-catenin along with inducing the expression of adiponectin (Fig. 6A, compare lanes 2, 3, and 11 with lane 1). These data suggest that changing E367 or K365 to alanine does not significantly alter PPARγ activity, whereas mutation of F372 to alanine alone (F) or in combination with E367A (EF) attenuates the ability of PPARγ to induce the degradation of β-catenin without significantly affecting its transcriptional activity (Fig. 6A, compare lanes 4 and 5 with lane 2). The data in Fig. 5C show that F372 is critical for the association of PPARγ with β-catenin, raising the possibility, therefore, that the CBD of PPARγ is important for the turnover of β-catenin. Mutation of D378 or D379 to alanine either alone (D1 or D2), together (D1D2), or in combination with F372A (FD1, FD2), however, completely blocks the ability of PPARγ to stimulate β-catenin degradation or induce adiponectin expression.
FIG. 6.
Identification of amino acids within helices 7 and 8 of PPARγ (CBD) responsible for regulating its activity. (A) Swiss fibroblasts expressing a control vector (C), WT PPARγ, or the following mutant PPARγ molecules were exposed to the adipogenic inducers in the presence of troglitazone for 4 days as outlined in Materials and Methods: E367A (E), F372A (F), E367A/F372A (EF), D378A (D1), D379A (D2), D378A/D379A (D1D2), F372A/D378A (FD1), F372A/D379A (FD2), and K365A (K). Total cellular proteins were then subjected to Western blot analysis of normal β-catenin, PPARγ, adiponectin, and actin. (B) Stable Swiss fibroblast cell lines expressing WT β-catenin and WT PPARγ (lanes 1 and 6) or tetracycline-responsive HA-tagged S37A β-catenin along with either WT PPARγ (lanes 2 and 7), E367A (E) PPARγ (lanes 3 and 8), E367A/F372A (EF) PPARγ (lanes 4 and 9), or D378A/D379A (D1D2) PPARγ (lanes 5 and 10) were cultured in the presence or absence of tetracycline and exposed to the adipogenic inducers with troglitazone for 4 days as outlined in Materials and Methods. Total cellular proteins were then subjected to Western blot analysis of PPARγ, normal β-catenin, HA-tagged S37A β-catenin, adiponectin, and actin.
To determine whether the CBD of PPARγ is involved in the inhibition of PPARγ target gene expression by oncogenic β-catenin, we ectopically expressed the WT and select mutants of PPARγ (E367A, E367A/F372A, and D378A/D379A) in Swiss fibroblasts containing a tetracycline-responsive HA-tagged S37A β-catenin gene (S37A cells). WT PPARγ was also expressed in Swiss fibroblasts expressing a WT β-catenin. The corresponding cell lines were exposed to adipogenic inducers, including troglitazone, in the presence or absence of tetracycline to control the expression of the ectopic S37A β-catenin. Figure 6B, lane 1, shows that WT PPARγ in cells expressing WT β-catenin stimulates adiponectin expression and induces the turnover of total cellular β-catenin as previously observed in Fig. 6A, lane 2. Activation of WT PPARγ or E367A PPARγ in S37A cells exposed to tetracycline induces the degradation of cellular β-catenin as well as stimulates adiponectin gene expression (Fig. 6B, lanes 2 and 3). The adipogenic activity of WT PPARγ, however, is significantly reduced in the S37A cells compared to cells containing a WT β-catenin (Fig. 6B, compare lane 2 with lane 1). This is possibly due to the fact that there is some leakage of S37A β-catenin expression in these cells even though they are exposed to tetracycline, as illustrated in the blot corresponding to the anti-HA antibody. This level of the HA-tagged S37A β-catenin is likely enough to attenuate PPARγ activity. The E367A PPARγ appears to be less sensitive to this inhibitory action of the oncogenic (S37A) β-catenin (Fig. 6B, compare lane 3 with lane 2). The E367A/F372A (EF) and D378A/D379A (D1D2) mutants of PPARγ are completely inactive in these cell lines (Fig. 6B, lanes 4 and 5). To assess the activity of each of the PPARγ molecules in cells expressing abundant amounts of the S37A β-catenin, the Swiss cell lines were exposed to the adipogenic inducers (plus troglitazone) in the absence of tetracycline. Activation of WT and E367A PPARγ by troglitazone is incapable of inducing the degradation of HA-tagged S37A β-catenin but does significantly reduce the total amount of β-catenin compared to cells expressing a defective PPARγ molecule (D1D2) (Fig. 6B, compare lanes 7 and 8 with lane 10). The ability of WT PPARγ to induce adiponectin gene expression, however, is completely blocked by the ectopic S37A β-catenin (Fig. 6B, lane 7). Interestingly, E367A PPARγ does retain some ability to activate adiponectin expression in the presence of the oncogenic β-catenin (Fig. 6B, compare lanes 7 and 8). The E367A/F372A (EF) and D378A/D379A (D1D2) mutants of PPARγ are completely inactive, since they are incapable of inducing adiponectin expression or enhancing the degradation of total cellular β-catenin (Fig. 6B, lanes 9 and 10). Taken together, these data suggest that S37A β-catenin inhibits the ability of WT PPARγ to induce adipogenic gene expression without inhibiting its ability to induce the degradation of normal cellular β-catenin. Furthermore, mutation of E367 to alanine appears to enhance PPARγ activity so that it can overcome to some extent the inhibitory action of S37A β-catenin. Mutation of F372 to alanine along with the E367A mutation produces a PPARγ molecule (EF PPARγ) that is inactive in the presence of small as well as abundant amounts of S37A β-catenin (Fig. 6B, lanes 4 and 9). Finally, as observed in Fig. 6A, mutation of D378 and D379 to alanines (D1D2) completely inactivates PPARγ.
Three-dimensional modeling of PPARγ interacting with β-catenin.
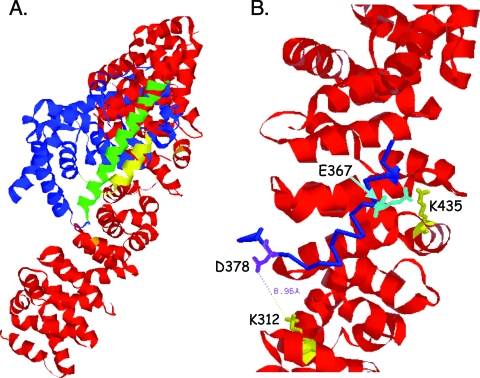
On the basis of the present data, we postulated that PPARγ directly interacts with β-catenin. Consequently, we obtained a predicted structure for the two proteins binding to each other via the TCF/LEF binding domain in β-catenin and the CBD in PPARγ using the protein-protein docking program ClusPro (8, 9) and the published crystal structures of each protein with Protein Data Bank (PDB) codes as follows: 2prg for PPARγ (38) and 1LUJ for β-catenin (18, 19). Figure 7A shows the predicted three-dimensional structure of the two interacting proteins, while Fig. 7B depicts the site of interaction between the TCF/LEF binding domain in β-catenin and the CBD in PPARγ. The charged buttons K312 and K435 in β-catenin are predicted to form salt bridges with D378 and E367, respectively, in PPARγ.
FIG. 7.
Predicted three-dimensional model of PPARγ interacting with β-catenin. (A) The ribbon model shows the entire structure of PPARγ (blue) and β-catenin (red). The model was generated using the protein-protein docking program (ClusPro) available at http://nrc.bu.edu/cluster. PDB codes 2prg (for PPARγ) and 1LUJ (for β-catenin) were downloaded from the PDB website (http://www.pdb.org). The yellow region (helices 7 and 8, aa 367 to 382) of PPARγ is shown in close proximity to the charged buttons, K435 to K312, of β-catenin and appears not to interfere with helix 10 (green), which is required for binding to RXRα. (B) A region of the model from panel A was enlarged to show the interaction of amino acids 367 to 382 of PPARγ (blue, backbone only shown) with the charged buttons K312 and K435 (yellow) of β-catenin (red). The side chains of two positively charged amino acids (K312 and K435) of β-catenin and two negatively charged amino acids (D378 in purple and E367 in cyan) of PPARγ are shown in stick drawing. The distance between the K312 (β-catenin) and D378 (of PPARγ) side chains is 8.96 Å, which is within the distance to form a “salt bridge.”
DISCUSSION
In earlier studies, we demonstrated that PPARγ can induce the proteasomal degradation of normal β-catenin but is incapable of degrading oncogenic β-catenin (S37A), which is resistant to phosphorylation by GSK3β (28). Our studies showed that S37A β-catenin inhibits PPARγ activity during the differentiation of mouse fibroblasts into adipocytes. In the present study, we questioned whether β-catenin and PPARγ interact with each other in a way that affects each of their activities. In fact, the data demonstrate an interaction between β-catenin and PPARγ that is facilitated by the TCF/LEF binding domain of β-catenin and a putative CBD within helices 7 and 8 of the ligand-binding domain of PPARγ. We show that mutation of the “charged buttons” K312 and K435 within the TCF/LEF binding domain significantly attenuates the ability of S37A β-catenin to interact with PPARγ as well as activate TCF/LEF target gene expression in Swiss mouse fibroblasts. Additionally, these mutations prevent S37A β-catenin from inhibiting PPARγ activity, and consequently, this oncogenic form of β-catenin is degraded in the proteasome in response to activation of PPARγ. Finally, mutation of E367 and F372 within the CBD of PPARγ prevents the association of PPARγ with β-catenin and, in so doing, reduces the ability of PPARγ to facilitate the degradation of normal β-catenin. On the basis of these data, we propose that in normal untransformed cells PPARγ can induce the proteasomal degradation of β-catenin through a mechanism that involves the CBD of PPARγ and the TCF binding domain of β-catenin. In transformed cells, oncogenic β-catenin escapes its PPARγ-associated degradation by inhibiting PPARγ activity, which appears to involve a functional TCF binding domain. This model suggests that the oncogenic form of β-catenin is dominant over PPARγ, thus explaining why PPARγ is capable only of suppressing tumorigenesis in cells (i.e., colon epithelial cells) that have a functional APC to facilitate the GSK3β inactivation of β-catenin (16).
What are the mechanisms by which oncogenic β-catenin inhibits PPARγ activity?
Figures 2 and 4C demonstrate that mutation of K312 and K435 to glutamic acids significantly reduces the capacity of β-catenin to interact with and inhibit the activity of PPARγ. Since these amino acids play a critical role in facilitating the association of β-catenin with TCF/LEF as well as PPARγ, one must consider the possibility that TCF/LEF signaling converges on PPARγ. In fact, recent studies have shown that cyclin D1, a well-recognized TCF/LEF target gene product, can inhibit PPARγ activity in a variety of cell types (15, 52). However, mutation of K312 or K435 reduces the inhibitory activity of S37A β-catenin on PPARγ but does not appear to reduce the expression of cyclin D1 (Fig. 2). In contrast, these single mutations do have a significant effect on the ability of S37A β-catenin to interact with PPARγ (Fig. 4C). Consequently, we propose that in response to the appropriate signal (Wnt activation or oncogenic mutation), β-catenin accumulates within the cell nucleus and associates with complexes containing PPARγ. It is likely that this interaction has no significant impact on β-catenin's ability to coactivate TCF/LEF, since the expression of S37A β-catenin in fibroblasts expressing abundant amounts of PPARγ can still activate cyclin D1 expression (Fig. 2). However, the interaction between PPARγ and β-catenin appears to affect PPARγ activity. It is generally accepted that activation of PPARγ involves its association with an appropriate ligand that stabilizes helix 12 and facilitates binding of coactivators, such as TIF2, to the AF-2 transactivation domain. It is possible that binding of β-catenin to helices 7 and 8 on the side of the ligand-binding domain opposite from helix 12 perturbs interactions with these coactivators or with RXRα. It is also conceivable that β-catenin blocks the binding of other factors that interact directly with helices 7 and 8 to regulate PPARγ activity by means that are independent of helix 12. In this regard, recent studies have shown that PPARγ can repress the expression of inflammatory genes such as inducible nitric oxide synthase through mechanisms that involve a ligand-dependent SUMOylation of PPARγ on K365 in helix 7, which induces the recruitment of the nuclear receptor corepressor and histone deacetylase complexes to the inducible nitric oxide synthase gene promoter (39). It is interesting that the consensus site for SUMOylation (PK365FE) overlaps with the putative CBD (PKFE367FAVKFNALELDD378; underlined amino acids are the CBD) within helices 7 and 8 of PPARγ (see Fig. S1 in the supplemental material). Consequently, it is possible that this region of PPARγ is subject to structural modification, such as SUMOylation on K365, which influences transcriptional activity that can also be altered in response to binding to β-catenin. It will be of interest to determine whether β-catenin affects the suppression of inflammation in response to PPARγ activation.
What are the mechanisms by which PPARγ enhances the proteasomal degradation of β-catenin?
In an earlier publication (28), we suggested that PPARγ enhances β-catenin degradation in the proteasome by stimulating GSK3β activity through a mechanism that could include the induction of PTEN expression by PPARγ (40). This notion was based on the fact that PTEN, in blocking phosphatidylinositol 3-kinase signaling, would lead to suppression of Akt/protein kinase B activity and consequently stimulate GSK3β. The present data suggest that alternative mechanisms must also exist, since PPARγ can enhance the proteasomal degradation of β-catenin molecules that are resistant to phosphorylation by GSK3β. Specifically, the data presented in Fig. 2 show that mutation of lysines 312 and 435 diminishes the inhibitory action of S37A β-catenin on PPARγ, and consequently these oncogenic β-catenin molecules become subject to proteasomal degradation in response to activation of PPARγ by troglitazone (Fig. 3). It is unlikely that PPARγ facilitates this degradation process through a direct interaction with β-catenin, since these mutations also prevent such an interaction (Fig. 4C). At present, we are considering the possibility that PPARγ induces the expression of other components of the ubiquitylation/proteasomal system that interact with β-catenin at sites other than those requiring GSK3β activity. In particular, studies have uncovered an alternative pathway of β-catenin degradation involving a distinct ubiquitin-ligase complex, Siah-1-SIP-Skp1-Ebi, in which the F-box protein Ebi binds to β-catenin at sites that do not require phosphorylation by GSK3β (29, 31). Future studies are aimed at investigating the effect of PPARγ on the expression and activity of these ubiquitylation/proteasomal system components. It is also important to mention that induction of β-catenin degradation by PPARγ involves F372 within helices 7 and 8 of PPARγ that appears to be independent of overall PPARγ transcriptional activity (Fig. 6A). Presently, we are pursuing the notion that this region of PPARγ (CBD) regulates the expression of select target genes, some of which are involved in the turnover of β-catenin. Other studies as well as these (Fig. 4A) support the notion that PPARγ and β-catenin coexist within larger complexes that contain TCF (23) as well as a variety of coactivators or corepressors. Such complexes could possibly be involved in regulating the transcription of select target genes, as is the case for the interaction of LRH-1 and β-catenin (6). Additionally, PPARγ and β-catenin might exist in a complex whose function is to facilitate their degradation in the 26S proteasome.
The interaction between β-catenin and PPARγ likely influences the function of each protein in controlling gene expression in a variety of cell types. Recent studies have demonstrated that canonical Wnt signaling mediated by β-catenin regulates the fate of mesenchymal stem cells. Specifically, activation of Wnt pathways or ectopic expression of oncogenic β-catenin inhibits adipogenesis in mesenchymal cells in favor of myogenesis (44). In fact, conditional deletion of β-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium (2). It appears, therefore, that mechanisms must exist within the adipogenic program to ensure suppression of β-catenin activity. These processes include a PPARγ-associated stimulation of β-catenin degradation in the proteasome as outlined in these and earlier studies as well as a downregulation of Wnt's and their frizzled receptors (3, 28, 32). β-Catenin and PPARγ are also proposed to influence each other's activities in colon epithelial cells. It is well established that oncogenic β-catenin contributes to the progression of colon cancer in humans as well as rodents (33, 41). Recent studies have also suggested that PPARγ has a role to play in the development of gut epithelial cells (12) and that activation of PPARγ in the colon can suppress carcinogenesis by reducing β-catenin levels in cells that express functional APC molecules (16). In conclusion, a greater understanding of the functional interaction between β-catenin and PPARγ should provide insight into the development of mesenchymal and epithelial stem cells as well as lead to potential therapeutics for cancer and obesity-related disorders.
Supplementary Material
Acknowledgments
We acknowledge the advice and assistance of Stephen Comeau and Michael Silberstein in the laboratory of Sandor Vajda in the Department of Biomedical Engineering, Boston University, and Pinglei Zhou in the Department of Molecular and Cellular Biology at Harvard University in developing the three-dimensional model of the interaction of PPARγ with β-catenin.
This work was supported by USPHS grants DK51586 and DK58825.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altiok, S., M. Xu, and B. M. Spiegelman. 1997. PPARgamma induces cell cycle withdrawal: inhibition of E2F/DP DNA-binding activity via down-regulation of PP2A. Genes Dev. 11:1987-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arango, N. A., P. P. Szotek, T. F. Manganaro, E. Oliva, P. K. Donahoe, and J. Teixeira. 2005. Conditional deletion of beta-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium. Dev. Biol. 288:276-283. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, C. N., S. E. Ross, K. A. Longo, L. Bajnok, N. Hemati, K. W. Johnson, S. D. Harrison, and O. A. MacDougald. 2002. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277:30998-31004. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Ze'ev, A. 1997. Cytoskeletal and adhesion proteins as tumor suppressors. Curr. Opin. Cell Biol. 9:99-108. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Ze'ev, A. 1999. The dual role of cytoskeletal anchor proteins in cell adhesion and signal transduction. Ann. N. Y. Acad. Sci. 886:37-47. [DOI] [PubMed] [Google Scholar]
- 6.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15:499-509. [DOI] [PubMed] [Google Scholar]
- 7.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 8.Comeau, S. R., D. W. Gatchell, S. Vajda, and C. J. Camacho. 2004. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32:W96-W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comeau, S. R., D. W. Gatchell, S. Vajda, and C. J. Camacho. 2004. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20:45-50. [DOI] [PubMed] [Google Scholar]
- 10.Conacci-Sorrell, M., J. Zhurinsky, and A. Ben-Ze'ev. 2002. The cadherin-catenin adhesion system in signaling and cancer. J. Clin. Investig. 109:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels, D. L., and W. I. Weis. 2002. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol. Cell 10:573-584. [DOI] [PubMed] [Google Scholar]
- 12.Drori, S., G. D. Girnun, L. Tou, J. D. Szwaya, E. Mueller, K. Xia, R. A. Shivdasani, and B. M. Spiegelman. 2005. Hic-5 regulates an epithelial program mediated by PPARgamma. Genes Dev. 19:362-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, R. M., G. D. Barish, and Y. X. Wang. 2004. PPARs and the complex journey to obesity. Nat. Med. 10:355-361. [DOI] [PubMed] [Google Scholar]
- 14.Fajas, L., M. B. Debril, and J. Auwerx. 2001. PPAR gamma: an essential role in metabolic control. Nutr. Metab. Cardiovasc. Dis. 11:64-69. [PubMed] [Google Scholar]
- 15.Fu, M., M. Rao, T. Bouras, C. Wang, K. Wu, X. Zhang, Z. Li, T. P. Yao, and R. G. Pestell. 2005. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J. Biol. Chem. 280:16934-16941. [DOI] [PubMed] [Google Scholar]
- 16.Girnun, G. D., W. M. Smith, S. Drori, P. Sarraf, E. Mueller, C. Eng, P. Nambiar, D. W. Rosenberg, R. T. Bronson, W. Edelmann, R. Kucherlapati, F. J. Gonzalez, and B. M. Spiegelman. 2002. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc. Natl. Acad. Sci. USA 99:13771-13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girnun, G. D., and B. M. Spiegelman. 2003. PPARgamma ligands: taking Ppart in chemoprevention [sic]. Gastroenterology 124:564-567. [DOI] [PubMed] [Google Scholar]
- 18.Graham, T. A., W. K. Clements, D. Kimelman, and W. Xu. 2002. The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT. Mol. Cell 10:563-571. [DOI] [PubMed] [Google Scholar]
- 19.Graham, T. A., C. Weaver, F. Mao, D. Kimelman, and W. Xu. 2000. Crystal structure of a beta-catenin/Tcf complex. Cell 103:885-896. [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber, A. H., W. J. Nelson, and W. I. Weis. 1997. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90:871-882. [DOI] [PubMed] [Google Scholar]
- 22.Huber, A. H., and W. I. Weis. 2001. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105:391-402. [DOI] [PubMed] [Google Scholar]
- 23.Jansson, E. A., A. Are, G. Greicius, I. C. Kuo, D. Kelly, V. Arulampalam, and S. Pettersson. 2005. The Wnt/beta-catenin signaling pathway targets PPARgamma activity in colon cancer cells. Proc. Natl. Acad. Sci. USA 102:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kliewer, S. A., H. E. Xu, M. H. Lambert, and T. M. Willson. 2001. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog. Horm. Res. 56:239-263. [DOI] [PubMed] [Google Scholar]
- 25.Krey, G., O. Braissant, F. L'Horset, E. Kalkhoven, M. Perroud, M. G. Parker, and W. Wahli. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 11:779-791. [DOI] [PubMed] [Google Scholar]
- 26.Lazar, M. A. 2005. PPAR gamma, 10 years later. Biochimie 87:9-13. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., and D. I. Beller. 2003. Distinct pathways for NF-kappa B regulation are associated with aberrant macrophage IL-12 production in lupus- and diabetes-prone mouse strains. J. Immunol. 170:4489-4496. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., and S. R. Farmer. 2004. Regulating the balance between peroxisome proliferator-activated receptor gamma and beta-catenin signaling during adipogenesis. A glycogen synthase kinase 3beta phosphorylation-defective mutant of beta-catenin inhibits expression of a subset of adipogenic genes. J. Biol. Chem. 279:45020-45027. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927-936. [DOI] [PubMed] [Google Scholar]
- 30.Lu, D., H. B. Cottam, M. Corr, and D. A. Carson. 2005. Repression of beta-catenin function in malignant cells by nonsteroidal antiinflammatory drugs. Proc. Natl. Acad. Sci. USA 102:18567-18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 32.Moldes, M., Y. Zuo, R. F. Morrison, D. Silva, B. H. Park, J. Liu, and S. R. Farmer. 2003. Peroxisome-proliferator-activated receptor gamma suppresses Wnt/beta-catenin signalling during adipogenesis. Biochem. J. 376:607-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, R. F., and S. R. Farmer. 1999. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 274:17088-17097. [DOI] [PubMed] [Google Scholar]
- 35.Mueller, E., P. Sarraf, P. Tontonoz, R. M. Evans, K. J. Martin, M. Zhang, C. Fletcher, S. Singer, and B. M. Spiegelman. 1998. Terminal differentiation of human breast cancer through PPAR gamma. Mol. Cell 1:465-470. [DOI] [PubMed] [Google Scholar]
- 36.Mulholland, D. J., S. Dedhar, G. A. Coetzee, and C. C. Nelson. 2005. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr. Rev. 26:898-915. [DOI] [PubMed] [Google Scholar]
- 37.Nelson, W. J., and R. Nusse. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303:1483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 39.Pascual, G., A. L. Fong, S. Ogawa, A. Gamliel, A. C. Li, V. Perissi, D. W. Rose, T. M. Willson, M. G. Rosenfeld, and C. K. Glass. 2005. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 437:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel, L., I. Pass, P. Coxon, C. P. Downes, S. A. Smith, and C. H. Macphee. 2001. Tumor suppressor and anti-inflammatory actions of PPARgamma agonists are mediated via upregulation of PTEN. Curr. Biol. 11:764-768. [DOI] [PubMed] [Google Scholar]
- 41.Peifer, M. 1997. Beta-catenin as oncogene: the smoking gun. Science 275:1752-1753. [DOI] [PubMed] [Google Scholar]
- 42.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 43.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 44.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950-953. [DOI] [PubMed] [Google Scholar]
- 45.Sarraf, P., E. Mueller, W. M. Smith, H. M. Wright, J. B. Kum, L. A. Aaltonen, A. de la Chapelle, B. M. Spiegelman, and C. Eng. 1999. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol. Cell 3:799-804. [DOI] [PubMed] [Google Scholar]
- 46.Schoonjans, K., L. Dubuquoy, J. Mebis, E. Fayard, O. Wendling, C. Haby, K. Geboes, and J. Auwerx. 2005. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA 102:2058-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spiegelman, B. M. 1998. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes 47:507-514. [DOI] [PubMed] [Google Scholar]
- 48.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPARg, a lipid-activated transcription factor. Cell 79:1147-1156. [DOI] [PubMed] [Google Scholar]
- 50.Truica, C. I., S. Byers, and E. P. Gelmann. 2000. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 60:4709-4713. [PubMed] [Google Scholar]
- 51.von Kries, J. P., G. Winbeck, C. Asbrand, T. Schwarz-Romond, N. Sochnikova, A. Dell'Oro, J. Behrens, and W. Birchmeier. 2000. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 7:800-807. [DOI] [PubMed] [Google Scholar]
- 52.Wang, C., N. Pattabiraman, J. N. Zhou, M. Fu, T. Sakamaki, C. Albanese, Z. Li, K. Wu, J. Hulit, P. Neumeister, P. M. Novikoff, M. Brownlee, P. E. Scherer, J. G. Jones, K. D. Whitney, L. A. Donehower, E. L. Harris, T. Rohan, D. C. Johns, and R. G. Pestell. 2003. Cyclin D1 repression of peroxisome proliferator-activated receptor γ expression and transactivation. Mol. Cell. Biol. 23:6159-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willert, K., and R. Nusse. 1998. Beta-catenin: a key mediator of Wnt signaling. Curr. Opin. Genet. Dev. 8:95-102. [DOI] [PubMed] [Google Scholar]
- 54.Willson, T. M., M. H. Lambert, and S. A. Kliewer. 2001. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem. 70:341-367. [DOI] [PubMed] [Google Scholar]
- 55.Wu, Z., N. L. R. Bucher, and S. R. Farmer. 1996. Induction of peroxisome proliferator-activated receptor γ during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol. Cell. Biol. 16:4128-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yap, A. S., W. M. Brieher, and B. M. Gumbiner. 1997. Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol. 13:119-146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.