Abstract
The binding of Brf1 to the tetratricopeptide repeat (TPR)-containing transcription factor IIIC (TFIIIC) subunit (Tfc4) represents a rate-limiting step in the ordered assembly of the RNA polymerase III initiation factor TFIIIB. Tfc4 contains multiple binding sites for Brf1 within its amino terminus and adjacent TPR arrays, but the access of Brf1 to these sites is limited by autoinhibition. Moreover, the Brf1 binding sites in Tfc4 overlap with sites important for the subsequent recruitment of another TFIIIB subunit, Bdp1, implying that repositioning of Brf1 is required after its initial interaction with Tfc4. As a starting point for dissecting the steps in TFIIIC-directed assembly of TFIIIB, we conducted yeast two-hybrid screens of Brf1 peptide libraries against different TPR-containing Tfc4 fragments. Short, biochemically active peptides were identified in three distinct regions of Brf1. Two peptides defined conserved but distal regions of Brf1 that participate in stable binding of Brf1 to TFIIIC-DNA. Remarkably, a third peptide that binds specifically to TPR6-9 of Tfc4 was found to promote the formation of both TFIIIC-DNA and Brf1-TFIIIC-DNA complexes and to reduce the mobility of these complexes in native gels. The data are consistent with this peptide causing a conformational change in TFIIIC that overcomes Tfc4 autoinhibition of Brf1 binding and suggest a structural model for the Brf1-Tfc4 interaction.
Transcription factor IIIC (TFIIIC) is an essential multisubunit assembly factor for RNA polymerase (Pol) III transcription of 5S rRNA, tRNA, and other genes that have similar promoter architectures. Upon binding to the promoters of these genes (with TFIIIA in the case of 5S genes), TFIIIC directs the assembly of the heterotrimeric initiation factor TFIIIB to the DNA upstream of the transcription start site (reviewed in references 11 and 32). The subunits of TFIIIB are loosely associated in the absence of DNA and are assembled sequentially by TFIIIC, beginning with the TFIIB-related factor Brf1. Subsequent binding of the TATA-binding protein, TBP, is followed by the SANT domain-containing protein Bdp1 to form an exceptionally stable TFIIIB-DNA complex in which the DNA is kinetically trapped (5, 14, 15, 21). The recruitment of both the Brf1 and Bdp1 subunits of TFIIIB is directed by the tetratricopeptide repeat (TPR)-containing subunit of TFIIIC, Tfc4 (8, 23, 27).
The interaction between Tfc4 and Brf1 represents a rate-limiting step in preinitiation complex assembly, as activating mutations in Tfc4 that increase Brf1 recruitment also facilitate TFIIIB complex assembly and Pol III transcription in vitro and increase the expression of a specific tRNA reporter gene in vivo (25, 27). The binding of Brf1 to Tfc4 is known to involve the N-terminal half of Tfc4 (Nt-TPR9), which contains two tandem TPR arrays (TPR1-5 and TPR6-9) separated by a 134-amino-acid intervening region (3, 28). Each TPR array can bind independently to Brf1 in vitro, and the significance of these interactions in the context of TFIIIC has been demonstrated genetically and biochemically (23, 25, 27).
TPR motifs are found in functionally diverse proteins and have a general role as protein-protein interaction domains and in multisubunit complex assembly (7). Each TPR motif is composed of a degenerate sequence of 34 amino acids arranged as a pair of antiparallel α helices (designated A and B). The known structures of tandem TPRs, either free or in complex with ligands, reveal a similar superhelical architecture with a concave groove that provides or supports a site for ligand binding. Peptides as short as five amino acids bind in the superhelical groove in an extended conformation with moderate to low micromolar affinity (10, 31). This mode of binding together with the large number of TPR sequences in public databases suggests that the TPR protein family is likely to recognize many different peptides. At present, however, relatively few peptide ligands have been identified and the basis of their sequence-specific recognition has been determined in only a few cases (4, 10, 22, 31, 36). Guided by the available structures of TPR proteins, our genetic and biochemical studies of Tfc4 suggest that the superhelical groove of each TPR array provides binding sites for Brf1 (23, 25, 27, 28). Moreover, as there is little sequence similarity between the two TPR arrays in Tfc4 (apart from the residues that are important for the overall fold), each TPR array is predicted to interact with different sites in Brf1.
Besides the binding sites in the TPR arrays, the N-terminal 128 amino acids of Tfc4 (Nt), which are essential for viability in Saccharomyces cerevisiae (8), are part of a high-affinity binding site for Brf1 (Nt-TPR5). Interestingly, the binding affinity of this site for Brf1, as well as the site present in TPR6-9, is significantly higher than that of a larger Tfc4 fragment (Nt-TPR9) that contains both of these sites (26). This has led to the suggestion that access of Brf1 to its binding sites in Tfc4 is limited by competing interactions within Nt-TPR9. This interpretation is supported by the behavior of a dominant mutation (the PCF1-1 mutation, H190Y) which maps to the convex surface of the first TPR array (25). The PCF1-1 mutation increases the affinity of the Nt-TPR9-Brf1 interaction but does not affect the interaction of Nt-TPR5 or TPR1-5 with Brf1. Thus, the mutation is thought to facilitate Brf1 binding by an indirect mechanism. Previous work has proposed that the PCF1-1 mutation relieves autoinhibition by stabilizing an alternative conformation of Tfc4 (25).
Brf1 plays multiple roles in Pol III transcription. In addition to its participation in initiation complex assembly, which involves associations with Tfc4 and the other TFIIIB subunits, Brf1 functions in polymerase recruitment through interactions with Rpc34 and Rpc17 and is also important for promoter melting (1, 9, 18, 20, 35). These functions are mediated by multiple conserved domains in the TFIIB-related N terminus of Brf1 (Nt-Brf1) and in the Brf1-specific C-terminal half of the protein (Ct-Brf1). The two halves of Brf1 appear to be structurally and functionally independent, as TFIIIB-DNA complexes formed with Brf1 that is split into two fragments at residue 283 are transcriptionally active and are indistinguishable by footprinting from complexes formed with intact Brf1 (17). Moreover, a triple fusion protein that inserts the conserved core of TBP between the N- and C-proximal domains of Brf1 is able to replace Brf1 and TBP in transcription in vitro and can support viability of a BRF1 deletion strain (19). Interestingly, whereas each half of Brf1 is able to support TATA box-directed TFIIIB complex formation in the absence of TFIIIC, both halves of Brf1 are required for proper TFIIIC-directed complex assembly (17), indicating that the structural requirements on Brf1 are more stringent for TFIIIC-dependent reactions.
Brf1 is not the only ligand for the TPRs of Tfc4; Bdp1 can also bind to Tfc4 fragments containing both TPR arrays (TPR1-9 or Nt-TPR9). A mutation (L469K) in TPR7 of Tfc4, which affects the recruitment of Brf1, also inhibits the direct binding of Bdp1 to Tfc4 and decreases the affinity of Bdp1 for the TBP-Brf1-TFIIIC-DNA complex, suggesting that Brf1 and Bdp1 share an overlapping binding site in Tfc4 (23). However, unlike Brf1, Bdp1 does not bind detectably to the individual TPR arrays or to TFIIIC-DNA. These data suggest that TFIIIC-dependent assembly of TFIIIB involves dynamic protein-protein interactions that likely include the repositioning of Brf1 to allow subsequent recruitment of Bdp1 (28).
To better understand the molecular interactions between Brf1 and Tfc4 and the concerted binding of TFIIIB subunits to TFIIIC-DNA, we constructed Brf1 peptide libraries and used a yeast two-hybrid assay to identify peptides that bind to the high-affinity (Nt-TPR5) and autoinhibited (Nt-TPR9) binding sites of Tfc4. Two peptides identified against Nt-TPR5 were found to inhibit Brf1 recruitment by TFIIIC-DNA, and the corresponding regions in Brf1 were shown to be critical for this function. In contrast, a peptide identified against Nt-TPR9 increased Brf1-TFIIIC-DNA complex formation and affected complex mobility, consistent with a role in relieving autoinhibition within Tfc4. The results suggest a model for the dynamic interaction between Tfc4 and Brf1.
MATERIALS AND METHODS
Cloning of Brf1 domains into pACTII.
A pACTII clone containing the entire BRF1 coding sequence was used as the starting plasmid for PCR and restriction analysis in order to construct activation domain fusions of smaller Brf1 fragments, including the N- and C-terminal halves, which are conveniently separated by a StuI site. The amino acid boundaries of pACTII clones containing repeat II and other conserved Brf1 domains are given below and were engineered at a filled-in BglII site. All of the clones were validated by DNA sequence analysis before transformation into yeast strain Y190 for two-hybrid assays.
Construction and screening of Brf1 peptide libraries in a yeast two-hybrid system.
Peptide libraries were constructed by cloning randomly digested fragments of the Brf1 coding sequence into the plasmid pACTII. Briefly, a 1.8-kb coding sequence fragment was cut with DNase I in the presence of 10 mM MnCl2 to obtain double-stranded DNA fragments ranging in size from approximately 30 to 150 bp. The digest was fractionated on a 2% agarose gel, and DNA fragments were purified by electroelution and filled in by T4 DNA polymerase before ligation into the filled-in XhoI site of pACTII. A library of ∼6,000 unique transformants was generated by transformation into Escherichia coli DH5α, and the pooled cells were used to prepare plasmid DNA for screening against the Nt-TPR5 fragment of Tfc4 in pASCYH2 (23, 26). The peptide library used to screen the Nt-TPR9 fragment of Tfc4 was constructed similarly but contained Brf1 fragments ranging in size from 100 to 400 bp that were cloned into pACTII at a filled-in BglII site. These DNA-based Brf1 peptide libraries were transformed into yeast strain Y190 containing the respective Tfc4 constructs in pASCYH2 and plated on synthetic complete (SC) medium lacking His, Leu, and Trp and containing 35 mM 3-aminotriazole to select for the plasmids and expression of the HIS3 reporter gene. His+ transformants were then screened for expression of the second reporter gene (lacZ) by use of a β-galactosidase colony lift filter assay with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as the substrate (23, 34). Positive colonies were picked from the original plates and streaked onto SC medium lacking His, Leu, and Trp (SC−His-Leu-Trp medium) and subsequently onto SC−Leu medium containing 2.5 μg/ml cycloheximide to evict the pASCYH2 plasmid. These cells were then mated with transformants of yeast strain Y187 containing pASCYH2 plasmids expressing Nt-TPR5, Nt-TPR9, or unrelated fusion proteins (p53 and Snf2) to determine the reproducibility and specificity of the initial interaction. Plasmid DNA was recovered from the validated colonies and transformed into E. coli, and the insert DNA was sequenced and translated. Individual candidate Brf1 sequences were recloned into pACTII at a filled-in BglII site and transformed into strain Y190 to confirm their interaction with Nt-TPR5 or Nt-TPR9. Fivefold serial dilutions of these transformants were spotted onto 3-aminotriazole-containing medium (see above) to monitor expression of the HIS3 reporter gene.
Mutagenesis of BRF1.
Mutagenesis of BRF1 was performed on pET11d BRF1ΔSD (24) and/or pRS313-BRF1 by using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations. The pRS313-BRF1 template used for mutagenesis is truncated at an Eag1 site 190 bp upstream of the initiating methionine and confers conditional growth at 37°C. Primers were used to generate the following Brf1 amino acid substitutions: R238A-R239A, N235A-N236A, M234A, L237A, R233A-N235A, N236A, R233A, R238A, and K501A-R502A. The K501A-R502A substitutions were also introduced into a BRF1 background containing the R238A-R239A mutations. All of the mutations were confirmed by sequencing, and mutant BRF1 alleles were transformed into yeast strain YSB108 (33). Single colonies were picked and resuspended in SC−His-Ura medium at a concentration of 2 × 107 cells/ml. Fivefold serial dilutions of each strain were spotted onto SC−His-Ura and 5-fluoroorotic acid-containing media to monitor growth in the presence and absence, respectively, of wild-type BRF1. Growth phenotypes were assayed at 16, 30, and 34°C.
Proteins and peptides.
Yeast TFIIIC, yeast RNA polymerase III, yeast B′′, recombinant TBP, and recombinant Bdp1 were prepared as reported previously (26, 27). Wild-type and mutant Brf1 were expressed in E. coli and purified under denaturing conditions before refolding, as previously described (24). The concentration of Brf1 was determined by image analysis of Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gels using bovine serum albumin as a standard and then corrected for differential staining of bovine serum albumin versus a gel-purified Brf1 standard. Seven peptides (101 and 103 to 108) were synthesized by Mimotopes Corp. and were judged to be about 80% pure by high-performance liquid chromatography and mass spectrometry. Chemical derivatives of peptide 101 (containing p-benzoylphenylalanine [Bpa] at the C terminus and/or biotin at the N terminus) and mutants of peptide 104 (99% pure), along with their-wild type counterparts, were synthesized by the proteomic center of Rockefeller University. All peptides were dissolved in H2O at a concentration of 10 mM.
Complex assembly and transcription assays.
TFIIIC-DNA complexes were preassembled for 10 min on a 32P-labeled sup3-eST tRNA gene-containing probe (27). Brf1, TBP, and Bdp1 were added as required in specific experiments and incubated at room temperature for 45 min before native gel electrophoresis (27). Peptides, unless otherwise indicated, were added to the preassembled TFIIIC-DNA complexes prior to the addition of Brf1. Brf1-TFIIIC-DNA and TFIIIB-TFIIIC-DNA complex formation was quantified and analyzed as previously described (26). Briefly, digital images collected on phosphor storage screens were quantified with ImageQuant software. Individual lines, one lane wide, were analyzed by peak fitting to calculate peak areas corresponding to the Brf1-TFIIIC-DNA or TFIIIB-TFIIIC-DNA and TFIIIC-DNA complexes. For each lane, assembly of the complex of interest was calculated as a percentage of the total number of TFIIIC-containing complexes. These values were then compared in the presence versus the absence of added Brf1 peptide or between wild-type and mutant Brf1. TFIIIC-dependent transcription was carried out using a sup4 tRNATyr gene (0.5 μg) under multiple-round conditions (1 h at 25°C) as previously described, with recombinant Brf1 (6 pmol), recombinant TBP (250 fmol), yeast-purified TFIIIC (5 fmol DNA-binding activity), B′′ fraction (125 fmol Bdp1), and Pol III (0.6 μg).
Photo-cross-linking of peptide 101 to Tfc4.
Peptide 101 analogs were incubated in the dark at 20°C with recombinant Tfc4 fragments (26) or TBP (each at 5 μM final concentration) for 10 min in Costar polystyrene 96-round-well plates. The solution conditions were the same as those used for complex assembly assays except that the salt concentration was raised to 150 mM NaCl. The plate without its lid was then held on a circulating ice-water bath and irradiated at 365 nm for 20 min by placing the UV lamp (115 V, model UVL-56, Blak-Ray lamp) directly on top of the plate. In competition experiments, a 50-fold molar excess of unmodified peptide 101 was preincubated with the Tfc4 fragments prior to the addition of the derivatized peptide. The irradiated samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane, and blotted with ECL streptavidin-horseradish peroxidase (HRP) conjugate (GE Healthcare) to detect the biotin signal on the proteins. The membrane was then stained with Gelcode blue (Pierce) by following the manufacturer's recommendations to demonstrate that equivalent amounts of Tfc4 fragments were present in all of the reactions. The preparations of recombinant Nt-TPR9, TPR1-5, and TPR6-9 have been described previously (26).
RESULTS
Identification of Brf1 peptides that interact with Tfc4 by use of a yeast two-hybrid assay.
Previous two-hybrid studies have shown that the TFIIB-related domain of Brf1 (Nt-Brf1) (Fig. 1) is able to bind Tfc4 (3), but the important sequences within this region of Brf1 have not been identified. Conversely, sequences in Ct-Brf1 are not known to interact with Tfc4, and yet TFIIIC can assemble higher-order complexes containing this fragment (3, 17). To look more closely at the interactions between Tfc4 and Brf1, we expressed several conserved domains of Brf1 as Gal4 activation domain fusion proteins and tested their interactions with a Gal4 DNA binding domain fusion to the high-affinity Brf1-binding region of Tfc4 (Nt-TPR5) (26) in a typical two-hybrid assay (Fig. 1B and C). As shown previously (3, 26), Nt-Brf1 (amino acids [aa] 1 to 286) itself suffices for a strong interaction with Nt-TPR5. Within this region of Brf1, the second direct repeat (repeat II, aa 174 to 286) (Fig. 1B) is sufficient for an interaction with Nt-TPR5. In contrast, the C-terminal half of Brf1 (aa 287 to 596) and a fragment containing conserved domains I and II (aa 287 to 561) did not detectably interact with Nt-TPR5. However, a smaller fragment encompassing conserved domain II (aa 435 to 561) (Fig. 1B) was able to interact efficiently with Nt-TPR5. The existence of a masked Tfc4 interaction site in domain II of Brf1 suggests that a conformational transition in this region of Brf1 may occur during its recruitment by TFIIIC. Interestingly, conformational changes in the C-terminal region of Brf1 have previously been proposed to expose a cryptic DNA-binding activity (12).
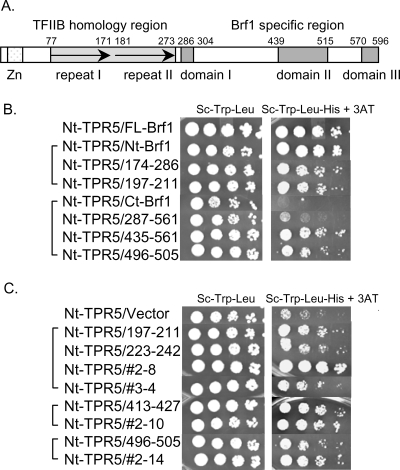
FIG. 1.
Delimiting the regions of Brf1 that interact with Nt-TPR5 in Tfc4 by two-hybrid assay. (A) Schematic representation of yeast Brf1 showing the zinc ribbon domain (Zn), the two imperfect repeats of the TFIIB homology region (arrows), and domains I, II, and III in the Brf1-specific C terminus. (B) Two-hybrid interactions of large Brf1 fragments with Nt-TPR5. Interactions are compared for full-length Brf1 (FL-Brf1), the N-terminal TFIIB homology region (Nt-Brf1), repeat II (aa 174 to 286 [174-286]), the C-terminal half of Brf1 (Ct-Brf1), and subfragments in this region (287-561 and 435-561). Shorter Brf1 peptide fusions (197-211 and 496-505) are included as a reference for the interactions shown in panel C. (C) Two-hybrid interactions between Nt-TPR5 and Brf1 peptide library-selected or engineered fusions (Table 1) are compared with empty pACTII (vector). In panels B and C, fivefold serial dilutions of the transformants were plated on the indicated media to compare cell numbers (left panels) and expression of the HIS3 reporter gene (right panels). 3-aminotriazole (3AT), a competitive inhibitor of the HIS3 gene product, was added to His− medium at a concentration of 10 mM. All of the colonies shown in panel C are from the same plate and were rearranged to facilitate comparison.
To more precisely define the regions of Brf1 that interact with Tfc4, we constructed a Brf1 peptide library by cloning randomly digested fragments of the coding sequence into pACTII (to create C-terminal fusions with the Gal4 activation domain) and then screened this library by using the yeast two-hybrid system against the high-affinity binding site in Tfc4 (Nt-TPR5). This approach has been used successfully to identify peptides that interact with the retinoblastoma protein (37) and is well suited to identify TPR ligands which are generally short and bind in an extended conformation (7). Positive interactions were selected for HIS3 expression and confirmed by expression of a second reporter gene, lacZ. From ∼6,000 yeast transformants, about 1% were His+ and ∼30% of these were also LacZ+. The specificity of the interactions was demonstrated by reversion of the His+ and LacZ+ phenotypes after eviction of the plasmid encoding Nt-TPR5 from positive strains and by the phenotypes obtained after reintroducing Nt-TPR5 or negative-control proteins. DNA sequence analysis of positive clones identified two conserved regions within the TFIIB homology domain and two regions within the unique C terminus of Brf1 as potential sites of interaction with Nt-TPR5 (Table 1). In the TFIIB homology domain, two noncontiguous regions in repeat II (aa 207 to 211 and aa 222 to 238) were recovered from a single clone (no. 2-8), as a result of multiple fragment insertions into the vector. The identification of aa 222 to 238 as a potential ligand for Nt-TPR5 was further supported by the recovery of aa 232 to 238 from an independent positive clone (no. 3-4). The two regions within the C terminus of Brf1 map to domain II (aa 497 to 505) and to a less conserved region between domains I and II (aa 413 to 427). The sequences of other positive clones showed that they contained a stop codon immediately adjacent to the insertion site or contained out-of-frame inserts that could be translated to give single or double arginine residues at or near the C terminus of the fusion protein. These sequences were not studied further. However, as will be discussed later, the basic character of these peptides may reflect a feature of the interaction with Nt-TPR5.
TABLE 1.
Identification of Brf1 peptides that bind to Nt-TPR5 in a two-hybrid assay
| Positive clone no. | Insert sequencea | Location in Brf1 (aa)b | Engineered sequencec (aa) | Synthetic peptide identification no., sequence (aa) |
|---|---|---|---|---|
| 2-8 | QRMSK | Nt repeat II (207-211) | KVVKDAVKLAQRMSK (197-211) | 106, KKIKVVKDAVKLAQRMSKDW (194-213) |
| 2-8 | GIAGACILLACRMNNLR | Nt repeat II (222-238) | IAGACILLACRMNNLRRTHT (223-242) | 107, IAGAAILLAARMNNLRRTHTW (223-243) |
| 3-4 | CRMNNLR | Nt repeat II (232-238) | IAGACILLACRMNNLRRTHT (223-242) | 107, IAGAAILLAARMNNLRRTHTW (223-243) |
| 2-10 | EEKKENESGHFQDAI | Ct (413-427) | EEKKENESGHFQDAI (413-427) | 105, EEKKENESGHFQDAIDGY (413-430) |
| 2-14 | EQESKRLKQ | Ct domain II (497-505) | LEQESKRLKQ (496-505) | 104, FLLEQESKRLKQEAD (494-508) |
Sequences shown occur after -IRIRAR. Randomly digested Brf1 fragments were cloned into pACTII at a filled-in XhoI site.
Peptide-containing regions in the amino terminus (Nt) and carboxy terminus (Ct) correspond to regions shown in Fig. 1.
Sequences shown occur after -PNPKKEI. The indicated sequences were engineered into pACTII at a filled-in BglII site.
To test individual Brf1 peptide sequences as well as the effect of fusion protein context on the interaction with Nt-TPR5, we cloned each candidate Brf1 sequence into the pACTII vector at a different insertion site (Table 1). The resulting engineered clones along with the original clones were tested against Nt-TPR5 for expression of the HIS3 reporter gene. Positive interactions with Nt-TPR5 were confirmed for all four Brf1 peptides (Fig. 1C). Notably, the strain bearing clone no. 2-8, which contains two Brf1 peptides (aa 207 to 211 and aa 222 to 238 [Table 1]), grew more robustly than strains containing the individual engineered peptides (aa 197 to 211 and aa 223 to 242), reflecting a contribution of both sequences in the overall interaction. Moreover, the strain expressing the latter peptide (aa 223 to 242) grew equally as well as the original clone (no. 3-4), which contains a single small insert (aa 232 to 238 [Table 1]) (Fig. 1C). Thus, this seven-amino-acid sequence in repeat II of Brf1 is likely to define an important site of interaction with Tfc4. The two engineered clones in the C-terminal half of Brf1 (aa 413 to 427 and aa 496 to 505) interacted with Nt-TPR5 similarly to their original isolates (no. 2-10 and no. 2-14, respectively), indicating that these interactions are not dependent on the context of the Brf1 peptide fusion with the Gal4 activation domain. Each of the four engineered Brf1 sequences was also tested for interactions with a Tfc4 fragment containing only the N-terminal TPR array (TPR1-5). Biochemical experiments have previously measured a low-affinity interaction between this TPR array and Brf1 (26). Consistent with this observation, Nt-Brf1 interacted weakly with TPR1-5 in a yeast two-hybrid assay. A similar interaction was seen for the sequences derived from the N-terminal half of Brf1. Sequences derived from the C terminus of Brf1 did not bind to TPR1-5 (Y. Liao, unpublished data).
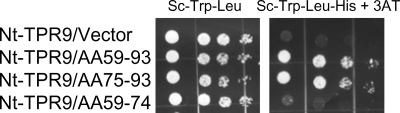
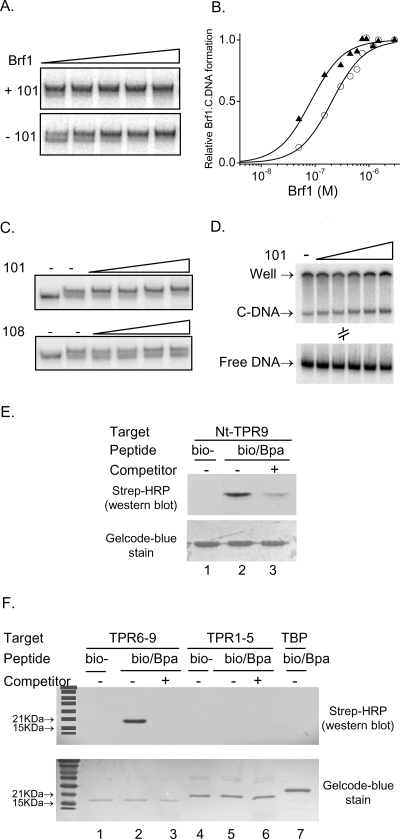
A fragment of Tfc4 extending from the N terminus to the end of TPR9 (Nt-TPR9) binds to Brf1 with reduced affinity relative to the two independent Brf1 binding sites (Nt-TPR5 and TPR6-9) contained within this region (26). To probe the Brf1 binding site in this larger Tfc4 fragment, we screened a Brf1 peptide library against Nt-TPR9. Four novel Brf1 fragments were identified (Table 2 and Fig. 2): three of these mapped to an overlapping region (aa 59 to 93) in the N terminus, while the fourth mapped to conserved domain II (aa 440 to 449) in the C terminus. Based on the homology with TFIIB, amino acids 75 to 93 of Brf1 correspond to the α1 helix of repeat I and adjacent upstream sequences, while amino acids 59 to 74 of Brf1 correspond to the linker region of TFIIB that connects the B finger to the TFIIB core (2). We cloned the sequences encoding these regions in Brf1 into pACTII and tested which of the two segments contributes to the interaction with Nt-TPR9. The region including the α1 helix of repeat I (aa 75 to 93) interacted with Nt-TPR9 with a strength similar to that of the parental clone (aa 59 to 93), whereas no binding was detected for amino acids 59 to 74 (Fig. 2).
TABLE 2.
Identification of Brf1 peptides that bind to Nt-TPR9 in a two-hybrid assay
| Positive clone no.a | Insert sequenceb | Location in Brf1 (aa)c | Locations (aa) of engineered sequencesd | Synthetic peptide identification no., sequence(s) (aa) |
|---|---|---|---|---|
| 45 | GAGQSHAAFYGSSALESREATLNNARRKLRAVSYA | Nt (59-93) | 59-74, 75-93 | 101, SREATLNNARRKLRAVSYA (75-93); 108, GAGQSHAAFYGSSALE (59-74)e |
| 4 | MPVCKN---YITDAAF | Nt (1-105) | ||
| 41 | MPVCKN---ACRKEKTH | Nt (1-140) | ||
| 50 | RNLHLLPTTDTYLSKVSDD | Ct (440-458) | 103, RNLHLLPTTDTYLSKVSDD (440-458) |
Clones 4, 41, and 45 contain overlapping sequences in the amino terminus of Brf1.
Sequences shown occur after -PNPKKEI. Randomly digested Brf1 fragments were cloned into pACTII at a filled-in BglII site.
Peptide-containing regions in the amino terminus (Nt) and carboxy terminus (Ct) correspond to regions shown in Fig. 1.
Locations shown are those of sequences occurring after -PNPKKEI. These sequences were engineered into pACTII at a filled-in BglII site.
Peptide no. 108 was the control.
FIG. 2.
Two-hybrid interactions between Nt-TPR9 of Tfc4 and Brf1 peptides. A Brf1 library-selected clone (no. 45, aa 59 to 93) and engineered fusions (Table 2) were compared with empty pACTII (vector) for interaction with Nt-TPR9. Serial dilutions of the transformants were plated on the indicated media to compare cell numbers (left panel) and expression of the HIS3 reporter gene (right panel) as described in the legend for Fig. 1.
Synthetic Brf1 peptides affect TFIIIC-mediated assembly of Brf1 and TFIIIB.
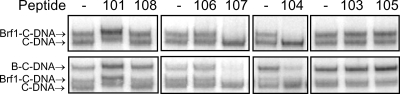
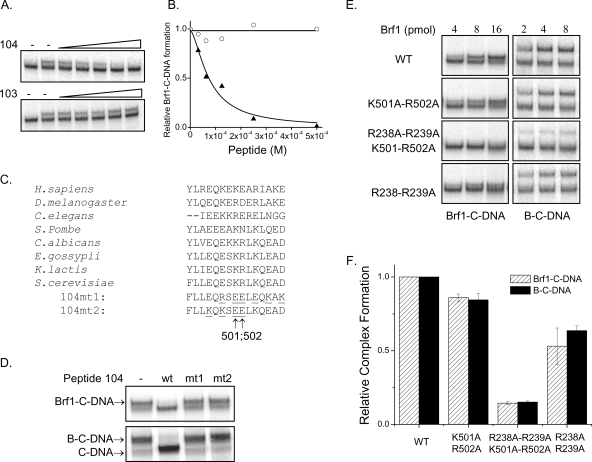
Six peptides, ranging from 15 to 21 amino acids in length, were synthesized based on the positive yeast two-hybrid interactions with Tfc4 fragments (Tables 1 and 2). These peptides were examined for their ability to perturb both the recruitment of Brf1 to TFIIIC-DNA complexes and the subsequent assembly of a complete TFIIIB complex. As a negative control for these experiments, we synthesized a 16-amino-acid peptide, corresponding to Brf1 amino acids 59 to 74, which did not interact with Nt-TPR9 in the two-hybrid assay (no. 108 [Table 2]) (Fig. 2). The peptides were added individually to preassembled TFIIIC-DNA prior to the addition of Brf1 and the two other TFIIIB components. Two of the peptides (no. 104 and no. 107) identified using the high-affinity Tfc4 fragment (Nt-TPR5) inhibited the recruitment of Brf1 to TFIIIC-DNA as well as the formation of the TFIIIB-TFIIIC-DNA complex (Fig. 3). In contrast, a peptide (no. 101) identified using the autoinhibited Nt-TPR9 fragment increased the formation of Brf1 and TFIIIB complexes and reduced the mobility of these complexes in the native gel. Given the small size of the peptide relative to the complexes, the unique effect of peptide 101 on complex mobility implies that it alters their conformation. Three other peptides (no. 103, no. 105, and no. 106), despite positive interactions with Tfc4 fragments in the yeast two-hybrid assay, did not affect complex assembly relative to reactions performed with the negative-control peptide (no. 108) or no added peptide. These Brf1 peptides do not play an important role in transcription complex assembly under the current experimental conditions and were not studied further. Subsequent experiments focused on the three biochemically active peptides.
FIG. 3.
Biochemical effects of synthetic Brf1 peptides on the formation of Brf1-TFIIIC-DNA (Brf1-C-DNA) and TFIIIB-TFIIIC-DNA (B-C-DNA) complexes. TFIIIC-DNA (C-DNA) complexes were assembled for 10 min on a 32P-labeled sup3-eST tRNA gene. Peptides (Tables 1 and 2) were then added to a final concentration of 500 μM prior to the addition of Brf1 without (top panels) or with (bottom panels) the other TFIIIB components, TBP and Bdp1. Reaction mixtures were incubated for 45 min at 20°C, and the Brf1-containing complexes (Brf1-C-DNA and B-C-DNA) were resolved from C-DNA complexes by native gel electrophoresis.
A loop region in repeat II of the TFIIB homology domain is important for Brf1 binding to TFIIIC-DNA.
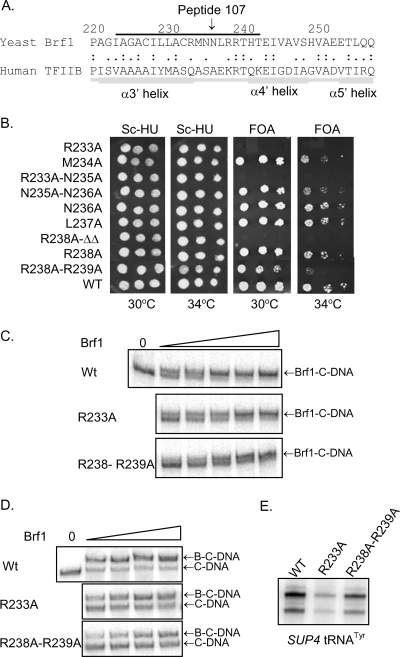
Structural modeling of Brf1 sequences using human TFIIB as a template shows that inhibitory peptide 107 corresponds to the α3′ helix-loop-α4′ helix region of repeat II (Fig. 4A). This region of TFIIB in the TFIIB-TBP-DNA ternary complex is solvent exposed and lies opposite the interaction interface with the C-terminal stirrup of TBP (29). The positive two-hybrid interaction of this peptide with TPR1-5 (Liao, unpublished) suggests that it may bind in the superhelical groove of this TPR array. Since ligand binding in TPR grooves typically involves an extended peptide conformation, we focused on the predicted loop region residues of the peptide (RMNNLRRT, aa 233 to 240) as a potential interaction site with Tfc4. To examine the significance of the amino acids within this region of Brf1, alanine substitutions were generated individually or in pairs and were evaluated for effects on Brf1 function in vivo (Fig. 4B). Two of the mutants (the R233A and R238A-R239Δ-T240Δ mutants) were lethal, while three others (the M234A, N235A-N236A, and R238A-R239A mutants) had a temperature-sensitive phenotype. Thus, the residues in peptide 107 that map to the α3′, α4′ loop in repeat II have clear biological significance.
FIG. 4.
Brf1 peptide 107 defines a functionally important loop in repeat II of the TFIIB homology region. (A) Alignment of repeat II sequences from the TFIIB homology region of yeast Brf1 with those of human TFIIB. The alignment is based on a multiple mapping method combining ClustalW and Muscle alignment with structural modeling (30). The region defined by peptide 107 (line above the sequence) corresponds to the α3′ helix-loop-α4′ helix region of TFIIB. (B) In vivo phenotypes of BRF1 mutants. BRF1 residues in the loop connecting helices α3′ and α4′ were mutagenized to alanine, and the ability of the mutants to support growth of a brf1Δ strain (YSB108) at 30°C and 34°C was tested after eviction of the rescuing plasmid on 5-fluoroorotic acid (FOA)-containing media. Normalization for cell number was confirmed by spotting serial dilutions of the transformants on SC−His-Ura (Sc-HU) medium. (C) Recombinant Brf1 proteins carrying the R233A or R238A-R239A mutation were compared with wild-type (WT) Brf1 for their abilities to assemble Brf1-TFIIIC-DNA (Brf1-C-DNA) complexes. Reaction mixtures contained equal amounts of TFIIIC-DNA and 0, 750, 1,500, 3,000, 6,000 or 12,000 fmol of Brf1. (D) Brf1 proteins from panel C were compared for their abilities to assemble TFIIIB-TFIIIC-DNA (B-C-DNA) complexes. Reaction mixtures contained equal amounts of TFIIIC-DNA (C-DNA) and no Brf1 or 182.5, 375, 750, and 1,500 fmol of Brf1 together with TBP and Bdp1. (E) Brf1 mutations affect TFIIIC-dependent SUP4 tRNA gene transcription. Reactions were carried out under multiple-round transcription conditions with equal amounts of Brf1 and other components as specified in Materials and Methods.
To examine the biochemical function of the α3′, α4′ loop mutants, recombinant Brf1 carrying the R233A or R238A-R239A mutation was prepared in parallel with wild-type Brf1 and analyzed for TFIIIC-mediated complex assembly. As expected, both of the mutant proteins were less efficient than the wild type in forming Brf1-TFIIIC-DNA and TFIIIB-TFIIIC-DNA complexes, consistent with a defect of the Brf1 mutants in binding to TFIIIC-DNA (Fig. 4C and D). All together, the in vivo and in vitro data support a role for the α3′, α4′ loop that is encompassed by Brf1 peptide 107 in binding to Tfc4 during preinitiation complex assembly. Additional data supporting this conclusion are presented below.
The modest effect of the Brf1 R233A mutation on complex assembly in vitro contrasts with the lethal phenotype of this mutant in vivo and suggests that additional defects may be associated with the mutation. To investigate this possibility, we compared in vitro transcription levels on a SUP4 tRNA gene in reactions performed with wild-type or mutant (R233A and R238A-R239A mutant) Brf1 (Fig. 4E). Transcription with the conditional mutant, the R238A-R239A mutant, was reduced by about 50%, consistent with its modest complex assembly defect (Fig. 4C to E). In comparison, transcription with the lethal R233A mutant was reduced to about 14% of the wild-type level. The more severe effect of the R233A mutation on transcription relative to complex assembly implies a defect in the function of the mutant TFIIIB complex. This is consistent with the suggestion that repeat II of Brf1 makes an important interaction with the Rpc17 subunit of Pol III (9).
Domain II in the C terminus of Brf1 participates with the N-terminal α3′, α4′ loop region in binding to TFIIIC-DNA.
As described above for peptide 107, the inhibitory effect of peptide 104 in the formation of Brf1-TFIIIC-DNA complexes (Fig. 3) suggests that conserved domain II in the C-terminal half of Brf1 (Table 1 and Fig. 1A) also plays a role in the recruitment of Brf1 by TFIIIC. To further characterize the function of this region in Brf1, TFIIIC-DNA complexes were assembled and peptide 104 was titrated in the presence of a fixed amount of Brf1. This resulted in a concentration-dependent decrease in the number of Brf1-TFIIIC-DNA complexes (Fig. 5A). In contrast, titration of another C-terminal Brf1 peptide (no. 103 [Table 2]) had no effect (Fig. 5A). Quantitation of Brf1-TFIIIC-DNA complex assembly as a function of peptide concentration followed by curve fitting to the Hill equation (26) yielded an apparent binding affinity for the peptide-TFIIIC-DNA complex of 75 ± 14.2 μM (Fig. 5B). Phylogenetic analysis of the Brf1 region represented in peptide 104 reveals that it is highly conserved and very hydrophilic, with a high content of acidic and basic residues (Fig. 5C). To further test the specificity of the peptide in inhibiting Brf1 recruitment, we generated two mutant peptides (104mt1 and 104mt2) in which different pairs of basic and acidic residues were switched in order to maintain the net charge of the peptides (Fig. 5C). The mutant peptides did not inhibit Brf1 recruitment by TFIIIC-DNA or the formation of TFIIIB-TFIIIC-DNA complexes under conditions where the wild-type peptide completely inhibited the assembly of these complexes (Fig. 5D). A common feature of the mutant peptides is the substitution of lysine 501 and arginine 502 (Fig. 5C). To directly test the importance of these residues, we prepared recombinant Brf1 carrying a K501A-R502A mutation and compared its ability to assemble complexes with that of wild-type Brf1. In addition, to test whether the regions defined by peptides 104 and 107 in domain II and repeat II, respectively, contribute together in the binding of Brf1 to TFIIIC-DNA, we also introduced the K501A-R502A substitutions into a Brf1 background containing the R238A-R239A mutations (Fig. 4). Each of the mutant proteins was prepared and assayed together with wild-type Brf1. Similarly to the mutation in repeat II (R238A-R239A) (Fig. 4C and D), the K501A-R502A mutation in domain II showed a modest but reproducible decrease in the binding of Brf1 to TFIIIC-DNA and a similarly modest reduction in the assembly of TFIIIB (Fig. 5E and F). Strikingly, the combination of mutations in domain II and repeat II led to a significant reduction in the assembly of the Brf1-TFIIIC-DNA and TFIIIB-TFIIIC-DNA complexes (Fig. 5E and F). This indicates that both sites participate in Brf1 binding to TFIIIC-DNA.
FIG. 5.
Role for conserved domain II of Brf1 in binding to TFIIIC-DNA (C-DNA). (A) Peptide 104 competes with Brf1 for binding to C-DNA. Preformed C-DNA complexes were incubated in the absence (−) or the presence of a fixed concentration of Brf1 and various amounts of peptide 104 or peptide 103, as indicated, from 0 to 500 μM (final concentration). (B) Brf1-TFIIIC-DNA (Brf1-C-DNA) complex formation in the presence of peptide 104 (triangles) or peptide 103 (circles) as shown in panel A was quantified and plotted as a function of peptide concentration. The data for peptide 104 were fit to the Hill equation (26) to obtain an apparent Kd of 75 μM. (C) Comparison of the peptide 104 amino acid sequences among Brf1 orthologs. The sequences of mutant peptides (104mt1 and 104mt2) that were synthesized are shown, with the mutated residues underlined. The net charge of these peptides is the same as that of the wild-type peptide. Amino acids 501 and 502 in Saccharomyces cerevisiae (S. cerevisiae) Brf1 are indicated by arrows below the mutant peptide sequences. H. sapiens, Homo sapiens; D. melanogaster, Drosophila melanogaster; C. elegans, Caenorhabditis elegans, S. pombe, Schizosaccharomyces pombe; C. albicans, Candida albicans; E. gossypii, Eremothecium gossypii; K. lactis, Kluyveromyces lactis. (D) Mutant peptides do not inhibit Brf1 binding to C-DNA. Wild-type (WT) and mutant (mt1, 104mt1; mt2, 104mt2) peptides (500 μM final concentration) were added to preassembled C-DNA complexes to monitor the inhibition of Brf1-C-DNA (upper panel) and TFIIIB-TFIIIC-DNA (B-C-DNA, lower panel) complex formation as described in the legend for Fig. 3. (E) Wild-type and mutant Brf1 proteins were added to preassembled C-DNA complexes to compare their abilities to form Brf1-C-DNA and B-C-DNA complexes. (F) Triplicate experiments of the type shown in panel E were quantified by peak fitting analysis (Microcal Origin software), and the complexes formed with mutant Brf1 were expressed relative to that formed with the wild type. The plot shows the data obtained with 8,000 fmol of Brf1.
A Brf1 peptide that binds to TPR6-9 of Tfc4 stimulates transcription complex formation.
Peptide 101 was identified by virtue of its interactions with a large Tfc4 fragment (Nt-TPR9) whose ability to bind Brf1 is autoinhibited (26). This peptide maps to the N terminus of repeat I in the TFIIB homology region. Initial analysis of this peptide revealed properties that are distinct from those of the other biochemically active peptides in two respects: peptide 101 promotes the formation of Brf1 and TFIIIB complexes by TFIIIC, and it slows the migration of these complexes in native gels (Fig. 3, leftmost panel). To examine these properties in greater detail, Brf1 was titrated onto TFIIIC-DNA in the presence or absence of the peptide. The different concentration dependences of forming Brf1-TFIIIC-DNA complexes are evident from a representative native gel (Fig. 6A). Quantitation and analysis of several such experiments revealed that the peptide increased the apparent affinity of Brf1 for TFIIIC-DNA by 2.6-fold (Fig. 6B). The ability of the peptide to drive the recruitment of Brf1 by TFIIIC was also apparent from a peptide titration experiment. In contrast to titration of the negative-control peptide (no. 108), which had no effect, titration of peptide 101 not only increased Brf1-TFIIIC-DNA formation but also caused an upward shift in the mobility of all of the complexes (Fig. 6C). Quantitative and qualitative changes in complex assembly and mobility, respectively, were also independently confirmed when the peptide was titrated against TFIIIC-DNA alone (Fig. 6D). The results suggest that the binding of peptide 101 to TFIIIC promotes a conformational change in the factor that facilitates its binding to DNA and increases its affinity for Brf1.
FIG. 6.
Peptide 101 promotes Brf1 binding to TFIIIC-DNA (C-DNA). (A) Peptide 101 increases the affinity of Brf1 for C-DNA. Preassembled C-DNA complexes were incubated for 45 min at 20°C with Brf1 (750, 1,500, 3,000, 6,000, and 12,000 fmol) in the presence (+101) or absence (−101) of peptide 101 (500 μM) prior to native gel electrophoresis. (B) Multiple experiments of the type shown in panel A were quantified by peak fitting analysis (Microcal Origin) to determine the relative amounts of Brf1-TFIIIC-DNA (Brf1-C-DNA) complex assembly. These data were fit to the Hill equation (26) to obtain Brf1 binding isotherms in the presence (triangles) or absence (circles) of peptide 101. (C) Peptide 101 facilitates Brf1-C-DNA complex formation and retards the mobility of Brf1-C-DNA complexes. Preassembled C-DNA complexes were incubated in the absence (−) or presence of Brf1 (750 fmol) and increasing concentrations (0, 125, 250, 500, and 1,000 μM) of peptide 101 or 108. (D) Peptide 101 increases TFIIIC binding to DNA and retards the mobility of C-DNA. C-DNA complexes were incubated with peptide 101 at concentrations of 0, 125, 250, 500, 1,000, and 1,500 μM. The region of the gel between the complex and the free DNA was cropped to conserve space. (E) Photo-cross-linking of peptide 101 to Nt-TPR9. Peptide 101 was synthesized with an N-terminal biotin group (bio) and then conjugated, or not, via its C terminus to Bpa. The modified peptides were incubated in the dark at 20°C for 10 min with Nt-TPR9 (each at 5 μM final concentration) in the presence (+) or absence (−) of a 50-fold molar excess of unmodified peptide 101 (competitor). After UV irradiation at 365 nm on ice for 20 min, the samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were transferred to a nitrocellulose membrane and blotted with streptavidin (Strep)-HRP to detect the biotin moiety of the cross-linked peptide. The membrane was then stained with Gelcode blue to demonstrate that equivalent amounts of Nt-TPR9 were present in all reactions. (F) Photo-cross-linking of peptide 101 to TPR6-9 of Tfc4. Reaction conditions were the same as those described for panel E except that the TPR6-9 or TPR1-5 fragments of Tfc4 or TBP were incubated with the modified peptides. Western blot detection of the cross-linked, biotinylated peptide and Gelcode blue staining of the membrane are shown. Molecular size markers are shown in the leftmost lane.
To identify the region of Tfc4 that is bound by peptide 101, peptide analogs were synthesized with a C-terminal photoreactive label (Bpa) and/or an N-terminal biotin group. Bpa enables the covalent attachment of the peptide to its target upon UV irradiation, while the biotin group allows detection of the photo-cross-link by use of streptavidin-HRP. The peptide analogs demonstrated the same effect as the unmodified peptide on Brf1 binding to TFIIIC-DNA (data not shown). Subsequently, the Bpa- and biotin-labeled peptide was incubated with bacterially expressed and purified Nt-TPR9 prior to UV irradiation and Western blot analysis. As expected, a cross-linked band of the appropriate size for Nt-TPR9 was detected (Fig. 6E, lane 2), and this band was specifically competed by a 50-fold excess of the unmodified peptide (Fig. 6E, lane 3). Importantly, no signal was observed in reactions performed with Nt-TPR9 and the biotinylated peptide lacking the Bpa moiety (Fig. 6E, lane 1), indicating that cross-linking was dependent on the Bpa group. Similarly, no cross-linked products were observed with samples that were not UV irradiated (Liao, unpublished). Next, we used this photo-cross-linking methodology to determine whether the binding of the peptide to Nt-TPR9 could be mapped to either of the TPR arrays. Recombinant TPR1-5 or TPR6-9 fragments along with TBP as a negative-control protein were separately irradiated in the presence of the double-labeled peptide (i.e., Bpa and biotin). A biotinylated product of ∼20 kDa was detected in the reaction performed with TPR6-9, consistent with the expected size of cross-linked protein (18.4-kDa TPR6-9 plus 2-kDa peptide) (Fig. 6F). This band disappeared in the presence of a 50-fold excess of unmodified peptide. In contrast, no signal was detected in reactions performed with the double-labeled peptide and TPR1-5 or TBP (Fig. 6F). These results indicate that peptide 101 binds to Nt-TPR9 through its interaction specifically with TPR6-9.
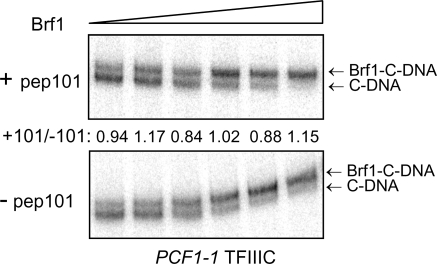
The stimulatory effect of peptide 101 on Brf1 binding to TFIIIC-DNA is reminiscent of the effect of a dominant mutation, the PCF1-1 mutation, in TPR2 of Tfc4. As noted earlier, this mutation increases the affinity of Brf1 for Nt-TPR9 without affecting its interaction with Nt-TPR5 or TPR1-5 and is thought to facilitate Brf1 binding to TFIIIC by stabilizing an alternative conformation of Tfc4 in which autoinhibition is relieved (25). We considered the possibility that peptide 101 might drive a similar conformational change in Tfc4 by functioning as a competitor of the proposed autoinhibitory interaction. This mechanistic similarity predicts that the peptide would not stimulate Brf1 binding to TFIIIC complexes harboring the dominant mutation. To test this hypothesis, Brf1 titrations were performed in the presence or absence of peptide 101 by use of TFIIIC purified from a PCF1-1 mutant strain. In contrast to the results obtained with wild-type TFIIIC (Fig. 6), addition of the peptide had no significant effect on the fraction of TFIIIC-DNA that was converted into Brf1-TFIIIC-DNA across the range of Brf1 concentrations tested (Fig. 7). At limiting Brf1 concentrations, levels of Brf1-TFIIIC-DNA formation in the presence and absence of peptide 101 differed by less than 17%. We conclude that peptide 101 and the PCF1-1 mutation operate by similar mechanisms to promote Brf1 binding to TFIIIC-DNA.
FIG. 7.
The stimulatory effect of peptide 101 on Brf1 binding is not seen with a dominant activating mutation (the PCF1-1 mutation) in TFIIIC. Preassembled PCF1-1 TFIIIC-DNA (C-DNA) complexes were incubated for 45 min at 20°C with Brf1 (125, 250, 500, 750, 1,000, and 2,000 fmol) in the presence (+pep101) or absence (−pep101) of peptide 101 (125 μM) prior to native gel electrophoresis. The amount of Brf1-TFIIIC-DNA (Brf1-C-DNA) complex relative to the total amount of TFIIIC-containing complexes per lane was quantified by peak fitting (Microcal Origin). For each Brf1 concentration, the ratio of these values in the presence and absence of peptide 101 was determined and is shown between the two panels.
DISCUSSION
By screening Brf1 peptide libraries against Tfc4 fragments with unique binding properties, we have uncovered two sites in Brf1 that are important for its high-affinity interaction with the Nt-TPR5 region of Tfc4 and with TFIIIC-DNA. In addition, we identified a third site in Brf1 that can function in trans to promote TFIIIC interactions with DNA and Brf1 by binding directly to the second TPR array (TPR6-9) of Tfc4. The function of this region in Brf1 supports an earlier proposal that the structure of Tfc4 limits Brf1 binding and illustrates how this autoinhibition of Tfc4 can be relieved. The results provide molecular evidence of the dynamic nature of the Tfc4-Brf1 interaction and help to define the early steps in the concerted assembly of TFIIIB by TFIIIC.
Distal sites in Brf1 are required for high-affinity binding to TFIIIC-DNA.
Two-hybrid interactions between fragments of Brf1 and its high-affinity binding site (Nt-TPR5) in Tfc4 initially identified repeat II in the TFIIB-related N terminus and domain II in the C-terminal Brf1-specific region as important sites of interaction with Tfc4 (Fig. 1B). Consistent with these observations, we subsequently identified short conserved peptides within repeat II and domain II by two-hybrid screening of a DNA-based Brf1 peptide library (Fig. 1 and Table 1) and demonstrated that these regions are biochemically important for the interaction of Brf1 with TFIIIC (Fig. 3 to 5). Synthetic peptides (104 and 107) encompassing either of these regions blocked the recruitment of Brf1 to TFIIIC-DNA (Fig. 3), indicating that binding to both sites is required for a high-affinity interaction. The same conclusion was reached based on the striking decrease in Brf1-TFIIIC-DNA complex assembly obtained with a Brf1 protein carrying adjacent double point mutations at each site (Fig. 5E). Although the latter result could in principle be explained by an effect of the mutations on the global structure of Brf1, this is considered unlikely since the mutations reside in two structurally independent halves of Brf1 (17). Moreover, the structures of Brf1 domain II and of TFIIB bound to TBP-DNA show that the mutated residues are solvent exposed and do not appear to make important intramolecular contacts (13, 29). In contrast to these large effects on complex assembly, Brf1 proteins carrying mutations in one binding site or the other were only modestly defective in their interaction with TFIIIC-DNA (Fig. 4C and 5E). This reflects the multiplicity of contacts between Brf1 and Tfc4; the loss of one or two contacts does not significantly impact the interaction. Accordingly, it seems likely that other residues within the region defined by Brf1 peptides 104 and 107 interact directly with Tfc4.
Different domains in the Nt-TPR5 fragment of Tfc4 interact with Brf1.
The segment of Brf1 corresponding to the α3′, α4′ loop in repeat II (peptide 107 [Table 1]) was able to bind to TPR1-5 in a yeast two-hybrid assay, while the segment corresponding to the C-terminal end of domain II (peptide 104) interacted only with Nt-TPR5. This suggests that the site of interaction of peptide 104 resides in the N terminus of Tfc4 (Fig. 8).
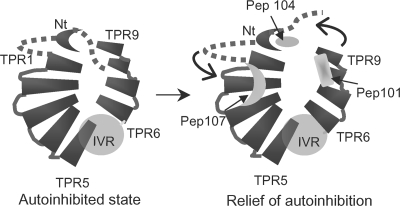
FIG. 8.
Model of Brf1 binding to Nt-TPR9 and relief of autoinhibition. Nt-TPR9 is proposed to adopt a PEX5-like structure (see text). Putative binding sites for Brf1 peptides (Pep) 101, 104, and 107 are indicated in the model on the right. In the absence of Brf1, a region in the N terminus of Tfc4 (dotted line) is hypothesized to interact with TPR9, thereby limiting access of Brf1 to its binding site in this TPR. Binding of the Brf1 region equivalent to peptide 101 promotes a conformational change (denoted by arrows) in Tfc4 that frees the N terminus from TPR6-9. A similar effect can be achieved by the dominant PCF1-1 mutation in TPR2 that has been proposed previously to stabilize the higher-affinity conformation. Further conformational changes, perhaps driven by TBP, are thought to be required for repositioning of Brf1 in order to expose Bdp1 binding sites in Tfc4 (23, 28). IVR, intervening region.
Previous genetic and biochemical studies of dominant mutations in TPR1-5 together with structural modeling have suggested that the superhelical groove of this array serves as a binding site for Brf1 (26). From the studies reported here, we propose that the α3′, α4′ loop in repeat II of Brf1 is a ligand for the TPR1-5 array (Fig. 8). Further studies are obviously needed to test the potential complementarity of this interface. However, several points distinguish this putative ligand-TPR interaction from other examples: (i) as described previously (28), the proposed interaction sites in TPR1-5 traverse the superhelical groove rather than proceeding along it, as seen in the structures of several TPR-ligand complexes (4, 31, 36), and (ii) the sequence of the Brf1 ligand is rich in polar and basic residues and thus is distinct from the acidic peptides that bind via a carboxylate clamp to the TPR arrays of the cochaperone Hop and protein phosphatase 5 (4, 31). Interestingly, with the yeast two-hybrid screen against Nt-TPR5, we recovered numerous positive clones with stop codons immediately adjacent to the insertion site (IRIRAR [Table 1]) or with out-of-frame inserts yielding short peptide fusions with double arginine residues at or near the C terminus (Liao, unpublished). These observations together with the defects associated with mutation of the arginine residues in the α3′, α4′ loop (Fig. 4B to D and 5E and F) suggest that basic residues provide an important recognition element for peptides that bind to TPR1-5. Thus, the Brf1 α3′, α4′ loop interaction with TPR1-5 exhibits novel specificity and a potentially novel mode of interaction among known TPR-ligand complexes.
Domain II in the C-terminal half of Brf1 has been shown to interact with high affinity to both TBP and Bdp1 (6, 17). In the crystal structure of the domain II-TBP-DNA complex, Brf1 is remarkably extended and makes interactions across the convex upper surface of TBP and down its N-terminal stirrup (13). Recent work has shown that segments of domain II interacting with both of these regions in TBP are also critical for Bdp1 binding (16). For example, Brf1 helix H24, which interacts with the N-terminal stirrup of TBP, serves as a two-sided adhesive surface with the side chains projecting away from TBP engaged in interactions with Bdp1. Our studies show that domain II is also important for Brf1 binding to Tfc4. Peptide 104 lies at the C-terminal end of domain II and maps to the last resolved helix (H25) in the domain II-TBP-DNA crystal structure. Helix H25 makes no significant contacts with TBP and is extended away from TBP towards the downstream DNA. This juxtaposes Brf1 residues important for the interaction with Tfc4 and Brf1 residues that interact with the C-terminal SANT domain of Bdp1 (16). Since Bdp1 also interacts with Tfc4, it seems likely that the binding sites in Tfc4 for domain II of Brf1 and the SANT domain of Bdp1 are adjacent or may overlap (requiring repositioning of Brf1 for Bdp1 binding), as suggested previously (23, 28).
Brf1 peptide 101 promotes complex assembly by relieving autoinhibition in Nt-TPR9.
The Nt-TPR9 fragment of Tfc4 has been investigated previously as a model for autoinhibition of Brf1 binding following initial biochemical studies of wild-type and dominant mutant TFIIIC (25, 28). In the current work, Brf1 peptide 101 was identified as an interacting partner of Nt-TPR9 and was shown to photo-cross-link specifically to TPR6-9. The effects of peptide 101 on complex formation are indicative of its role in relieving the autoinhibition of TFIIIC binding to Brf1. In addition to increasing TFIIIC-DNA, Brf1-TFIIIC-DNA, and TFIIIB-TFIIIC-DNA complex formation, the peptide slows the migration of these complexes in native gels (Fig. 6), consistent with a peptide-induced conformational change in Tfc4. The increased affinity of wild-type but not mutant (PCF1-1 mutant) TFIIIC-DNA complexes for Brf1 in the presence of the peptide further supports this interpretation: as already noted, previous studies have indicated that the PCF1-1 mutation does not make direct contacts with Brf1 (25). This mutation is thought to facilitate Brf1 binding to Nt-TPR9 and to TFIIIC-DNA complexes by stabilizing an alternative conformation of Tfc4 in which autoinhibition is relieved.
What is the nature of the autoinhibitory interaction in Tfc4, and how is it perturbed by Brf1 peptide 101 and the PCF1-1 mutation? Some insight into these questions is provided by the properties of mutations in the superhelical groove of TPR6-9 (23). A loss-of-function mutation in TPR7 (L469K) of Tfc4 was previously shown to decrease Brf1 (and Bdp1) binding to TFIIIC. However, this reduced binding is still stimulated by Brf1 peptide 101, suggesting that the L469K mutation does not interfere with peptide binding (Liao, unpublished). Similarly, a TPR6-9 fragment containing the L469K mutation was efficiently photo-cross-linked to the derivatized peptide (Liao, unpublished). These results suggest that the binding site for peptide 101 resides elsewhere in this array. Likely sites of interaction have been mapped to TPR9, where loss-of-function mutations (S541I and L542G) can be suppressed by Brf1 overexpression (23) (Fig. 8).
The mechanistic similarity of Brf1 peptide 101 and the PCF1-1 mutation in TPR2 suggests that these binding/interaction sites in Tfc4 may be in relatively close proximity (although it is not possible to exclude the propagation of structural changes between distal sites). With this in mind and with the likelihood that peptide 101 may bind to TPR9 (Fig. 8), it is possible to consider autoinhibition in Tfc4 in terms of a TPR structural model similar to the peroxisomal importer PEX5 model (10, 28). In this model, the two TPR arrays are oriented antiparallel to one another (Fig. 8). This places the N terminus of Tfc4 and TPR2 considerably closer to TPR9 than would be the case for an extended superhelical array of the type found in p67phox (28, 31). A PEX5-like structure for Tfc4 predicts that autoinhibition may result from an intramolecular interaction between N-terminal sequences and TPR6-9. Binding of Brf1 peptide 101 would prevent this interaction and potentially enable additional interactions between Tfc4 and Brf1 and/or repositioning of Brf1, as proposed previously (23, 28). The PCF1-1 mutation, on the other hand, would stabilize an intramolecular interaction with the N terminus of Tfc4 that is mutually exclusive with N terminus binding in the autoinhibited conformation. Experiments to address these possibilities are in progress.
Acknowledgments
We thank Andras Fiser and members of his laboratory, Rai Brajesh Kumar and Fernandez-Fuentes Narcis, for help with TFIIB homology modeling and docking. We also thank Karen Puglia for providing the Tfc4 fragments and for experimental support in these studies.
This work was supported by National Institutes of Health grant GM42728.
REFERENCES
- 1.Brun, I., A. Sentenac, and M. Werner. 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16:5730-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushnell, D. A., K. D. Westover, R. E. Davis, and R. D. Kornberg. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303:983-988. [DOI] [PubMed] [Google Scholar]
- 3.Chaussivert, N., C. Conesa, S. Shaaban, and A. Sentenac. 1995. Complex interactions between yeast TFIIIB and TFIIIC. J. Biol. Chem. 270:15353-15358. [DOI] [PubMed] [Google Scholar]
- 4.Cliff, M. J., R. Harris, D. Barford, J. E. Ladbury, and M. A. Williams. 2006. Conformational diversity in the TPR domain-mediated interaction of protein phosphatase 5 with Hsp90. Structure 14:415-426. [DOI] [PubMed] [Google Scholar]
- 5.Cloutier, T. E., M. D. Librizzi, A. K. Mollah, M. Brenowitz, and I. M. Willis. 2001. Kinetic trapping of DNA by transcription factor IIIB. Proc. Natl. Acad. Sci. USA 98:9581-9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colbert, T., S. Lee, G. Schimmack, and S. Hahn. 1998. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol. Cell. Biol. 18:1682-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 8.Dumay-Odelot, H., J. Acker, R. Arrebola, A. Sentenac, and C. Marck. 2002. Multiple roles of the τ131 subunit of yeast transcription factor IIIC (TFIIIC) in TFIIIB assembly. Mol. Cell. Biol. 22:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferri, M. L., G. Peyroche, M. Siaut, O. Lefebvre, C. Carles, C. Conesa, and A. Sentenac. 2000. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol. 20:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatto, G. J., Jr., B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091-1095. [DOI] [PubMed] [Google Scholar]
- 11.Geiduschek, E. P., and G. A. Kassavetis. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310:1-26. [DOI] [PubMed] [Google Scholar]
- 12.Huet, J., C. Conesa, C. Carles, and A. Sentenac. 1997. A cryptic DNA binding domain at the COOH terminus of TFIIIB70 affects formation, stability, and function of preinitiation complexes. J. Biol. Chem. 272:18341-18349. [DOI] [PubMed] [Google Scholar]
- 13.Juo, Z. S., G. A. Kassavetis, J. Wang, E. P. Geiduschek, and P. B. Sigler. 2003. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422:534-539. [DOI] [PubMed] [Google Scholar]
- 14.Kassavetis, G. A., B. Bartholomew, J. A. Blanco, T. E. Johnson, and E. P. Geiduschek. 1991. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc. Natl. Acad. Sci. USA 88:7308-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235-245. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis, G. A., R. Driscoll, and E. P. Geiduschek. 2006. Mapping the principal interaction site of the Brf1 and Bdp1 subunits of Saccharomyces cerevisiae TFIIIB. J. Biol. Chem. 281:14321-14329. [DOI] [PubMed] [Google Scholar]
- 17.Kassavetis, G. A., A. Kumar, E. Ramirez, and E. P. Geiduschek. 1998. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol. 18:5587-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassavetis, G. A., G. A. Letts, and E. P. Geiduschek. 2001. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J. 20:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kassavetis, G. A., E. Soragni, R. Driscoll, and E. P. Geiduschek. 2005. Reconfiguring the connectivity of a multiprotein complex: fusions of yeast TATA-binding protein with Brf1, and the function of transcription factor IIIB. Proc. Natl. Acad. Sci. USA 102:15406-15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoo, B., B. Brophy, and S. P. Jackson. 1994. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 8:2879-2890. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, A., A. Grove, G. A. Kassavetis, and E. P. Geiduschek. 1998. Transcription factor IIIB: the architecture of its DNA complex, and its roles in initiation of transcription by RNA polymerase III. Cold Spring Harbor Symp. Quant. Biol. 63:121-129. [DOI] [PubMed] [Google Scholar]
- 22.Lapouge, K., S. J. Smith, P. A. Walker, S. J. Gamblin, S. J. Smerdon, and K. Rittinger. 2000. Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 6:899-907. [DOI] [PubMed] [Google Scholar]
- 23.Liao, Y., I. M. Willis, and R. D. Moir. 2003. The Brf1 and Bdp1 subunits of transcription factor TFIIIB bind to overlapping sites in the tetratricopeptide repeats of Tfc4. J. Biol. Chem. 278:44467-44474. [DOI] [PubMed] [Google Scholar]
- 24.Librizzi, M. D., R. D. Moir, M. Brenowitz, and I. M. Willis. 1996. Expression and purification of the RNA polymerase III transcription specificity factor IIIB70 from Saccharomyces cerevisiae and its cooperative binding with TATA-binding protein. J. Biol. Chem. 271:32695-32701. [DOI] [PubMed] [Google Scholar]
- 25.Moir, R. D., K. V. Puglia, and I. M. Willis. 2002. A gain-of-function mutation in the second tetratricopeptide repeat of TFIIIC131 relieves autoinhibition of Brf1 binding. Mol. Cell. Biol. 22:6131-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moir, R. D., K. V. Puglia, and I. M. Willis. 2002. Autoinhibition of TFIIIB70 binding by the tetratricopeptide repeat-containing subunit of TFIIIC. J. Biol. Chem. 277:694-701. [DOI] [PubMed] [Google Scholar]
- 27.Moir, R. D., I. Sethy-Coraci, K. Puglia, M. D. Librizzi, and I. M. Willis. 1997. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol. 17:7119-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moir, R. D., and I. M. Willis. 2004. Tetratricopeptide repeats of Tfc4 and a limiting step in the assembly of the initiation factor TFIIIB. Adv. Protein Chem. 67:93-121. [DOI] [PubMed] [Google Scholar]
- 29.Nikolov, D. B., H. Chen, E. D. Halay, A. A. Usheva, K. Hisatake, D. K. Lee, R. G. Roeder, and S. K. Burley. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119-128. [DOI] [PubMed] [Google Scholar]
- 30.Rai, B. K., and A. Fiser. 2006. Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins 63:644-661. [DOI] [PubMed] [Google Scholar]
- 31.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 32.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 33.Sethy-Coraci, I., R. D. Moir, A. Lopez-de-Leon, and I. M. Willis. 1998. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 26:2344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staudinger, J., M. Perry, S. J. Elledge, and E. N. Olson. 1993. Interactions among vertebrate helix-loop-helix proteins in yeast using the two-hybrid system. J. Biol. Chem. 268:4608-4611. [PubMed] [Google Scholar]
- 35.Werner, M., N. Chaussivert, I. M. Willis, and A. Sentenac. 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268:20721-20724. [PubMed] [Google Scholar]
- 36.Wu, B., P. Li, Y. Liu, Z. Lou, Y. Ding, C. Shu, S. Ye, M. Bartlam, B. Shen, and Z. Rao. 2004. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci. USA 101:8348-8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, M., Z. Wu, and S. Fields. 1995. Protein-peptide interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 23:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]